Abstract
The recruitment of transcriptional coactivators, including histone modifying enzymes, is an important step in transcription regulation. A typical activator is thought to interact with several cofactors, presumably in a sequential manner. The common use of several cofactors raises the question of how activators achieve both cofactor selectivity and diversity. Human STAGA is a multiprotein complex with the acetyltransferase GCN5L as the catalytic subunit. Here, we first show, through RNA interference-mediated knock-down and chromatin immunoprecipitation assays, that GCN5 plays a role in p53-dependent gene activation. We then employ p53 mutagenesis, in vitro binding, protein-protein cross-linking, and chromatin immunoprecipitation assays to establish a novel role for the second p53 activation subdomain (AD2) in STAGA recruitment and, further, to demonstrate that optimal binding of STAGA to p53 involves interactions of STAGA subunits TAF9, GCN5, and ADA2b, respectively, with AD1, AD2, and carboxy-terminal domains of p53. These results provide concrete evidence for mediation of transcription factor binding to coactivator complexes through multiple interactions. Based on our data, we propose a cooperative and modular binding mode for the recruitment of coactivator complexes to promoters.
The tumor suppressor p53, in large part through its action as a gene-specific transcriptional activator, mediates cell cycle arrest or apoptosis in mammalian cells in response to a variety of cellular stress conditions that include DNA damage, aberrant growth signals, and exposure to certain drugs. The p53 gene is the most frequent target of genetic alterations in cancer, and the majority of the observed p53 mutations map in its sequence-specific DNA-binding domain. The induction of p53 modifications and stabilization after genotoxic stimuli results in the activation of a large number of p53-dependent genes that include cell cycle regulatory genes such as GADD45 and p21 and proapoptotic genes such as PUMA and Noxa (63).
As a transcription factor, p53 has been shown to act through cofactors involved either in preinitiation complex formation (25) or covalent modification of chromosomal histones (reviewed in reference 24). In the latter case our laboratory and others have shown that, through direct interactions, p53 recruits a variety of histone modifying enzymes (including p300, PRMT1, and CARM1) to p53-dependent genes (1). While the importance of the histone acetyltransferase p300 or the related CBP for p53-dependent transcription in vivo (24) and in vitro (reviewed in reference 1) has long been established, there is mounting evidence that GCN5 and PCAF, two closely related acetyltransferases that are homologues of yeast GCN5 (yGCN5) (11, 71), also play roles in p53-dependent gene activation.
As first reported for yGCN5, which is found in the Saccharomyces cerevisiae SAGA complex (23), mammalian GCN5 and PCAF are found in large complexes. These include the GCN5-containing STAGA complex (50), the GCN5-containing TFTC complex (64), and the PCAF complex (52). The mammalian (human) complexes contain homologues of yeast SAGA subunits, as well as associated factors involved in DNA repair and RNA processing (8, 51). Furthermore, the diversity of the mammalian SAGA-like complexes is increased by the presence of not only two paralogues (GCN5 and PCAF) but also alternatively spliced forms of mammalian GCN5 (68) and two variant forms of the ADA2 subunit (4). Mice lacking PCAF develop normally and do not have a distinct phenotype, whereas GCN5 null embryos die during embryogenesis (67, 69).
Human STAGA has been shown to interact with the activation domains of VP16 (51) and c-Myc (46) and to affect Gal4-VP16-dependent transcription from a chromatinized template (51). This interaction appears to be conserved in yeast since yeast SAGA binds the Myc activation domain and since transactivation by a Myc-Pho4 fusion protein depends on the Gcn5, Ada2, and Ada3 components of yeast SAGA (21). A TFTC-type complex was reported to show ligand-dependent estrogen receptor interaction and recruitment of TRRAP and GCN5 subunits to the cathepsin D and c-fos promoters (70).
Recently, several proteins common both to STAGA and to other complexes have been functionally or physically linked to p53. Thus, TRRAP was found to act synergistically with p53 in vivo (2) and to be recruited to the p21 promoter after gamma irradiation (5). However, TRRAP is found in at least five different large multisubunit complexes implicated in chromatin modification. These include the STAGA (51), PCAF (61), TFTC (9), TIP60 (29), and p400 (22) complexes. In relation to these complexes, the TIP60 complex has been implicated in p53 transactivation (reviewed in reference 55), TRRAP and GCN5 have been shown to coimmunoprecipitate with p53 from nuclear extract (5), and coexpressed ADA3 has been shown to stabilize p53 (39).
The amino terminal activation domain (amino acids 1 to 80) of p53 consists of two subdomains, AD1 (residues 1 to 40) and AD2 (residues 41 to 80), that show independent functions in various assays (reviewed in reference 73). Interestingly, in an artificial yeast-based assay employing Gal4 fusion proteins, both the intact activation domain and the two subdomains were found to depend upon yeast Ada2 (yAda2), yAda3, and yGcn5 (12). Human p53 containing mutations in AD1 (L22Q W23S) is transcriptionally dysfunctional (44), and the corresponding L25Q W26S mutation in murine p53 is embryonic lethal (33). However, studies with a conditional p53 mutant knock-in mouse strain have revealed that although the mutant is impaired in the activation of several known p53-dependent genes, including the p21 and MDM2 genes, it is able to activate transcription of certain targets that include the proapoptotic BAX and APAF-1 genes (32, 33). AD1 has been shown to directly bind p300/CBP (27), the human TFIID TAF9 (hTAF9)/TAFII31 subunit (48, 59), and Mediator subunit TRAP80/MED17 (30), all of which are potentially responsible for transcription impairment by AD1 mutations.
Although early studies showed that subdomain AD2 also plays a role in p53-dependent transcription both in yeast (12) and in human cells (62), the underlying mechanism remains to be explained. AD2 previously has been shown to interact with replication protein A subunit 1 (6, 28) and with the p62 subunit of the general transcription factor TFIIH (17, 66). However, since replication protein A and TFIIH are also involved in DNA repair, the cellular events relevant to these interactions of p53 are presently unclear.
The present study shows that both the AD1 and AD2 activation subdomains interact with the STAGA complex in vitro and that both are needed for efficient recruitment of the complex in vivo, providing a novel explanation for the role of the second p53 activation subdomain. We show that the STAGA complex, through GCN5, is specifically involved in p53-dependent gene activation, and we document the recruitment of STAGA to promoters upon UV damage. We find that p53 binds the intact STAGA complex both in vivo and in vitro. Localizing direct physical interactions of the activator to various subunits of the complex, we find that unlike what was proposed for yeast SAGA (10), STAGA interaction with the activator is mediated by several distinct contacts. Our findings are indicative of a cooperative and modular activator-coactivator binding mechanism.
MATERIALS AND METHODS
Cell culture and transfection.
H1299 and U2OS cells were kept in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum. HeLa cells in spinner cultures were kept in phosphate-buffered Dulbecco's modified Eagle medium with 10% bovine serum for large-scale nuclear extract preparation. Transfections were conducted according to the manufacturer's instruction using FuGENE6 (Roche). Stable cell lines were created as described previously (51). H1299-derived cells, expressing p53 after tetracycline withdrawal, were kindly provided by Xinbin Chen (University of Alabama).
RNA interference.
H1299-derived Hμ cells were transfected with previously described small interfering RNAs (siRNAs) directed against GCN5 (54) or nontargeting control siRNAs (Dharmacon). At 48 h after transfection, cells were shifted to the permissive temperature (30°C) and harvested after 6 h for RNA isolation.
ChIP assays and reverse transcription-PCR (RT-PCR).
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (1). For ChIP studies with UV-treated cells, U2OS cells were irradiated with 80 J/m2. The antibodies against diacetylated histone H3K9K14 were from Upstate Biotechnology. Antibodies against GCN5 and p53 were from Santa Cruz Biotechnologies.
RNA was isolated from cells with Trizol (Invitrogen) and reverse transcribed with Superscript II RT (Invitrogen) according to the manufacturer's protocol. In RNA interference experiments, agarose gel bands after RT-PCR were quantified using the ImageJ program, version 1.34s, provided by the NIH. For the PCR of GADD45 shown in Fig. 1B, the 5′ and 3′ primers were located in the third and fourth exons, respectively (1). Alternatively, real-time PCR was used to quantify RNA levels. The primers for the real-time PCR were located in the 3′ untranslated region for GADD45, in the first and second exons for p21, and in the second and third exons for PUMA. Results and standard deviations refer to the outcome of at least three experiments.
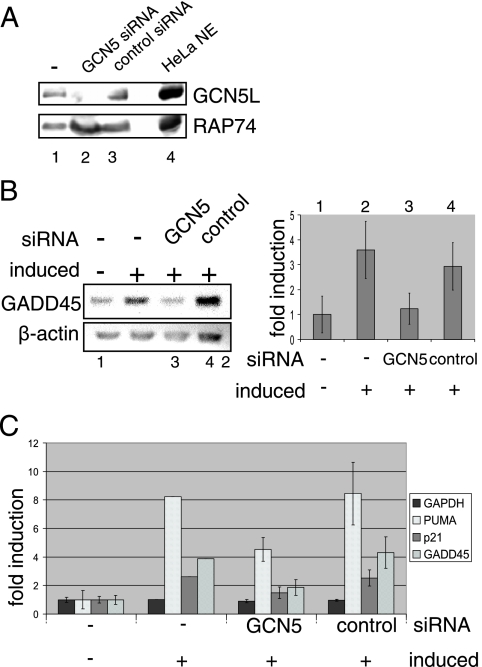
FIG. 1.
GCN5 is needed for efficient activation of p53-dependent genes after UV damage. (A) Knockdown of GCN5 in Hμ cells (H1299-derived cells expressing the temperature-sensitive p53 mutant p53V143A). Hμ cells were treated for 48 h with siRNA against GCN5 or nonspecific control siRNA. Equal amounts of whole-cell extract (based on protein content) were blotted and checked for GCN5. RAP74 and untransfected cells served as loading and positive controls. (B) GADD45 and β-actin levels in Hμ cells. Cells were transfected with siRNA against GCN5 or nonspecific control siRNA. At 48 h after siRNA treatment, cells were shifted to 30°C, and total RNA was isolated after 6 h. Untransfected cells and cells kept at 37°C served as controls. A representative ethidium bromide-stained agarose gel of RT-PCR products is shown. GADD45 RNA levels were quantified using ImageJ software (NIH) and normalized to β-actin levels. The plot shows the result of at least three independent experiments. (C) RNA levels of p53-dependent genes in Hμ cells. Cells were treated as described in panel B. GAPDH, PUMA, p21, and GADD45 RNA levels then were quantified by real-time PCR. The plot shows the result of at least three independent experiments.
Cloning of STAGA subunits.
STAF65γ and human ADA2b (hADA2b) were obtained from expressed sequence tags (clones HK04750 and BU860474). STAF42α was cloned from a fetal spleen cDNA library.
Antibodies.
Antibodies against human SPT3 (hSPT3), hTAF9/TAFII31, and hTAF12/TAFII20 were from previously reported laboratory stocks. Antibodies against hADA2b were raised in rabbits injected with bacterially purified His-tagged hADA2b. TRRAP and hGCN5 antibodies were from Santa Cruz. Antibodies against hADA3, TAF5L/PAF65β, and hTAF10/TAFII30 were kindly provided by Yoshihiro Nakatani (Dana-Farber Cancer Institute) and Laszlo Tora (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France).
GST pull-down assays.
Glutathione S-transferase (GST) fusion proteins were expressed in the Escherichia coli strain XA90 and purified as described previously (26). Proteins were labeled with [35S]methionine using TNT reticulate lysates, following the standard protocol provided by Promega. In vitro translated proteins were incubated with resin-bound proteins (10 μg) by rotating at 4°C for 3 to 4 h in BC buffer (10% glycerol, 20 mM Tris-HCl, pH 7.9, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol) containing 0.05% NP-40, 150 mM KCl, and 0.1 μg/μl bovine serum albumin. After washes with BC buffer containing 0.05% NP-40 and 200 mM KCl, the resin was either stripped with a mixture of 20 mM Tris-HCl, pH 7.9, 100 mM NaCl, and 0.2% N-lauroyl-sarcosine or directly boiled in sodium dodecyl sulfate (SDS) loading buffer before being subjected to SDS-polyacrylamide gel electrophoresis (PAGE). Gels were incubated in Amplify (Amersham Pharmacia) before drying and autoradiography steps to enhance the signal. The same resin preparation, washing, and elution conditions were used for HeLa nuclear extract and the purified STAGA complex when they were incubated with the resin-coupled GST-fusion proteins. Eluates were subjected to SDS-PAGE and blotted onto nitrocellulose membrane (Protran; Schleicher and Schüll) for immunoblotting.
Nuclear extract preparation and complex purification from stable cell lines.
Nuclear extract was prepared as described previously (16). The STAGA complex was purified from the nuclear extract of a cell line stably expressing FLAG-hemagglutinin (HA)-tagged SPT3 (51) by immunopurification with M2 agarose (Sigma). After successive washes with BC buffer containing 300 mM KCl-0.05% NP-40 and BC buffer containing 100 mM KCl-0.05% NP-40, the complex was eluted with FLAG peptide in BC buffer containing 100 mM KCl-0.05% NP-40.
In vivo interaction assays.
The FLAG-HA-tagged STAGA subunit STAF65γ or STAF42α was coexpressed with p53 in H1299 cells. At 24 h posttransfection the cells were washed and cross-linked with 1 mM dimethyl 3,3′-dithiobispropionimidate in phosphate-buffered saline (PBS) by shaking for 20 min at room temperature. After washes with PBS, the cells were lysed in stop-lysis wash (SLW) buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 5 mM EDTA, 0.5% NP-40, 0.5 mM phenylmethylsulfonyl fluoride) and sonicated. The lysate was cleared by centrifugation and incubated with M2 agarose for 8 h. After washes with SLW buffer, the beads were boiled in reducing SDS buffer for elution and reversal of the cross-link. Inputs and eluates were examined by immunoblotting with the monoclonal anti-HA (Santa Cruz) and polyclonal anti-p53 FL393 (Santa Cruz) antibodies. (The secondary antibody against mouse recognizes the band of the M2 heavy chain as well.) FLAG-HA-tagged glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a negative control. For the reverse immunoprecipitation of endogenous STAGA with tagged p53 (without cross-linking), whole-cell extracts from H1299 cells transfected with a vector expressing FLAG-tagged p53 were prepared using SLW buffer and incubated with M2 agarose. Bound material was washed and analyzed by immunoblotting against STAGA subunits. Untransfected cells served as a negative control.
Cross-linking experiments.
The STAGA complex was bound to GST fusion proteins on glutathione-Sepharose and washed as described for the GST pull-down experiments. After an additional two washes with PBS-0.05% NP-40 to eliminate traces of Tris, the resin was suspended in 600 μl of PBS-0.05% NP-40, and 55 μl of 25 mM dithiobis(succinimidylproprionate) (DSP) in dimethyl sulfoxide was added. After 2 min the supernatant was removed, and the reaction was quenched with 50 mM Tris-HCl, pH 7.5, for 30 min at room temperature. The proteins on the beads were denatured with 8 M urea for 1 h before being washed twice with 1 ml of BC buffer, 200 mM KCl, and 0.05% NP-40. Elution and reversal of cross-linking were effected by stripping with N-lauroyl-sarcosine in the presence of 0.1 M dithiothreitol at 37°C for 30 min or by boiling in reducing SDS buffer. The former elution method gave generally cleaner immunoblots.
RESULTS
GCN5 is needed for efficient activation of the p53-inducible GADD45 gene.
To investigate whether GCN5 is involved in p53-dependent gene expression, we derived a stable cell line (Hμ) expressing a temperature-sensitive p53 mutant (p53V143A) from p53-negative H1299 lung carcinoma cells. This p53 mutant activates a variety of target genes as effectively as wild-type p53 at 32°C but not at 37°C (72). Consistent with a report that GADD45a, cyclin G, p21, and HDM2 are induced by p53V143A at the permissive temperature (3), a shift from 37°C to 30°C led to increases of p21 and GADD45 RNA levels in Hμ cells but not in control H1299 cells (data not shown). Treatment of Hμ cells with GCN5 siRNA led to a greater than 90% decrease in the level of GCN5 (Fig. 1A). GCN5 knockdown in these cells resulted in a concomitant reduction in the accumulation of GADD45 RNA in response to the shift from 37°C to 30°C (Fig. 1B, lanes 1 to 3), whereas a control nontargeting siRNA had no significant effect on p53-dependent activation (lane 4). In an extension of these results, real-time PCR was used to quantify the effect of GCN5 siRNA on PUMA, p21, GADD45, and GAPDH RNA levels. GCN5 siRNA, but not control siRNA, resulted in impaired induction of the p53-dependent gene products PUMA, p21, and GADD45. GCN5 is thus required for the activation of several p53-dependent genes.
UV-induced recruitment of hGCN5 to promoter sites containing p53 response elements.
In yeast, Gcn5-containing complexes are recruited by transcription factors to promoters of several target genes (40, 42). To determine whether the hGCN5L-containing STAGA complex, an acetyltransferase complex with preference for histone H3 (50), is directly involved in p53 activation, a ChIP assay was employed to assess p53 and GCN5 binding to GADD45 and p21 promoters. The GADD45a gene contains a single p53 binding site 1.6 kb downstream of the core promoter in the third intron (35). The human p21 promoter contains two p53 response elements that are located 1.4 and 2.3 kb upstream of the transcription initiation site (18) (Fig. 2C). Both sites are at least partially bound by p53 in vivo (19, 34). p53-positive U2OS osteosarcoma cells showed, as expected, UV-induced increases in mRNAs from p53 target genes GADD45 and p21 (Fig. 2A). As reported for WS1 fibroblasts (43), UV irradiation of U2OS cells led to phosphorylation of p53 at serine 15 and to a slow, modest increase in the p53 protein level (Fig. 2B). The GCN5 protein level was not affected by UV irradiation.
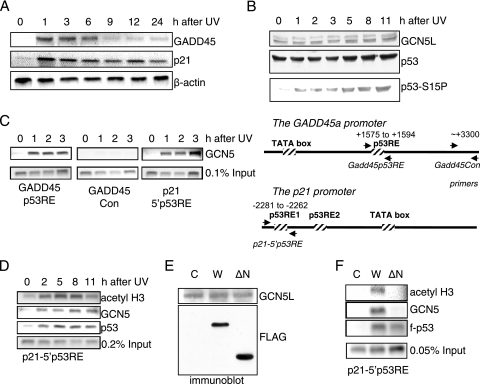
FIG. 2.
GCN5 is recruited to the p21 and GADD45a promoters. (A) RT-PCR analysis of p21 and GADD45 RNA levels in U2OS cells after UV irradiation. Total RNA of irradiated cells was purified after the indicated time points, and cDNA was prepared. Inverted ethidium bromide stains of PCR products are shown. β-Actin levels served as internal controls. (B) Immunoblot of UV-irradiated U2OS cells. p53-positive U2OS cells were UV irradiated and harvested after the indicated time intervals. Equal protein amounts of whole-cell extract were blotted and tested for the amounts of GCN5, p53, and serine 15-phosphorylated p53. (C and D) ChIP assays with antibodies against acetyl H3, GCN5, and p53 after UV irradiation. U2OS cells were UV irradiated and harvested after the indicated time intervals. The schematic representation of the GADD45 and p21 promoters shows the respective location of primers used for ChIP assays. (E) Immunoblot of transfected H1299 cells used for the ChIP assays in panel F. Equal amounts of whole-cell extracts were tested for GCN5 and exogenous tagged p53. (F) ChIP assays in p53-negative H1299 cells. Cells were transfected with expression vectors for wild-type p53 (W) or an amino-terminal deletion mutant of p53 (ΔN) and harvested after 30 h. Untransfected cells (C) served as controls. f, Flag tag.
As shown by ChIP analyses, UV irradiation resulted in recruitment of GCN5 to p53 response elements at both the GADD45a and p21 promoters in U2OS cells (Fig. 2C). A control site downstream of the p53 binding site in the GADD45 promoter was not bound by GCN5. GCN5 binding and histone H3 acetylation appeared to be concomitant with p53 binding to the p21 promoter (Fig. 2D), supporting the hypothesis that STAGA is recruited to the promoter by p53.
The acidic amino terminus (residues 1 to 73) of p53 encompasses a domain needed for transcription activation (20, 36, 53). This activation domain has been implicated in the recruitment of cofactors p300 and PRMT1 to the GADD45 promoter but is dispensable for CARM1 recruitment (1). To test whether GCN5 is recruited to the p21 promoter by p53 and, if so, whether the activation domain of p53 is needed for the recruitment, p53-negative H1299 lung fibroblasts were transiently transfected with either FLAG-tagged wild-type p53 or a FLAG-tagged mutant lacking the amino terminal 73 residues. An immunoblot showed that the wild-type (Fig. 2E, W) and the mutant protein (Fig. 2E, ΔN) were equally well expressed, and ChIP assays confirmed that both proteins bound to the p21 promoter (Fig. 2F). However, only wild-type p53 resulted in accumulation of GCN5, as well as acetylated H3, on the promoter, indicating that the amino-terminal activation domain of p53 is essential for the recruitment of GCN5 in vivo.
Binding of STAGA to p53 in vitro and in vivo.
GCN5 is a subunit of the human complex STAGA. To see whether the STAGA complex binds to p53, a bead-immobilized GST-full-length p53 fusion protein (GSTp53) was incubated with HeLa nuclear extract and washed. An immunoblot of bound proteins revealed binding of STAGA subunits TRRAP, GCN5L, TAF5L (PAF65β), TAF9 (TAFII31), and SPT3 to GSTp53 but not to GST alone (Fig. 3A). Similarly, an affinity-purified STAGA complex (51), derived from a cell line that expresses a FLAG-tagged SPT3, bound to GSTp53 but not to GST alone (Fig. 3B). Thus, STAGA binding to p53 is direct and does not require other cofactors for a strong interaction.
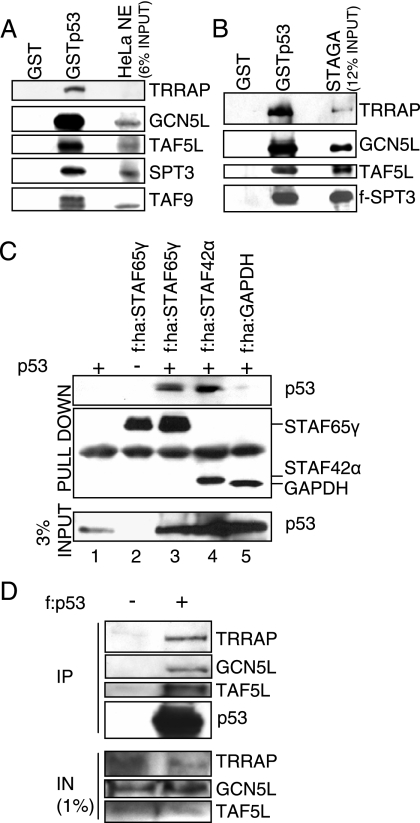
FIG. 3.
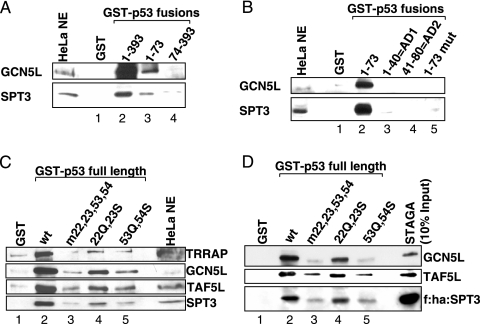
STAGA interacts with p53 in vitro and in vivo. (A) STAGA subunits in HeLa nuclear extract bind to p53. GST alone or GSTp53 was bound to glutathione-Sepharose and incubated with HeLa nuclear extract. The beads were washed, and bound proteins were analyzed by immunoblotting. (B) The STAGA complex purified from a tagged hSPT3 stable cell line binds to p53. GST and GSTp53 beads were incubated with purified STAGA, and bound proteins were analyzed as described in panel A. (C) In vivo interaction of STAGA with p53. Tagged STAGA subunit STAF65γ or STAF42α was coexpressed with p53 in H1299 cells. After in vivo cross-linking, whole-cell extracts were prepared and incubated with M2 agarose. Bound material was washed and analyzed by immunoblotting with antibodies against p53 and HA. Cells expressing tagged GAPDH, p53 only, or STAF65γ without p53 served as negative controls. (D) Immunoprecipitation of endogenous STAGA with tagged p53. Whole-cell extracts from H1299 cells transfected with a vector expressing FLAG-tagged p53 were prepared and incubated with M2 agarose. Bound material was washed and analyzed by immunoblotting. STAGA subunits were found to coprecipitate with tagged p53. Untransfected cells served as negative controls. f, Flag tag; ha, HA tag; IN, input; IP, immunoprecipitation.
We next determined whether STAGA also interacts with p53 in vivo. To this end we first characterized the purified STAGA complex with respect to STAF65γ and STAF42α, two associated proteins that are related, respectively, to the yeast SAGA subunits Spt7 and Ada1 (51). Immunoblotting for representative STAGA subunits in anti-FLAG (M2 agarose) immunoprecipitates from stable HeLa cell lines expressing FLAG-tagged STAF65γ and STAF42α confirmed that the two proteins are integral subunits of the STAGA complex (data not shown). Because their yeast counterparts are thought to play structural roles in the related SAGA complex (65), we chose these two subunits for in vivo interaction studies. Pull-down assays with in vitro translated proteins confirmed that STAF65γ and STAF42α do not directly interact with GSTp53 (see Fig. 5D below).
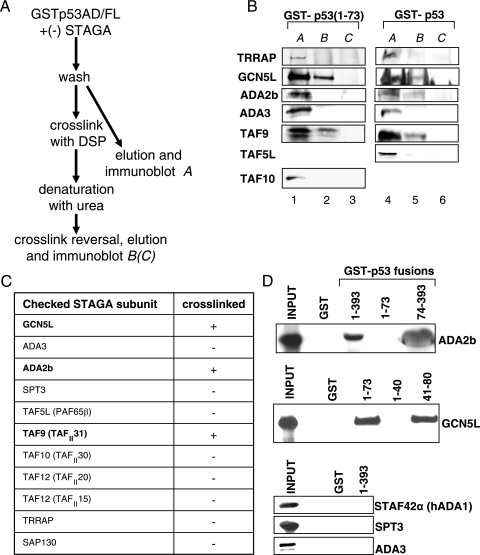
FIG. 5.
Direct targets of p53 within the STAGA complex. (A) Scheme for in vitro cross-linking. The purified STAGA complex was bound to glutathione-Sepharose immobilized GST fusion proteins containing full-length p53 or the p53 activation domain and washed. DSP was used to cross-link interacting proteins under conditions avoiding ternary cross-links. After the proteins were denatured on the beads and beads were washed to remove unbound proteins, the cross-links were reversed, and eluates were checked by immunoblotting (B). Interacting proteins eluted before cross-linking or beads without STAGA served as input (A) and control (C). (B) Representative immunoblots. Several STAGA subunits were checked for direct interaction with full-length p53 or the activation domain of p53. (C) List of STAGA subunits tested for direct interaction with p53 by cross-linking. Only TAF9, GCN5L, and ADA2b were found to cross-link under the tested conditions. ADA2b binding was only observed in the case of full-length p53. (D) Verification and mapping of binary p53-subunit interactions. GST alone or a GST fusion protein with full-length (residues 1 to 393) or truncated mutants of p53 were bound to glutathione-Sepharose and incubated with in vitro translated 35S-labeled STAGA subunits. Beads were washed, and eluted proteins were analyzed by autoradiography after SDS-PAGE. Twenty-five percent of actual input was loaded as Input. ADA2b binding to p53 is not mediated by the p53 activation domain. The interaction site of the p53 activation domain with GCN5L lies within the second subdomain (residues 41 to 80). STAF42α (hADA1), SPT3, and ADA3 served as negative controls.
Because the p53 pathway in HeLa cells is compromised by the endogenous human papillomavirus E6 protein (37, 57), we chose not to use the stable cell lines for in vivo interaction studies with p53. Instead, FLAG-HA-tagged STAF65γ and STAF42α were individually coexpressed with p53 in H1299 cells. The STAGA complex with bound proteins was immunoprecipitated with M2 agarose and subjected to immunoblotting. As shown in Fig. 3C, p53 was immunoprecipitated with FLAG-HA-tagged STAF65γ or STAF42α but not FLAG-HA-tagged GAPDH (lanes 3 to 5). We also performed the reverse immunoprecipitation of endogenous STAGA with tagged p53 (Fig. 3D). H1299 cells were transfected with a vector expressing FLAG-tagged p53. Whole-cell extracts from transfected and untransfected cells were prepared and incubated with M2 agarose. Bound material was washed and analyzed by immunoblotting for representative STAGA subunits. STAGA was found to coprecipitate with tagged p53, indicating that p53 also associates with the STAGA complex in vivo.
STAGA binds to distinct p53 domains that include the amino terminal activation domain of p53.
The requirement of the p53 amino terminus for GCN5 recruitment to endogenous target genes suggests that the STAGA complex binds directly to the activation domain of p53. To test this hypothesis, GST fusion proteins with full-length p53 [p53(1-393)], the p53 amino-terminal activation domain [p53(1-73)], or a p53 mutant lacking the amino terminus [p53(74-393)] were incubated with HeLa nuclear extract. As shown in Fig. 4A, endogenous STAGA, monitored by GCN5 and SPT3 subunits, showed only moderate (but significant) binding to the p53 activation domain (lane 3) compared to the intact p53 (lane 2) and only a very weak binding to the carboxy-terminal fragment (lane 4). These results indicate that while the amino-terminal activation domain of p53 is sufficient for a significant interaction of p53 with STAGA, a region(s) outside this domain contributes strongly to p53 binding to the complex.
FIG. 4.
Mapping the interaction of p53 with STAGA. (A) STAGA binding to full-length p53 and p53 truncation mutants. GST alone or GST fusion proteins with full-length p53, the p53 activation domain, or p53 lacking the amino-terminal activation domain were bound to glutathione-Sepharose and incubated with HeLa nuclear extract. The beads were washed, and bound proteins were analyzed by immunoblotting for the presence of SPT3 and GCN5. (B) Truncation or mutation (L22Q W23S) of the activation domain of p53 abolishes its interaction with STAGA. Beads with GST fusions of the p53 activation domain or various mutants thereof were incubated with HeLa nuclear extract and probed as described in panel A. (C and D) Double point mutations of the activation domain of p53 decrease the interaction of full-length p53 with STAGA. Beads with GST fusions of wild-type or mutant full-length p53 were incubated with HeLa nuclear extract (C) or purified STAGA (D) and probed as described in panel A. Mutants L22Q W23S (22Q, 23S; lanes 4) and W53Q F54S (53Q, 54S; lanes 5) are known to negatively affect p53-dependent transcription and cellular function, with a quadruple mutation, L22Q W23S W53Q F54S (m22, 23, 53, 53; lanes 3), having the most severe effect. f, Flag tag; ha, HA tag; wt, wild type; mut, mutant.
In a further analysis, we checked for interactions of isolated AD1 and AD2 domains with STAGA in nuclear extracts. As shown in Fig. 4B, the endogenous STAGA complex did not bind measurably to either of the two isolated subdomains of p53 (lanes 2 to 4) or to a p53 activation domain, p53(1-73), with the double point mutation L22Q W23S in subdomain AD1 (Fig. 4B, lane 5).
Point mutations in either of the two amino-terminal activation subdomains of p53 affect STAGA binding.
As mentioned, p53 regions containing amino acids 1 to 40 (AD1) and 41 to 80 (AD2) were proposed to function as independent activation domains. Interestingly, the mutations in either AD1 (L22Q W23S) or AD2 (W53Q F54S) negatively affect the transcription of a variety of p53-dependent genes, yet the mutants retain growth suppression activity, while a quadruple mutant completely loses transactivation and growth suppression activity (62). The deletion of p53 amino acids 1 to 23 or 63 to 91 leads to mutants with decreased, but still significant, activation of genes such as the p21 gene, whereas deletion of amino acids 1 to 63 or mutation of residues 22, 23, 53, and 54 completely abrogates activation (74). Other studies showed interactions of isolated hTAF9 and hTAF6 with the amino-terminal p53 activation domain (48, 59) and, further, that mutations either of residues L22 and W23 (48) or of residues F19 and W23 (60) in p53 interfere with binding of TAF9 to the p53 activation domain, whereas a fragment of p53 containing amino acids 9 to 25 is sufficient for an interaction (60). Although it was proposed that p53-dependent transcription activation involves recruitment of TFIID via the hTAF9 subunit, the fact that hTAF9 is also a subunit of STAGA (50) suggested that a TAF9 interaction could also facilitate binding of this complex to p53.
Based on these findings, we further tested the binding of STAGA, both from HeLa nuclear extract (Fig. 4C) and as a purified complex (Fig. 4D), to immobilized p53 mutants. Independent mutations of p53 in AD1 (L22Q W23S) and AD2 (W53Q F54S) weakened the binding of STAGA to p53, with the mutation in the second activation subdomain having a much more pronounced effect (Fig. 4C, lanes 4 and 5 versus lane 2). A quadruple mutation of the activation domain diminished the interaction most severely (lanes 3).
These results are in agreement with a binding mode involving interaction sites within both activation subdomains and at least one other interaction surface outside the amino terminus in p53. Consistent with these results, and indicative of a contribution of the first 40 amino acids of p53, a mutant lacking these residues leads to weaker binding of STAGA than intact p53 but stronger binding than p53(74-393) (data not shown).
Identification of STAGA subunits that are direct targets of p53.
In order to assess human STAGA subunits that directly interact with p53, purified STAGA was bound to immobilized GSTp53(1-393) or GST-p53(1-73) fusion proteins and subjected to cross-linking with DSP (15, 47) according to the protocol shown in Fig. 5A. This reversible homobifunctional cross-linker, which mainly targets lysines, precludes possible interference with target binding and altered mobilities that might complicate assays with other (e.g., covalently linked photoactivated cross-linking) agents. The cross-linking kinetics were chosen so as to restrict cross-linking to two adjacent partners and thus to avoid artificial positive signals due to ternary linkages. As a control, a GST fusion protein was cross-linked in the absence of STAGA (Fig. 5A).
In the analysis of Fig. 5B, in which representative STAGA subunits were scored by immunoblotting after cross-link reversal and elution, only GCN5L and hTAF9 were consistently found to directly interact with the isolated activation domain (residues 1 to 73) of p53 (lanes 1 to 3). In contrast, ADA2b, as well as GCN5L and TAF9, was found to interact with full-length p53, indicating that ADA2b binds p53 outside the amino terminus (lanes 4 to 6). Of note, we did not observe cross-linking to TRRAP or ADA3, despite previous reports of direct interactions of p53 with isolated ADA3 or a fragment of TRRAP (2, 39) and direct interactions of acidic activators with yeast SAGA through the TRRAP homologue Tra1 (10). In the case of TRRAP, detection limits of our immunoblot analysis could be responsible for a lack of observed binding. Of note regarding yeast SAGA, the proposed role of Tra1 as the sole subunit mediating interaction with acidic activators (10) has been challenged by more recent cross-linking data (38, 56).
As mentioned, the binding of (isolated) hTAF9 to p53 was shown to involve p53 residues 9 to 25 (60). To verify and map the observed interactions of ADA2b and GCN5L with p53, we tested the binding of in vitro translated ADA2b and GCN5L to GST-fused p53 deletion mutants. In agreement with the cross-linking data, ADA2b bound very efficiently to a carboxy terminal fragment, p53(74-393), but not detectably to the amino-terminal activation domain (Fig. 5D). Also in agreement with our cross-linking data, GCN5 bound efficiently to the intact AD of p53 and, importantly, almost equally well to AD2 (residues 41 to 80) but not to AD1 (residues 1 to 40) (Fig. 5D). Thus, GCN5 binds to a subdomain distinct from that reported to bind hTAF9 (48, 60). A further analysis with in vitro translated GCN5 showed that both hGCN5L and a shorter splice variant (68) not present in the STAGA complex (hGCN5S) bound p53 efficiently in vitro, indicating that the nonconserved amino terminus of hGCN5L is not needed for the interaction (data not shown). As a further control, and consistent with the cross-linking data (Fig. 5B and C), three other STAGA subunits (STAF42α, hADA3, and hSPT3) showed no detectable binding to p53 under the same conditions (Fig. 5D).
The results of the cross-linking and binding assays, indicating interactions of distinct p53 domains with three direct binding partners in STAGA, are in strong agreement with the observed affinities of STAGA for p53 mutants bearing various truncation and point mutations. Altogether, these data demonstrate, for the first time, that simultaneous interactions of different subunits with distinct activator regions contribute in a cooperative manner to activator-coactivator binding.
STAGA interactions with both p53 activation subdomains, AD1 and AD2, are important for STAGA recruitment in vivo.
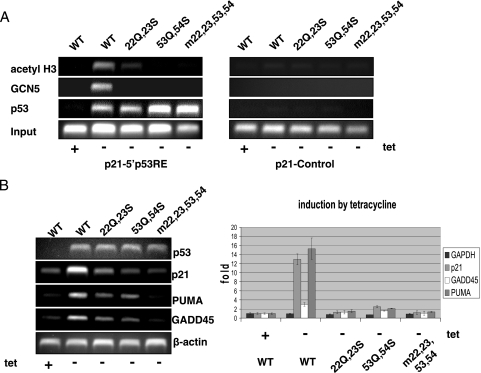
In order to test the in vivo relevance of the observed p53-STAGA subunit interactions, we used ChIP assays to determine whether AD1 (L22Q W23S) and AD2 (W53Q F54S) point mutations that impair in vitro binding of STAGA also impair GCN5 recruitment to endogenous p53 target genes. H1299 cells that express an inducible p53 were harvested 24 h after tetracycline withdrawal. These cells contain similar levels of overexpressed p53 (45) and showed comparable binding of p53 to the p21 promoter in a ChIP assay, indicating that the point mutations do not affect p53 binding to the regulatory elements under the present conditions (Fig. 6A). In contrast, the two sets of point mutations, alone and in combination, essentially eliminated GCN5 recruitment and drastically reduced H3 acetylation. This observation that an intact p53 activation domain is needed for detectable GCN5 binding to the p53 response element underscores the importance of the individual interactions of the p53 subdomains AD1 and AD2 with the STAGA complex, and these interactions appear responsible, at least in part, for the detrimental effects of the mutations on expression of p53 target genes (Fig. 6B). Residual transcription by the individual sets of mutants could be explained by residual interactions of AD1 or AD2 mutants with STAGA that are too weak to be detected by ChIP assay. It is therefore very likely that the same cooperative interactions that govern p53 binding to the complex in vitro are also involved in STAGA recruitment in vivo.
FIG. 6.
p53-dependent in vivo recruitment of STAGA involves both activation subdomains of p53. (A) ChIP assays in H1299-derived inducible p53 cell lines expressing full-length p53 or the L22Q W23S (22Q, 23S), W53Q F54S (53Q, 54S), and L22Q W23S W53Q F54S (m22, 23, 53, 53) mutants. At 24 h after tetracycline (tet) removal, induced wild-type (WT) and mutant p53 bind efficiently to the p21 promoter whereas GCN5 is differentially recruited by the mutants. (B) Mutations of the p53 activation domain affect GADD45, p21, and PUMA transcription. RNA levels in the p53 cell lines were checked by RT-PCR 24 h after tetracycline removal. Representative gels and quantitation by real-time PCR are shown. The RNA levels of actin or GAPDH served as controls.
DISCUSSION
This report presents a functional analysis of the human STAGA complex, which contains the histone acetyltransferase GCN5, in p53-dependent gene activation and a biochemical analysis of STAGA interactions with p53. It demonstrates that p53-dependent transcription of GADD45 requires GCN5, that p53 recruits GCN5 to the p53-dependent p21 and GADD45 promoters after UV damage, and that STAGA binding to p53 involves cooperative interactions of three distinct subunits of the complex with three distinct p53 domains. These results provide evidence that STAGA is directly involved in p53-dependent gene activation. They also provide evidence for a cooperative binding model of activators with multiprotein coactivator complexes, including clues on the possible regulation of cofactor recruitment and corresponding gene activation.
STAGA, through GCN5, acts as a coactivator of p53.
Studies of p53 function in yeast and reports that p53 interacts directly with several isolated proteins that are subunits of STAGA, as well as other complexes (2, 39, 48), have led to the speculation that the human STAGA complex itself might interact with p53 and contribute to p53 function. Here, we show through RNA interference-mediated knockdown and ChIP assays that GCN5 is important for p53-dependent gene activation. We further show that p53 can bind the STAGA complex specifically, both in vitro and in vivo, and that p53 effects GCN5 recruitment, concomitant with H3 acetylation, to endogenous p53-dependent promoters. These results are consistent with our previous demonstration (1) showing that, on the GADD45 gene, UV-induced recruitment of GCN5 correlates with enhanced acetylation of H3 (but not H4), whereas the UV-induced recruitment of p300 correlates with enhanced acetylation of H4 (but not H3). Similar to Drosophila GCN5, hGCN5 is found in the ADA2b-containing STAGA complex and also in a distinct complex containing ADA2a (A. M. Gamper, J. Kim, and R. G. Roeder, unpublished data). Our unpublished ChIP assays showing the recruitment of STAGA-specific ADA2b but not of ADA2a after UV irradiation to p53-dependent promoters strongly supports the idea that STAGA is recruited by p53. While STAGA is clearly involved in transcription of some p53 target genes, the extent to which it is generally required for transcription in mammalian cells remains to be determined.
Distinct domains of p53 interact with specific STAGA subunits.
By using a bifunctional cross-linker to analyze interactions of p53 or the p53 amino-terminal activation domain with bound STAGA, we identified subunits TAF9, ADA2b, and GCN5L as direct interaction partners of p53 in the context of the intact STAGA complex. These interactions were confirmed by the demonstration of direct interactions of p53 with isolated TAF9 (48, 59) and with isolated GCN5 and ADA2b proteins (this study). Further studies showed that the amino terminal p53 activation domain, comprised of the AD1 (residues 1 to 40) and AD2 (residues 41 to 80) subdomains, interacts with TAF9 and GCN5L, whereas the ADA2b interaction is mediated by an undefined domain(s) in the remaining p53 carboxy-terminal region, p53(74-393).
In relation to AD1, it has been suggested that the reduced transcription caused by AD1 mutations affecting the TAF9-p53 interaction reflects reduced p53-TFIID interaction (48, 59). However, our demonstration that TAF9 is involved in STAGA interactions with p53 and that mutations (L22Q W23S) in AD1 diminish p53 STAGA interactions in vitro and in vivo suggests decreased transcription due to reduced p53-STAGA interactions. Furthermore, AD1 also has been implicated in p53 interactions with the Mediator (30) and with p300 (27). Hence, the AD1 domain may well facilitate the recruitment and function of several coactivators.
In relation to AD2, we have demonstrated, first, that mutations (W53Q and F54S) previously shown to reduce p53-dependent transcription dramatically decrease p53-STAGA interactions and, second, that GCN5L binds strongly to the isolated AD2 subdomain. These results provide a novel explanation for the well-established physiological role of AD2 in transcription of various p53 target genes (62, 73, 74).
Beyond the p53 AD1 and AD2 interactions with STAGA through TAF9 and GCN5, respectively, we demonstrate an important role for a carboxy-terminal domain(s) in p53 interactions with STAGA through the ADA2b subunit. In an extension of the present work, our preliminary studies (data not shown) have shown ADA2b binding both to a central p53 fragment (residues 120 to 290) that contains the DNA binding domain and to a carboxy-terminal fragment (residues 290 to 393).
A cooperative and modular binding mode.
Despite the ability of the p53(1-40), p53(41-80), and p53(74-393) fragments to bind TAF9, GCN5, and ADA2b relatively well, none of these domains alone suffices for detectable interaction with the complete STAGA complex in vitro. However, certain combinations of two domains significantly increase the binding, with p53(1-73) and p53(41-393) showing easily detectable binding of STAGA (data not shown). These results underscore the cooperativity in the overall binding mechanism.
Interaction of a single activator such as p53 with multiple subunits of a coactivator complex such as STAGA, Mediator, or TFIID would make sense from energetic, evolutionary, and regulatory points of view. Thus, a combinatorial set of binding partners provides a means both for fine-tuning the strength of the overall interaction and for a wider range of control. It also allows the modular “construction” of activators from interacting (sub)domains during evolution—similar to the combinatorial use of activator binding sites in promoters—and the assembly of diverse coactivator complexes using common subunits. Importantly, individual activation domain-coactivator subunit interactions need not be strong, thus allowing for greater (perhaps stepwise) reversibility and for a dynamic interplay and exchange, potentially in a temporal sequence of multiple coactivators with distinct functions.
In the case of p53, it will be important to extend the current study of interaction multivalency to other relevant cofactor complexes. These include Mediator, TFIID, and the HDAC1 complex with demonstrated p53 interactions that involve, minimally, p53 AD1-MED17/TRAP80 (30; also S. Yamamura and R. G. Roeder, unpublished results), p53 AD1-TAF9 (48, 59), and p53 AD1-PID/MTA2 (49) interactions, respectively.
Regulation of p53-STAGA interactions.
p53 is subject to a number of constitutive and stress-induced posttranslational modifications in both the amino-terminal activation domain and the carboxy-terminal regulatory domain (reviewed in reference 7). The phosphorylation of specific p53 residues was shown to enhance p300/CBP interactions (41), and phosphorylation was also proposed to differentially regulate HDM2 and TAF9 binding to p53 (31), whereas mutations in a primary AD1 phosphorylation site were shown to reduce activation of some p53-dependent promoters (13, 58). This raises the interesting possibility that p53 phosphorylation events might affect STAGA binding to p53 as well. Also of possible relevance to p53-STAGA interactions, prior acetylation of full-length p53 or of a carboxy-terminal p53 fragment (residues 300 to 393) was reported to enhance interaction with TRRAP and GCN5 (possibly as part of a STAGA complex) in nuclear extract (5).
Finally, another important question is whether requirements for each of these multivalent p53-STAGA interactions show any target gene specificity. In this regard, gene-specific variations in AD1 and AD2 requirements have been noted (14, 33, 73, 74).
Acknowledgments
We thank Sohail Malik for valuable comments on the manuscript and Yoshihiro Nakatani, Xinbin Chen, and Laszlo Tora for providing reagents.
This work was supported in part by NIH Grants CA129325, CA113822, and DK071900. A.M.G. was a Burroughs Wellcome Fellow.
Footnotes
Published ahead of print on 4 February 2008.
REFERENCES
- 1.An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117735-748. [DOI] [PubMed] [Google Scholar]
- 2.Ard, P. G., C. Chatterjee, S. Kunjibettu, L. R. Adside, L. E. Gralinski, and S. B. McMahon. 2002. Transcriptional regulation of the mdm2 oncogene by p53 requires TRRAP acetyltransferase complexes. Mol. Cell. Biol. 225650-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baptiste, N., P. Friedlander, X. Chen, and C. Prives. 2002. The proline-rich domain of p53 is required for cooperation with anti-neoplastic agents to promote apoptosis of tumor cells. Oncogene 219-21. [DOI] [PubMed] [Google Scholar]
- 4.Barlev, N. A., A. V. Emelyanov, P. Castagnino, P. Zegerman, A. J. Bannister, M. A. Sepulveda, F. Robert, L. Tora, T. Kouzarides, B. K. Birshtein, and S. L. Berger. 2003. A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol. Cell. Biol. 236944-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 81243-1254. [DOI] [PubMed] [Google Scholar]
- 6.Bochkareva, E., L. Kaustov, A. Ayed, G. S. Yi, Y. Lu, A. Pineda-Lucena, J. C. Liao, A. L. Okorokov, J. Milner, C. H. Arrowsmith, and A. Bochkarev. 2005. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl. Acad. Sci. USA 10215412-15417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bode, A. M., and Z. Dong. 2004. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 4793-805. [DOI] [PubMed] [Google Scholar]
- 8.Brand, M., J. G. Moggs, M. Oulad-Abdelghani, F. Lejeune, F. J. Dilworth, J. Stevenin, G. Almouzni, and L. Tora. 2001. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 203187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 27418285-18289. [DOI] [PubMed] [Google Scholar]
- 10.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 2922333-2337. [DOI] [PubMed] [Google Scholar]
- 11.Candau, R., P. A. Moore, L. Wang, N. Barlev, C. Y. Ying, C. A. Rosen, and S. L. Berger. 1996. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol. Cell. Biol. 16593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Candau, R., D. M. Scolnick, P. Darpino, C. Y. Ying, T. D. Halazonetis, and S. L. Berger. 1997. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene 15807-816. [DOI] [PubMed] [Google Scholar]
- 13.Chao, C., M. Hergenhahn, M. D. Kaeser, Z. Wu, S. Saito, R. Iggo, M. Hollstein, E. Appella, and Y. Xu. 2003. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J. Biol. Chem. 27841028-41033. [DOI] [PubMed] [Google Scholar]
- 14.Cregan, S. P., N. A. Arbour, J. G. Maclaurin, S. M. Callaghan, A. Fortin, E. C. Cheung, D. S. Guberman, D. S. Park, and R. S. Slack. 2004. p53 activation domain 1 is essential for PUMA upregulation and p53-mediated neuronal cell death. J. Neurosci. 2410003-10012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuatrecasas, P., and I. Parikh. 1972. Adsorbents for affinity chromatography. Use of N-hydroxysuccinimide esters of agarose. Biochemistry 112291-2299. [DOI] [PubMed] [Google Scholar]
- 16.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 111475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Lello, P., L. M. Jenkins, T. N. Jones, B. D. Nguyen, T. Hara, H. Yamaguchi, J. D. Dikeakos, E. Appella, P. Legault, and J. G. Omichinski. 2006. Structure of the Tfb1/p53 complex: insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol. Cell 22731-740. [DOI] [PubMed] [Google Scholar]
- 18.el-Deiry, W. S., T. Tokino, T. Waldman, J. D. Oliner, V. E. Velculescu, M. Burrell, D. E. Hill, E. Healy, J. L. Rees, S. R. Hamilton, et al. 1995. Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer Res. 552910-2919. [PubMed] [Google Scholar]
- 19.Espinosa, J. M., R. E. Verdun, and B. M. Emerson. 2003. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol. Cell 121015-1027. [DOI] [PubMed] [Google Scholar]
- 20.Fields, S., and S. K. Jang. 1990. Presence of a potent transcription activating sequence in the p53 protein. Science 2491046-1049. [DOI] [PubMed] [Google Scholar]
- 21.Flinn, E. M., A. E. Wallberg, S. Hermann, P. A. Grant, J. L. Workman, and A. P. Wright. 2002. Recruitment of Gcn5-containing complexes during c-Myc-dependent gene activation. Structure and function aspects. J. Biol. Chem. 27723399-23406. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106297-307. [DOI] [PubMed] [Google Scholar]
- 23.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 111640-1650. [DOI] [PubMed] [Google Scholar]
- 24.Grossman, S. R. 2001. p300/CBP/p53 interaction and regulation of the p53 response. Eur. J. Biochem. 2682773-2778. [DOI] [PubMed] [Google Scholar]
- 25.Gu, W., S. Malik, M. Ito, C. X. Yuan, J. D. Fondell, X. Zhang, E. Martinez, J. Qin, and R. G. Roeder. 1999. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell 397-108. [DOI] [PubMed] [Google Scholar]
- 26.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90595-606. [DOI] [PubMed] [Google Scholar]
- 27.Gu, W., X. L. Shi, and R. G. Roeder. 1997. Synergistic activation of transcription by CBP and p53. Nature 387819-823. [DOI] [PubMed] [Google Scholar]
- 28.He, Z., B. T. Brinton, J. Greenblatt, J. A. Hassell, and C. J. Ingles. 1993. The transactivator proteins VP16 and GAL4 bind replication factor A. Cell 731223-1232. [DOI] [PubMed] [Google Scholar]
- 29.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102463-473. [DOI] [PubMed] [Google Scholar]
- 30.Ito, M., C. X. Yuan, S. Malik, W. Gu, J. D. Fondell, S. Yamamura, Z. Y. Fu, X. Zhang, J. Qin, and R. G. Roeder. 1999. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell 3361-370. [DOI] [PubMed] [Google Scholar]
- 31.Jabbur, J. R., A. D. Tabor, X. Cheng, H. Wang, M. Uesugi, G. Lozano, and W. Zhang. 2002. Mdm-2 binding and TAF(II)31 recruitment is regulated by hydrogen bond disruption between the p53 residues Thr18 and Asp21. Oncogene 217100-7113. [DOI] [PubMed] [Google Scholar]
- 32.Johnson, T. M., and L. D. Attardi. 2005. p53QS: an old mutant teaches us new tricks. Cell Cycle 4731-734. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, T. M., E. M. Hammond, A. Giaccia, and L. D. Attardi. 2005. The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat. Genet. 37145-152. [DOI] [PubMed] [Google Scholar]
- 34.Kaeser, M. D., and R. D. Iggo. 2004. Promoter-specific p53-dependent histone acetylation following DNA damage. Oncogene 234007-4013. [DOI] [PubMed] [Google Scholar]
- 35.Kastan, M. B., Q. Zhan, W. S. el-Deiry, F. Carrier, T. Jacks, W. V. Walsh, B. S. Plunkett, B. Vogelstein, and A. J. Fornace, Jr. 1992. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71587-597. [DOI] [PubMed] [Google Scholar]
- 36.Kaustov, L., G. S. Yi, A. Ayed, E. Bochkareva, A. Bochkarev, and C. H. Arrowsmith. 2006. p53 transcriptional activation domain: a molecular chameleon? Cell Cycle 5489-494. [DOI] [PubMed] [Google Scholar]
- 37.Kessis, T. D., R. J. Slebos, W. G. Nelson, M. B. Kastan, B. S. Plunkett, S. M. Han, A. T. Lorincz, L. Hedrick, and K. R. Cho. 1993. Human papillomavirus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proc. Natl. Acad. Sci. USA 903988-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein, J., M. Nolden, S. L. Sanders, J. Kirchner, P. A. Weil, and K. Melcher. 2003. Use of a genetically introduced cross-linker to identify interaction sites of acidic activators within native transcription factor IID and SAGA. J. Biol. Chem. 2786779-6786. [DOI] [PubMed] [Google Scholar]
- 39.Kumar, A., Y. Zhao, G. Meng, M. Zeng, S. Srinivasan, L. M. Delmolino, Q. Gao, G. Dimri, G. F. Weber, D. E. Wazer, H. Band, and V. Band. 2002. Human papillomavirus oncoprotein E6 inactivates the transcriptional coactivator human ADA3. Mol. Cell. Biol. 225801-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuo, M. H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 61309-1320. [DOI] [PubMed] [Google Scholar]
- 41.Lambert, P. F., F. Kashanchi, M. F. Radonovich, R. Shiekhattar, and J. N. Brady. 1998. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 27333048-33053. [DOI] [PubMed] [Google Scholar]
- 42.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 151946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Latonen, L., Y. Taya, and M. Laiho. 2001. UV-radiation induces dose-dependent regulation of p53 response and modulates p53-HDM2 interaction in human fibroblasts. Oncogene 206784-6793. [DOI] [PubMed] [Google Scholar]
- 44.Lin, J., J. Chen, B. Elenbaas, and A. J. Levine. 1994. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to Mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 81235-1246. [DOI] [PubMed] [Google Scholar]
- 45.Liu, G., T. Xia, and X. Chen. 2003. The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREB-binding protein. J. Biol. Chem. 27817557-17565. [DOI] [PubMed] [Google Scholar]
- 46.Liu, X., J. Tesfai, Y. A. Evrard, S. Y. Dent., and E. Martinez. 2003. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J. Biol. Chem. 27820405-20412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lomant, A. J., and G. Fairbanks. 1976. Chemical probes of extended biological structures: synthesis and properties of the cleavable protein cross-linking reagent [35S]dithiobis(succinimidyl propionate). J. Mol. Biol. 104243-261. [DOI] [PubMed] [Google Scholar]
- 48.Lu, H., and A. J. Levine. 1995. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc. Natl. Acad. Sci. USA 925154-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo, J., F. Su, D. Chen, A. Shiloh, and W. Gu. 2000. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408377-381. [DOI] [PubMed] [Google Scholar]
- 50.Martinez, E., T. K. Kundu, J. Fu, and R. G. Roeder. 1998. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J. Biol. Chem. 27323781-23785. [DOI] [PubMed] [Google Scholar]
- 51.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 216782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schiltz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 9435-44. [DOI] [PubMed] [Google Scholar]
- 53.O'Rourke, R. W., C. W. Miller, G. J. Kato, K. J. Simon, D. L. Chen, C. V. Dang, and H. P. Koeffler. 1990. A potential transcriptional activation element in the p53 protein. Oncogene 51829-1832. [PubMed] [Google Scholar]
- 54.Palhan, V. B., S. Chen, G. H. Peng, A. Tjernberg, A. M. Gamper, Y. Fan, B. T. Chait, A. R. La Spada, and R. G. Roeder. 2005. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc. Natl. Acad. Sci. USA 1028472-8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park, J. H., and R. G. Roeder. 2006. GAS41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation. Mol. Cell. Biol. 264006-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reeves, W. M., and S. Hahn. 2005. Targets of the Gal4 transcription activator in functional transcription complexes. Mol. Cell. Biol. 259092-9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 631129-1136. [DOI] [PubMed] [Google Scholar]
- 58.Sluss, H. K., H. Armata, J. Gallant, and S. N. Jones. 2004. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol. Cell. Biol. 24976-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thut, C. J., J. L. Chen, R. Klemm, and R. Tjian. 1995. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science 267100-104. [DOI] [PubMed] [Google Scholar]
- 60.Uesugi, M., and G. L. Verdine. 1999. The alpha-helical FXXΦΦ motif in p53: TAF interaction and discrimination by MDM2. Proc. Natl. Acad. Sci. USA 9614801-14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vassilev, A., J. Yamauchi, T. Kotani, C. Prives, M. L. Avantaggiati, J. Qin, and Y. Nakatani. 1998. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol. Cell 2869-875. [DOI] [PubMed] [Google Scholar]
- 62.Venot, C., M. Maratrat, V. Sierra, E. Conseiller, and L. Debussche. 1999. Definition of a p53 transactivation function-deficient mutant and characterization of two independent p53 transactivation subdomains. Oncogene 182405-2410. [DOI] [PubMed] [Google Scholar]
- 63.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408307-310. [DOI] [PubMed] [Google Scholar]
- 64.Wieczorek, E., M. Brand, X. Jacq, and L. Tora. 1998. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393187-191. [DOI] [PubMed] [Google Scholar]
- 65.Wu, P. Y., C. Ruhlmann, F. Winston, and P. Schultz. 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15199-208. [DOI] [PubMed] [Google Scholar]
- 66.Xiao, H., A. Pearson, B. Coulombe, R. Truant, S. Zhang, J. L. Regier, S. J. Triezenberg, D. Reinberg, O. Flores, C. J. Ingles, and et al. 1994. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol. Cell. Biol. 147013-7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu, W., D. G. Edmondson, Y. A. Evrard, M. Wakamiya, R. R. Behringer, and S. Y. Roth. 2000. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26229-232. [DOI] [PubMed] [Google Scholar]
- 68.Xu, W., D. G. Edmondson, and S. Y. Roth. 1998. Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol. Cell. Biol. 185659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamauchi, T., J. Yamauchi, T. Kuwata, T. Tamura, T. Yamashita, N. Bae, H. Westphal, K. Ozato, and Y. Nakatani. 2000. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 9711303-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yanagisawa, J., H. Kitagawa, M. Yanagida, O. Wada, S. Ogawa, M. Nakagomi, H. Oishi, Y. Yamamoto, H. Nagasawa, S. B. McMahon, M. D. Cole, L. Tora, N. Takahashi, and S. Kato. 2002. Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Mol. Cell 9553-562. [DOI] [PubMed] [Google Scholar]
- 71.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382319-324. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, W., X. Y. Guo, G. Y. Hu, W. B. Liu, J. W. Shay, and A. B. Deisseroth. 1994. A temperature-sensitive mutant of human p53. EMBO J. 132535-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu, J., S. Zhang, J. Jiang, and X. Chen. 2000. Definition of the p53 functional domains necessary for inducing apoptosis. J. Biol. Chem. 27539927-39934. [DOI] [PubMed] [Google Scholar]
- 74.Zhu, J., W. Zhou, J. Jiang, and X. Chen. 1998. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J. Biol. Chem. 27313030-13036. [DOI] [PubMed] [Google Scholar]