Abstract
The cell wall integrity mitogen-activated protein kinase (MAPK) cascade of Saccharomyces cerevisiae drives changes in gene expression in response to cell wall stress. We show that the MAPK of this pathway (Mpk1) and its pseudokinase paralog (Mlp1) use a noncatalytic mechanism to activate transcription of the FKS2 gene. Transcriptional activation of FKS2 was dependent on the Swi4/Swi6 (SBF) transcription factor and on an activating signal to Mpk1 but not on protein kinase activity. Activated (phosphorylated) Mpk1 and Mlp1 were detected in a complex with Swi4 and Swi6 at the FKS2 promoter. Mpk1 association with Swi4 in vivo required phosphorylation of Mpk1. Promoter association of Mpk1 and the Swi4 DNA-binding subunit of SBF were codependent but did not require Swi6, indicating that the MAPK confers DNA-binding ability to Swi4. Based on these data, we propose a model in which phosphorylated Mpk1 or Mlp1 forms a dimeric complex with Swi4 that is competent to associate with the FKS2 promoter. This complex then recruits Swi6 to activate transcription. Finally, we show that human ERK5, a functional ortholog of Mpk1, is similarly capable of driving FKS2 expression in the absence of protein kinase activity, suggesting that this mammalian MAPK may also have a noncatalytic function in vivo.
The cell wall of the budding yeast Saccharomyces cerevisiae is required to maintain cell shape and integrity (13, 41). The cell must remodel this rigid structure during vegetative growth and during pheromone-induced morphogenesis. Wall remodeling is monitored and regulated by the cell wall integrity (CWI) signaling pathway controlled by the Rho1p GTPase (reviewed in reference 43). Two essential functions have been identified for Rho1p. First, it serves as an integral regulatory subunit of the 1,3-β-glucan synthase (GS) complex stimulating GS activity in a GTP-dependent manner. A second essential function of Rho1 is to bind and activate protein kinase C, which is encoded by PKC1. Loss of Pkc1 function or of any of the components of the mitogen-activated protein kinase (MAPK) cascade under its control results in a cell lysis defect that is attributable to a deficiency in cell wall construction. The MAPK cascade is a linear pathway comprised of a MEKK (Bck1), a pair of redundant MEKs (Mkk1/2), and a MAPK (Mpk1/Slt2). Mpk1 is a functional homolog of human ERK5 (65), a MAPK that is activated in response to growth factors as well as physical and chemical stresses (1, 68).
CWI signaling is induced in response to a variety of cell wall stressors. First, signaling is activated persistently in response to growth at elevated temperatures (e.g., 37 to 39°C) (37), consistent with the finding that null mutants in many of the pathway components display cell lysis defects only when cultivated at high temperatures. Second, hypo-osmotic shock induces a rapid but transient activation of signaling (16, 37). Third, treatment with mating pheromone stimulates signaling at a time that is coincident with the onset of morphogenesis (12). Finally, CWI signaling is also stimulated by agents that interfere with cell wall biogenesis, such as the chitin antagonist calcofluor white (39), Congo red, caffeine, or zymolyase (18, 47).
One consequence of CWI signaling is activation of the Rlm1 transcription factor (19, 67) through phosphorylation by Mpk1 (35). Rlm1 regulates the transcription of a wide array of cell wall metabolism genes (23, 36, 55). However, the FKS2 gene, which encodes one of two alternative catalytic subunits of the GS complex, is an unusual transcriptional target of CWI signaling because it is induced independently of Rlm1 (36, 70).
A second transcription factor that plays a poorly defined role in CWI signaling is SBF (7, 44), a dimeric G1 regulator comprised of Swi4 and Swi6 (reviewed in reference 11). Swi4 is the sequence-specific DNA binding subunit (64), but Swi6 is required for DNA binding (4, 6, 58). SBF is essential to normal regulation of G1-specific transcription, but genetic and biochemical evidence suggests that SBF also participates in CWI signaling as a target of Mpk1. First, the cell lysis defect of an mpk1Δ mutant is suppressed by overexpression of Swi4 (44). Second, both swi4Δ and swi6Δ mutants are hypersensitive to calcofluor white, supporting a role for SBF in cell wall biogenesis (33). Third, Mpk1 associates with SBF in vivo, as judged by coprecipitation experiments (44), and with Swi4 (but not Swi6) in vitro (7). Fourth, Swi6 is phosphorylated in vivo and in vitro by Mpk1 in response to cell wall stress (44).
Here, we find that CWI signaling drives expression of the FKS2 gene through SBF. Although Mpk1 must be in the active (phosphorylated) conformation to induce FKS2 expression, its protein kinase activity is not required. Indeed, the pseudokinase paralog of Mpk1, Mlp1 (for Mpk-like protein), is also capable of inducing FKS2 expression. This novel noncatalytic function involves stable association of Mpk1 or Mlp1 with SBF at the FKS2 promoter. Transcriptional activity of FKS2 has been conserved in mammalian ERK5, suggesting that this MAPK may also have noncatalytic functions in metazoan cells.
MATERIALS AND METHODS
Strains, growth conditions, and transformations.
The S. cerevisiae strains used in this study are listed in Table 1. Yeast cultures were grown in YEPD medium (1% Bacto yeast extract, 2% Bacto peptone, 2% glucose) with or without 10% sorbitol, or in SD medium (0.67% yeast nitrogen base, 2% glucose) supplemented with the appropriate nutrients to select for plasmids and gene replacements. Escherichia coli DH5α was used to propagate all plasmids. E. coli cells were cultured in Luria broth medium (1% Bacto tryptone, 0.5% Bacto yeast extract, 1% NaCl) and transformed to carbenicillin resistance by standard methods. Two-hybrid tests were conducted using yeast strain SFY526 (Clontech Laboratories) in cultures grown to mid-log phase. β-Galactosidase assays for promoter-lacZ fusion expression experiments and for two-hybrid analyses were conducted as described previously (70).
TABLE 1.
S. cerevisiae strains
| Strain | Relevant genotypea | Reference or source |
|---|---|---|
| 1788 | MATa/MATα EG123 leu2-3,112 trp1-1 ura3-52 his4 can1r | I. Herskowitz |
| DL454 | MATa EG123 mpk1Δ::TRP1 | 42 |
| DL3145 | MATa/MATα EG123 swi4Δ::TRP1/swi4Δ::TRP1 | This study |
| DL3148 | MATa/MATα EG123 swi6Δ::LEU2/swi6Δ::LEU2 | This study |
| DL3166 | MATa/MATα EG123 mlp1Δ::URA3/mlp1Δ::URA3 mpk1Δ::TRP1/mpk1Δ::TRP1 | This study |
| DL3183 | MATa S288c mpk1Δ::G418 mlp1Δ::G418 | This study; Research Genetics |
| DL3187 | MATa S288c (BY4741) his3Δ leu2Δ ura3Δ lys2Δ | Research Genetics |
| DL3193 | MATa/MATα S288c (BY4743) | Research Genetics |
| DL3194 | MATa/MATα S288c mlp1Δ::G418/mlp1Δ::G418 | This study; Research Genetics |
| DL3195 | MATa/MATα S288c mpk1Δ::G418/mpk1Δ::G418 | This study; Research Genetics |
| DL3196 | MATa/MATα S288c mpk1Δ::G418/mpk1Δ::G418 mlp1Δ::G418/mlp1Δ::G418 | This study; Research Genetics |
| DL3233 | MATa S288c swi6Δ::G418 | Research Genetics |
| DL3405 | MATa S288c swi4Δ::G418 | Research Genetics |
| DL3529 | MATa/MATα EG123 bck1Δ::G418/bck1Δ::G418 | Research Genetics |
| DL3541 | MATa/α S288c mkk1Δ::G418/mkk1Δ::G418 mkk2Δ::G418/mkk2Δ::G418 | This study; Research Genetics |
| SFY526 | MATagal4-542 gal80-538 ura3-52 ura3::GAL1-lacZ ade2-101 leu2-3,112 his3-200 trp1-905 lys2-801 canR | Clontech |
Strain background EG123 is described by Siliciano and Tatchell (61).
Plasmids.
Plasmids used in this study are listed in Table 2. A plasmid consisting of FKS2 residues −540 to −375 [FSK2(−540 to −375)]-CYC1-lacZ (plasmid p2052) was constructed by replacing the 134-bp SmaI/XhoI fragment of the CYC1 promoter of pLGΔ-312 (26) with a 165-bp PCR-amplified FKS2 fragment, resulting in fusion of the FKS2 promoter sequences to the CYC1 minimal promoter. Three SBF-binding site mutant alleles of FKS2 were constructed by PCR overlap mutagenesis (32) of p2052: an A to T at residue −386 (plasmid p2053), a G to C at residue −388 (p2054), and a C to G at residue −389 (p2055). A plasmid consisting of FKS2(−706 to −360)-CYC1-lacZ (plasmid p1070), which bears a LEU2 marker, was created by replacement of the URA3 marker in plasmid p916 (70) by the method of Cross (15). A plasmid consisting of CLN2(−600 to −400)-CYC1-lacZ (plasmid p2066) was constructed by replacing the 134-bp SmaI/XhoI fragment of the CYC1 promoter of pLGΔ-312 (26) with a 200-bp PCR-amplified CLN2 fragment, resulting in fusion of the CLN2 promoter sequences containing multiple SBF-binding sites to the CYC1 minimal promoter.
TABLE 2.
Plasmids
| Plasmid | Description | Reference or source |
|---|---|---|
| pRS315 | LEU2-based centromeric plasmid | 60 |
| YEp351 | LEU2-based 2μm plasmid | 31 |
| YEp352 | URA3-based 2μm plasmid | 31 |
| p740 | YEp351[MPK1] | 37 |
| p777 | YEp351[MPK1-3xHA] | 37 |
| p778 | YEp351[mpk1(T190 Y192F)-3xHA] | 37 |
| p903 | pLGΔ-312; CYC1-lacZ reporter vector | 26 |
| p904 | pLGΔ-178; CYC1-lacZ reporter vector | 26 |
| p916 | FKS2(−706 to −360)-CYC1-lacZ in p903 | 70 |
| p950 | YEp352[FKS2] | 70 |
| p1070 | FKS2(−706 to −360)-CYC1-lacZ with LEU2 marker; derived from p916 | This study |
| p1150 | YEp24(SWI4) | Joseph Gray |
| p1172 | pGBT9; 2-hybrid vector with Gal4 DBD | Clontech |
| p1173 | pGAD424; 2-hybrid vector with Gal4 AD | Clontech |
| p1366 | PRM5(YIL117c)-lacZ | 35 |
| p2022 | YEp351[MLP1-3xHA] | This study |
| p2024 | YEp351[mlp1(Y192F)-3xHA] | This study |
| p2052 | FKS2(−540 to −375)-CYC1-lacZ with URA3 marker | This study |
| p2053 | FKS2(−540 to −375)-CYC1-lacZ with SBF-binding site mutation at −386T | This study |
| p2054 | FKS2(−540 to −375)-CYC1-lacZ with SBF-binding site mutation at −388C | This study |
| p2055 | FKS2(−540 to −375)-CYC1-lacZ with SBF-binding site mutation at −389G | This study |
| p2066 | CLN2(−600 to −400)-CYC1-lacZ | This study |
| p2119 | YEp351[mpk1(K54R)-3xHA] | This study |
| p2188 | pRS315[MPK1-3xHA] | This study |
| p2190 | pRS315[mpk1(T190A Y192F)-3xHA] | This study |
| p2193 | pRS315[mpk1(K54R)-3xHA] | This study |
| p2248 | pGBT9 MPK1(1-328) | 62 |
| p2313 | YEp351[MPK1-FLAG] | 40 |
| p2316 | YEp351[mpk1(T190A Y192F)-FLAG] | This study |
| p2317 | YEp351[mpk1(K54R)-FLAG] | This study |
| p2339 | BG1805-GAL1-SWI4-ZZ-HA-6HIS | 24 |
| p2341 | BG1805-GAL1-SWI6-ZZ-HA-6HIS | 24 |
| p2344 | pRS304[swi4Δ::TRP1] | This study |
| p2345 | pRS305[swi6Δ::LEU2] | This study |
| p2346 | pRS315[MLP1-3xHA] | This study |
| p2347 | pRS315[mlp1(Y192F)-3xHA] | This study |
| p2348 | pUT34; HIS3-based MET25 promoter | 49 |
| p2349 | pUT34[ERK5-6HIS] | This study |
| p2350 | pUT34[ERK5(T219A Y221F)-6HIS] | This study |
| p2351 | pUT34[ERK5(K84R)-6HIS] | This study |
| p2352 | pGAD424 SWI4(1-1093) | This study |
| p2415 | pUT36; URA3-based MET25 promoter | 49 |
| p2418 | pUT36 (SWI4-6HIS) | This study |
PCR overlap mutagenesis was used to construct point mutants in MPK1-3xHA (where HA is hemagglutinin) and MLP1-3xHA. These alleles (mpk1-K54R-3xHA and mlp1-Y192F-3xHA), their wild-type counterparts, and mpk1-T190A Y192F-3xHA (mpk1-TA/YF-3xHA) (37) were subcloned from a URA3-based multicopy plasmid (YEp352) (31) into LEU2-based multicopy (YEp351) (31) and centromeric (pRS315) (60) plasmids, using SalI/EcoRI for MPK1 alleles and XbaI/SacI for MLP1 alleles. Mutant alleles of MPK1 (mpk1-TA/YF and mpk1-K54R) were tagged with the FLAG epitope by subcloning a SacI/SacII fragment bearing the mutant regions from the appropriate YEp351 clones into YEp351(MPK1-FLAG) (40).
SWI4-6HIS was created by PCR amplification of the SWI4 coding region using primers that include a SpeI site in the upstream primer and an Xhol site in the downstream primer. The downstream primer also included a six copies of the HIS tag adjacent to the termination codon. This fragment was cloned into pUT36 (plasmid p2415) (49), creating pUT36(SWI4-6HIS) (p2418), which expresses Swi4-6HIS under the control of the MET25 promoter and is selectable by the URA3 marker.
ERK5-6HIS was subcloned from pUT36(ERK5-6HIS) (49) (gift of P. Piper) using BamHl and Sall into vector pUT34 (plasmid p2348), creating pUT34(ERK5-6HIS) (p2349), which expresses ERK5-6HIS under the control of the MET25 promoter and is selectable by the HIS3 marker. QuikChange site-directed mutagenesis (66) was used to create pUT34(ERK5-T219A Y221F-6HIS) (p2350) and pUT34(ERK5-K84R-6HIS) (p2351).
A Gal4 activation domain (AD) fusion to Swi4 for two-hybrid analysis was constructed in pGAD424 (Clontech Technologies) (plasmid p1173) by PCR amplification of the coding region using primers that include an SalI site in the upstream primer and a BglII site in the downstream primer. The Gal4 DNA-binding domain (DBD) fusion to the catalytic domain of Mpk1 residues 1 to 328 [pGBT9-Mpk1(1-328)]; plasmid (p2248) was the gift of M. Molina. All PCR-amplified sequences were confirmed by DNA sequence analysis across the entire amplified region.
Genomic deletions of SWI4 and SWI6.
To delete the genomic copy of SWI4, 595 bp of sequence 5′ to the SWI4 start codon and 614 bp of sequence 3′ of the SWI4 stop codon were amplified in separate PCRs from genomic DNA of yeast strain 1788. The 5′ fragment was amplified with primers that placed an NotI site at the end adjacent to the SWI4 coding sequence and a PstI site at the opposite end. The 3′ fragment was amplified with primers that placed an SalI site adjacent to the SWI4 coding sequence and a PstI site at the opposite end. These fragments were ligated in a three-molecule reaction to the NotI and SalI sites of the integrative plasmid pRS304 (60) to create a unique PstI site between the fragments. The resulting plasmid, pRS304(swi4Δ::TRP1) (p2344), was linearized with PstI and used to transform yeast strains to tryptophan prototrophy.
To delete the genomic copy of SWI6, 615 bp of sequence 5′ to the SWI6 start codon and 611 bp of sequence 3′ of the SWI6 stop codon were amplified in separate PCRs from genomic DNA of yeast strain 1788. The 5′ fragment was amplified with primers that placed an NotI site at the end adjacent to the SWI6 coding sequence and a PstI site at the opposite end. The 3′ fragment was amplified with primers that placed an SalI site adjacent to the SWI6 coding sequence and a PstI site at the opposite end. These fragments were ligated in a three-molecule reaction to the NotI and SalI sites of the integrative plasmid pRS305 (60) to create a unique PstI site between the fragments. The resulting plasmid, pRS305(swi6Δ::LEU2) (p2345), was linearized with PstI and used to transform yeast strains to leucine prototrophy.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were conducted as described by Hecht and Grunstein (30), with the following modifications. Yeast cells expressing HA-tagged proteins of interest and multicopy plasmids containing various alleles of FKS2 were grown in YEPD medium at room temperature or at 39°C to an A600 of 1.5. DNA was cross-linked to proteins by the addition of formaldehyde to 1%, followed by incubation at 30°C for 1 h. Chromatin was sheared using a Branson Sonifier 450 (output setting 5) in 10-s bursts for a total of 90 s so as to generate fragments of an approximately mean length of 500 bp. Mouse monoclonal antibody 12CA5 (BabCo) was used for immunoprecipitation of 500 μg of protein. PCR (35 cycles) was used for detection of coprecipitated DNA sequences. For FKS2-CYC1-lacZ promoter association, one primer of the pair was homologous to CYC1 sequence to avoid interference from the endogenous FKS2 promoter. This was critical for experiments testing Mpk1-HA and Mlp1-HA association to the SBF-site mutant allele of the FKS2 promoter. A primer pair for amplification of part of the DYN1 gene was used as a negative control for genomic ChIP experiments. For primer sequences, see Table S1 in the supplemental material.
Coimmunoprecipitation.
Protein extractions and coimmunoprecipitation of yeast strain DL454 (mpk1Δ) expressing Swi4-His6 with Mpk1-FLAG (from 300 μg of protein) were conducted as described previously (37) using mouse monoclonal anti-FLAG (M2) affinity beads (Sigma) or protein A affinity beads (no antibody controls; Sigma). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (on 7.5% gels) of samples representing 100 μg of initial protein was followed by immunoblotting to detect either Mpk1-FLAG (M2 anti-FLAG antibody; Sigma) or Swi4-His6 (mouse monoclonal tetra-His antibody; Qiagen). Input controls represented 50 μg of initial protein.
RESULTS
Cell wall stress induction of FKS2 requires SBF.
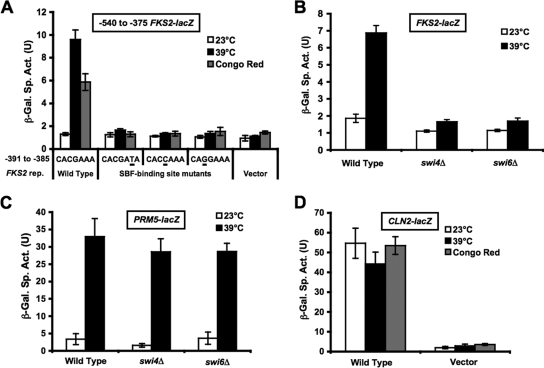
Expression of the FKS2 gene is induced in response to a variety of cell wall stressing agents including elevated growth temperature, Congo red, and calcofluor white (70; also K.-Y. Kim, unpublished data). A lacZ transcriptional reporter bearing the FKS2 promoter region between residues −706 and −360 fused to the basal CYC1 promoter (FKS2-lacZ) is responsive to thermal stress (70) and to expression of a constitutive form of the CWI MEK, Mkk1, but its induction is independent of the Rlm1 transcription factor (36). A consensus SBF-binding site (SCB; CACGAAA) resides within the responsive region of the FKS2 promoter (−385 to −391). To assess the contribution of this site to cell wall stress-induced activation of the FKS2 promoter, we mutated three residues predicted to abolish SBF binding (5). Mutant promoters were tested in the context of a shorter FKS2-lacZ reporter plasmid (with residues −540 to −375). All three mutations abolished induction of this reporter at increased temperatures or following Congo red treatment (Fig. 1A), indicating that the SCB is necessary for the response. Because elevated growth temperature was the most effective of several cell wall stressors for induction of FKS2 expression, subsequent experiments were conducted using that stress. Thermal induction of FKS2-lacZ was similarly abolished in swi4Δ and swi6Δ mutants (Fig. 1B), indicating the requirement for both subunits of SBF in this response. Because the swi4Δ mutant has an osmotically remedial growth defect at elevated temperatures (44, 50), it was necessary to conduct this experiment in the presence of sorbitol for osmotic support, which diminished somewhat the stress response in the wild-type strain. In contrast to these results, a reporter that is activated by cell wall stress through the Rlm1 transcription factor (PRM5-lacZ) was induced normally at an elevated temperature in swi4Δ and swi6Δ mutants (Fig. 1C), indicating that Rlm1-mediated transcription is not dependent on these factors and that the mutant strains are not generally impaired for transcriptional induction at high temperature.
FIG. 1.
Cell wall stress induction of FKS2 expression is dependent on an SBF-binding site and SBF. (A) The SBF-binding site in the FKS2 promoter is required for cell wall stress-induced activation. FKS2-lacZ reporter plasmids bearing point mutations in a predicted SBF-binding site (wild-type, p2052; −386T, p2053; −388C, p2054; and −389G, p2055) were transformed into wild-type yeast strain 1788. Transformants were grown to saturation at 23°C in SD-uracil medium. Cultures were diluted into 3 ml of YEPD medium so that subsequent incubation at 23°C, 39°C, or 23°C with 50 μg/ml Congo red for 15 h resulted in mid-log-phase cultures (A600 of 1.0 to 1.5). β-Galactosidase (β-Gal) activity was measured in crude extracts. (B) Mutants in SWI4 and SWI6 are defective for thermal activation of FKS2 expression. An FKS2-lacZ reporter plasmid (p2052) was transformed into wild-type (1788), swi4Δ (DL3145), or swi6Δ (DL3148) strains. Transformants were treated as described in panel A, except that cultures were diluted into YEPD medium containing 10% sorbitol for osmotic support. (C) Mutants in SWI4 and SWI6 are competent for thermal activation of PRM5 expression. A PRM5-lacZ reporter plasmid (p1366) was transformed into wild-type (1788), swi4Δ (DL3145), or swi6Δ (DL3148) strains. Transformants were treated as described in panel A, except that cultures were diluted into YEPD medium containing 10% sorbitol for osmotic support. (D) A cell cycle-regulated SCB reporter is not induced in response to cell wall stress. A CLN2-lacZ reporter plasmid (p2066) and its parent vector (p904) were transformed into the wild-type strain (1788). Transformants were treated as described in panel A. Each value represents the mean and standard deviation from three independent transformants. Sp. Act., specific activity; U, unit.
It has been demonstrated previously that a pkc1 mutation does not affect expression of an SBF-driven cell cycle-regulated promoter (33). Consistent with this, we found that an SBF-driven reporter that is regulated periodically through the cell cycle (CLN2-lacZ) showed no activation in response to cell wall stress induced either by high temperatures or by Congo red (Fig. 1D), revealing that the SCB in FKS2 functions differently than the SCBs in cell cycle-regulated genes.
Thermal induction of FKS2 requires either the Mpk1 MAPK or its pseudokinase paralog, Mlp1.
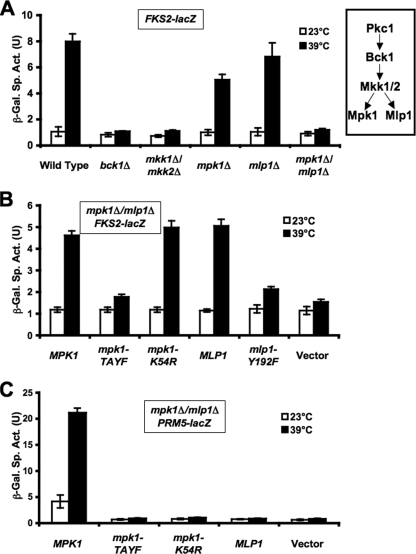
To determine which elements of the CWI signaling pathway are required for thermal induction of FKS2 through SBF, we examined mutants in the MAPK (mpk1Δ), the MEK (mkk1Δ/mkk2Δ), and the MEKK (bck1Δ) of this pathway. As above, these mutants were exposed to thermal stress in the presence of sorbitol for osmotic support. Although the bck1Δ and mkk1Δ/mkk2Δ mutants were defective for FKS2 induction, we were surprised to find that the mpk1Δ mutant was only modestly impaired (Fig. 2A). This was unexpected because the MAPK branch of the CWI signaling pathway has been thought to be linear, with Mpk1 being the sole MAPK activated through this pathway (43).
FIG. 2.
CWI requirements for thermal induction of FKS2 and PRM5. (A) Induction of FKS2 expression is dependent on either Mpk1 or its pseudokinase paralog, Mlp1. An FKS2-lacZ reporter plasmid (p2052) was transformed into the wild-type (DL3193), bck1Δ (DL3529), mkk1Δ mkk2Δ (DL3541), mpk1Δ (DL3195), mlp1Δ (DL3194), or mpk1Δ mlp1Δ (DL3196) strain. Transformants were cultured as described in the legend of Fig. 1 in the presence of YEPD medium containing 10% sorbitol for osmotic support. β-Galactosidase (β-Gal) activity was measured in crude extracts. (B) Mpk1 does not require catalytic activity for thermal induction of FKS2, but Mpk1 and Mlp1 must be phosphorylated. An FKS2-lacZ reporter plasmid (p2052) was cotransformed with centromeric plasmids bearing MPK1 (p2188), mpk1-TA/YF (p2190), mpk1-K54R (p2193), MLP1 (p2346), mlp1-Y192F (p2347), or vector (pRS315) into an mpk1Δ mlp1Δ strain (DL3196). Transformants were treated as above. (C) Mpk1 catalytic activity is required for thermal induction of PRM5. A PRM5-lacZ reporter plasmid (p1366) was cotransformed with the indicated plasmids from panel B into an mpk1Δ mlp1Δ strain (DL3196). Transformants were treated as above, except that culture time was 5 h rather than 15 h. Each value represents the mean and standard deviation from three independent transformants. Sp. Act., specific activity; U, unit.
A paralog of Mpk1, encoded by the MLP1 gene (67), exists within the yeast genome as a consequence of an ancestral whole-genome duplication (38). Mlp1 shares 53% amino acid sequence identity with Mpk1. However, Mlp1 lacks two catalytic domain residues recognized to be critical for protein kinase activity. First, a universally conserved Lys residue within subdomain II of all protein kinases, which is important for ATP positioning (28), is mutated to an Arg residue in Mlp1 (residue 54). This mutation is often engineered to create catalytically inactive forms of protein kinases. Second, a universally conserved Asp residue within the triplet DFG in subdomain VII is mutated to an Asn residue in Mlp1 (residue 171). This residue normally coordinates a magnesium ion that is also critical for ATP positioning (28), further suggesting that Mlp1 does not have the capability to bind ATP in a catalytically productive manner. Additionally, we have been unable to detect protein kinase activity associated with immunoprecipitated Mlp1 using in vitro substrates of Mpk1 (unpublished results), supporting the conclusion that Mlp1 is a pseudokinase. In addition to the differences in the ATP-binding site, a conserved Thr residue within the dual phosphorylation site of MAPK activation loops (TXY) is mutated to a Lys residue in Mlp1 (residue 190), further indicating that it is not a true MAPK. However, because phosphorylation of the activation loop Tyr residue by MEKs precedes that of the Thr residue (21, 29), it is possible that Mlp1 is phosphorylated on Tyr192 by Mkk1/2.
We tested an mlp1Δ mutation individually and in combination with mpk1Δ for thermal induction of FKS2 expression (Fig. 2A). Although the mlp1Δ mutant was not compromised, the double mutant mlp1Δ mpk1Δ was incapable of inducing FKS2 expression, indicating that these proteins serve an overlapping function for activation of transcription through SBF. This surprising result also indicated that Mlp1 drives FKS2 expression through a noncatalytic mechanism.
Because protein kinase activity is not required for Mlp1 to induce FKS2 expression, we asked if Mpk1 requires its protein kinase activity for this function. To address this, two mutant forms of Mpk1 were constructed. The first was mpk1-K54R, a mutation within the ATP-binding site, which blocks catalytic activity by interfering with ATP positioning and is analogous to Arg54 found naturally in MLP1. This allele fails to complement the cell lysis defect of an mpk1Δ mutant (46) and is devoid of detectable protein kinase activity (44, 69). The second mutant form, mpk1-TA/YF, eliminates the two phosphorylation sites within the activation loop of Mpk1 that are normally phosphorylated by Mkk1/2. This allele similarly fails to complement the cell lysis defect of an mpk1Δ mutant (42) and is devoid of detectable protein kinase activity (37). These forms were introduced into an mpk1Δ mlp1Δ double mutant on centromeric plasmids. Figure 2B shows that although the mpk1-TA/YF mutant was blocked for thermal induction of FKS2, the mpk1-K54R mutant was not, indicating that Mpk1 must be in the active (phosphorylated) conformation to drive transcription through SBF but need not be catalytically active. A mutant form of Mlp1 that eliminates its lone potential phosphorylation site (Y192F) was similarly blocked for FKS2 induction (Fig. 2B), suggesting that Mlp1 must also be in the active conformation to drive transcription. These results are in contrast to cell wall stress induction of PRM5 transcription, which requires phosphorylation of the Rlm1 transcription factor by Mpk1 (35). Mutations in Mpk1 that either block its phosphorylation by Mkk1/2 or interfere with its catalytic activity block thermal induction of PRM5 (Fig. 2C). Additionally, although Mlp1 can associate with Rlm1 (67), it cannot substitute for Mpk1 in the activation of this transcription factor (Fig. 2C).
Mpk1 and Mlp1 associate with the FKS2 promoter through Swi4.
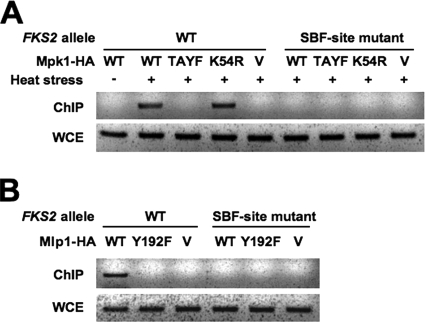
Because Mpk1 and Mlp1 must be in the active conformation but do not require catalytic activity to drive FKS2 expression through SBF, we considered a model in which these proteins act by stable association with SBF on the FKS2 promoter. We used ChIP analysis to test this model. Cells expressing epitope-tagged forms of Mpk1 were first exposed to mild heat stress to activate CWI signaling (37). Proteins were then cross-linked to DNA with formaldehyde, followed by cell lysis and DNA fragmentation. Mpk1-HA was immunoprecipitated from extracts, and coprecipitating FKS2 promoter sequence was detected by PCR. Wild-type Mpk1-HA was detected in a complex with the FKS2 promoter only after its activation by heat stress (Fig. 3A). The ATP-binding site mutant form of Mpk1 (Mpk1-K54R-HA) was also detected in a promoter complex. However, we failed to detect the phosphorylation site mutant form of Mpk1 (Mpk1-TA/YF-HA) bound to the FKS2 promoter even though this mutant protein is maintained at normal levels (37). To determine if this association was dependent on the SBF-binding site, the plasmid bearing the wild-type FKS2 promoter was replaced with one bearing the mutant SBF-binding site form described above. Mpk1-HA was not detected in association with this mutant promoter. Similarly to Mpk1-HA, wild-type Mlp1-HA was detected in a complex with the FKS2 promoter, but the phosphorylation site mutant of this protein (Mlp1-Y192F-HA) failed to bind the promoter (Fig. 3B). These results are consistent with the transcriptional data presented above and indicate that phosphorylated Mpk1 and Mlp1 drive transcription of FKS2 through association with the SCB at the promoter.
FIG. 3.
Phosphorylated Mpk1 and Mlp1 bind to the FKS2 promoter. (A) ChIP analysis of the FKS2 promoter with Mpk1-HA. A wild-type yeast strain (1788) was cotransformed with FKS2-lacZ reporter plasmids (p2052, wild-type; or p2053, SBF-site mutant −386T) and multicopy plasmids expressing the indicated HA-tagged MPK1 allele (wild-type, p777; mpk1-TA/YF, p778; mpk1-K54R, p2119; or vector, YEp351). Transformants were cultivated in YEPD medium at 23°C or subjected to thermal stress at 39°C for 15 h prior to ChIP analysis using primers designed to detect only the plasmid-borne FKS2 promoter. PCRs from whole-cell extracts (WCE) are also shown. (B) ChIP analysis of the FKS2 promoter with Mlp1-HA. Wild-type yeast strain 1788 was cotransformed with FKS2-lacZ reporter plasmids from panel A and multicopy plasmids expressing the indicated HA-tagged MLP1 allele (wild-type, p2022; mlp1-Y192F, p2024; or vector, YEp351). Transformants were cultivated and subjected to thermal stress prior to ChIP analysis, as above. WT, wild type; TAYF, T190A Y192F.
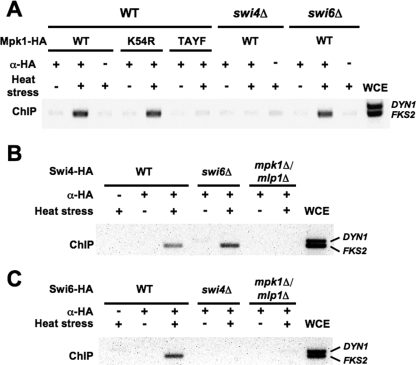
To determine the specific requirements for complex formation on the native FKS2 promoter, we conducted genomic ChIP analysis in swi4Δ, swi6Δ, and mpk1Δ/mlp1Δ mutants. As was observed using the plasmid-borne FKS2 promoter, wild-type Mpk1-HA and its ATP-binding site mutant form bound to the genomic FKS2 promoter in response to activation by heat stress (Fig. 4A). By contrast, the phosphorylation site mutant form of Mpk1-HA failed to bind FKS2. Mpk1-HA did not associate with the FKS2 promoter in an swi4Δ mutant but was still capable of association in the swi6Δ mutant (Fig. 4A). The latter result was somewhat surprising because Swi4 requires Swi6 to bind DNA at cell cycle-regulated promoters (4, 6, 58), and this suggests that Mpk1 can replace that function of Swi6 in this context. Swi4-HA associated with the FKS2 promoter in an swi6Δ mutant but not in an mpk1Δ mlp1Δ mutant (Fig. 4B), supporting the conclusion that Mpk1 (or Mlp1), but not Swi6, confers DNA-binding ability to Swi4 in this promoter context. Thus, Mpk1/Mlp1 and Swi4 promoter binding is codependent. Finally, because Swi6 is required for thermal induction of FKS2 expression, we examined the requirements for Swi6-HA promoter binding. We found that Swi6 association with the FKS2 promoter requires both Mpk1/Mlp1 and Swi4 (Fig. 4C). Taken together, these results support a model in which phosphorylated Mpk1 (or Mlp1) in stable complex with Swi4 is competent to bind the FKS2 promoter at the SBF binding site. However, transcriptional activation of FKS2 requires the further recruitment of Swi6 to the complex.
FIG. 4.
Interdependent recruitment of Mpk1 and Swi4 to the FKS2 promoter. (A) Genomic ChIP demonstrates that association of Mpk1-HA with the FKS2 promoter is dependent on Swi4 but not Swi6. Yeast strains (wild-type, DL3187; swi4Δ, DL3405; or swi6Δ, DL3233) were transformed with a multicopy plasmid bearing the indicated MPK1-HA allele and subjected to thermal stress prior to ChIP analysis as described in the legend of Fig. 3, except that strains were cultivated in the presence of 10% sorbitol. An additional primer pair for PCR amplification DYN1 sequence was included as a negative control. (B) Association of Swi4-HA with the FKS2 promoter is dependent on Mpk1/Mlp1 but not Swi6. Yeast strains (wild-type, DL3187; swi6Δ, DL3233; or mpk1Δ mlp1Δ, DL3183) were transformed with a multicopy plasmid bearing SWI4-HA (p2339) and treated as above. (C) Association of Swi6 with the FKS2 promoter is dependent on both Swi4 and Mpk1/Mlp1. Yeast strains (wild-type, DL3187; swi4Δ, DL3405; or mpk1Δ mlp1Δ, DL3183) were transformed with a multicopy plasmid bearing SWI6-HA (p2341) and treated as above. WCE, whole-cell extract; WT, wild type; α, anti; TAYF, mpk1-T190A Y192F.
Association of Mpk1 with Swi4 is regulated by the phosphorylation state of Mpk1.
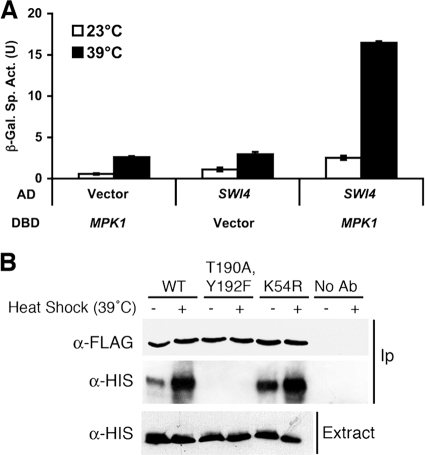
Phosphorylation of Mpk1 might regulate its association with Swi4, indicating that formation of the heterodimeric complex is the key step controlling DNA binding. Alternatively, Mpk1 might reside constitutively in complex with Swi4 and induce a conformational shift in response to activation that allows DNA binding of the complex. Therefore, we examined the regulation of the interaction of these proteins by two-hybrid analysis and by coimmunoprecipitation. Because Mpk1 possesses a transcriptional activation region within its C-terminal domain (62) that would interfere with two-hybrid analysis, we used a truncated form [Gal4DBD-Mpk1(1-328)] that possesses only the Mpk1 catalytic domain. A full-length Swi4 fusion (Gal4AD-Swi4) displayed interaction with Mpk1 but only in response to thermal stress (Fig. 5A), revealing that the interaction is regulated by the activation state of Mpk1.
FIG. 5.
Physical association of activated Mpk1 with Swi4. (A) Two-hybrid association of Mpk1 with Swi4 is dependent on activation of Mpk1. A Gal4DBD fusion to the catalytic domain of Mpk1, Mpk1(1-328) (plasmid p2248), was tested for interaction with a Gal4AD fusion to Swi4 (p2352) in yeast two-hybrid strain SFY526. Transformants were cultivated in selective medium for 15 h either at 23°C or 39°C. Vector controls (Gal4DBD vector, p1172; Gal4AD vector, p1173) are included for each fusion. Each value represents the mean and standard deviation from three independent transformants. (B) Coimmunoprecipitation (IP) of Swi4 with Mpk1 requires phosphorylation of Mpk1 but not its protein kinase activity. Yeast strain DL454 (mpk1Δ) was cotransformed with multicopy plasmids bearing the indicated FLAG-tagged MPK1 allele (wild-type, p2313; mpk1-TA/YF, p2316; mpk1-K54R, p2317) and His-tagged SWI4 allele (p2418). Transformants were cultivated to mid-log phase in selective medium lacking methionine (to induce expression of Swi4-His6) and either subjected to heat stress for 1 h at 39°C or maintained at 25°C. Cell extracts (Input) and immunoprecipitates with anti-FLAG M2 affinity gel (Sigma) or protein A affinity gel (Sigma) or no-antibody controls were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by immunoblotting with anti-FLAG or anti-His (Qiagen) antibodies. α, anti; WT, wild type; β-Gal, β-galactosidase; Sp. Act., specific activity; U, unit; Ab, antibody.
We next tested for coimmunoprecipitation of Mpk1 and Swi4. Epitope-tagged forms of Mpk1 were immunoprecipitated from extracts of cells that had been either exposed to thermal stress or grown under nonstress conditions. Epitope-tagged Swi4 (Swi4-6His) coprecipitated with either wild-type Mpk1 (Mpk1-FLAG) or the ATP-binding site mutant form of Mpk1 (Mpk1-K54R-FLAG) with an increased signal under thermal stress conditions (Fig. 5B). However, Swi4 failed to associate with the phosphorylation site mutant form of Mpk1 (Mpk1-TA/YF-FLAG) under either condition, confirming that Mpk1 must be in the active (phosphorylated) state but does not require protein kinase activity to bind Swi4. Thus, we propose that Mpk1 (or Mlp1), activated by cell wall stress, associates with Swi4. This dimeric complex is competent to bind the SCB of the FKS2 promoter. Finally, Swi6 associates with this ternary complex to drive transcription initiation (Fig. 6).
FIG. 6.
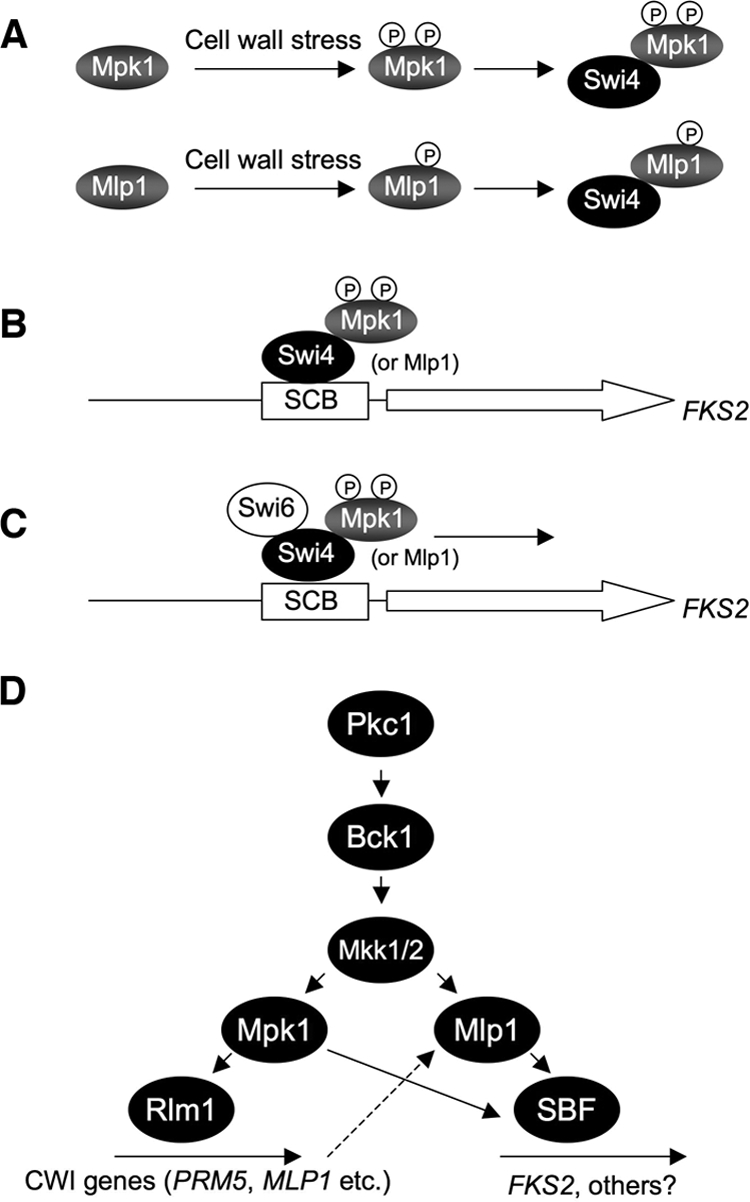
Model for CWI-regulated induction of FKS2 expression. (A) Cell wall stress results in phosphorylation and activation of the Mpk1 MAPK and the Mlp1 pseudokinase. Activated Mpk1 or Mlp1 binds Swi4. (B) The dimeric complex binds to the SBF-binding site within the FKS2 promoter. (C) Swi6 engages the ternary complex to initiate transcription. (D) Model for activation of transcription by CWI signaling. Cell wall stress activates the Mpk1 MAPK and the Rlm1 transcription factor. MLP1 expression is under the transcriptional control of Rlm1. Stress-induced Mlp1 is phosphorylated by MEKs (Mkk1/2) and stimulates FKS2 transcription redundantly with Mpk1 by noncatalytic activation of SBF. P, phosphate.
Human ERK5 drives FKS2 expression through a noncatalytic mechanism.
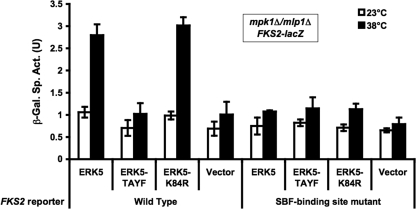
When expressed in yeast, the human ERK5 MAPK is activated in response to cell wall stress and suppresses the phenotypic defects of an mpk1Δ mutant (65). Additionally, ERK5 is capable of activating the Rlm1 transcription factor. We therefore asked if the noncatalytic transcriptional activation of SBF by Mpk1 and Mlp1 is also conserved in ERK5. Human ERK5 variants were expressed in an mpk1Δ mlp1Δ mutant and tested for their ability to activate FKS2 transcription in response to thermal stress. Both the wild-type and ATP-binding site mutant (K84R) alleles of ERK5 were capable of activating FKS2 reporter expression (Fig. 7). By contrast, a nonphosphorylatable form of ERK5 (T219A Y221F) failed to show an increase over the vector control. As was seen for Mpk1 and Mlp1, induction of FKS2 by ERK5 was dependent on the SCB. We conclude that the noncatalytic transcriptional function of Mpk1 and Mlp1 is conserved in ERK5.
FIG. 7.
The noncatalytic transcriptional function of Mpk1 and Mlp1 is conserved in human ERK5. FKS2-lacZ reporter plasmids (wild-type, p2052; SBF-binding site mutant, p2053) were cotransformed with ERK5 expression plasmids into an mpk1Δ mlp1Δ strain (DL3196). ERK5 plasmids were wild-type (ERK5; p2349), ERK5-T219A Y221F (TAYF; p2350), ERK5-K84R (p2351), or vector (pUT34; p2348). Transformants were cultured as described in the legend of Fig. 1 in the presence of YEPD medium containing 10% sorbitol for osmotic support. β-Galactosidase (β-Gal) activity was measured in crude extracts. Each value represents the mean and standard deviation from three independent transformants. Sp. Act., specific activity; U, unit.
DISCUSSION
A common mechanism by which MAPKs regulate gene expression in response to environmental stresses is through phosphorylation of transcription factors. However, protein kinases can also regulate transcription through the formation of stable interactions on the DNA. For example, the yeast Kss1 and Fus3 MAPKs function as repressors of transcription in the unphosphorylated state through association with the Ste12/Tec1 transcription factor complex (8, 14, 45). Inactive Kss1 binds directly to Ste12, recruiting the Dig1 and Dig2 transcriptional repressors to the DNA. Phosphorylation of Kss1 destabilizes this complex, causing derepression through a mechanism that does not require Kss1 protein kinase activity. Additionally, recent reports have revealed that some MAPKs can drive gene expression as integral components of transcription complexes (2, 3, 17, 51, 52, 53). This has been studied most thoroughly in the yeast Hog1 stress-activated protein kinase, which is activated by osmotic stress and uses several distinct mechanisms for inducing gene expression including recruitment of transcription factors, chromatin remodelers, and RNA polymerase to responsive promoters (reviewed in reference 20). All of these mechanisms require Hog1 protein kinase activity. In this study, we investigated the mechanism by which the yeast Mpk1 MAPK and its pseudokinase paralog, Mlp1, regulate gene expression.
Mpk1 and its pseudokinase paralog, Mlp1, function as transcriptional coactivators through a noncatalytic mechanism.
The majority of transcriptional regulation through the CWI signaling pathway results from phosphorylation and activation of the Rlm1 transcription factor by Mpk1 (23, 35, 36, 55). We demonstrated that Mpk1 additionally uses a novel mechanism that does not require its protein kinase activity to drive expression of the FKS2 gene in response to cell wall stress. This mechanism, which involves stable association with the FKS2 promoter, was revealed through the discovery that Mpk1 is redundant with its catalytically inactive paralog Mlp1 for this specific function. Mpk1 and Mlp1 must nevertheless be activated through phosphorylation by their cognate MEKs (Mkk1/2).
In addition to phosphorylated Mpk1 or Mlp1, we found that FKS2 transcription in response to wall stress requires the Swi4/Swi6 (SBF) transcription factor. SBF regulates G1 cyclin transcription in a cell cycle-periodic manner (11) but has been suggested to serve another function in the response to cell wall stress (7, 44). We found that association of phosphorylated Mpk1 and Mlp1 with the FKS2 promoter is dependent on an SBF-binding site within the promoter. Moreover, promoter association of Mpk1 and Swi4 was codependent but independent of Swi6. This is in contrast to the promoters of SBF-regulated cell cycle genes, in which association of Swi4 with Swi6 by their C termini renders Swi4 competent to engage SBF-binding sites (4, 6, 58). Intramolecular association of the Swi4 C terminus with its DBD interferes with its ability to bind DNA, which is relieved by Swi6 association. Our results indicate that in the context of the FKS2 promoter, Mpk1 (or Mlp1) replaces the function of Swi6 in allowing Swi4 to bind DNA. However, it is likely that Swi4 possesses separate binding sites for Mpk1 and Swi6 because Mpk1 forms a ternary complex with SBF in vivo and in vitro (7, 44). The difference between the SBF-binding site in the FKS2 promoter, which is not regulated periodically through the cell cycle (48, 63), and the sites found in promoters of genes subject to G1-specific transcriptional regulation must be defined outside of the core SBF-binding site, which is identical in FKS2 and cell cycle-regulated genes. Perhaps the presence of a single SBF-binding site in FKS2, rather than the 2 to 10 sites found in cell cycle-regulated genes (5), contributes to this differential regulation.
Mpk1 has been shown previously to associate directly with Swi4 in vitro but not with Swi6 (7). We found that Mpk1 must be in the active conformation to associate with Swi4. MAPKs are activated by dual phosphorylation of neighboring Thr and Tyr residues in a TXY motif within their so-called activation loops, which is thought to relieve steric inhibition of protein substrate binding (28). The requirement for Mpk1 phosphorylation to bind Swi4 therefore suggests that the kinase engages Swi4 as though it is a substrate. Nevertheless, catalytic activity is not required for either DNA binding of the resulting complex or for transcriptional activation.
Although Swi6 is not required for Mpk1 or Swi4 to bind the FKS2 promoter, we found that it is required for transcriptional activation and that Swi6 associates with the FKS2 promoter in an Mpk1- and Swi4-dependent manner. Mpk1 is known to phosphorylate Swi6 in response to activation by cell wall stress (44). However, the consequence of phosphorylation has not been investigated. Our results demonstrate that this modification is not required for the involvement of Swi6 in FKS2 induction. Indeed, loss of Mpk1 catalytic activity results in a small, but reproducible enhancement in transcriptional activation of FKS2 (Fig. 2B). Mpk1 phosphorylation of Swi6 may be a negative regulatory modification, similar to phosphorylation of Swi6 by Cdc28, which results in its nuclear egress (25, 59). To our knowledge, this noncatalytic mode of transcriptional activation by a protein kinase (or a pseudokinase) is unprecedented.
The Mlp1 pseudokinase.
Mlp1 is a paralog of Mpk1 that arose as a consequence of an ancestral whole-genome duplication (38). It was identified initially as a protein that interacts with the Rlm1 transcription factor (67). However, we demonstrated that Mlp1 neither contributes to Rlm1-driven transcription (Fig. 2C) nor interferes appreciably (K.-Y. Kim, unpublished). Although Mlp1 is not a catalytically active protein kinase, it possesses a Tyr residue within its activation loop that, as in MAPKs, is predicted to be phosphorylated in response to an activating signal. Mutation of this residue to Phe blocked the ability of Mlp1 to drive FKS2 transcription and to associate with the FKS2 promoter in response to cell wall stress. This result strongly suggests that tyrosine phosphorylation induces a conformational change in Mlp1 that allows it to bind Swi4 in a manner similar to activated Mpk1. As noted above, MAPKs are dually phosphorylated on neighboring Thr and Tyr residues. However, the activation loop Thr is replaced by a Lys residue in Mlp1. The significance of this change is not clear, but it evidently allows Mlp1 to be activated through a single phosphorylation event. This may alter its kinetics of activation by Mkk1/2 (34) or inactivation by the Msg5 and Sdp1 dual-specificity protein phosphatases (22, 27).
It is interesting that the MLP1 gene is transcriptionally induced in response to cell wall stress (35, 36). Its induction results from phosphorylation and activation of the Rlm1 transcription factor by Mpk1. Thus, in the absence of Mpk1 or cell wall stress, very little Mlp1 is expressed (unpublished results). Nevertheless, it is evident that sufficient Mlp1 exists in the absence of Mpk1 to activate FKS2 transcription to nearly normal levels (Fig. 2A). Figure 6C depicts a revised model of the CWI MAPK cascade based on these results.
Other characterized pseudokinases fall into two categories. Some, including mammalian STRAD (10), the ErbB3 epidermal growth factor receptor (9), and kinase suppressor of Ras (56), form complexes with active protein kinases (LKB1, ErbB, and Raf, respectively) and are critical to the regulation of their associated kinases. Others exist as pseudokinase domains within active protein kinases, which also serve to regulate kinase activity. This group includes the yeast Gcn2 (54) and mammalian JAK kinases (57). Although we have not tested for a physical interaction between Mpk1 and Mlp1, our results clearly demonstrate that their functions are not interdependent.
What is the consequence of the absence of protein kinase activity in Mlp1? This is not yet clear, but it appears that mutational loss of catalytic activity in Mpk1 paralogs has evolved independently at least twice. Saccharomyces castellii is a yeast species which, like S. cerevisiae, arose after an ancestral whole-genome duplication event that was followed by deletion of most of the paralogous genes (38). Also like S. cerevisiae, S. castellii retained both copies of the MPK1 gene and appears to have allowed one protein to evolve to a catalytically inactive state (Scas_678.13 [wolfe.gen.tcd.ie/browser]) but through a different set of mutations than those that inactivated S. cerevisiae Mlp1. This convergent evolution suggests that loss of catalytic activity in Mpk1 paralogs was the product of selection.
The noncatalytic mechanism for activation of FKS2 expression is conserved in human ERK5.
The human ERK5 MAPK complements loss of Mpk1 with respect to its cell lysis defect and transcriptional activation through Rlm1 (65). We found that this complementation extends to the noncatalytic transcriptional activation of FKS2 through SBF. Although no Swi4 or Swi6 homologs are evident in metazoan genomes, our results raise the possibility that ERK5, or perhaps other MAPKs, possess previously unappreciated noncatalytic functions that require a signal from upstream kinases.
Supplementary Material
Acknowledgments
We thank Andrew Sobering and Un Sung Jung for early studies with Mlp1; Brenda Andrews, Fred Cross, Val Culotta, Jean François, Joe Gray, Eric Grote, María Molina, and Peter Piper for strains and plasmids; Alan Hinnebusch, Humberto Martín, and María Molina for valuable discussions; Eric Grote for comments on the manuscript; and an anonymous reviewer for valuable suggestions.
This work was supported by a grant from the NIH (GM48533) to D.E.L.
Footnotes
Published ahead of print on 11 February 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abe, J., M. Kusuhara, R. J. Ulevitch, B. C. Berk, and J. D. Lee. 1996. Big mitogen-activated protein kinase 1 (BMK1) is a redox-sensitive kinase. J. Biol. Chem. 27116586-16590. [DOI] [PubMed] [Google Scholar]
- 2.Alepuz, P. M., E. de Nadal, M. Zapater, G. Ammerer, and F. Posas. 2003. Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. EMBO J. 222433-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alepuz, P. M., A. Jovanovic, V. Reiser, and G. Ammerer. 2001. Stress-induced MAP kinase Hog1 is part of transcription activation complexes. Mol. Cell 7767-777. [DOI] [PubMed] [Google Scholar]
- 4.Andrews, B. J., and L. A. Moore. 1992a. Interaction of the yeast Swi4 and Swi6 cell cycle regulatory proteins in vitro. Proc. Natl. Acad. Sci. USA 8911852-11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews, B. J., and L. A. Moore. 1992b. Mutational analysis of a DNA sequence involved in linking gene expression to the cell cycle. Biochem. Cell Biol. 701073-1080. [DOI] [PubMed] [Google Scholar]
- 6.Baetz, K., and B. Andrews. 1999. Regulation of the cell cycle transcription factor Swi4 through auto-inhibition of DNA binding. Mol. Cell. Biol. 196729-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baetz, K., J. Moffat, J. Haynes, M. Chang, and B. Andrews. 2001. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol. Cell. Biol. 216515-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardwell, L., J. G. Cook, D. Voora, D. M. Baggott, A. R. Martinez, and J. Thorner. 1998. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 122887-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger, M. B., J. M. Mendrola, and M. A. Lemmon. 2004. ErbB3/HER3 does not homodimerize upon neuregulin binding at the cell surface. FEBS Lett. 569332-336. [DOI] [PubMed] [Google Scholar]
- 10.Boudreau, J., J. W. Scott, N. Resta, M. Deak, A. Kieloch, D. Komander, D. G. Hardie, A. R. Prescott, D. M. F. van Aalten, and D. R. Alessi. 2004. Analysis of the LKB1-STRAD-MO25 complex. J. Cell Sci. 1176365-6375. [DOI] [PubMed] [Google Scholar]
- 11.Breeden, L. L. 2003. Periodic transcription: a cycle within a cycle. Curr. Biol. 13R31-38. [DOI] [PubMed] [Google Scholar]
- 12.Buehrer, B. M., and B. Errede. 1997. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol. Cell. Biol. 176517-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cid, V. J., A. Duran, F. Rey, M. P. Snyder, C. Nombela, and M. Sanchez. 1995. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol. Rev. 59345-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook, J. G., L. Bardwell, and J. Thorner. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous growth signaling pathway. Nature 39085-88. [DOI] [PubMed] [Google Scholar]
- 15.Cross, F. R. 1997. “Marker swap” plasmids: convenient tools for budding yeast molecular genetics. Yeast 13647653. [DOI] [PubMed] [Google Scholar]
- 16.Davenport, K. R., M. Sohaskey, Y. Kamada, D. E. Levin, and M. C. Gustin. 1995. A second osmosensing signal transduction pathway in yeast. Hypotonic shock activates the PKC1 protein kinase-regulated cell integrity pathway. J. Biol. Chem. 2730157-30161. [DOI] [PubMed] [Google Scholar]
- 17.de Nadal, E., M. Zapater, P. M. Alepuz, L. Sumoy, G. Mas, and F. Posas. 2004. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427370-374. [DOI] [PubMed] [Google Scholar]
- 18.de Nobel, H., C. Ruiz, H. Martin, W. Morris, S. Brul, M. Molina, and F. M. Klis. 2000. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 1462121-2132. [DOI] [PubMed] [Google Scholar]
- 19.Dodou, E., and R. Treisman. 1997. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 171848-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edmunds, J. W., and L. C. Mahadevan. 2004. MAP kinases as structural adaptors and enzymatic activators in transcription complexes. J. Cell Sci. 1173715-3723. [DOI] [PubMed] [Google Scholar]
- 21.Ferrell, J. E., and R. R. Bhatt. 1997. Mechanistic studies of the dual phosphorylation of Mitogen-activated protein kinase. J. Biol. Chem. 27219008-19016. [DOI] [PubMed] [Google Scholar]
- 22.Flandez, M., I. C. Cosano, C. Nombela, H. Martin, and M. Molina. 2004. Reciprocal regulation between Slt2 MAPK and isoforms of Msg5 dual-specificity protein phosphatase modulates the yeast cell integrity pathway. J. Biol. Chem. 27911027-11034. [DOI] [PubMed] [Google Scholar]
- 23.Garcia, R., C. Bermejo, C. Grau, R. Perez, J. M. Rodriquez-Pena, J. Francois, C. Nombela, and J. Arroyo. 2004. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 27915183-15195. [DOI] [PubMed] [Google Scholar]
- 24.Gelperin, D. M., M. A. White, M. L. Wilkinson, Y. Kon, L. A. Kung, K. J. Wise, N. Lopez-Hoyo, L. Jiang, S. Piccirillo, H. Yu, M. Gerstein, M. E. Dumont, E. M. Phizicky, M. Snyder, and E. J. Grayhack. 2005. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 192816-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geymonat, M., A. Spanos, G. P. Wells, S. J. Smerdon, and S. G. Sedgwick. 2004. Clb6/Cdc28 and Cdc14 regulate the phosphorylation status and cellular localization of Swi6. Mol. Cell. Biol. 242277-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guarente, L., and T. Mason. 1983. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell 321279-1286. [DOI] [PubMed] [Google Scholar]
- 27.Hahn, J.-S., and D. J. Thiele. 2002. Regulation of the Saccharomyces cerevisiae Slt2 kinase pathway by the stress-inducible Sdp1 dual specificity phosphatase. J. Biol. Chem. 27721278-21284. [DOI] [PubMed] [Google Scholar]
- 28.Hanks, S. K., and T. Hunter. 1995. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9576-596. [PubMed] [Google Scholar]
- 29.Haystead, T. A. J., P. Dent, J. Wu, M. M. Haystead, and T. W. Sturgill. 1992. Ordered phosphorylation of p42mapk by MAP kinase kinase. FEBS Lett. 30617-22. [DOI] [PubMed] [Google Scholar]
- 30.Hecht, A., and M. Grunstein. 1999. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 304399-414. [DOI] [PubMed] [Google Scholar]
- 31.Hill, J. E., A. M. Muers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2163-167. [DOI] [PubMed] [Google Scholar]
- 32.Ho, S.-N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 7751-59. [DOI] [PubMed] [Google Scholar]
- 33.Igual, J. C., A. L. Johnson, and L. H. Johnston. 1996. Coordinated regulation of gene expression by the cell cycle transcription factor SWI4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 155001-5013. [PMC free article] [PubMed] [Google Scholar]
- 34.Irie, K., M. Takase, K. S. Lee, D. E. Levin, H. Araki, K. Matsumoto, and Y. Oshima. 1993. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase kinase homologs, function in the pathway mediated by protein kinase C. Mol. Cell. Biol. 133076-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung, U. S., A. K. Sobering, M. J. Romeo, and D. E. Levin. 2002. Regulation of the yest Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46781-789. [DOI] [PubMed] [Google Scholar]
- 36.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 341049-1057. [DOI] [PubMed] [Google Scholar]
- 37.Kamada, Y., U. S. Jung, J. Piotrowski, and D. E. Levin. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 91559-1571. [DOI] [PubMed] [Google Scholar]
- 38.Kellis, M., B. W. Birren, and E. S. Lander. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428617-624. [DOI] [PubMed] [Google Scholar]
- 39.Ketela, T., R. Green, and H. Bussey. 1999. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J. Bacteriol. 1813330-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, K.-Y., I. C. Cosano, D. E. Levin, M. Molina, and H. Martin. 2007. Dissecting the transcriptional activation function of the cell wall integrity MAP kinase. Yeast 24335-342. [DOI] [PubMed] [Google Scholar]
- 41.Klis, F. M. 1994. Review: cell wall assembly in yeast. Yeast 10851-869. [DOI] [PubMed] [Google Scholar]
- 42.Lee, K. S., K. Irie, Y. Gotoh, Y. Watanabe, H. Araki, E. Nishida, K. Matsumoto, and D. E. Levin. 1993. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signaling by protein kinase C. Mol. Cell. Biol. 133067-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levin, D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Micro. Mol. Biol. Rev. 69262-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madden, K., Y. J. Sheu, K. Baetz, B. Andrews, and M. Snyder. 1997. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 2751781-1784. [DOI] [PubMed] [Google Scholar]
- 45.Madhani, H. D., C. Styles, and G. R. Fink. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91673-684. [DOI] [PubMed] [Google Scholar]
- 46.Martin, H., J. Arroyo, M. Sanchez, M. Molina, and C. Nombela. 1993. Activity of the yeast MAP kinase homologue Slt2 is critically required for cell integrity at 37 degrees C. Mol. Gen. Genet. 241177-184. [DOI] [PubMed] [Google Scholar]
- 47.Martin, H., J. M. Rodriguez-Pachon, C. Ruiz, C. Nombela, and M. Molina. 2000. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2751511-1519. [DOI] [PubMed] [Google Scholar]
- 48.Mazur, P., N. Morin, W. Baginsky, M. el-Sherbeini, J. A. Clemas, J. B. Nielsen, and F. Foor. 1995. Differential expression and function of two homologous subunits of yeast 1,3-beta-d-glucan synthase. Mol. Cell. Biol. 155671-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Millson, S. H., A. W. Truman, V. King, C. Prodromou, L. H. Pearl, and P. W. Piper. 2005. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p). Eukaryot. Cell 4849-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogas, J., B. J. Andrews, and I. Herskowitz. 1991. Transcriptional activation of CLN1, CLN2 and a putative new G1 cyclin (HCS26) by Swi4, a positive regulator of G1 specific transcription. Cell 661015-1026. [DOI] [PubMed] [Google Scholar]
- 51.Pokholok, D. K., J. Zeitlinger, N. M. Hannett, D. B. Reynolds, and R. A. Young. 2006. Activated signal transduction kinases frequently occupy target genes. Science 313533-536. [DOI] [PubMed] [Google Scholar]
- 52.Proft, M., G. Mas, E. de Nadal, A. Vendrell, N. Noriega, K. Struhl, and F. Posas. 2006. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol. Cell 23241-250. [DOI] [PubMed] [Google Scholar]
- 53.Proft, M., and K. Struhl. 2002. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 91307-1317. [DOI] [PubMed] [Google Scholar]
- 54.Qiu, H., M. T. Garcia-Barrio, and A. G. Hinnebusch. 1998. Dimerization by translation initiation factor 2 kinase GCN2 is mediated by interactions in the C-terminal ribosome-binding region and the protein kinase domain. Mol. Cell. Biol. 182697-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAP kinase pathways revealed by a matrix of global gene expression profiles. Science 287873-880. [DOI] [PubMed] [Google Scholar]
- 56.Roy, F., G. Laberge, M. Douziech, D. Ferland-McCollough, and M. Therrien. 2002. KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev. 16427-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saharinen, P., M. Vihinen, and O. Silvennoinen. 2003. Autoinhibition of Jak2 tyrosine kinase is dependent on specific regions in its pseudokinase domain. Mol. Cell. Biol. 141448-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sidorova, J. M., and L. L. Breeden. 1993. Analysis of the SWI4/SWI6 protein complex, which directs G1/S-specific transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 131069-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sidorova, J. M., G. E. Mikesell, and L. L. Breeden. 1995. Cell cycle-regulated phosphorylation of Swi6 controls its nuclear localization. Mol. Biol. Cell 61641-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siliciano, P. G., and K. Tatchell. 1984. Transcription and regulatory signals at the mating type locus in yeast. Cell 37969-978. [DOI] [PubMed] [Google Scholar]
- 62.Soler, M., A. Plovins, H. Martin, M. Molina, and C. Nombela. 1995. Characterization of domains in the yeast MAP kinase Slt2 (Mpk1) required for functional activity and in vivo interaction with protein kinases Mkk1 and Mkk2. Mol. Microbiol. 17833-842. [DOI] [PubMed] [Google Scholar]
- 63.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 93273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taylor, I. A., P. B. McIntosh, P. Pala, M. K. Treiber, S. Howell, A. N. Lane, and S. J. Smerdon. 2000. Characterization of the DNA-binding domains from the yeast cell-cycle transcription factors Mbp1 and Swi4. Biochemistry 393943-3954. [DOI] [PubMed] [Google Scholar]
- 65.Truman, A. W., S. H. Millson, J. M. Nuttall, V. King, M. Mollapour, C. Prodromou, L. H. Pearl, and P. W. Piper. 2006. Expressed in the yeast Saccharomyces cerevisiae, human ERK5 is a client of the Hsp90 chaperone that complements loss of the Slt2 (Mpk1) cell integrity stress-activated protein kinase. Euk. Cell 51914-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, W., and B. A. Malcolm. 1989. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange Site-Directed Mutagenesis. BioTechniques 26680-682. [DOI] [PubMed] [Google Scholar]
- 67.Watanabe, Y., G. Takaesu, M. Hagiwara, K. Irie, and K. Matsumoto. 1997. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 172615-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan, C., H. Luo, J. D. Lee, J. Abe, and B. C. Berk. 2001. Molecular cloning of mouse ERK5/BMK1 splice variants and characterization of ERK5 functional domains. J. Biol. Chem. 27610870-10878. [DOI] [PubMed] [Google Scholar]
- 69.Zarzov, P., C. Mazzoni, and C. Mann. 1996. The SLT2 (MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 1583-91. [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao, C., U. S. Jung, P. Garrett-Engele, T. Roe, M. S. Cyert, and D. E. Levin. 1998. Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol. Cell. Biol. 181013-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.