Abstract
The development of a highly branched dendritic tree is essential for the establishment of functional neuronal connections. The evolutionarily conserved immunoglobulin superfamily member, the protein dendrite arborization and synapse maturation 1 (Dasm-1) is thought to play a critical role in dendrite formation of dissociated hippocampal neurons. RNA interference-mediated Dasm-1 knockdown was previously shown to impair dendrite, but not axonal, outgrowth and branching (S. H. Shi, D. N. Cox, D. Wang, L. Y. Jan, and Y. N. Jan, Proc. Natl. Acad. Sci. USA 101:13341-13345, 2004). Here, we report the generation and analysis of Dasm-1 null mice. We find that genetic ablation of Dasm-1 does not interfere with hippocampal dendrite growth and branching in vitro and in vivo. Moreover, the absence of Dasm-1 does not affect the modulation of dendritic outgrowth induced by brain-derived neurotrophic factor. Importantly, the previously observed impairment in dendrite growth after Dasm-1 knockdown is also observed when the Dasm-1 knockdown is performed in cultured hippocampal neurons from Dasm-1 null mice. These findings indicate that the dendrite arborization phenotype was caused by off-target effects and that Dasm-1 is dispensable for hippocampal dendrite arborization.
Neurons are polarized cells that often grow highly branched dendrites that serve as the input compartment and an axon that mediates the output. Proper development of the dendritic tree is essential for establishing connections between neurons and for receiving and computing their signals (20). Dendritic arborization and synaptic partner choice are controlled by intrinsic and extrinsic factors. Among the latter, cell surface molecules appear particularly important. The Down syndrome-related cell adhesion molecule (Dscam), which in the fly is expressed in thousands of different isoforms, promotes repulsive interactions between the dendrites of olfactory projection neurons and thus ensures proper spacing of dendrites and complete coverage of the dendritic field (14-16, 19, 25). The homophilic cell adhesion molecule, N-cadherin, mediates dendro-dendritic interactions between olfactory projection neurons and thus helps to refine their dendrites to single glomeruli (28). Sidekicks (Sdks) are immunoglobulin superfamily (IgSF) members that mediate homophilic adhesion and synaptic connectivity between retinal ganglion cell dendrites and their presynaptic partner neurons (27). Other extrinsic factors include brain-derived neurotrophic factor (BDNF), which stimulates dendritic growth of cultured hippocampal and cortical neurons and maintains cortical dendrites in vivo (4, 5, 13, 18). Despite recent progress, the molecular cues and pathways that regulate dendrite arborization and network formation are still poorly understood.
The Drosophila transmembrane IgSF protein Turtle (tutl) had previously been shown to be essential for establishing neuronal circuits capable of executing coordinated motor output. Specifically, tutl mutants were unable to regain an upright position when inverted (hence, the name “turtle”), and they were unable to fly in adulthood (2). The overall morphology of the nervous system, basal synaptic transmission, and locomotor movements were normal in tutl mutants, raising a number of questions regarding the mechanisms by which tutl mediates complex behaviors. Based on the initial report, tutl apparently does not play a role in axon pathfinding or nervous system morphogenesis.
The mammalian homologue of tutl was originally cloned and named IgSF9 (7); the protein was recently renamed dendrite arborization and synapse maturation protein 1 (Dasm-1) (22, 23). Dasm-1 was shown to be expressed in the developing nervous system and more specifically in the dendrites of cultured rat hippocampal neurons (23). Suppression of Dasm-1 expression by RNA interference (RNAi) impaired dendrite but not axonal growth in vitro (23). In a parallel study the same authors showed that Dasm-1 was localized at excitatory synapses of hippocampal neurons and controlled excitatory synapse maturation in hippocampal organotypic slice cultures (22). Dasm-1 was shown to regulate synaptic α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) via its C-terminal PDZ interaction site, which interacted with synaptic PDZ domain-containing proteins. The current view is therefore that Dasm-1 acts as a neuronal cell surface receptor (10). The identity and the source of the Dasm-1 ligand are, however, unknown. The molecular mechanism by which Dasm-1 regulates dendrite development and/or synapse maturation also remains to be established. Moreover, the in vivo implications of the effects of Dasm-1 on dendrite growth displayed in culture assay need to be understood.
To begin investigating Dasm-1's function in vivo, we generated knockout mice. We found no defects in dendrite arborization in Dasm-1−/− hippocampal neurons in vitro or in vivo. In addition Dasm-1 seems also not to be important for the accelerated dendritic growth observed in the presence of BDNF. This is in contrast to the previously reported Dasm-1 RNAi experiments (22, 23), raising the question of functional compensation in the knockouts compared to a more acute knock-down by RNAi or off-target effects of the previously reported RNAi study. The data presented here clearly demonstrate off-target effects of the RNAi constructs used and suggest that Dasm-1 is either not required for dendrite arborization or that the lack of Dasm-1 can be compensated by other outgrowth and branching factors.
MATERIALS AND METHODS
Construction of the Dasm-1 targeting vector and generation of mutant mice.
A 6-kb Dasm-1 genomic fragment upstream of exon 4 and a 2.4-kb fragment downstream of exon 9 were amplified from 129/Sv mouse DNA by PCR. Primers were designed to incorporate restriction enzyme cleavage sites (NotI for the 6-kb fragment; SalI and KpnI for the 2.4-kb fragment) for ligation into the targeting vector. After amplification by PCR, all exons and exon-intron borders were sequenced. The 6-kb and 2.4-kb fragments were inserted separately as long and short homology arms, respectively, into a modified pBluescript vector, which contained a loxP site-flanked neomycin cassette. The targeting vector was linearized with PvuI and electroporated into embryonic day 14 (E14) embryonic stem (ES) cells. Resistant cells were selected in the presence of G418, DNA was isolated, and homologous recombinants were screened by Southern blotting. Genomic DNA was digested with SpeI and identified with probe 1, a specific PCR fragment of 600 bases located downstream of the targeted Dasm-1 locus. Two clones were injected in C57BL/6 blastocysts, which were subsequently transferred into pseudo-pregnant females to generate chimeric offspring. The chimeras were crossed into C57BL/6 mice for germ line transmission. The null mutants (Dasm-1−/−) were generated by intercrossing heterozygous Dasm-1 mice (Dasm-1+/−). To visualize individual hippocampal neurons, Dasm-1+/− mice were crossed with transgenic mice expressing green fluorescent protein (GFP) under the Thy1 promoter (11). All animal use was carried out in compliance with institutional and appropriate governmental policies.
Genotyping of mutant mice.
The genotypes of mutant mice were determined by PCR and confirmed by Southern blot analysis of genomic DNA prepared from tail biopsy samples. Routine genotyping through PCR was performed with genomic DNA using specific oligonucleotide primers for the Dasm-1 wild-type allele (F1, 5′-ACTAC TGTTTGTCACCTGGACCAAAGACGG-3′; and R1, 5′-CAATCAACTCGGAATG AGGTCATGTTAAGC-3′). For the targeted allele, a primer was designed from within the neomycin resistance gene (N1, 5′-TTATTAGGTCCCTCGACCTGCAGCCCAAGC-3′). The three primers were used in a multiplex PCR with the following amplification conditions: 94°C for 3 min, and 30 cycles of 94°C for 30 s, 61°C for 30 s, and 72°C for 40 s. Amplification products were resolved on a 2% agarose gel. The PCR product was 582 bp for the wild-type allele and 450 bp for the targeted allele. The presence of the Thy1-GFP transgene was detected by using specific primers for GFP (F, 5′-GCGGATCTTGAAGTTCACCTTGATGCC-3′; R, 5′-GCACGACTTCTTCAAGTCCGCCATGCC-3′) with the following amplification conditions: 94°C for 2 min and 35 cycles of 94°C for 1 min, 68°C for 1 min, and 72°C for 1 min.
Reverse transcription-PCR analysis.
Total cellular RNA was isolated from brains of postnatal day 0 (P0) mice with RNAzol reagent (Invitrogen). Two micrograms of this RNA was transcribed into cDNA (Superscript II; Invitrogen) before being amplified with primers specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (forward, 5′-AGTATGACTCCACTCACGGCA-3′; reverse, 5′-GAGGGGCCATCCACAGTCT-3′), EphA4 (forward, 5′-GAGAAACATTTGAATCAAGGTGTTAGAACTTAT-3′; reverse, 5′-CTCCATGTACTCCGTTATGATCATTACC-3′), and Dasm-1 (forward, 5′-AGACAGGAGCATCTCTCCAGATTGAGG-3′; reverse, 5′-AGCTGCAAGGCCTGTCCGTCTTTGG-3′). The following amplification protocol was used: 95°C for 3 min and 35 cycles of 95°C for 30 s plus 1 min at 60°C (GAPDH), 67°C (EphA4), or 62°C (Dasm-1), followed by 1 min at 72°C.
Antibody production.
To generate Dasm-1-specific antibodies the C-terminal part of the Dasm-1 cDNA encoding the last 100 amino acids (C100) of Dasm-1 was subcloned into pET-28 plasmid vector from Novagen. The fusion protein His6-Dasm-1(C100) was purified and used to immunize rabbits. The IgG-enriched fraction of the polyclonal rabbit antiserum was used to detect Dasm-1 protein by Western blotting.
Western blot analysis.
Protein extracts for immunoblotting were obtained by homogenizing brain tissue or HeLa cells in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% Triton X-100) with protease inhibitors (Complete; Roche). Lysates were loaded (50 or 25 μg/lane) and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted to nylon membranes, and developed using enhanced chemiluminescence (Amersham) following manufacturer's instructions. For the lectin pull-down, lysates (500 μg) were first incubated with lectin-agarose beads (Sigma) for 3 h at 4°C. Beads were washed, boiled under denaturing conditions, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The following antibodies were used for immunodetection: anti-Map2 (1:1,000; Sigma), anti-α-tubulin (1:20,000; Sigma), anti-Dasm-1 (1:1,000), anti-FLAG (1:1,000; Sigma), and horseradish peroxidase-conjugated goat anti-mouse and goat anti-rabbit IgG (Jackson ImmunoResearch).
In situ hybridization.
For in situ hybridization, P0 and adult mice were deeply anesthetized, and cardiac perfusion with 4% paraformaldehyde was performed. Whole brains were removed, postfixed overnight, sectioned using a vibratome (75-μm thickness), and used for hybridization. The N-terminal Dasm-1 PCR fragment (1 kb) was subcloned into pCRII-TOPO plasmid vector (Invitrogen). Digoxigenin (DIG)-labeled Dasm-1 RNA antisense/sense probes were transcribed from EcoRV/HindIII-linearized constructs in vitro using SP6/T7 RNA polymerases, respectively (DIG RNA labeling kit; Boehringer Mannheim), according to the manufacturer's instructions. Hybridized RNA was detected using alkaline phosphatase-conjugated anti-DIG antibody (Roche Diagnostics).
Histology and immunohistochemistry.
For histology, mice were perfused with phosphate-buffered saline (PBS) and 4% paraformaldehyde. Subsequently, brains were removed from the skull and postfixed overnight in the same fixative. Immunostaining was performed on 60-μm thick, coronal, free-floating, vibratome sections. The sections were blocked for 1 h in 5% bovine serum albumin and 5% donkey serum-0.3% Triton X-100 in PBS and incubated with the first antibody (anti-Map2; Sigma) diluted 1:200 in the blocking solution at 4°C overnight. The sections were washed three times in PBS with 0.1% Triton X-100 for 15 min and incubated in biotinylated secondary antibody (1:300 anti-mouse; Vectastain) for 2 h at room temperature. For diaminobenzidine detection of the secondary antibody, a Vectastain ABC kit (Vector Laboratories) was used according to the provider's instructions.
To visualize hippocampal neurons in vivo, Dasm-1+/− mice were crossed with a transgenic mouse line (GFP-M) to give Dasm-1+/−; GFP-M mice that when back-crossed with Dasm-1+/− mice, produced GFP-M control and Dasm-1−/−; GFP-M littermates. Mice were perfused with PBS and 4% paraformaldehyde, brains were dissected, and 200-μm thick, sagittal vibratome sections were collected and mounted using aqueous mounting medium with antifading reagent (Biomedia). Fluorescence images of individual CA1 pyramidal neurons and groups of dentate gyrus granule neurons were acquired with a TCS SP2 confocal microscope (Leica Microsystems) using a 40× objective with a numerical aperture of 1.25 (HCX Plan-Apochromat CS oil; Leica Microsystems).
Golgi-Cox staining was performed on freshly dissected brains using an FD Rapid GolgiStain Kit (FD NeuroTechnologies, Inc.) following the manufacturer's instructions. Dentate gyrus granule neurons were traced using Neurolucida software (MBF Bioscience).
RNAi and dominant negative constructs.
Three different Dasm-1 RNAi plasmids against different regions of Dasm-1 were tested for a strong and specific effect on suppressing Dasm-1 protein expression in HeLa cells. Dasm-1 RNAi plasmids were constructed as follows. Two oligonucleotides that form a hairpin structure corresponding to Dasm-1 coding nucleotides (NCBI accession number AF317839) 69 to 87 (5′-GATCCCCgcctgaggtggtctctgtgTTCAAGAGAcacagagaccacctcaggcTTTTTA-3′ and 5′-AGCTTAAAAAgcctgaggtggtctctgtgTCTCTTGAAcacagagaccacctcaggcGGG-3′), 868 to 886 (5′-GATCCCCgccactcagcctgatgacgTTCAAGAGAcgtcatcaggctgagtggcTTTTTA-3′ and 5′-AGCTTAAAAAgccactcagcctgatgacgTCTCTTGAAcgtcatcaggctgagtggcGGG-3′) and 3341 to 3359 (5′-GATCCCCcttgagccagagttaggtgTTCAAGAGAcacctaactctggctcaagTTTTTA-3′ and 5′-AGCTTAAAAActtgagccagagttaggtgTCTCTTGAAcacctaactctggctcaagGGG-3′) were annealed together, and these fragments were individually inserted into pSUPER plasmid vector (OligoEngine) (uppercase letters indicate sequence from this plasmid; lowercase letters indicate residues homologous to Dasm-1). The nucleotide BLAST search did not show any significant similarity between the Dasm-1 short hairpin RNA (shRNA) sequences and IgSF9b, a close homolog of Dasm-1, showing that the selected sequences are not conserved in the two proteins. As previously reported (22), the cytoplasmic tail deletion mutant (Dasm-1delC-EGFP) (where EGFP is enhanced GFP) was generated by amplifying the first 795 amino acids by PCR and cloning into pEGFP-N1 (Clontech). The cDNA encoding the last 200 amino acids (979 to 1178) of the Dasm-1 cytoplasmic tail was amplified by PCR and cloned into p3×FLAG-CMV-9 (Sigma) with FLAG tags fused to its N terminus (FLAG-C200) to preserve the type I PDZ domain-binding motif at the C terminus. All of the constructs were confirmed by sequencing.
Hippocampal neuron cultures and transfections.
Hippocampi from E18 mice were separated from diencephalic structures and digested with 0.25% trypsin containing 1 mM EDTA for 20 min at 37°C. Neurons were plated in 24-well plates (40,000 per well) on coverslips coated with poly-d-lysine (1 mg/ml) and laminin (5 μg/ml) in neurobasal medium supplemented with B27 containing 0.5 mM glutamine (all reagents are from Invitrogen). Neurons were cultured for 5 or 10 days in vitro (DIV), after which they were fixed and stained with anti-Map2 antibody. Transfection of dissociated neurons in culture with different Dasm-1 RNAi plasmids, FLAG-C200, and EGFP was carried out using a calcium phosphate-DNA precipitation procedure after 2 DIV. Neurons were fixed after 8 DIV and stained with anti-Map2 and anti-GFP antibodies. Each experiment was repeated at least three times.
Immunocytochemistry.
Neurons were fixed with 4% paraformaldehyde and 4% sucrose in PBS for 20 min at 4°C, rinsed twice with PBS, and then incubated with 50 mM NH4Cl in PBS for 10 min at 4°C and rinsed twice again before permeabilization for 5 min with ice-cold 0.1% Triton X-100 in PBS. After a washing step, cells were blocked for 30 min at room temperature (25°C) with 2% bovine serum albumin and 4% donkey serum, 4% goat serum, or both (Jackson ImmunoResearch) and incubated with primary and secondary antibodies for 60 and 30 min, respectively. Samples were mounted using the Gel Mount antifading medium (Biomeda). The following antibodies were used: mouse anti-Map2 (1:1,000; Sigma), rabbit anti-GFP (1:2,000; RDI), Alexa Fluor 488-conjugated goat anti-rabbit (1:500; Molecular Probes), and Cy3-conjugated donkey anti-mouse (1:500; Jackson ImmunoResearch) antibodies. Images were acquired using a digital camera (SpotRT; Diagnostic Instruments) attached to an epifluorescence microscope (Zeiss) equipped with a 40× objective (Plan-Apochromat; Zeiss).
Image analysis and quantification.
All quantitative measurements were performed using Neurolucida, Neuroexplorer (MBF Bioscience), and MetaMorph software (Molecular Devices). The maximum projection of the confocal stacks of hippocampal neurons in the CA1 and the dentate gyrus region were used for the analysis. Apical dendrites of individual CA1 pyramidal neurons, Golgi-stained dentate gyrus granule neurons, and in vitro cultured hippocampal neurons were traced using Neurolucida software. Neuronal tracings were subjected to Sholl analysis using Neuroexplorer software. In order to quantitate the area covered by fluorescent dendrites, the images of denate gyrus granule neurons were first thresholded by subtracting the background fluorescence to improve the signal-to-noise ratio. The part of the total area (marked by dashed lines in Fig. 2L and M) that was covered by the fluorescent dendrites was calculated using Metamorph software and was divided by the number of cell bodies to give the percent dendrite area per granule neuron in each image. This was used as a measure of the dendrite arbor elaboration for granule neurons (see Fig. 2N). All neuronal tracings and image analyses were done by an investigator blind to the genotype. Statistical analysis was performed using Microsoft Excel. Dendritic parameters from different groups of neurons were compared using a Student's t test. Values are represented as means ± standard error of means.
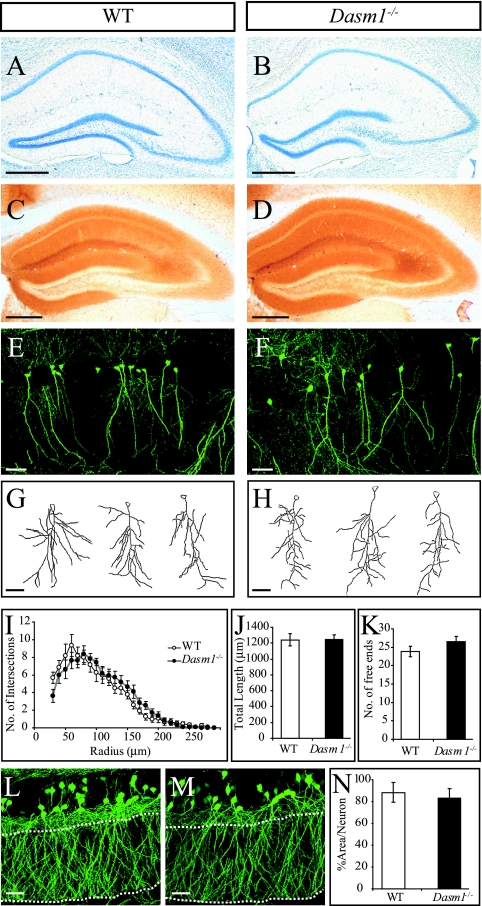
FIG. 2.
Normal dendritic arborization in Dasm-1−/− hippocampal neurons in vivo. (A and B) Nissl-stained adult (P45) hippocampi of wild-type (WT) and Dasm-1−/− mice. (C and D) Map2 immunostaining to visualize overall dendrite density and organization in P45 hippocampus. (E and F) GFP-fluorescent CA1 pyramidal neurons in the P45 hippocampi of GFP-M control and Dasm-1−/−; GFP-M mice. (G and H) Representative tracings of the apical dendrites of GFP-M control and Dasm-1−/−; GFP-M CA1 pyramidal neurons. (I) Sholl analysis of GFP-M control (WT) and Dasm-1−/−; GFP-M (Dasm-1−/−) CA1 neurons revealing similar dendrite organization. The total dendritic length (J) and the number of free dendritic ends of CA1 neurons as a measure of the complexity of the dendritic arbor (K) are comparable in the GFP-M control and Dasm-1−/−; GFP-M mice. Representative photomicrographs of the dentate gyrus region of the hippocampus in the GFP-M control (L) and Dasm-1−/−; GFP-M (M) mice are shown. (N) Quantitative measurement comparing the percentage of area covered by the fluorescent dendrites of a granule neuron in the GFP-M control and Dasm-1−/−; GFP-M mice (analysis was done in the region delineated by the dashed lines). Scale bars, 500 μm (A to D) and 50 μm (E to H, L, and M). The error bars indicate standard errors of the mean.
RESULTS
Gene targeting of Dasm-1.
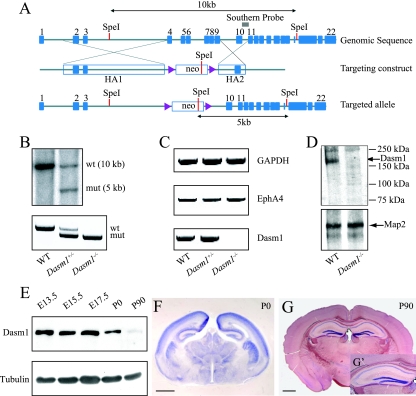
To generate Dasm-1-deficient mice (Dasm-1−/−), we used a targeting construct carrying a deletion of 2.7 kb of genomic DNA including exons 4 to 9 of the mouse Dasm-1 gene (Fig. 1A) which partially encode Ig domains 1 and 4 and completely encode Ig domains 2 and 3 of the Dasm-1 ectodomain. We screened targeted ES cell clones by Southern blot analysis and established a genotyping PCR (Fig. 1B). We then confirmed the absence of Dasm-1 mRNA in P0 brain lysates of Dasm-1−/− mice (Fig. 1C) and of Dasm-1 protein in E15 brain extracts of Dasm-1−/− mice (Fig. 1D). The use of an antiserum specific for the most C-terminal 100 amino acids also confirmed that Dasm-1−/− mice did not produce detectable amounts of a truncated Dasm-1 protein lacking most of its ectodomain (expected size of ∼89 kDa). A developmental profile of Dasm-1 expression in brain protein lysates from E13.5 to P90 showed high expression of Dasm-1 during mid- to late gestation and a decrease in postnatal stages (Fig. 1E). The spatial distribution of Dasm-1 expression was revealed by in situ hybridization. Dasm-1 mRNA expression was widespread in the newborn brain (Fig. 1F) and highest in hippocampal CA1 and dentate gyrus in adult brain (Fig. 1G and G′). Labeling with the corresponding Dasm-1 sense probe did not show any staining (data not shown).
FIG. 1.
Gene targeting and expression analysis of Dasm-1. (A) Schematic representation of the targeting strategy to generate Dasm-1−/− mice. At top is the wild-type locus. Exons (blue boxes) are shown. The diagram in the middle represents the targeting strategy. A loxP PGKneobpA-loxP cassette (indigo blue triangles represent the loxP sites) was inserted by homologous recombination replacing the region spanning exons 4 to 9. In the targeting construct, flanking homology arms (HA) were included that extended ∼6.0 kb 5′ (HA1) and ∼2.4 kb 3′ (HA2) to the PGKneobpA cassette. The bottom diagram represents the targeted allele. (B) Homologous recombination was verified by Southern hybridization with ES cell genomic DNA after digestion with SpeI. A radioactive Southern probe (indicated by the gray box in panel A) detected a wild-type (wt) band (∼10.0 kb) and a mutant (mut) band (∼5.0 kb) in a clone of transfected ES cells (+/−). The bottom panel shows results of a genotyping PCR. The wild-type allele is identified by a ∼600-bp band, and the mutant allele is detected as a ∼450-bp band using a three-primer PCR. (C) Loss of Dasm-1 mRNA in Dasm-1−/− mice. Reverse transcription-PCR from P0 brains shows expression of GAPDH (top), Dasm-1 (bottom), and another unrelated gene, the EphA4 gene (middle), in the wild-type (WT), heterozygous (Dasm-1+/−), and homozygous (Dasm-1−/−) mutants. (D) Lectin pull-down and Western blot analysis of E15 brain extracts show a protein band of the expected size (≥130 kDa) for Dasm-1 in the wild-type mice that was absent in the Dasm-1−/− mice (top). A 280-kDa protein band corresponding to Map2 was present both in the wild-type and Dasm-1−/− mice (bottom). (E) Developmental profile of Dasm-1 expression in the brain is shown by Western blot analysis (top). An equal amount of protein (∼50 μg) was loaded for each lane, and the same sample was reprobed with antitubulin antibody (bottom). (F and G) In situ hybridization analysis of wild-type newborn (P0) and adult (P90) brains using a Dasm-1 antisense probe corresponding to the N-terminal region. Panel G′ is the view of hippocampus at a higher magnification. In the adult, strongest expression is found in the hippocampus including CA1 and dentate gyrus. Scale bar, 1 mm.
Dasm-1−/− mice have normal dendrite arborization patterns in the hippocampus.
Dasm-1−/− mice are viable and fertile and do not show any obvious abnormalities in different genetic backgrounds (C57BL/6 or CD1). We focused our phenotypic analysis on the adult hippocampus where Dasm-1 is highly expressed. Overall, hippocampal architecture and dendritic pattern as revealed by Nissl staining and immunostaining with the dendritic marker Map2 were indistinguishable from controls (Fig. 2A to D). To investigate dendrite arborization of individual hippocampal neurons in vivo, we crossed Dasm-1−/− mice with a transgenic mouse line (GFP-M) which expresses GFP in small neuronal subsets (11). We prepared 200-μm-thick hippocampal slices from P45 brains, acquired fluorescence images with a confocal microscope (Fig. 2E and F), and traced individual CA1 neurons using Neurolucida software (Fig. 2G and H). Traced images were further analyzed for dendritic complexity using Sholl analysis (24). Contrary to our expectations, apical dendrites of wild-type and Dasm-1−/− CA1 neurons were indistinguishable with respect to dendrite branching complexity (Fig. 2I), overall length (1,240.4 ± 76.9 versus 1,248.1 ± 59.2 μm; P > 0.4) (Fig. 2J), and the number of free ends (23.8 ± 1.5 versus 26.5 ± 1.5; P > 0.1 (Fig. 2K). Similar results were obtained from the dentate gyrus. Because individual neurons were difficult to trace, we obtained images from the dendritic regions of several granule neurons, thresholded the images to increase the signal-to-noise ratio, and quantified the area that was covered by fluorescent dendrites, taking into account the number of cell bodies in that area (Fig. 2L and M). The percentages of thresholded area per neuron were not different in the wild-type and Dasm-1−/− hippocampus (88.4% ± 9.1% versus 82.9% ± 9.0%; P > 0.3) (Fig. 2N). We also performed Golgi staining on P90 brains, traced the dendritic trees of wild-type and Dasm-1−/− dentate gyrus granule neurons, and analyzed the degree of branching by Sholl analysis. Dasm-1−/− hippocampal neurons were not impaired in dendritic branching compared to their littermate controls (see Fig. S1 in the supplemental material). These results indicate that Dasm-1 expression is not required for proper dendrite morphogenesis in the hippocampus in vivo.
Normal dendrite organization in Dasm-1−/− hippocampal neurons in culture.
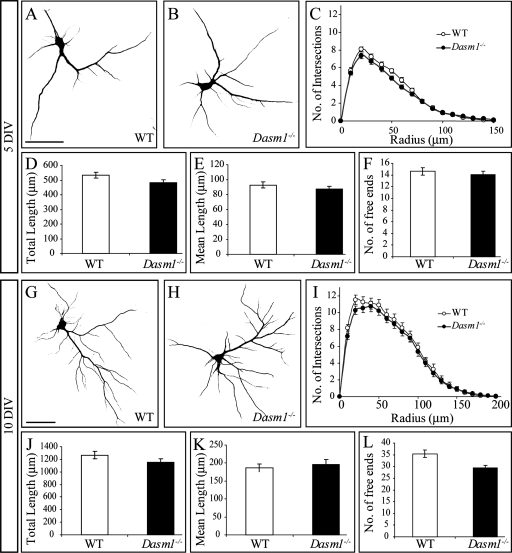
Previous evidence of Dasm-1's function in dendrite organization was largely based on experiments performed in rat dissociated hippocampal neuron cultures (23). To test the possibility that the lack of significant defects in Dasm-1−/− mice was due to compensatory mechanisms in vivo, we investigated dendrite organization of Dasm-1-deficient dissociated mouse hippocampal neurons. Cultures were established from E18 embryos, and neurons were cultured for 5 and 10 DIV. To specifically label dendrites, we stained the neurons with Map2 and used Sholl analysis to measure various dendrite parameters. We found that after 5 DIV, Dasm-1−/− neurons (Fig. 3B) were indistinguishable from wild-type littermate neurons (Fig. 3A) with respect to the number of intersections of dendrites with concentric rings drawn around the cell body (Fig. 3C), to total dendritic branch length (482.5 ± 18.9 versus 532.9 ± 20.0 μm; P > 0.05) (Fig. 3D), to mean dendritic branch length (87.4 ± 4.1 versus 93.0 ± 4.2 μm; P > 0.1) (Fig. 3E), and to the number of free ends (14.0 ± 0.6 versus 14.6 ± 0.6; P > 0.2) (Fig. 3F). Similar results were obtained in older cultures (10 DIV) (Fig. 3G to K) with no significant difference in the total dendritic branch length (1265.4 ± 59.3 versus 1149.1 ± 54.6 μm; P > 0.05) (Fig. 3J) and mean dendritic branch length (186.3 ± 10.6 versus 195.8 ± 13.9 μm; P > 0.2) (Fig. 3K) values between the wild-type and Dasm-1−/− neurons. There was a small (16.9%) yet significant reduction in the number of free ends in Dasm-1−/− neurons compared to wild-type littermate neurons (29.4 ± 1.1 versus 35.4 ± 1.6; P < 0.01) (Fig. 3L). This was mostly due to the shortest dendrites close to the cell body (Fig. 3I). However, in a subsequent series of experiments using 8 DIV neurons, this result was not confirmed (Fig. 4, compare panels L and M), and the numbers of free ends were similar in the wild-type and the Dasm-1−/− neurons (30.9 ± 1.8 versus 31.1 ± 1.5; P > 0.4). Together, these findings indicate that genetic ablation of Dasm-1 does not impair dendrite arborization of cultured hippocampal neurons.
FIG. 3.
Normal dendrite organization in dissociated Dasm-1−/− hippocampal neurons in vitro. (A and B) Representative photomicrographs of Map2-labeled dissociated wild-type (A) and Dasm-1−/− (B) neurons cultured for 5 DIV. (C to F) Morphometric analysis of the dendritic architecture using Sholl analysis. (C) The number of intersections of dendrites with concentric circles drawn around the cell bodies are plotted in relation to their radii (n = 46 for wild-type and n = 59 for Dasm-1−/− neurons). None of the parameters shows significant changes in the mutants. (D) Total dendrite length (P > 0.05). (E) Mean dendrite length (P > 0.1). (F) Number of free ends (P > 0.2). (G and H) Representative photomicrographs of Map2-labeled dissociated wild-type (G) and Dasm-1−/− (H) neurons cultured for 10 DIV. (I) Number of intersections of dendrites with concentric circles in relation to their radii (n = 33 for wild-type and n = 33 for Dasm-1−/− neurons). (J) Total dendrite length (P > 0.05). (K) Mean dendrite length (P > 0.2). (L) Number of free ends (P < 0.01). The number of free ends shows significant changes in this experiment, but this alteration is not consistently observed (Fig. 4). Scale bar, 50 μm. The error bars indicate standard errors of the mean. WT, wild type.
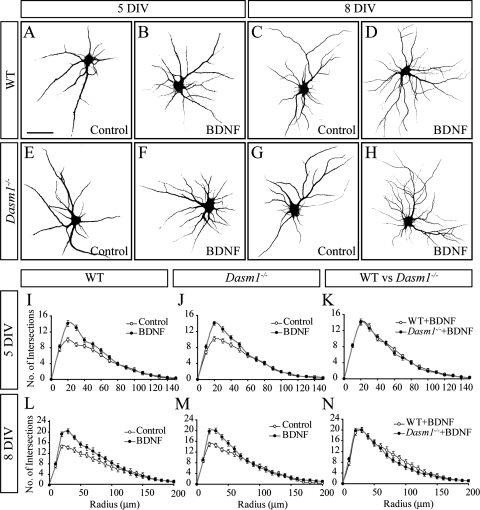
FIG. 4.
Normal BDNF-induced dendritic arborization in Dasm-1−/− hippocampal neurons in vitro. (A to H) Representative photomicrographs are shown of Map2-labeled dissociated wild-type (A to D) and Dasm-1−/− (E to H) neurons in the absence (A, C, E, and G) and presence (B, D, F, and H) of BDNF. (I to N) Sholl analysis of the dendritic complexity of neurons cultured for 5 DIV (I to K) and 8 DIV (L to N). For wild type, n = 24 neurons; for Dasm-1−/− mice, n = 24 neurons. The effects of BDNF are highly significant (for wild-type and Dasm-1−/− neurons, without versus with BDNF, after 5 and 8 DIV, P is <0.0001), whereas the differences between wild-type and Dasm-1−/− neurons are not (for wild-type versus Dasm-1−/− neurons without BDNF, after 5 and 8 DIV, P values are >0.1 and >0.2, respectively; for wild-type versus Dasm-1−/− neurons with BDNF, after 5 and 8 DIV, P values are >0.1 and >0.3, respectively). Values corresponding to radii of 20, 30, and 40 μm are pooled for statistics. Scale bar, 50 μm. The error bars indicate standard errors of the mean.
Normal dendrite organization in BDNF-stimulated Dasm-1−/− hippocampal neurons.
The mechanism of action of Dasm-1 is currently unknown. It seemed plausible that Dasm-1 helps in recruiting or stabilizing the machinery necessary for dendrite branch formation under conditions of accelerated growth, e.g., in the presence of BDNF. We therefore followed the development of cultured hippocampal neurons from Dasm-1−/− embryos and control littermates in the presence of BDNF. In both wild-type and Dasm-1−/− neurons cultured for either 5 or 8 DIV, BDNF caused a significant increase in dendrite complexity close to the cell body (Fig. 4). There was no difference in the pattern (Fig. 4A to H) or magnitude (Fig. 4I to N) of dendrite complexity between wild-type and Dasm-1−/− neurons at either time point, indicating that lack of Dasm-1 did not change the ability of the neurons to respond to BDNF with enhanced dendritic growth or branching.
The effects on dendrite arborization by Dasm-1 RNAi knockdown are due to off-target effects.
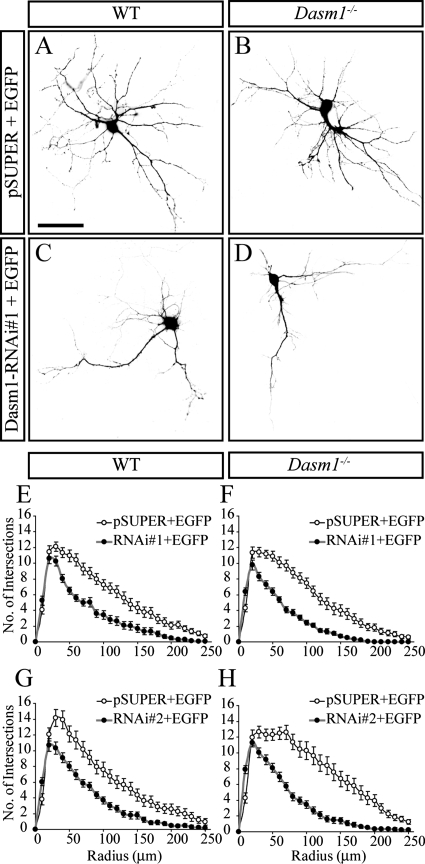
Phenotypic differences between RNAi knockdown and genetic knockout experiments can result from compensations that occur in vivo, e.g., the upregulation of structurally related proteins, and/or from nonspecific (off-target) effects of the RNAi constructs. To distinguish between these possibilities, we decided to perform Dasm-1 knockdowns using shRNA constructs that were originally used to establish a role for Dasm-1 in dendrite arborization in rat hippocampal neurons (23). We showed that these RNAi constructs suppressed expression of Dasm-1 in transfected HeLa cells and confirmed that RNAi 1 strongly reduced dendrite arborization in rat hippocampal neurons (see Fig. S2 in the supplemental material). We then performed Dasm-1 knockdowns using RNAi 1 and 2 constructs in hippocampal neurons derived from wild-type and Dasm-1−/− mice (Fig. 5). Both RNAi constructs reduced dendrite complexity to similar degrees in wild-type neurons, as previously reported (Fig. 5E and G). Surprisingly, identical effects were seen when these RNAi constructs were expressed in Dasm-1−/− neurons (Fig. 5F and H). The RNAi 2 construct was directed against a portion of the open reading frame that had been deleted by homologous recombination in Dasm-1−/− neurons. These results firmly established that the reduction in dendrite arborization was due to off-target effects and was not the result of loss of Dasm-1 expression. The Dasm-1 knockdown using RNAi 3 did not affect dendritic arborization of wild-type and Dasm-1−/− neurons (see Fig. S3 in the supplemental material), although the reduction of Dasm-1 protein was similar to results with RNAi 1 and 2 (see Fig. S2 in the supplemental material). This result excludes the possibility that Dasm-1−/− neurons were somehow sensitized to nonspecific effects of RNAi.
FIG. 5.
Off-target effects of Dasm-1 RNAi knockdown. (A to D) Representative photomicrographs of hippocampal neurons transfected with either control pSUPER plasmid (A and B) or Dasm-1 shRNA plasmid (C and D) together with a plasmid expressing EGFP. Compared to neurons transfected with pSUPER/EGFP, transfection with Dasm-1-RNAi/EGFP resulted in reduced dendritic development in the wild-type (A and C) and Dasm-1−/− (B and D) neurons. Quantification by Sholl analysis showed a strong reduction in the dendritic arbors of neurons expressing Dasm-1-RNAi 1 and Dasm-1-RNAi 2 in the wild type (E and G) as well as Dasm-1−/− (F and H) compared to neurons transfected with pSUPER (n > 25 neurons quantified per condition). Neurons were dissected from E18 embryos, transfected at 2 DIV, and fixed after an additional 6 DIV. Scale bar, 50 μm. The error bars indicate standard errors of the mean.
Dendrite development in neurons expressing putative dominant negative forms of Dasm-1.
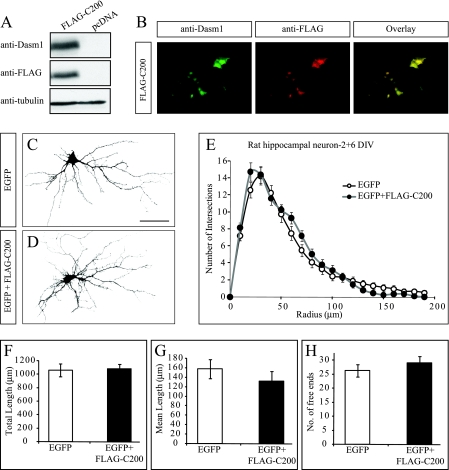
A previous report had shown that transfection of neurons with a truncated form of Dasm-1, lacking the entire cytoplasmic tail (Dasm-1delC-EGFP), resulted in impaired dendrite outgrowth (23). Repeating the same experiment, we observed a similar reduction in dendrite arbor of neurons expressing Dasm-1delC-EGFP compared to those expressing EGFP only (data not shown). Another potential dominant negative mutant consisting of the last 200 amino acids of the Dasm-1 cytoplasmic tail (C200) had previously been shown to affect synaptic transmission of hippocampal neurons (22). We investigated if an N-terminally FLAG-tagged version (FLAG-C200), which was well expressed in HeLa cells (Fig. 6A and B), would affect dendrite arborization in neurons. Neurons transfected with FLAG-C200 developed normally and showed similar dendritic complexity as EGFP-expressing neurons (Fig. 6C to E) or untransfected neurons (data not shown). There were no significant differences in the total dendritic branch length (1,061.2 ± 94.9 versus 1,080.9 ± 69.7 μm; P > 0.4) (Fig. 6F), mean dendritic length (158.1 ± 14.1 versus 132.4 ± 9.4 μm; P > 0.05) (Fig. 6G), and number of free ends (26.4 ± 2.2 versus 29.2 ± 2.3; P > 0.1) (Fig. 6H) between the EGFP- and FLAG-C200/EGFP-transfected neurons. These data indicate that the effects on dendrite arborization of dominant negative Dasm-1 constructs are not easy to interpret and that the underlying mechanisms are not understood.
FIG. 6.
Overexpression of the most C-terminal 200 amino acids of the Dasm-1 cytoplasmic tail (FLAG-C200), a putative dominant negative Dasm-1 mutant, does not impair dendrite outgrowth. (A) Western blot analysis showing the expression of FLAG-C200 in HeLa cells. A unique band of expected size (∼25 kDa) was detected using anti-Dasm-1 antibody. An equal amount of protein (∼25 μg) was loaded in each lane, and the same sample was probed with anti-Dasm-1 (top), anti-FLAG (middle), and antitubulin (bottom) antibodies. (B) Immunostaining of HeLa cells overexpressing FLAG-C200 showed coexpression with anti-Dasm-1 (left) and anti-FLAG (middle) antibody as seen in the overlay of the two images (right). Representative photomicrographs of dissociated rat hippocampal neurons transfected with EGFP (C) or FLAG-C200/EGFP (D). (E) Quantification by Sholl analysis showed no significant difference in the dendritic arbors of neurons expressing FLAG-C200 (n = 20) compared to neurons transfected with EGFP (n = 20). None of the parameters shows significant changes in the FLAG-C200-transfected neurons. (F) Total dendrite length (P > 0.4). (G) Mean dendrite length (P > 0.05). (H) Number of free ends (P > 0.1). Scale bar, 50 μm. The error bars indicate standard errors of the mean.
DISCUSSION
The closest known relative of mouse Dasm-1 is the Drosophila melanogaster gene tutl, which is essential for coordinated motor control (2). Larvae from tutl mutants show defective exploratory behavior, escape response to tactile stimulation, and righting behavior. Adult tutl mutants walk normally and have well-developed and innervated flight muscles but fail to perform flight of any kind. Although the Turtle protein has structural similarity to the neogenin/deleted in colorectal carcinoma/Frazzled and Roundabout families, which regulate axon pathfinding (6), tutl mutants do not display any detectable morphological alterations (2). It was speculated that Turtle may play a role in axon pathfinding of a very small subset of neurons or that it functions in a novel way unrelated to other IgSF family members. Our analysis of Dasm-1 null mutant mice is in agreement with the view that Dasm-1 is not required for morphogenesis of the developing or adult hippocampus. Both CA1 and dentate gyrus neurons in Dasm-1 null mice develop normal dendritic arborization patterns in vivo. Dissociated hippocampal neurons from Dasm-1 null mice also show normal dendritic differentiation in vitro. Moreover, the density of dendritic spines (based on the Thy-GFP marker) and the numbers of synapses in CA1 neurons (using transmission electron microscopy) were not altered in Dasm-1−/− mice compared to age-matched controls (data not shown). Major neuronal subtypes outside the hippocampus, such as cortical and Purkinje neurons, appeared normal in Golgi-stained mutant brains (data not shown), but a more thorough analysis including markers for neuronal subtypes will be required in future analyses. It therefore remains a possibility that a small subset of Dasm-1-positive neurons requires Dasm-1 for dendrite morphogenesis in vivo.
We also investigated the possibility that Dasm-1 regulates the fine-tuning of dendritic growth and branching in response to BDNF. BDNF expression is induced by neuronal activity, and only electrically active neurons seem able to respond to BDNF with an increase in dendritic arborization (17). Our observation that Dasm-1−/− neurons were not impaired in BDNF-induced neurite outgrowth in vitro suggests that Dasm-1 might not be necessary for activity-dependent structural plasticity in hippocampal neurons. Alternatively, the absence of Dasm-1 may be functionally compensated by other related molecules. Another immunoglobulin superfamily member, IgSF9b, a close homologue of Dasm-1 (3) that is coexpressed in the developing hippocampus and whose expression is unaltered in Dasm-1−/− brain (see Fig. S4 in the supplemental material), could be a putative substitute for Dasm-1.
In contrast to the tutl mutants, Dasm-1−/− mice do not show any overt impairment in coordinated motor control. Dasm-1−/− mice show normal spatial learning in the Morris water maze (A. Mishra, R. Klein, D. Wolfer, and H.-P. Lipp, unpublished observations). In agreement with the tutl mutants, which display normal basal synaptic transmission at the neuromuscular junction, Dasm-1−/− mice are not impaired in basal synaptic transmission in the hippocampus including input-output relationships of the field excitatory postsynaptic currents and paired pulse facilitation at CA1 synapses (M. Traut, V. Stein, A. Mishra, and R. Klein, unpublished observations). Whether Dasm-1 plays a role in synaptic maturation in the hippocampus, as previously suggested (22), remains to be investigated.
The normal development and morphogenesis of hippocampal neurons in the Dasm-1−/− mice were surprising in light of the reported in vitro effects of Dasm-1 RNAi knockdown in hippocampal neurons (23). The lack of defects in genetically targeted neurons compared to an RNAi knockdown suggests functional compensation in the knockout neurons. Such compensation has recently been demonstrated for membrane-associated guanylate kinases for their role in targeting AMPARs to the synaptic membrane and allowing AMPAR-mediated synaptic transmission. The knockout of PSD-95 had no effect, while acute knockdown of PSD-95 by RNAi led to the removal of all AMPA receptors at about half of excitatory synapses. Expression of shRNAs in the corresponding knockouts did not affect AMPAR-mediated synaptic currents, indicating that the effects of the knockdown were real and not due to off-target effects (9). This led to the suggestion to interpret the results from knockout animals with great caution due to compensation that can happen (12). In contrast to the PSD-95 knockdown, the Dasm-1-specific RNAis 1 and 2 produced similar impairments in dendritic growth in wild-type and Dasm-1−/− neurons, strongly indicating that the Dasm-1 knockdown phenotype is entirely due to off-target effects. One of the RNAis (RNAi 2) is even directed against the exons that were deleted by the gene targeting event, and absence of these exons in Dasm-1−/− mice was confirmed by reverse transcription-PCR.
More recent work involving RNAi has shown that the expression of shRNAs against an exogenous protein such as Photinus pyralis luciferase can trigger severe morphological and functional perturbations in cultured neurons in part by activating the cellular antiviral response pathway (1). In neurons, neurite growth and dendritic spine morphology are regulated by endogenous microRNAs (21, 26). Thus, siRNAs may compete for the endogenous machinery and indirectly alter neuron morphology. Because false-positive results are common in RNAi-based analyses, general recommendations have been offered for minimizing the risk that an observed phenotype arises from off-target effects (8). One important control which was omitted from the study of Shi et al. (23) was a rescue experiment with a Dasm-1 cDNA carrying silent mutations that would render the mRNA insensitive to the shRNA. Such a control would have probably invalidated the effects as being nonspecific. One control that was included in the study of Shi et al. was the transfection of a putative dominant negative Dasm-1 construct encoding Dasm-1 lacking the intracellular part (Dasm-1delC). Expression of a Dasm-1delC-EGFP fusion protein also impaired dendrite development of cultured hippocampal neurons (23). We have performed similar experiments and obtained similar outcomes (data not shown). However, this finding was not consistent with the results acquired by expressing another potential dominant negative Dasm-1 mutant, C200. Expression of C200 in neurons produced no difference in dendrite elaboration although it was previously shown to reduce synaptic function (22). The reason behind this discrepancy and the mechanism of Dasm-1delC-mediated impairment of dendrite growth are unclear. The subcellular localization of overexpressed Dasm-1delC was not analyzed; it is therefore possible that Dasm-1delC is targeted to aberrant subcellular sites and that it recruits endogenous proteins to those sites, thereby inhibiting normal development of the neurons. Without knowing the physiological ligand and downstream interaction partners of Dasm-1, we cannot determine if Dasm-1delC specifically inhibits endogenous Dasm-1. It is also possible that Dasm-1delC interacts with IgSF9b, thereby inhibiting both Dasm-1 and IgSF9b. We are currently investigating this possibility. Independently of our ongoing studies involving IgSF9b, our genetic data strongly indicate that Dasm-1 is dispensable for dendrite arborization of hippocampal neurons in vitro and in vivo.
Supplementary Material
Acknowledgments
This work was supported by the Max-Planck Society. S.P. was supported by a postdoctoral fellowship from FCT, Portugal, cofinanced by POCI 2010 and FSE.
We thank J. R. Sanes for GFP-M mice, J. Schultz for help with identifying the Dasm-1 cDNA as a candidate guidance receptor in a bioinformatics search, and J. Bailey, R. Sanchez, L. Meyn, M. Boesl, and M. Rodriguez for expert technical assistance.
Footnotes
Published ahead of print on 11 February 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alvarez, V. A., D. A. Ridenour, and B. L. Sabatini. 2006. Retraction of synapses and dendritic spines induced by off-target effects of RNA interference. J. Neurosci. 267820-7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodily, K. D., C. M. Morrison, R. B. Renden, and K. Broadie. 2001. A novel member of the Ig superfamily, turtle, is a CNS-specific protein required for coordinated motor control. J. Neurosci. 213113-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carninci, P., T. Kasukawa, S. Katayama, J. Gough, M. C. Frith, N. Maeda, R. Oyama, T. Ravasi, B. Lenhard, C. Wells, R. Kodzius, K. Shimokawa, V. B. Bajic, S. E. Brenner, S. Batalov, A. R. Forrest, M. Zavolan, M. J. Davis, L. G. Wilming, V. Aidinis, J. E. Allen, A. Ambesi-Impiombato, R. Apweiler, R. N. Aturaliya, T. L. Bailey, M. Bansal, L. Baxter, K. W. Beisel, T. Bersano, H. Bono, A. M. Chalk, K. P. Chiu, V. Choudhary, A. Christoffels, D. R. Clutterbuck, M. L. Crowe, E. Dalla, B. P. Dalrymple, B. de Bono, G. Della Gatta, D. di Bernardo, T. Down, P. Engstrom, M. Fagiolini, G. Faulkner, C. F. Fletcher, T. Fukushima, M. Furuno, S. Futaki, M. Gariboldi, P. Georgii-Hemming, T. R. Gingeras, T. Gojobori, R. E. Green, S. Gustincich, M. Harbers, Y. Hayashi, T. K. Hensch, N. Hirokawa, D. Hill, L. Huminiecki, M. Iacono, K. Ikeo, A. Iwama, T. Ishikawa, M. Jakt, A. Kanapin, M. Katoh, Y. Kawasawa, J. Kelso, H. Kitamura, H. Kitano, G. Kollias, S. P. Krishnan, A. Kruger, S. K. Kummerfeld, I. V. Kurochkin, L. F. Lareau, D. Lazarevic, L. Lipovich, J. Liu, S. Liuni, S. McWilliam, M. Madan Babu, M. Madera, L. Marchionni, H. Matsuda, S. Matsuzawa, H. Miki, F. Mignone, S. Miyake, K. Morris, S. Mottagui-Tabar, N. Mulder, N. Nakano, H. Nakauchi, P. Ng, R. Nilsson, S. Nishiguchi, S. Nishikawa, et al. 2005. The transcriptional landscape of the mammalian genome. Science 3091559-1563. [DOI] [PubMed] [Google Scholar]
- 4.Cheung, Z. H., W. H. Chin, Y. Chen, Y. P. Ng, and N. Y. Ip. 2007. Cdk5 is involved in BDNF-stimulated dendritic growth in hippocampal neurons. PLoS Biol. 5e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danzer, S. C., K. R. Crooks, D. C. Lo, and J. O. McNamara. 2002. Increased expression of brain-derived neurotrophic factor induces formation of basal dendrites and axonal branching in dentate granule cells in hippocampal explant cultures. J. Neurosci. 229754-9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickson, B. J. 2002. Molecular mechanisms of axon guidance. Science 2981959-1964. [DOI] [PubMed] [Google Scholar]
- 7.Doudney, K., J. N. Murdoch, C. Braybrook, C. Paternotte, L. Bentley, A. J. Copp, and P. Stanier. 2002. Cloning and characterization of Igsf9 in mouse and human: a new member of the immunoglobulin superfamily expressed in the developing nervous system. Genomics 79663-670. [DOI] [PubMed] [Google Scholar]
- 8.Echeverri, C. J., P. A. Beachy, B. Baum, M. Boutros, F. Buchholz, S. K. Chanda, J. Downward, J. Ellenberg, A. G. Fraser, N. Hacohen, W. C. Hahn, A. L. Jackson, A. Kiger, P. S. Linsley, L. Lum, Y. Ma, B. Mathey-Prevot, D. E. Root, D. M. Sabatini, J. Taipale, N. Perrimon, and R. Bernards. 2006. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods 3777-779. [DOI] [PubMed] [Google Scholar]
- 9.Elias, G. M., L. Funke, V. Stein, S. G. Grant, D. S. Bredt, and R. A. Nicoll. 2006. Synapse-specific and developmentally regulated targeting of AMPA receptors by a family of MAGUK scaffolding proteins. Neuron 52307-320. [DOI] [PubMed] [Google Scholar]
- 10.Falls, D. L. 2005. Dasm-1: a receptor that shapes neuronal dendrites and turns on silent synapses? Sci. STKE 2005pe10. [DOI] [PubMed] [Google Scholar]
- 11.Feng, G., R. H. Mellor, M. Bernstein, C. Keller-Peck, Q. T. Nguyen, M. Wallace, J. M. Nerbonne, J. W. Lichtman, and J. R. Sanes. 2000. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 2841-51. [DOI] [PubMed] [Google Scholar]
- 12.Fitzjohn, S. M., A. J. Doherty, and G. L. Collingridge. 2006. Promiscuous interactions between AMPA-Rs and MAGUKs. Neuron 52222-224. [DOI] [PubMed] [Google Scholar]
- 13.Gorski, J. A., S. R. Zeiler, S. Tamowski, and K. R. Jones. 2003. Brain-derived neurotrophic factor is required for the maintenance of cortical dendrites. J. Neurosci. 236856-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hattori, D., E. Demir, H. W. Kim, E. Viragh, S. L. Zipursky, and B. J. Dickson. 2007. Dscam diversity is essential for neuronal wiring and self-recognition. Nature 449223-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes, M. E., R. Bortnick, A. Tsubouchi, P. Baumer, M. Kondo, T. Uemura, and D. Schmucker. 2007. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron 54417-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthews, B. J., M. E. Kim, J. J. Flanagan, D. Hattori, J. C. Clemens, S. L. Zipursky, and W. B. Grueber. 2007. Dendrite self-avoidance is controlled by Dscam. Cell 129593-604. [DOI] [PubMed] [Google Scholar]
- 17.McAllister, A. K., L. C. Katz, and D. C. Lo. 1996. Neurotrophin regulation of cortical dendritic growth requires activity. Neuron 171057-1064. [DOI] [PubMed] [Google Scholar]
- 18.McAllister, A. K., L. C. Katz, and D. C. Lo. 1997. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron 18767-778. [DOI] [PubMed] [Google Scholar]
- 19.Millard, S. S., J. J. Flanagan, K. S. Pappu, W. Wu, and S. L. Zipursky. 2007. Dscam2 mediates axonal tiling in the Drosophila visual system. Nature 447720-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parrish, J. Z., K. Emoto, M. D. Kim, and Y. N. Jan. 2007. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu. Rev. Neurosci. 30399-423. [DOI] [PubMed] [Google Scholar]
- 21.Schratt, G. M., F. Tuebing, E. A. Nigh, C. G. Kane, M. E. Sabatini, M. Kiebler, and M. E. Greenberg. 2006. A brain-specific microRNA regulates dendritic spine development. Nature 439283-289. [DOI] [PubMed] [Google Scholar]
- 22.Shi, S. H., T. Cheng, L. Y. Jan, and Y. N. Jan. 2004. The immunoglobulin family member dendrite arborization and synapse maturation 1 (Dasm-1) controls excitatory synapse maturation. Proc. Natl. Acad. Sci. USA 10113346-13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi, S. H., D. N. Cox, D. Wang, L. Y. Jan, and Y. N. Jan. 2004. Control of dendrite arborization by an Ig family member, dendrite arborization and synapse maturation 1 (Dasm-1). Proc. Natl. Acad. Sci. USA 10113341-13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sholl, D. A. 1953. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 87387-406. [PMC free article] [PubMed] [Google Scholar]
- 25.Soba, P., S. Zhu, K. Emoto, S. Younger, S. J. Yang, H. H. Yu, T. Lee, L. Y. Jan, and Y. N. Jan. 2007. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron 54403-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vo, N., M. E. Klein, O. Varlamova, D. M. Keller, T. Yamamoto, R. H. Goodman, and S. Impey. 2005. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. USA 10216426-16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamagata, M., J. A. Weiner, and J. R. Sanes. 2002. Sidekicks: synaptic adhesion molecules that promote lamina-specific connectivity in the retina. Cell 110649-660. [DOI] [PubMed] [Google Scholar]
- 28.Zhu, H., and L. Luo. 2004. Diverse functions of N-cadherin in dendritic and axonal terminal arborization of olfactory projection neurons. Neuron 4263-75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.