Abstract
ATP-dependent chromatin remodelers of the CHD family play important roles during differentiation and development. Three CHD proteins, dMi-2, dChd1, and Kismet, have been described for Drosophila melanogaster. Here, we study dCHD3, a novel member of the CHD family. dCHD3 is related in sequence to dMi-2 but lacks several domains implicated in dMi-2 function. We demonstrate that dCHD3 is a nuclear protein and that expression is tightly regulated during fly development. Recombinant dCHD3 remodels mono- and polynucleosomes in an ATP-dependent manner in vitro. Its chromodomains are critical for nucleosome binding and remodeling. Unlike dMi-2, dCHD3 exists as a monomer. Nevertheless, both proteins colocalize with RNA polymerase II to actively transcribed regions on polytene chromosomes, suggesting that both remodelers participate in the process of transcription.
Dynamic regulation of chromatin structure is an important mechanism for modulating genome activity in eukaryotes. Two major groups of proteins with enzymatic activities directed toward chromatin are histone-modifying enzymes and ATP-dependent chromatin remodelers (2, 18). The former modify histones by attaching or removing acetyl, methyl, phosphate, ADP-ribose, ubiquitin, or SUMO groups. These modifications alone do not lead to major changes of chromatin structure. The histone code hypothesis postulates that these “marks” are specifically bound by factors which in turn influence chromatin structure by mechanisms that are not well understood (19, 33, 35). ATP-dependent chromatin remodelers can directly change nucleosome structure by altering interactions between DNA and the histone octamer. This results in nucleosome assembly or eviction, incorporation of histone variants, altered DNA-to-octamer contacts within an existing nucleosome, or lateral movement of the octamer along DNA (6, 23, 38). As a result, the accessibility of nucleosomal DNA is changed.
Most ATP-dependent chromatin remodelers are multisubunit complexes containing an ATPase of the SNF2 superfamily (12). These enzymes share a conserved ATPase domain but can differ in additional domains. Accordingly, they are subdivided into several families, including the DDM1, the SWI/SNF, the ISWI, the INO80, and the CHD family (3, 15, 44).
CHD proteins are characterized by the presence of two tandemly arranged chromodomains (30). The mammalian CHD family has nine members, only a few of which have been subjected to a detailed molecular analysis. The Chd3/Chd4 subfamily is characterized by the presence of a pair of PHD fingers preceding the chromodomains. These proteins exist in multisubunit complexes with nucleosome remodeling and deacetylation (NuRD) activity (7). In addition to Chd3 or Chd4, NuRD complexes contain histone deacetylase 1 (HDAC1) and HDAC2, the histone binding proteins RbAp46 and RbAp48, proteins of the MTA and p66 families (MTA1, MTA2, or MTA3 and p66α or p66β), and subunits with methylated DNA binding domains (MBD3). It has been suggested that these complexes couple ATP-dependent chromatin remodeling and deacetylation to effect transcriptional repression.
Complexes related to NuRD exist in Drosophila melanogaster and Caenorhabditis elegans and play important roles during differentiation and cell fate determination (3, 13). This function is conserved in mammals: an MTA3-containing NuRD complex regulates B-lymphocyte differentiation and is indispensable for BCL-6-mediated repression of plasma cell-specific genes in B lymphocytes (16).
Two different modes of targeting NuRD complexes have been discussed. First, recruitment via interaction with DNA bound transcriptional repressors, such as BCL-6, Hunchback, or Tramtrack 69 (16, 17, 20, 34). Second, direct binding of stoichiometric (MBD3) or associated (MBD2A) MBD proteins to methylated DNA (31, 45, 50).
The ATP-dependent chromatin remodeling functions of mammalian Chd4 and Drosophila dMi-2 have been analyzed in vitro. Whereas Chd4 ATPase activity is stimulated by both DNA and nucleosomes, the stimulation of dMi-2 is nucleosome specific (9, 46). Recombinant dMi-2 mobilizes nucleosomes in the absence of other NuRD subunits in vitro (9). The chromodomains of dMi-2 are critical for this activity and appear to mediate the interaction between enzyme and nucleosome substrate (5). Experiments with mammalian Chd4 suggest that the PHD fingers are required for binding the HDAC1 subunit of NuRD (49).
In addition to dMi-2, the D. melanogaster genome encodes a second putative protein belonging to the Chd3/Chd4 subfamily: dCHD3 shares one of the conserved PHD fingers, both chromodomains, and the ATPase domain with dMi-2. By contrast, regions that are important for interaction with other proteins are missing (C terminus) or incomplete (PHD fingers). In addition, a region in the N terminus of dMi-2 that plays a role in regulating ATPase activity and is phosphorylated by casein kinase 2 is absent in dCHD3 (4).
Here, we show that recombinant dCHD3 is a nucleosome-stimulated ATPase and remodels mono- and polynucleosomal substrates in an ATP- and chromodomain-dependent manner in vitro. dCHD3 is expressed as a nuclear protein during development and in adult females. dCHD3 and dMi-2 colocalize with active RNA polymerase II on polytene chromosomes. However, unlike dMi-2, dCHD3 exists as a monomer and cannot functionally replace dMi-2.
MATERIALS AND METHODS
Recombinant proteins.
dCHD3 cDNA was obtained from BDGP (clone no. RE55932). A premature stop codon was repaired by PCR. This sequence was used for the generation of Flag-tagged dCHD3 and mutants by PCR using appropriate sets of primers (see the supplemental material). All PCR products were cloned into pVL1392 and verified by sequencing. Baculovirus production and protein purification have previously been described (9).
Antibodies and immunoprecipitations.
Mouse monoclonal RBF1 (DX3) and rabbit polyclonal dMi-2 (anti-dMi-2-N) and dRPD3 antibodies have previously been described (9, 22). Antibodies against RNA polymerase II (H5) were purchased from Covance, anti-Flag antibody was from Sigma, and secondary antibodies were from Invitrogen and GE Healthcare.
The generation of antibodies against dCHD3 and dMi-2 is described in the supplemental material. Immunoprecipitations were performed using standard procedures (see the supplemental material).
Histone octamer purification, nucleosome assembly, and mononucleosome mobilization assay.
Histone octamers were isolated from 100 g dechorionated embryos as described previously (9). Poly- and mononucleosomes were assembled on pUC18 plasmid and 200-bp fragments, respectively, by stepwise salt dialysis in siliconized Eppendorf tube lids (Biozym) (see the supplemental material). Mononucleosome mobilization assays are described in the supplemental material.
Enzymatic assays.
ATPase and HDAC assays were performed as described previously (5, 9).
Immunofluorescence.
The preparation of polytene chromosomes was described previously (51). DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole) (0.5 μg/μl). Antibody dilutions were anti-dCHD3 (rat 7A11) at 1:2, anti-dMi-2 (rabbit, N terminal) at 1:200, anti-dMi-2 (rat 4D8) at 1:2, and anti-pol II (mouse H5 or H14) at 1:50. As secondary antibodies, Alexa Fluor 488 goat anti-rabbit (1:200), Alexa Fluor 546 goat anti-rat (1:200; Invitrogen), or fluorescein isothiocyanate-conjugated goat anti-mouse (1:200; Jackson Laboratory) antibody was used.
Embryo immunostaining was performed as described previously (32). Briefly, wild-type Oregon R embryos were washed, dechorionated, fixed, and permeabilized. Primary antibodies used were anti-dCHD3 (rat 7A11) undiluted and anti-dMi-2-N (rabbit) at 1:200. Embryos were analyzed by confocal microscopy.
RNAi.
Double-stranded RNA was prepared using primers designed according to the GenomeRNAi database (DKFZ, Heidelberg, Germany [www.dkfz.de/signaling2/rnai/]). RNA interference (RNAi) was prepared with the MEGAscript T7 kit (Ambion) according to the manufacturer's instructions. A total of 1 × 106 KC cells were incubated with 12 μg of double-stranded RNA in serum-free medium for 40 min at 25°C. Complete medium with serum was added, and cells were incubated for 5 to 7 days at 25°C. Knockdown was verified by Western blot analysis.
Gel filtration.
Two hundred microliters of embryo or KC cell nuclear extract was applied to a Superose 6 HR 10/30 gel filtration column (Amersham Pharmacia) and resolved in 10 mM HEPES, pH 7.6, 300 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, and 10% glycerol on an Äkta purifier system (GE Healthcare) according to the manufacturer's instructions.
Biocomputing.
Multiple sequence alignments and phylogenetic trees were generated with ClustalW (www.ebi.ac.uk) and Jalview.
RESULTS
Sequence analysis of dCHD3.
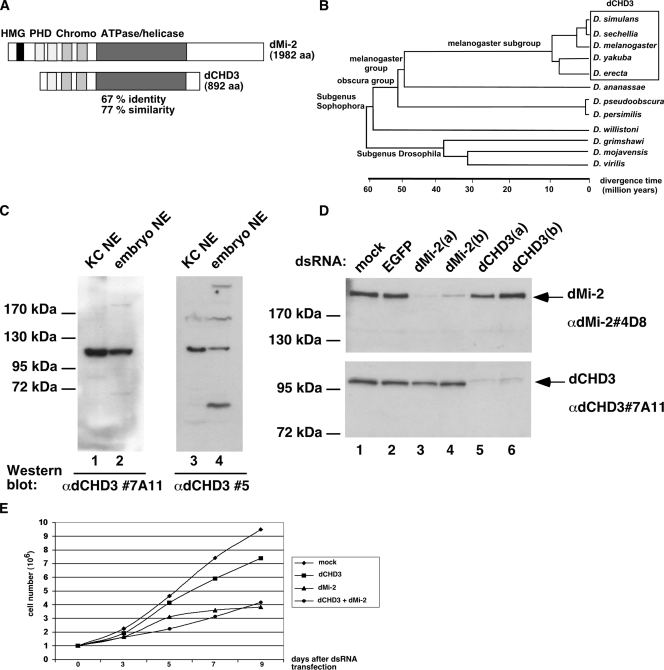
The D. melanogaster genome encodes four CHD family members: dChd1, Kismet, dMi-2, and dCHD3 (3). Of these four, dMi-2 and dCHD3 contain the highest amino acid sequence similarity (67% identity and 77% similarity) (Fig. 1A). We searched for dMi-2 and dCHD3 in the genomes of different Drosophila species. As expected, dMi-2 was found in all genomes examined (data not shown). dCHD3 genes were identified in Drosophila simulans, Drosophila sechellia, Drosophila yakuba, and Drosophila erecta (D. melanogaster subgroup). By contrast, no dCHD3 sequences were found in Drosophila ananassae (D. melanogaster group) or in more distantly related Drosophila species (Fig. 1B). The evolutionary relationship suggests that the event giving rise to the dCHD3 gene took place more than 10 million years ago (Fig. 1B).
FIG. 1.
dCHD3 is expressed in D. melanogaster (A) Schematic representation of dMi-2 and dCHD3. HMG, high-mobility-group box; PHD, PHD finger; Chromo, chromodomain; aa, amino acids. (B) Phylogenetic tree of drosophilae. Species with dCHD3 genes are boxed. (C) Nuclear extracts derived from KC cells and embryos were subjected to Western blot analysis by using rat monoclonal (left panel) and rabbit polyclonal (right panel) antibodies. α, anti. (D) KC cells were treated with double-stranded RNAs directed against enhanced green fluorescent protein (EGFP), dMi-2 [dMi-2(a) and dMi-2(b)], and dCHD3 [dCHD3(a) and dCHD3(b)]. Nuclear extracts were prepared 6 days after treatment and subjected to Western blot analysis using dMi-2 (upper panel) and dCHD3 antibodies (lower panel). Mock, nuclear extract derived from mock-treated cells. Positions of dMi-2 and dCHD3 are indicated on the right. (E) KC cells were treated with double-stranded RNA (dsRNA) directed against dCHD3, dMi-2, or a combination of the two. Cell numbers were determined after 3, 5, 7, and 9 days. Mock, treatment omitting double-stranded RNA.
Sequence alignment of dCHD3, dMi-2, dChd1, Kismet, ISWI, and Brahma revealed the conservation of important ATPase domain motifs in dCHD3 (see Fig. S1 in the supplemental material). This result is consistent with the view that the enzymatic activity of the putative dCHD3 protein has been maintained through evolution.
Expression of dCHD3 in the fly embryo.
dCHD3-specific RNA has been detected by microarray analysis in embryos and adult flies, suggesting that the dCHD3 gene is expressed, at least at the RNA level (www.fruitfly.org/cgi-bin/ex/insitu.pl). We generated rabbit polyclonal and rat monoclonal antibodies to analyze dCHD3 protein expression. When nuclear extracts of KC cells were subjected to Western blot analysis, both dCHD3 antibodies reacted strongly with a 100-kDa polypeptide, closely corresponding to dCHD3's predicted molecular mass of 103 kDa (Fig. 1C, lanes 1 and 3). Similar results were obtained when S2 cells and embryo extracts were probed (Fig. 1C, lanes 2 and 4, and data not shown).
We used RNAi to assess the specificity of dCHD3 and dMi-2 antibodies (Fig. 1D). Treatment of KC cells with two independent double-stranded RNAs directed against different regions of dCHD3 mRNA strongly decreased the detection of the 100-kDa polypeptide by dCHD3 antibody (Fig. 1D, compare lanes 5 and 6 with lanes 1 to 4). By contrast, the recognition of dMi-2 by dMi-2-specific antibodies was not affected. Conversely, RNAi against dMi-2 weakened the dMi-2 signal but did not affect the dCHD3 signal (compare lanes 3 and 4 with lanes 1, 2, 5, and 6). In addition, we tested antibody specificity using recombinant dMi-2 and dCHD3. We failed to detect cross-reactivity even when microgram amounts of dMi-2 and dCHD3 were used (see Fig. S2 in the supplemental material). We conclude that dCHD3 protein is expressed in embryos and cell lines. Furthermore, these experiments establish the specificity of the dCHD3 and dMi-2 antibodies used in this study.
Nonredundant functions of dCHD3 and dMi-2.
dMi-2 is required for the viability of germ cells (20). We used RNAi to determine whether this requirement also applied to KC cells and to test for functional redundancy between dMi-2 and dCHD3 (Fig. 1E). The depletion of dMi-2 had a significant effect on cell growth and viability, suggesting that dMi-2 is essential for KC cell survival. By contrast, the depletion of dCHD3 had no significant effect on cell growth. The codepletion of both factors affected cell growth to a degree similar to that of the depletion of dMi-2 alone. This result indicates that dCHD3 cannot substitute for the loss of dMi-2, arguing that dMi-2 and dCHD3 perform nonredundant functions in KC cells.
dCHD3 is a nucleosome-stimulated ATPase.
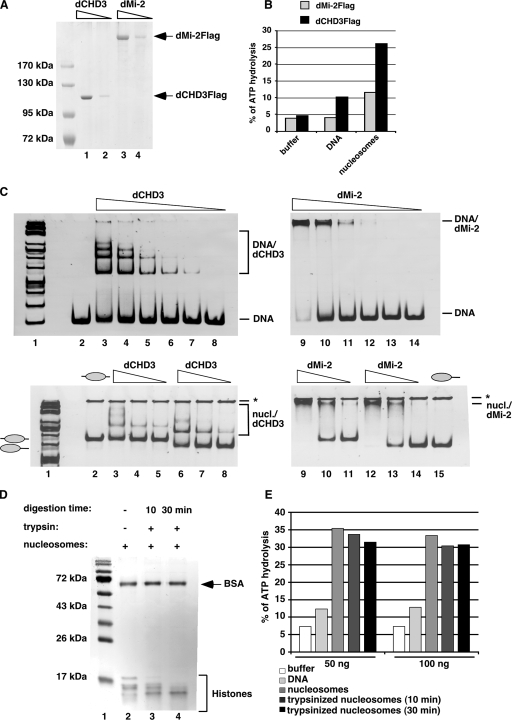
We generated a baculovirus expressing dCHD3 to test whether it was an active enzyme. Flag-tagged dCHD3 and dMi-2 were affinity purified from extracts of baculovirus-infected Sf9 cells (Fig. 2A). We determined the ATPase activity of both proteins in the absence and presence of DNA and polynucleosomes (Fig. 2B). As observed previously, dMi-2 displayed low ATPase activity in the absence of effector, no significant stimulation by DNA, and a robust activation by nucleosomes (9). dCHD3 had low basal ATPase activity. Stimulation by DNA was clearly detectable (twofold) but weaker than the activation by nucleosomes (fivefold). Increasing the DNA concentration did not significantly increase the stimulation of dMi-2 and dCHD3 ATPase activity, indicating that DNA was not limiting (see Fig. S3 in the supplemental material). We conclude that dCHD3 is an ATPase that is activated by DNA but is preferentially stimulated by nucleosomes.
FIG. 2.
dCHD3 has DNA and nucleosome-stimulated ATPase activity in vitro. (A) Coomassie-stained gel showing recombinant proteins used for assays. Lanes 1 and 2, Flag-tagged dCHD3 (36 and 9 pmol); lanes 3 and 4, Flag-tagged dMi-2 (36 and 9 pmol). The molecular mass marker is shown on the left. (B) 2.7 pmol of dMi-2 and dCHD3 was incubated in the absence or presence of 100 ng DNA or nucleosomes and [γ-32P]ATP as indicated. The percentage of hydrolyzed ATP was determined by thin-layer chromatography and quantified by PhosphorImager analysis. (C) Upper panels, decreasing amounts of dCHD3 (lane 3, 18 pmol; lane 4, 9 pmol; lane 5, 3.6 pmol; lane 6, 1.8 pmol; lane 7, 0.9 pmol; and lane 8, 0.36 pmol) and dMi-2 (lane 9, 18 pmol; lane 10, 9 pmol; lane 11, 3.6 pmol; lane 12, 1.8 pmol; lane 13, 0.9 pmol; and lane 14, 0.36 pmol) were incubated with a 200-bp DNA fragment, and complexes were resolved by native agarose gel electrophoresis. The positions of DNA/protein complexes and unbound DNA are indicated on the right. Lower panels, decreasing amounts of dCHD3 (lanes 2 and 6, 18 pmol; lanes 3 and 7, 9 pmol; and lanes 4 and 8, 3.6 pmol) and dMi-2 (lanes 9 and 12, 18 pmol; lanes 10 and 13, 9 pmol; and lanes 11 and 14, 3.6 pmol) were incubated with centrally (lanes 2 to 5 and 9 to 11) or distally positioned (lanes 6 to 8 and 12 to 15) mononucleosomes. Complexes were resolved by native agarose gel electrophoresis. The positions of complexes are indicated on the right, and the positions of unbound mononucleosomes are indicated on the left. Asterisks denote assembled plasmid backbone that remains in the well. nucl., nucleosome. (D) Polynucleosomes were digested with trypsin for 10 (lane 3) or 30 min (lane 4) as indicated. Histones were visualized by Coomassie staining following sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Positions of bovine serum albumin (BSA) and histones are indicated on the right. −, absence of (for trypsin) or time zero (for digestion time); +, presence of. (E) Fifty and 100 ng of trypsinized nucleosomes were incubated with dCHD3, and ATPase activity was determined as described for panel B.
dCHD3 binds DNA and mononucleosomes.
We compared the binding of dCHD3 and dMi-2 to 200-bp DNA fragments and mononucleosomes (Fig. 2C). dCHD3 formed distinct complexes with DNA that were stable during electrophoresis through native polyacrylamide gels (Fig. 2C, upper panel, lanes 3 to 8). Increasing concentrations of dCHD3 led to the formation of up to four complexes of decreasing mobility, suggesting that several dCHD3 molecules can simultaneously bind the probe. As shown previously, dMi-2 displayed DNA binding affinity (5, 9). However, dMi-2 complexes had a low mobility and did not enter the gel (Fig. 2C, upper panel, lanes 9 to 14).
To assay nucleosome binding, we used two mononucleosomes that differ in the position of the octamer relative to the end of a 200-bp DNA fragment (see below). dCHD3 formed complexes with both nucleosomes (Fig. 2C, lower panel, lanes 3 to 8). Again, multiple dCHD3 complexes were detected when higher dCHD3 concentrations were used. dMi-2 also bound both mononucleosomes, leading to the formation of complexes of slow mobility (Fig. 2C, lower panel, lanes 9 to 14).
dCHD3 does not require histone tails in vitro.
N-terminal histone tails are critical for nucleosome-stimulated ATPase and nucleosome mobilization activities of ISWI (2). By contrast, dMi-2 remodeling activity does not require histone tails in vitro (9). We assessed the importance of histone tails for dCHD3 ATPase activity. We progressively removed histone tails from polynucleosomes by partial trypsin digestion (Fig. 2D). We then assessed the activation of dCHD3 by trypsinized nucleosomes (Fig. 2E). No significant differences in nucleosome-stimulated ATPase activities were observed between the different nucleosome samples tested. We conclude that histone tails are not critical for dCHD3 ATPase activation in vitro.
dCHD3 mobilizes mononucleosomes in vitro.
We subjected dCHD3 and dMi-2 to nucleosome sliding assays (5). We used two 200-bp DNA fragments bearing the so-called 601 nucleosome binding sequence either near the center or near the end (28). Assembly onto these DNA fragments produced two mononucleosomes that differ in the relative position of the histone octamer and in electrophoretic mobility (Fig. 2C, lower panels, compare lanes 2 and 15).
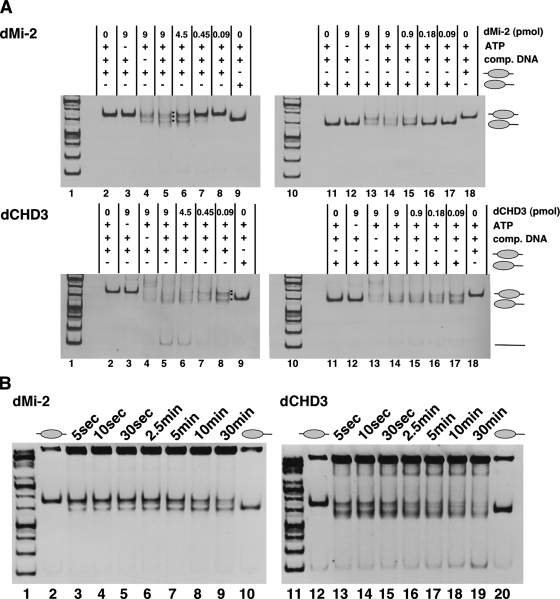
As reported previously, recombinant dMi-2 displayed ATP-dependent nucleosome mobilization activity (Fig. 3A, upper panels). Both types of nucleosomes were mobilized. Like dMi-2, recombinant dCHD3 showed ATP-dependent nucleosome mobilization activity on both substrates (Fig. 3A, lower panels). Lower concentrations of dCHD3 than dMi-2 were required to effect nucleosome mobilization.
FIG. 3.
dCHD3 and dMi-2 remodel mononucleosomes in vitro. (A) dMi-2 (upper panels) and dCHD3 (lower panels) were incubated with positioned mononucleosomes (left panels show centrally positioned mononucleosomes, and right panels show distally positioned mononucleosomes) as indicated on top. Lanes 3 to 5 and 12 to 14, 9 pmol protein; lane 6, 4.5 pmol; lane 15, 0.9 pmol; lane 7, 0.45 pmol; lane 16, 0.18 pmol; lanes 8 and 17, 0.09 pmol; lanes 2, 9, 11, and 18, no protein. Nucleosome mobilization was visualized by ethidium bromide staining following agarose gel electrophoresis. Positions of free DNA (straight line) and mononucleosomes (ovals) are indicated on the right. Comp. DNA, competitor DNA used to stop the reaction. Nucleosome substrates used are indicated as symbols on the right. −, absence of; +, presence of. (B) dMi-2 (9 pmol; left panel) and dCHD3 (9 pmol; right panel) were incubated with centrally positioned mononucleosomes. Reactions were stopped at the times indicated at the top of the panels.
In several reactions, multiple products were generated, likely reflecting differently positioned nucleosomes (Fig. 3A, upper panel, lane 5, and lower panel, lane 8). In some cases, a product migrated slightly faster than the end-positioned nucleosome did. It is conceivable that here, the histone octamer has been pushed over the edge and is no longer making contact with all 146 bp of DNA.
In contrast to dMi-2, dCHD3 produced a significant amount of free DNA when the centrally positioned mononucleosome was used as a substrate (Fig. 3A, left lower panel), suggesting that dCHD3 action removes octamers from DNA fragments.
Next, we assessed the kinetics of dMi-2- and dCHD3-catalyzed reactions (Fig. 3B). Within 10 min, approximately 50% of nucleosomes had been mobilized by dMi-2 (left panel). In striking contrast, dCHD3 required only 5 s to move 50% of nucleosomes (right panel). Further nucleosome mobilization occurred for 30 min, at which time most nucleosomes had been repositioned. An increase in the amount of free DNA became detectable after 10 min.
We conclude that recombinant dMi-2 and dCHD3 display different kinetics in the mobilization assay. Moreover, nucleosome mobilization and octamer removal reactions can be separated kinetically, with nucleosome movement occurring earlier than octamer removal.
dCHD3 remodels polynucleosomes in vitro.
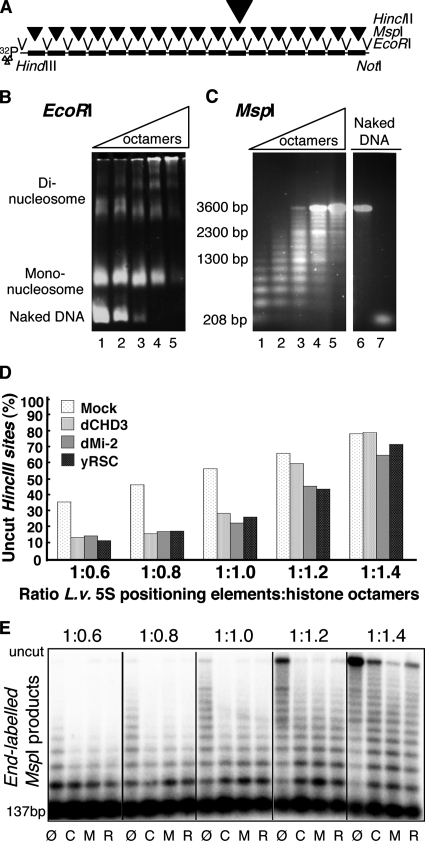
We next investigated whether dMi-2 and dCHD3 can remodel polynucleosomal substrates. We utilized a linear array bearing 17 repeats of the 5S ribosomal DNA (rDNA) nucleosome positioning sequence (Fig. 4A) (25). Repeats are separated by EcoRI restriction sites, and each contains an MspI restriction site within the nucleosome positioning sequence. In addition, repeat 11 harbors a single HincII site. The presence of nucleosomes on this array decreases the accessibility of MspI and HincII to their cognate sites (27).
FIG. 4.
dCHD3 and dMi-2 remodel polynucleosomes in vitro. (A) Schematic representation of polynucleosome substrate used. Black boxes, 5S rDNA nucleosome positioning elements; thin arrows, EcoRI sites; filled arrowheads, MspI sites; bold arrow, HincII site; 32P, labeled end of fragment. (B) EcoRI digests of polynucleosomes assembled with increasing histone octamer-to-DNA ratios (lane 1, 0.6:1; lane 2, 0.8:1; lane 3, 1:1; lane 4, 1.2:1; and lane 5, 1.4:1). DNA was visualized by ethidium bromide staining following native polyacrylamide gel electrophoresis (26). (C) MspI digests of polynucleosomes, analyzed by agarose gel electrophoresis of naked DNA after phenol extraction. Lanes 1 to 5 are as described for panel B. Lane 6, uncut DNA; lane 7, MspI cut DNA. (D) Polynucleosome arrays (1 nM) were incubated with ATP, HincII, and the indicated remodelers for 1 h. The proportion of cut HincII sites was determined by PhosphorImager analysis. (E) Polynucleosomes with indicated saturation levels were incubated with remodelers and ATP for 1 h. Remodeling was stopped by the addition of apyrase. Samples were then digested with MspI and extracted with phenol, and DNA was resolved by agarose gel electrophoresis and visualized by autoradiography. Ø, mock; R, RSC; C, dCHD3; M, dMi-2.
We generated a series of subsaturated, fully saturated, and oversaturated polynucleosomal arrays and monitored assembly by EcoRI and MspI digestion (Fig. 4B and C). We then incubated arrays with HincII in the absence or presence of remodeling enzymes and ATP (Fig. 4D). Since neither dCHD3 nor dMi-2 had been tested on polynucleosomal substrates before, we included purified yeast RSC complex as a positive control (8). Depending on the saturation level of the array, between 30% and 80% of DNA molecules were not cut by HincII in the absence of remodeler. This result is in good agreement with previous work and attributable to occlusion of the HincII site (27). The incubation of subsaturated and fully saturated arrays with remodelers and ATP significantly decreased the percentage of uncut DNA molecules. This result suggests that RSC, dMi-2, and dCHD3 all can increase the accessibility of the HincII site to a similar extent by nucleosome remodeling. By contrast, oversaturated nucleosomal arrays were largely refractory to remodeler action.
We also tested whether the three enzymes were able to generate a stably remodeled polynucleosomal product in the absence of restriction enzyme (Fig. 4E). End-labeled arrays were incubated with remodelers, remodeling was terminated by the addition of apyrase, and the accessibility of MspI sites was tested by restriction digest. The three remodelers significantly increased the production of shorter restriction fragments. Thus, all three enzymes were able to produce stably remodeled nucleosomal arrays.
Chromodomains are essential for dCHD3 remodeling activity.
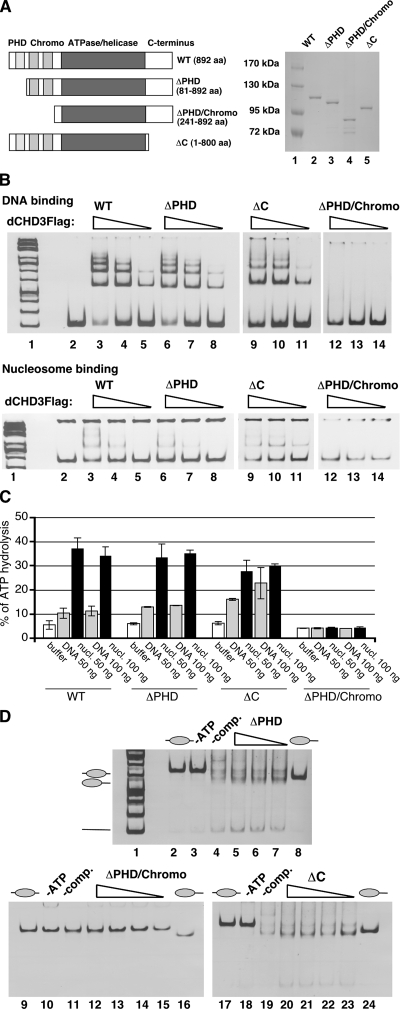
We created deletion mutants of dCHD3 to define domains important for nucleosome remodeling (Fig. 5A). Flag-tagged mutants were expressed and purified using the baculovirus system (Fig. 5A, right panel).
FIG. 5.
The chromodomains are critical for nucleosome remodeling in vitro. (A) Left panel, schematic representation of dCHD3 deletion mutants. Right panel, Coomassie-stained gel showing recombinant dCHD3 mutants. (B) DNA (upper panels) and nucleosome (lower panels) binding by dCHD3 mutants. WT, wild type. Eighteen picomoles (lanes 3, 6, 9 and 12), 9 pmol (lanes 4, 7, 10 and 13), or 1.8 pmol (lanes 5, 8, 11 and 14) of protein was incubated with DNA and mononucleosomes as indicated. Complexes were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. Lanes 1, molecular weight marker; lanes 2, DNA (upper panel) and nucleosome (lower) probes. (C) Recombinant dCHD3 proteins were incubated in the absence or presence of DNA or nucleosomes (nucl.) and [γ-32P]ATP as indicated. ATPase activity was determined as described above. Error bars indicate standard deviations. (D) Recombinant dCHD3 deletion mutants were incubated with centrally positioned mononucleosomes as indicated. Nucleosome mobilization was visualized by ethidium bromide staining following agarose gel electrophoresis. Positions of free DNA (straight line) and mononucleosomes (ovals) are indicated on the left. Lanes 3 to 5, 10 to 12, and 18 to 20, 9 pmol of recombinant protein; lanes 6, 13, and 21, 4.5 pmol; lanes 7, 14, and 22, 2.25 pmol; lanes 15 and 23, 0.45 pmol; lanes 2, 9, 16, centrally positioned nucleosome; lanes 8, 16, and 24, distally positioned nucleosome. −ATP, ATP was omitted; −comp., no competitor DNA was used to stop the reaction.
The deletion of the PHD finger or the C terminus did not abrogate DNA or nucleosome binding or ATPase activity (Fig. 5B and C). However, the deletion of the C terminus led to a noticeable increase of DNA-stimulated ATPase activity (from twofold to fourfold). The mutant lacking both PHD finger and chromodomains failed to bind DNA and nucleosomes. This mutant retained basal ATPase activity but was no longer stimulated by DNA or nucleosomes.
Finally, we assessed nucleosome mobilization activity of dCHD3 mutants (Fig. 5D). Again, the activity of mutants retaining the chromodomains was comparable to that of the wild type. By contrast, the mutant lacking the chromodomains displayed no detectable nucleosome mobilization activities.
These results are in agreement with the view that the chromodomains of dCHD3 are essential for DNA and nucleosome binding and nucleosome-stimulated ATPase and ATP-dependent nucleosome mobilization activities.
dCHD3 and dMi-2 expression in embryos, larvae, and flies.
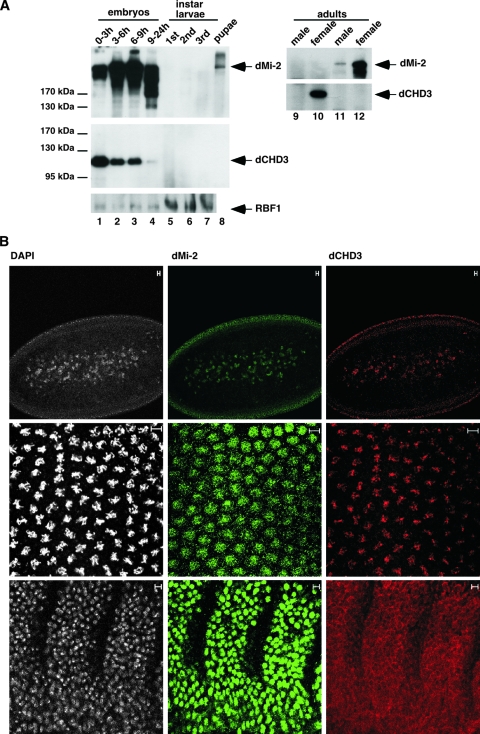
We probed whole-cell extracts from embryos, larvae, and pupae to determine the developmental expression patterns of dCHD3 and dMi-2 (Fig. 6A).
FIG. 6.
dCHD3 expression in embryos. (A) Extracts derived from different developmental stages were immunoprecipitated and subjected to Western blot analysis as indicated using the following antibodies: for dMi-2, anti-dMi-2-N; for dCHD3, 5; and for RBF1, DX3. Antibodies used for immunoprecipitation were, for lanes 1 to 8, anti-dMi-2 (4D8, upper panel), anti-dCHD3 (7A11, middle panel), and anti-RBF1 (DX3, bottom panel); for lanes 9 and 10, anti-dCHD3 (7A11); and for lanes 11 and 12, anti-dMi-2 (4D8). Antibodies used to probe the Western blots are indicated on the right. (B) Immunofluorescence. Early preblastoderm (upper panels), late preblastoderm (middle panels), and postgastrulation (lower panels) embryos were stained with DAPI (white; left panels), dMi-2 (green; middle panels), and dCHD3 antibody (red; right panels). Scale bars, 5 μm.
Highest dMi-2 levels were detected in embryos, with expression levels peaking around 6 to 9 h after egg deposition. dMi-2 levels then declined sharply and became undetectable at the first larval stage. dMi-2 remained undetectable until the pupa stage, when weak dMi-2 expression was observed. dCHD3 was most strongly expressed in early embryos (0 to 3 h after egg deposition). Expression then became weaker, and no dCHD3 was detected in larval and pupal stages. As a control, extracts were probed with an antibody recognizing RBF1. In agreement with published work, RBF1 levels remain relatively constant throughout embryonic and larval development (42). Female and male adult flies showed sharp differences in dMi-2 and dCHD3 expression. No dCHD3 and little dMi-2 was detectable in extracts prepared from male flies, whereas strong expression of both proteins was apparent in extracts from female flies. This finding is consistent with the expression of both proteins in ovaries (data not shown). These results demonstrate that dMi-2 and dCHD3 expression is tightly regulated during development, is sex specific in adult flies, and suggests a significant maternal deposition of dMi-2 and dCHD3.
We used immunofluorescence and confocal microscopy to assess the subcellular localization of dMi-2 and dCHD3 during embryogenesis (Fig. 6B). Expression of dMi-2 and dCHD3 was detected in nuclei before their migration to the membrane of the preblastoderm embryo (upper panel). Both proteins remained nuclear after migration (middle panel). Interestingly, dCHD3 remained associated with condensed chromosomes in mitotic nuclei where dMi-2 displayed a diffuse nuclear staining. At later stages, dMi-2 was still detectable in nuclei of the epidermis (lower panels). dCHD3 immunofluorescence, however, had declined to background levels. These findings are consistent with the results of Western blot analysis (Fig. 6A). They demonstrate that both chromatin remodelers are expressed in nuclei of the embryo.
dCHD3 and dMi-2 localize to overlapping sites on polytene chromosomes.
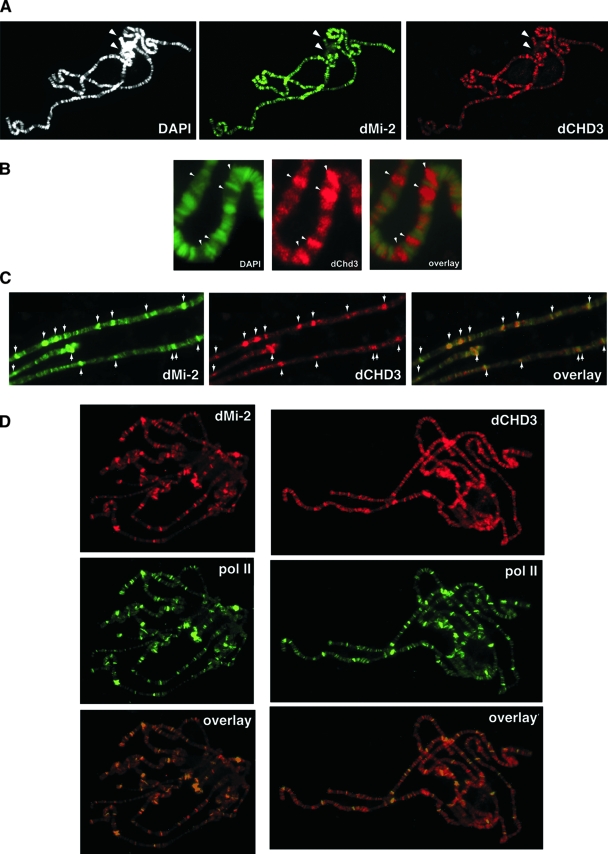
We immunostained polytene chromosomes of third instar larvae with dMi-2 and dCHD3 antibodies. Despite the absence of robust dCHD3 and dMi-2 Western blot signals in larval extracts, the immunostaining of salivary glands confirmed nuclear expression of both factors (Fig. 6A; see Fig. S4 in the supplemental material). In agreement with previous work, dMi-2 localized to multiple bands (Fig. 7A, middle panel) (34). Comparatively weak dMi-2 staining was apparent at the highly condensed chromocenter and the fourth chromosome (Fig. 7A, upper panel). The dCHD3 antibody likewise revealed multiple bands and low staining of the chromocenter and fourth chromosome (Fig. 7A, bottom panel). dCHD3 staining localized mostly to interbands and was largely excluded from DAPI-dense regions (Fig. 7B).
FIG. 7.
dCHD3 and dMi-2 colocalize to sites of active transcription. (A) Polytene chromosome preparations (right panels) were stained with DAPI (white; top), dMi-2 (green; middle), and dCHD3 antibody (red; bottom). Arrows indicate the chromocenter and the fourth chromosome. (B) Close-up of polytene chromosome stained with DAPI (green; left) and dCHD3 antibody (red; middle). An overlay of both signals is shown on the right. Arrows indicate bands of strong dCHD3 and weak DAPI staining. (C) Polytene chromosomes were stained with DAPI (white; top), dMi-2 (green; middle), and dCHD3 antibody (red; middle). An overlay of dMi-2 and dCHD3 signals is shown on the bottom. Arrows indicate overlapping dMi-2 and dCHD3 bands. (D) Polytene chromosomes were stained with dMi-2, dCHD3, and RNA polymerase II antibodies as indicated. Bottom panels show an overlay of RNA polymerase II and dMi-2 (left) and dCHD3 (right) signals.
Costaining with dMi-2 and dCHD3 antibodies revealed a remarkable overlap of binding sites (Fig. 7C). These data suggest that dMi-2 and dCHD3 are recruited to the same chromosomal regions on polytene chromosomes.
To determine whether the dMi-2 and dCHD3 binding sites correspond to sites of active transcription, we employed antibodies recognizing active RNA polymerase II. An antibody binding phosphorylated serine 2 of the heptad repeats of the carboxy-terminal domain highlighted multiple sites of active transcription (Fig. 7D, middle panels). Costaining with antibodies directed against dMi-2 and dCHD3 revealed a high degree of colocalization with elongating RNA polymerase II. We also carried out costaining experiments by using an antibody recognizing the serine 5 phosphorylation in the carboxyterminal domain. Again, dMi-2 and dCHD3 colocalized with active RNA polymerase II (data not shown). No colocalization of either dMi-2 or dCHD3 with RBF2, a component of the repressive dREAM complex which is excluded from actively transcribed chromatin was detected (data not shown) (22).
dCHD3 exists as a monomer.
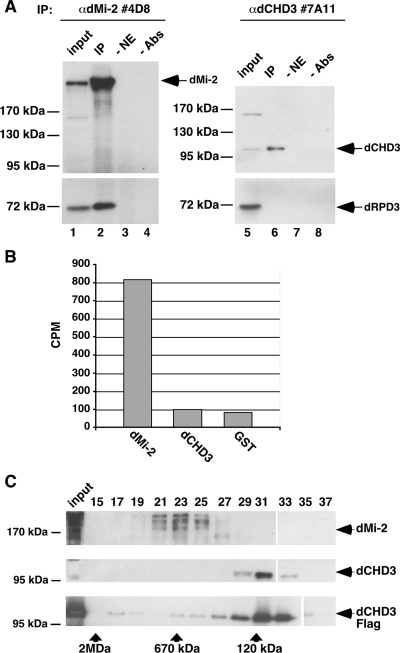
We next investigated whether dCHD3 is part of a dNuRD-like complex. As expected, the immunoprecipitation of dMi-2 coprecipitated dRPD3 (Fig. 8A). By contrast, no dRPD3 was detected in dCHD3 immunoprecipitates. In agreement with this result, dMi-2 immunoprecipitates displayed robust HDAC activity in vitro (Fig. 8B). However, no HDAC activity was associated with dCHD3 immunoprecipitates. Moreover, in contrast to dMi-2, dCHD3 was not detectable in dRPD3 immunoprecipitates obtained from cells overexpressing Flag-tagged dRPD3 (see Fig. S5 in the supplemental material). In addition, we failed to detect an interaction between recombinant dCHD3 and dRPD3 overexpressed in baculovirus infected Sf9 cells (data not shown).
FIG. 8.
dCHD3 exists as a monomer. (A) Embryo nuclear extracts were immunoprecipitated with dMi-2 (left panels) and dCHD3 (right panels) antibodies. Immunoprecipitates were analyzed by Western blot using antibodies against dMi-2 (upper, left), dCHD3 (upper, right), and dRPD3 (lower panels). IP, immunoprecipitates; −NE, IP without extract; −Abs, mock IP without antibody. (B) Embryo nuclear extracts were immunoprecipitated with dMi-2, dCHD3, and glutathione S-transferase (GST) antibodies as indicated. Immunoprecipitates were subjected to HDAC assays by using 3H-labeled histone octamers. (C) Embryo nuclear extracts (upper and middle panels) and recombinant dCHD3 (bottom panel) were applied to a Superose 6 column. Fractions were analyzed by Western blotting using appropriate antibodies as indicated. Gel filtration size markers are shown below (arrows); positions of dMi-2 and dCHD3 are shown on the right.
Next, we fractionated nuclear extracts from embryos by gel filtration and assayed the fractions for the presence of dMi-2 and dCHD3 (Fig. 8C). dMi-2 eluted with an apparent molecular mass of larger than 670 kDa, confirming that it is part of a multisubunit complex (Fig. 8C, upper panel). By contrast, dCHD3 eluted with an apparent molecular mass of 120 kDa (Fig. 8C, middle panel). dCHD3 eluted in similar fractions when KC nuclear extracts or purified recombinant dCHD3 protein was applied to the column (Fig. 8C, lower panel and data not shown).
Taken together, these data suggest that unlike dMi-2, dCHD3 exists as a monomer in vivo.
DISCUSSION
Saccharomyces cerevisiae has a single CHD family member, CHD1. In metazoans, the family has expanded to four CHD proteins in D. melanogaster and nine in Homo sapiens (30). In this study, we have analyzed dCHD3, a novel chromatin remodeler of D. melanogaster. By analyzing genomes of different Drosophila species, we have traced the gene duplication event giving rise to dCHD3. We postulate that the dCHD3 gene originated from integration of a truncated, reverse-transcribed dMi-2 mRNA. This hypothesis is supported by the fact that the dCHD3 gene lacks introns.
We have performed a detailed analysis of the nucleosome remodeling activities of dCHD3 in vitro. Like dMi-2, dCHD3 binds DNA and nucleosomes and is a nucleosome-stimulated ATPase. However, dCHD3 and dMi-2 differ in the extent to which their ATPases are activated by DNA. Whereas the activation of dMi-2 by DNA is insignificant, the activation of dCHD3 is more pronounced. Interestingly, DNA stimulation of dMi-2 is increased in a deletion mutant lacking the C-terminal 700 residues (5). dCHD3 lacks most of this region, and when the remaining C-terminal residues are removed, the stimulation of dCHD3 ATPase activity by DNA is further increased. These results strengthen the hypothesis that the C-terminal sequences of dMi-2 and dCHD3 have a negative regulatory effect on DNA-stimulated ATPase activity (5).
dCHD3 mobilizes mononucleosomes in an ATP-dependent manner. Mobilization is effected from both a position close to one end of the DNA fragment to a more central one and a central position to more distal ones. Recombinant dMi-2 displays a strong preference for distal-to-central mobilization when a nucleosome assembled on the mouse 5S rDNA fragment is used (9). In the current study, mononucleosomes were assembled on DNA fragments containing the high-affinity 601 sequence (28). Neither dCHD3 nor dMi-2 displayed a significant preference for the direction of nucleosome movement. This demonstrates that direction preference is not an invariant property of the remodeling machine but is influenced by the underlying DNA sequence.
dCHD3 removes a fraction of histone octamers from DNA during the mobilization assay. It is possible that some histone octamers are pushed over the edge during mobilization and become unstable. The kinetics of the mobilization reaction are in agreement with this view: mobilized nucleosomes are produced within seconds, whereas free DNA becomes detectable only after 10 min, when most nucleosomes have already been relocalized.
We have tested the ability of recombinant dCHD3 and dMi-2 to remodel polynucleosomal substrates in vitro. Both factors increased the accessibility of nucleosomal restriction sites. Interestingly, this effect was obtained when the restriction enzyme was present during the remodeling reaction as well as when it was added after remodeling had taken place. This result indicates that both factors can create stably remodeled polynucleosomes in the absence of other proteins.
Removal of histone tails by partial proteolysis does not affect nucleosome stimulation of the dCHD3 ATPase. Likewise, dMi-2 is stimulated by nucleosomes assembled from recombinant histones lacking all N-terminal tails and NuRD complex purified from Xenopus laevis is activated by trypsinized nucleosomes (8, 9). These results suggest that histone tail interactions do not play a critical role during remodeling catalyzed by this CHD subfamily, despite the fact that both factors contain chromodomains and PHD fingers. Both of these domains have recently been demonstrated to specifically bind methylated histone tails (10, 33). For example, the chromodomains of human CHD1 recognize histone H3 methylated at K4 (14, 40). Moreover, human CHD4 (Mi2β) has been suggested to bind histone H3 methylated at K36 (33). There is no data suggesting that chromodomains and PHD fingers of dCHD3 and dMi-2 bind methylated histone tails. Indeed, based on sequence differences between chromodomains of CHD3/CHD4 and CHD1, it has been suggested that CHD3/CHD4-type chromodomains do not bind methylated histone tails (36). Our results indicate that the chromodomains of dCHD3 are critical for nucleosome remodeling by providing an interaction surface for DNA. These findings are in agreement with results that we have previously reported for the chromodomains of dMi-2 (5). However, we cannot formally rule out the possibility that PHD fingers and chromodomains of dCHD3 carry out redundant functions. Moreover, it remains possible that specific tail modifications positively or negatively influence the interactions of these remodelers with chromatin in vivo.
Most ATP-dependent chromatin remodelers exist in stable complexes with one or more nonenzymatic subunits implicated in targeting and regulation of enzymatic activity. Given the similarity between dCHD3 and dMi-2, it was conceivable that dCHD3 would interact with NuRD subunits. However, our results strongly suggest that dCHD3 exists as a monomer in vivo. In this respect, dCHD3 resembles the CHD1 protein which has been reported both to exist in a complex and as a monomer in S. cerevisiae and has been shown to be largely monomeric in D. melanogaster (29, 37, 43).
dCHD3 and dMi-2 antibodies stain the same bands on polytene chromosomes. We have ruled out that this is due to antibody cross-reactivity (see Fig. S2 in the supplemental material). The high degree of colocalization of dCHD3 and dMi-2 suggests a common way of targeting. However, the targeting mechanisms that have been proposed for dMi-2 do not seem to be applicable to dCHD3. dMi-2 interacts with DNA bound transcriptional repressors via a C-terminal domain that is missing in dCHD3 (20, 34). An alternative targeting mechanism of NuRD complexes is provided by MBD-domain-containing subunits which could guide the complex to regions of methylated DNA (31). However, since dCHD3 exists as a monomer, such a mechanism appears unlikely. Of course, we cannot exclude the possibility that transient interactions might be involved in dCHD3 recruitment. Given that dCHD3 and dMi-2 colocalize with RNA polymerase II at transcriptionally active regions, it is conceivable that targeting involves binding to components of the transcription apparatus. Indeed, such interactions have been demonstrated for yeast CHD1 and SWI2/SNF2 (39, 47). However, the immunoprecipitation of dCHD3 and dMi-2 from nuclear extracts did not coprecipitate RNA polymerase II and vice versa (data not shown). Like dMi-2 and dCHD3, dChd1, Kismet, and BRM associate with actively transcribed regions on polytene chromosomes, although no physical interactions with RNA polymerase II can be detected (1, 41). This result suggests that these ATP-dependent chromatin remodelers recognize an as-yet-unidentified component of actively transcribed regions.
It is important to note that dMi-2 and dCHD3 do not colocalize under all circumstances. Our analysis of early embryos suggests different affinities for binding to condensed chromosomes during mitosis. dCHD3 remains associated with condensed chromosomes, whereas dMi-2 is distributed more evenly throughout the nucleus.
Their similar expression patterns, remodeling properties, and colocalization on polytene chromosomes raise the question of whether dMi-2 and dCHD3 carry out redundant functions. dMi-2-deficient flies are generally nonviable (although escapers have been reported) and die during larval stages (20, 48). Their survival to larval stage has been attributed to maternal contribution of dMi-2. Indeed, our immunofluorescence and Western blot analysis results show a high level of dMi-2 protein in embryos prior to the onset of zygotic transcription. dMi-2-deficient germ cells are not viable (20). The depletion of dMi-2 by RNAi in KC cells decreases cell growth, supporting the view that dMi-2 is required for viability on a cell autonomous level. By contrast, we have not detected adverse effects of dCHD3 depletion. In addition, no dCHD3 mutant flies have been reported to date. These results suggest that dCHD3 and dMi-2 are not fully redundant and that dCHD3 cannot compensate for the loss of dMi-2. However, we cannot formally exclude the possibility that the differential response of cells to dMi-2 and dCHD3 depletion is due to incomplete knockdown of dCHD3 or a similar dosage effect.
It is remarkable that all four CHD family members colocalize with RNA polymerase II to sites of active transcription on polytene chromosomes (41; this study). This finding is not easily reconcilable with the proposed role of dMi-2 and dNuRD in transcriptional repression. However, it has recently been demonstrated that HDAC complexes play important roles in silencing cryptic promoters in transcribed genes in S. cerevisiae (11, 24). Srinivasan and colleagues have used mutants to demonstrate that Kismet likely acts at an early stage of the transcription process (41). CHD1 mutants are deficient in the replication-independent incorporation of the histone H3.3 variant both during the restructuring of the male pronucleus and at later stages following the onset of zygotic transcription (21). Suitable dmi-2 or dchd3 mutants are not yet available to perform similar studies. Whether the four CHD remodelers are all involved at different stages of transcription and what their molecular function is remain to be analyzed.
Supplementary Material
Acknowledgments
We are grateful to R. Renkawitz-Pohl and R. Jacob for access to microscopes. We are indebted to A. Sapetschnig for RNAi constructs and advice. We thank G. Suske for critical reading of the manuscript and all members of the Brehm lab for support and discussion.
M.M. was supported by a fellowship from the International Graduate School GRK1384. J.V.V., C.L., and A.B. acknowledge support from EuroDYNA (J.V.V., C.L. and A.B.) and the Deutsche Forschungsgemeinschaft (DFG BR2102-5/1 [A.B.]).
Footnotes
Published ahead of print on 4 February 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Armstrong, J. A., O. Papoulas, G. Daubresse, A. S. Sperling, J. T. Lis, M. P. Scott, and J. W. Tamkun. 2002. The Drosophila BRM complex facilitates global transcription by RNA polymerase II. EMBO J. 215245-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71247-273. [DOI] [PubMed] [Google Scholar]
- 3.Bouazoune, K., and A. Brehm. 2006. ATP-dependent chromatin remodeling complexes in Drosophila. Chromosome Res. 14433-449. [DOI] [PubMed] [Google Scholar]
- 4.Bouazoune, K., and A. Brehm. 2005. dMi-2 chromatin binding and remodeling activities are regulated by dCK2 phosphorylation. J. Biol. Chem. 28041912-41920. [DOI] [PubMed] [Google Scholar]
- 5.Bouazoune, K., A. Mitterweger, G. Langst, A. Imhof, A. Akhtar, P. B. Becker, and A. Brehm. 2002. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 212430-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulard, M., P. Bouvet, T. K. Kundu, and S. Dimitrov. 2007. Histone variant nucleosomes: structure, function and implication in disease. Subcell. Biochem. 4171-89. [PubMed] [Google Scholar]
- 7.Bowen, N. J., N. Fujita, M. Kajita, and P. A. Wade. 2004. Mi-2/NuRD: multiple complexes for many purposes. Biochim. Biophys. Acta 167752-57. [DOI] [PubMed] [Google Scholar]
- 8.Boyer, L. A., C. Logie, E. Bonte, P. B. Becker, P. A. Wade, A. P. Wolffe, C. Wu, A. N. Imbalzano, and C. L. Peterson. 2000. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem. 27518864-18870. [DOI] [PubMed] [Google Scholar]
- 9.Brehm, A., G. Langst, J. Kehle, C. R. Clapier, A. Imhof, A. Eberharter, J. Muller, and P. B. Becker. 2000. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J. 194332-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brehm, A., K. R. Tufteland, R. Aasland, and P. B. Becker. 2004. The many colours of chromodomains. Bioessays 26133-140. [DOI] [PubMed] [Google Scholar]
- 11.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123581-592. [DOI] [PubMed] [Google Scholar]
- 12.Eisen, J. A., K. S. Sweder, and P. C. Hanawalt. 1995. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 232715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fay, D. S., and M. Han. 2000. The synthetic multivulval genes of C. elegans: functional redundancy, Ras-antagonism, and cell fate determination. Genesis 26279-284. [DOI] [PubMed] [Google Scholar]
- 14.Flanagan, J. F., L. Z. Mi, M. Chruszcz, M. Cymborowski, K. L. Clines, Y. Kim, W. Minor, F. Rastinejad, and S. Khorasanizadeh. 2005. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 4381181-1185. [DOI] [PubMed] [Google Scholar]
- 15.Flaus, A., D. M. Martin, G. J. Barton, and T. Owen-Hughes. 2006. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 342887-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujita, N., D. L. Jaye, C. Geigerman, A. Akyildiz, M. R. Mooney, J. M. Boss, and P. A. Wade. 2004. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell 11975-86. [DOI] [PubMed] [Google Scholar]
- 17.Fujita, N., D. L. Jaye, M. Kajita, C. Geigerman, C. S. Moreno, and P. A. Wade. 2003. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell 113207-219. [DOI] [PubMed] [Google Scholar]
- 18.Imhof, A. 2006. Epigenetic regulators and histone modification. Brief. Funct. Genomics Proteomics. 5222-227. [DOI] [PubMed] [Google Scholar]
- 19.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 20.Kehle, J., D. Beuchle, S. Treuheit, B. Christen, J. A. Kennison, M. Bienz, and J. Muller. 1998. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 2821897-1900. [DOI] [PubMed] [Google Scholar]
- 21.Konev, A. Y., M. Tribus, S. Y. Park, V. Podhraski, C. Y. Lim, A. V. Emelyanov, E. Vershilova, V. Pirrotta, J. T. Kadonaga, A. Lusser, and D. V. Fyodorov. 2007. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science 3171087-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korenjak, M., B. Taylor-Harding, U. K. Binne, J. S. Satterlee, O. Stevaux, R. Aasland, H. White-Cooper, N. Dyson, and A. Brehm. 2004. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119181-193. [DOI] [PubMed] [Google Scholar]
- 23.Längst, G., and P. B. Becker. 2004. Nucleosome remodeling: one mechanism, many phenomena? Biochim. Biophys. Acta 167758-63. [DOI] [PubMed] [Google Scholar]
- 24.Li, B., M. Gogol, M. Carey, D. Lee, C. Seidel, and J. L. Workman. 2007. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 3161050-1054. [DOI] [PubMed] [Google Scholar]
- 25.Logie, C., and C. L. Peterson. 1997. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 166772-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Logie, C., and C. L. Peterson. 1999. Purification and biochemical properties of yeast SWI/SNF complex. Methods Enzymol. 304726-741. [DOI] [PubMed] [Google Scholar]
- 27.Logie, C., C. Tse, J. C. Hansen, and C. L. Peterson. 1999. The core histone N-terminal domains are required for multiple rounds of catalytic chromatin remodeling by the SWI/SNF and RSC complexes. Biochemistry 382514-2522. [DOI] [PubMed] [Google Scholar]
- 28.Lowary, P. T., and J. Widom. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 27619-42. [DOI] [PubMed] [Google Scholar]
- 29.Lusser, A., D. L. Urwin, and J. T. Kadonaga. 2005. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 12160-166. [DOI] [PubMed] [Google Scholar]
- 30.Marfella, C. G., and A. N. Imbalzano. 2007. The Chd family of chromatin remodelers. Mutat. Res. 61830-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marhold, J., K. Kramer, E. Kremmer, and F. Lyko. 2004. The Drosophila MBD2/3 protein mediates interactions between the MI-2 chromatin complex and CpT/A-methylated DNA. Development 1316033-6039. [DOI] [PubMed] [Google Scholar]
- 32.Marhold, J., M. Zbylut, D. H. Lankenau, M. Li, D. Gerlich, E. Ballestar, B. M. Mechler, and F. Lyko. 2002. Stage-specific chromosomal association of Drosophila dMBD2/3 during genome activation. Chromosoma 11113-21. [DOI] [PubMed] [Google Scholar]
- 33.Mellor, J. 2006. It takes a PHD to read the histone code. Cell 12622-24. [DOI] [PubMed] [Google Scholar]
- 34.Murawsky, C. M., A. Brehm, P. Badenhorst, N. Lowe, P. B. Becker, and A. A. Travers. 2001. Tramtrack69 interacts with the dMi-2 subunit of the Drosophila NuRD chromatin remodelling complex. EMBO Rep. 21089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nightingale, K. P., L. P. O'Neill, and B. M. Turner. 2006. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr. Opin. Genet. Dev. 16125-136. [DOI] [PubMed] [Google Scholar]
- 36.Okuda, M., M. Horikoshi, and Y. Nishimura. 2007. Structural polymorphism of chromodomains in Chd1. J. Mol. Biol. 3651047-1062. [DOI] [PubMed] [Google Scholar]
- 37.Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates III, and P. A. Grant. 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433434-438. [DOI] [PubMed] [Google Scholar]
- 38.Saha, A., J. Wittmeyer, and B. R. Cairns. 2006. Mechanisms for nucleosome movement by ATP-dependent chromatin remodeling complexes. Results Probl. Cell Differ. 41127-148. [DOI] [PubMed] [Google Scholar]
- 39.Simic, R., D. L. Lindstrom, H. G. Tran, K. L. Roinick, P. J. Costa, A. D. Johnson, G. A. Hartzog, and K. M. Arndt. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 221846-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sims, R. J. III, C. F. Chen, H. Santos-Rosa, T. Kouzarides, S. S. Patel, and D. Reinberg. 2005. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 28041789-41792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srinivasan, S., J. A. Armstrong, R. Deuring, I. K. Dahlsveen, H. McNeill, and J. W. Tamkun. 2005. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA polymerase II. Development 1321623-1635. [DOI] [PubMed] [Google Scholar]
- 42.Stevaux, O., D. Dimova, M. V. Frolov, B. Taylor-Harding, E. Morris, and N. Dyson. 2002. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J. 214927-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran, H. G., D. J. Steger, V. R. Iyer, and A. D. Johnson. 2000. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 192323-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Vugt, J. J., M. Ranes, C. Campsteijn, and C. Logie. 2007. The ins and outs of ATP-dependent chromatin remodeling in budding yeast: biophysical and proteomic perspectives. Biochim. Biophys. Acta 1769153-171. [DOI] [PubMed] [Google Scholar]
- 45.Wade, P. A., A. Gegonne, P. L. Jones, E. Ballestar, F. Aubry, and A. P. Wolffe. 1999. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 2362-66. [DOI] [PubMed] [Google Scholar]
- 46.Wang, H. B., and Y. Zhang. 2001. Mi2, an auto-antigen for dermatomyositis, is an ATP-dependent nucleosome remodeling factor. Nucleic Acids Res. 292517-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson, C. J., D. M. Chao, A. N. Imbalzano, G. R. Schnitzler, R. E. Kingston, and R. A. Young. 1996. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell 84235-244. [DOI] [PubMed] [Google Scholar]
- 48.Yamasaki, Y., and Y. Nishida. 2006. Mi-2 chromatin remodeling factor functions in sensory organ development through proneural gene repression in Drosophila. Dev. Growth Differ. 48411-418. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95279-289. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Y., H. H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 131924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zink, B., and R. Paro. 1989. In vivo binding pattern of a trans-regulator of homoeotic genes in Drosophila melanogaster. Nature 337468-471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.