Abstract
Histone modifications occur in precise patterns and are proposed to signal the recruitment of effector molecules that profoundly impact chromatin structure, gene regulation, and cell cycle events. The linked modifications serine 10 phosphorylation and lysine 14 acetylation on histone H3 (H3S10phK14ac), modifications conserved from Saccharomyces cerevisiae to humans, are crucial for transcriptional activation of many genes. However, the mechanism of H3S10phK14ac involvement in these processes is unclear. To shed light on the role of this dual modification, we utilized H3 peptide affinity assays to identify H3S10phK14ac-interacting proteins. We found that the interaction of the known phospho-binding 14-3-3 proteins with H3 is dependent on the presence of both of these marks, not just phosphorylation alone. This is true of mammalian 14-3-3 proteins as well as the yeast homologues Bmh1 and Bmh2. The importance of acetylation in this interaction is also seen in vivo, where K14 acetylation is required for optimal Bmh1 recruitment to the GAL1 promoter during transcriptional activation.
Chromatin plays an important role not only in DNA packaging but also in regulating cell cycle progression and gene expression. The core histones are subject to a wide variety of posttranslational modifications that include lysine acetylation, lysine and arginine methylation, serine and threonine phosphorylation, ubiquitination, sumoylation, and ADP-ribosylation (reviewed in references 4, 18, and 26). The signal these modifications put forth can be read at the level of a singular modification as well as in the context of unique patterns of multiple modifications. It has been proposed that the different combinations of modification patterns are recognized and read by specific effector molecules that carry out the precise downstream function encoded (reviewed in references 13, 14, 17, 18, 26, and 34). The roles of these posttranslational modifications in the regulation of disparate cellular events have been the subject of intense investigation and are becoming increasingly clear. Moreover, specific domains in effector molecules that recognize acetylated and methylated histones have been and continue to be identified (reviewed in reference 18). Very little is known, however, about the involvement of some modifications, such as phosphorylation, in cellular processes, and domains that read the phosphorylation signal remain elusive.
Phosphorylation of serine 10 on histone H3 (H3S10ph) is involved in transcriptional activation, chromatin condensation, and mitotic progression (28, 30). During interphase, H3S10ph affects only a subset of genes, those that are transcriptionally activated. Mitogens stimulate H3S10ph within immediate-early response genes by the kinases Msk1 and Msk2 (mitogen- and stress-activated kinases 1 and 2) (36) in a time course consistent with the expression of these genes. In addition, H3S10ph has been shown to increase during activation of cyclic AMP-dependent protein kinase A responsive genes (10), and cytokines are known to trigger inflammatory responses that lead to H3S10ph at NF-κB-regulated promoters by the ΙκB kinase α (3, 51). In addition to the kinases described above, our previous research has identified the well-studied transcriptionally linked kinase Snf1 as an H3S10 kinase in Saccharomyces cerevisiae (20, 21).
Interestingly, H3S10ph has been linked to another modification, lysine 14 acetylation (K14ac) on the same histone tail, and the doubly modified H3S10phK14ac tail is important for transcriptional activation of several genes. Our work in Saccharomyces cerevisiae has found a mechanistic linkage between the two modifications at the INO1 gene where H3S10ph precedes and promotes K14ac on histone H3 (H3K14ac) (9, 20, 21). The histone kinase and histone acetyltransferase pair in these studies was Snf1 and Gcn5, respectively. However, this does not appear to be the case for other genes, such as GAL1, where H3K14ac exists prior to H3S10ph (22). Nevertheless, both of these modifications are essential for complete transcriptional activation of these genes (20, 21, 22). This pattern also occurs in mammals during proto-oncogene induction involving the Msk1 and Rsk2 kinases (7, 33) as well as during beta interferon activation (1). However, during these pathways of induction, it is not entirely clear whether these modifications work in concert with one another or are uncoupled in establishing complete transcriptional activation (8, 37).
Many phospho-specific binding domains have been identified for nonhistone proteins, such as WW, FHA, WD40, LRR, Polo box, SH2, PTB, BRCT, and 14-3-3 (38, 49, 50). 14-3-3 is a family of well-characterized phospho-Ser/Thr-binding proteins that function as adaptors/chaperones (48). These approximately 30-kDa acidic proteins form homo- and heterodimers and bind to other proteins to alter their conformation, enzymatic activity, interaction with other proteins, or intracellular localization (47). 14-3-3 proteins are highly conserved from yeast to mammals (32, 44) and are even interchangeable among species (40, 42). In mammals, there are at least seven 14-3-3 isoforms, and they are known to interact with over 200 proteins (reviewed in reference 2). S. cerevisiae has only two isoforms, Bmh1 and Bmh2, which are most closely related to mammalian 14-3-3ɛ (39, 41). Deletion of either of the BMH genes alone has little effect on the cell (43). Disruption of both genes, however, results in lethality for most laboratory strains (15, 40). The double deletion is known to be viable only in the Σ1278b strain background, and this results in severe growth phenotypes and increased sensitivity to a variety of stresses (31).
It has long been established that 14-3-3 proteins bind to chromatin-modifying proteins and transcriptional regulators, such as histone acetyltransferase 1 (16), histone deacetylases (5), p53 (45), and TATA-binding protein (29). They have also been found to bind to histones (6, 25), although it was not previously known whether any specific histone modification promoted 14-3-3 binding. During the course of our studies, another group reported that 14-3-3 binds to H3S10ph and that this binding occurs during gene activation in mammalian cells (24). However, this analysis did not report increased relative binding to the dual modification H3S10phK14ac as we show here, nor was there an examination of the binding in S. cerevisiae, which, as we describe here, allows for examination of binding to the combinatorial modification pattern in vivo. Our results demonstrate a novel feature of an effector molecule in that 14-3-3 proteins are able to specifically recognize a dual modification pattern apparently within a single domain in 14-3-3.
MATERIALS AND METHODS
Peptides.
The amino acid sequences of the histone peptides are listed in Table 1. These were synthesized by several sources: Abgent (San Diego, CA), Open Biosystems (Huntsville, AL), and the Baylor College of Medicine Protein Chemistry Core (Houston, TX). Peptides were attached to SulfoLink coupling gel (Pierce, Rockford, IL) via the C-terminal cysteine at a concentration of 1 mg/ml per the manufacturer's recommendation.
TABLE 1.
Peptides used in this study
| Peptide | Amino acid sequencea |
|---|---|
| H3 (unmodified) | ARTKQTARKSTGGKAPRKQLASKAAR-C |
| H3S10ph | ARTKQTARK(pS)TGGKAPRKQLASKAAR-C |
| H3K9acS10ph | ARTKQTAR(Kac)(pS)TGGKAPRKQLASKAAR-C |
| H3K9acS10phK14ac | ARTKQTAR(Kac)(pS)TGG(Kac)APRKQLASKAAR-C |
| H3S10phK14ac | ARTKQTARK(pS)TGG(Kac)APRKQLASKAAR-C |
| H3K14ac | ARTKQTARKSTGG(Kac)APRKQLASKAAR-C |
| H4 (unmodified) | SGRGKGGKGLGKGGAKRHR-C |
| H4S1ph | (pS)GRGKGGKGLGKGGAKRHR-C |
The C-terminal cysteine is indicated by -C at the right end of the sequence. pS, phosphorylated serine; Kac, lysine (K) acetylation.
Peptide affinity pull-down assays.
HeLa cell nuclear extract was prepared as described previously (12), diluted to 4 mg/ml in IPH buffer (50 mM Tris [pH 8], 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40 [vol/vol]) as described previously (52), and precleared with an equal volume of cysteine-blocked, peptide-free SulfoLink coupling gel. Sodium butyrate (Sigma-Aldrich, St. Louis, MO) was added to a final concentration of 250 mM, and phosphatase inhibitor cocktail was added per the manufacturer's recommendation (Sigma-Aldrich, St. Louis, MO). Affinity assays were performed by incubating 5 to 10 μg of immobilized peptide with 0.6 to 1.2 mg of nuclear extract for 1.5 h on a rotator at 4°C. Resin was washed five times in 1 ml IPH300 buffer (containing 300 mM NaCl [final concentration]) and boiled in sodium dodecyl sulfate (SDS) sample buffer, and proteins were resolved on Tris-glycine gels (Invitrogen, Carlsbad, CA) and detected by Silver Stain Plus (Bio-Rad, Hercules, CA) or colloidal blue staining (Invitrogen, Carlsbad, CA). Yeast whole-cell extract (WCE) was prepared from cells grown to mid-log phase. Strains used in this study are listed in Table 2. Pellets were washed once in EB350 (40 mM HEPES [pH 7.5], 350 mM NaCl, 0o.1% Tween 20, and 10% glycerol), resuspended in 1 ml EB350 containing sodium butyrate and phosphatase inhibitors as described above, and lysed with a mini bead beater three times for 1.25 min each time with 2-min rest intervals on ice between pulses. The lysate was separated from beads and cleared by centrifugation at 4°C for 15 min at 12,000 rpm. Affinity assays were performed as described above except that 1 to 2 mg of WCE was incubated with the peptide resin. Recombinant protein (0.5 to 1.0 μg) was diluted in IPH or EB containing the indicated NaCl concentration and incubated with peptide resin and analyzed as described above. Mass spectrometry was performed by the proteomics facility at The Wistar Institute (Philadelphia, PA).
TABLE 2.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference(s) or source |
|---|---|---|
| LPY8056 | matahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128 htt1-hhfΔ::LEU2 hht2-hhf2::HIS3 pRM204 [HHT2-HHF2-CEN-TRP1] | 20, 21, 22 |
| LPY8058 | matahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128 htt1-hhfΔ::LEU2 hht2-hhf2::HIS3 pRM204 [HHT2(S10A)-HHF2-CEN-TRP1] | 20, 21, 22 |
| WWY75 | matahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128 htt1-hhfΔ::LEU2 hht2-hhf2::HIS3 pRM204 [HHT2(K14R)-HHF2-CEN-TRP1] | This study |
| WWY76 | matahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128d htt1-hhfΔ::LEU2 hht2-hhf2::HIS3 pRM204 [HHT2(S10AK14R)-HHF2-CEN-TRP1] | This study |
| YKI155 | matahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128d htt1-hhfΔ::LEU2 hht2-hhf2::HIS3 pRM204 [FLAG-HHT2-HHF2-CEN-TRP1] | This study |
| DBY111 | matahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128d htt1-hhfΔ::LEU2 hht2-hhf2::HIS3 pRM204 [FLAG-HHT2(K14R)-HHF2-CEN-TRP1] | This study |
| WWY5 | matahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128d htt1-hhfΔ::LEU2 hht2-hhf2::HIS3 pRM204 [HHT2-HHF2-CEN-TRP1] three-FLAG-Bmh1 kanMX6 | This study |
| WWY69 | matahis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128d htt1-hhfΔ::LEU2 hht2-hhf2::HIS3 pDM9 [HHT1-HHF1-CEN-URA3] three-HA-Bmh1 kanMX6 | This study |
| RRY1217 | MATabmh1::HIS3 bmh2::HIS3 ura3-52 his3 leu2 trp1 | 31 |
| 10560-4a | MATaura3-52 leu2 trp1 his3 | 31 |
Western blots and antibodies.
Affinity assays were performed as described above, but proteins resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) were transferred to nitrocellulose and probed with the following antibodies: anti-14-3-3ɛ (T-16; Santa Cruz Biotechnology, Santa Cruz, CA), anti-Hsp70 (Hsp72; Stressgen, Ann Arbor, MI), anti-FLAG M2-peroxidase (conjugated to horseradish peroxidase) (Sigma-Aldrich, St. Louis, MO), antibody to glutathione S-transferase (anti-GST) (Upstate/Millipore, Charlottesville, VA), and antihemagglutinin (anti-HA) conjugated to horseradish peroxidase (clone 3F10; Roche, Indianapolis, IN).
Yeast strains.
S. cerevisiae strains used in this study are described in Table 2. C-terminal epitope tagging of endogenous proteins was performed as described previously (23). Following selection on appropriate media, correct integration was tested by PCR of genomic DNA, and tagged proteins were also immunoprecipitated and analyzed by Western blotting. Strains containing histone mutations were created as described previously (20, 21, 22).
Plasmids and recombinant proteins.
The BMH1 and BMH2 genes were amplified from wild-type (IPY36 background) yeast genomic DNA using the following primers: for BMH1, 5′-GGAATTCATATGATGTCAACCAGTCGTGAAGAT-3′ and 5′-CGCGGATCCTTACTTTGGTGCTTCACCTTCG-3′; and for BMH2, 5′-GGAATTCATATGATGTCCCAAACTCGTGAAGA-3′ and 5′-CGCGGATCCTTATTTGGTTGGTTCACCTTG-3′.
PCR products were digested with NdeI and BamHI (New England Biolabs, Ipswich, MA) and cloned into pET28a+ vector (Novagen, San Diego, CA) digested with the same enzymes. Recombinant protein was expressed in Escherichia coli BL21(DE3) cells (Stratagene, La Jolla, CA), purified using Ni2+-nitrilotriacetic acid agarose resin (Qiagen, Valencia, CA) per the manufacturer's recommendations, and concentrated to 12 mg/ml. Recombinant GST-tagged human 14-3-3ɛ was purchased from Biomol (Plymouth Meeting, PA). In addition, BMH2 was amplified using the primers 5′-CGCGGATCCATGTCCCAAACTCGTGAAGAT-3′ and 5′-CCGGAATTCTTATTTGGTTGGTTCACCTTG-3′. The PCR product was digested with BamHI and EcoRI (New England Biolabs, Ipswich, MA) and cloned into the pGEX-4T-1 vector (GE Healthcare, Piscataway, NJ) digested with the same enzymes. The protein was expressed in E. coli BL21(DE3) cells and purified using glutathione Sepharose resin (GE Healthcare, Piscataway, NJ) per the manufacturer's recommendations.
Affinity assay with GST-Bmh2.
About 10 mg WCE prepared from S. cerevisiae strains YKI155 and DBY111 as described above was incubated with 10 μg of glutathione Sepharose-immobilized GST-Bmh2 for 3 h on a rotator at 4°C. Resin was washed five times for 5 min each time in EB350 containing sodium butyrate and phosphatase inhibitors and analyzed as described above.
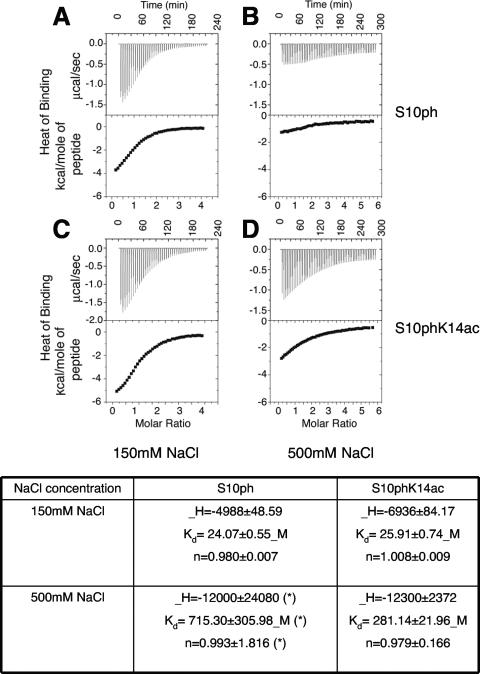
ITC studies.
All isothermal titration calorimetry (ITC) measurements were carried out using a MicroCal VP-ITC isothermal titration calorimeter (MicroCal, Inc.). Bmh1 was dialyzed against phosphate-buffered saline (PBS) at 4°C overnight and diluted to a final concentration of 90 μM, and the H3S10ph and H3S10phK14ac peptides (Abgent) were resuspended to 3.0 mM in the resultant dialysis buffer. About 1.4 ml of Bmh1 protein was added into the sample cell, and about 300 μl of the peptides was loaded into the injection syringe with the desired dilution. For each titration experiment, a 300-second delay at the start of the experiment was followed by selected number of injections (at 300-second intervals), with the sample cell stirred at 270 rpm throughout and maintained at 25°C. For titrations in PBS buffer, H3 peptides were titrated into Bmh1 at 2.4 mM with 42 injections (5 μl/each). In another round of titrations, NaCl was added to both protein and peptide solutions to a final concentration of 500 mM, and peptides were titrated at 2.4 mM with 56 injections (5 μl/each). Titration data were analyzed using the Origin 5.0 software supplied by MicroCal, Inc., and data sets were corrected for baseline heats of dilutions from control runs as appropriate. The corrected data were then fit to a theoretical titration curve describing one binding site per titrant. The area under each peak of the resultant heat profile was integrated and plotted against the molar ratio of Bmh1 to H3 peptide. A nonlinear best-fit binding isotherm for the data was used to calculate Bmh1/H3 peptide stoichiometry, dissociation constant, and standard enthalpy change. Other details are similar to those previously described (35).
ChIP.
Chromatin immunoprecipitation (ChIP) was performed based on a method previously described (27). Cells were grown in 50 ml of yeast extract-peptone-dextrose (YEPD) to a density of ∼1.5 × 107 cells/ml followed by growth for 2 h in YEP containing 2% raffinose. Galactose was then added to a final concentration of 2%, and the cells were allowed to grow for a further 3 h before cross-linking for 30 min by the addition of formaldehyde (1% final concentration) (Fisher Chemicals). Cross-linking was stopped by the addition of 2.5 ml of 2.5 M glycine, and cells were harvested and resuspended in 500 μl FA buffer [0.1% SDS, 1% Triton X-100, 10 mM HEPES, 0.1% deoxycholate, 150 mM NaCl, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), 0.8 μg/ml pepstatin A, 0.6 μg/ml leupeptin, 1 μl/10 ml Z-Leu-Leu-Leu-Al] following two PBS washes. Lysis of the yeast cells was performed by grinding with acid-washed glass beads. Fixed chromatin was sonicated using a bioruptor (Cosmo Bio) for 30 min at 3°C (1-min pulses with 20-second intervals) to a fragment size of less than 500 bp. Immunoprecipitations were performed using 200 μl of the sonicated chromatin solution and 3 μl anti-Bmh1 (kind gift from Paul van Heusden), 3 μl anti-histone H3 overnight at 4°C with rotation. Purification of the immunoprecipitated protein/DNA complexes was achieved by adding 40 μl of 50% slurry protein A-Sepharose beads (CL-4B Amersham) for a minimum of 90 min. Following binding, beads were washed with TSE-150/500 (1% Triton X-100, 0.1% SDS, 2 mM EDTA, 20 mM Tris HCl [pH 8.0], 150/500 mM NaCl), LiCl wash (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris HCl [pH 8.0]) and Tris-EDTA. Elution of bound protein/DNA was achieved by 30-min incubation with 100 μl elution buffer (1% SDS, 0.1 M NaHCO3) prewarmed to 65°C. DNA was uncross-linked for a minimum of 6 h by incubation at 65°C followed by ethanol precipitation. All samples were treated with RNase A and proteinase K (Roche) prior to purification with QIAquick PCR purification columns (Qiagen). DNA was quantified using real-time PCR with the Rotorgene-3000 system (Corbett Robotics). PCR was performed using Sensimix (Quantance SYBR green) and the following primers: for GAL1, 5′-GAACGAGAAAAATCCATCCA-3′ and 5′-CAAACCTTTCCGGTGCAAGT-3′; and for MET16, 5′-GCCAGCAAAGGTATCAACCC-3′ and 5′-GCGTTTCCAGCTTGATTAGT-3′. Quantitation of the DNA was calculated on the basis of a standard curve generated from serially diluted sonicated yeast genomic DNA. The percent input was calculated from three replicates of immunoprecipitation (IP), control IP (no antibody), and input DNA using the following calculation: [(Σ IP signals/Σ IP samples) − (Σ no-antibody signals/Σ no-antibody samples)]/(Σ input signals/Σ input samples). Binding of anti-Bmh1 was normalized by taking a ratio of Bmh1 percent input/histone H3 percent input).
Northern blotting.
Induction of GAL1 was performed as described above for ChIP. Extraction of RNA was performed using hot phenol extraction. Cells were resuspended in 400 μl TES (10 mM Tris HCl [pH 7.5], 5 mM EDTA [pH 8.0], 1% SDS) followed by the addition of an equivalent volume of acid phenol (pH 4.5) (Qbiogene) and incubated for 1 h at 65°C with shaking. RNA was purified by phenol-chloroform treatment and ethanol precipitated. Northern blotting was performed using standard technique and a probe that spanned the coding region from −401 to +1573 relative to the ATG.
RESULTS
14-3-3 binds to H3 specifically with the pattern S10phK14ac.
With the aim of gaining insight into the role of the linked modifications H3S10phK14ac in transcriptional activation, we performed peptide affinity assays to identify effector molecules that recognize these marks in combination. Peptides corresponding to the N-terminal tail of H3 histones were synthesized such that they contained no modifications, a single modification (H3S10ph or H3K14ac), or both modifications on the same tail (H3S10phK14ac). In addition, H4 peptides (unmodified and S1ph) were synthesized for use as a control, and a C-terminal cysteine was incorporated into all peptides for coupling onto SulfoLink (Pierce) resin (Table 1).
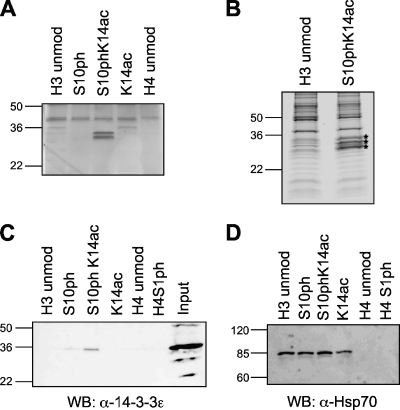
Peptide-linked resin was incubated in the presence of HeLa cell nuclear extract for 1.5 h, washed extensively with buffer containing 300 mM NaCl, resuspended in SDS-PAGE loading buffer, and boiled. Resolution of the bound proteins was achieved by SDS-PAGE, and silver staining revealed that a group of proteins around 30 kDa in size specifically bound to the H3 tail peptides in a pattern-specific manner. This binding was not detectable with the unmodified peptides or in the presence of either single modification but was observed only when both modifications occurred together (Fig. 1A).
FIG. 1.
14-3-3 proteins bind specifically to H3S10phK14ac peptides. (A) Proteins bound to differentially modified or unmodified (unmod) H3 and H4 peptides after incubation with HeLa cell nuclear extract, washing in the presence of 300 mM NaCl, resolution by SDS-PAGE (16% gel), and silver staining. (B) Proteins bound and resolved as described above for panel A but visualized by colloidal blue staining. Bands analyzed by mass spectrometry are indicated by asterisks. (C) Western blot (WB) of proteins bound to peptides as described above for panel A using an antibody against 14-3-3ɛ (α-14-3-3ɛ). (D) Western blot analysis of proteins bound to peptides as described above for panel A (except with resolution on 8% gel) with antibody against Hsp70 (α-HSP70). The numbers to the left of the gels are the sizes (in kilodaltons) of the proteins.
In order to identify these proteins, the binding reaction was scaled up, and the corresponding bands were subjected to digestion and mass spectrometry (Fig. 1B). The proteins were identified as the 14-3-3 family proteins, specifically, the α, β, ɛ, γ, and ζ isoforms. The presence of 14-3-3ɛ was confirmed by Western blotting (Fig. 1C). In order to confirm that there were equal loads in the lanes, we additionally probed for Hsp70. We had previously identified Hsp70 as a H3-binding protein that is not dependent on the modifications studied in this assay (data not shown), and therefore, the Western blot with antibody against Hsp70 confirms comparable loads of peptides and input proteins for each H3-bound sample (Fig. 1D).
Yeast 14-3-3 homologues Bmh1 and Bmh2 bind to H3S10 phK14ac.
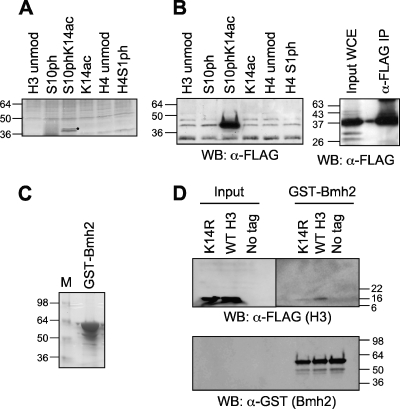
There are two 14-3-3 homologues in S. cerevisiae, the major isoform Bmh1 and the minor isoform Bmh2. Given that Bmh1 and Bmh2 are most closely related to the human 14-3-3ɛ isoform (39, 41), the fact that human 14-3-3ɛ bound specifically to H3S10phK14ac prompted us to test whether the yeast proteins also exhibited this pattern-specific recognition of the H3 tail. As described for the human nuclear extract above, yeast WCE was incubated with peptide-linked resin and washed extensively with buffer containing 350 mM NaCl. Bound proteins were analyzed by SDS-PAGE and silver staining (Fig. 2A). Once again, proteins around 30 kDa in size were detected in the sample containing the phosphoacetylated H3S10phK14ac peptide. This binding was specific to the dual-modified H3 peptide, as the proteins did not bind to the unmodified H3 peptide or to the singly modified H3 peptides containing S10ph or K14ac. Moreover, this binding was specific to H3; no binding was visualized in the presence of the unmodified H4 peptide or even in the presence of the phosphorylated H4 peptide H4S1ph. Once again, the samples were scaled up, and the bound proteins were identified by mass spectrometry to be the 14-3-3 family proteins (Bmh1 and Bmh2 [data not shown]).
FIG. 2.
Yeast 14-3-3 proteins also bind specifically to the phosphoacetylated H3 peptide. (A) Proteins bound to modified or unmodified (unmod) H3 and H4 peptides after incubation with yeast whole-cell extract (WCE), washing in the presence of 350 mM NaCl, resolution by SDS-PAGE (4 to 12% gel), and silver staining. (B) (Left) Western blot (WB) of bound proteins as described above for panel A, but incubating with WCE containing three-FLAG-tagged Bmh1 and probing with antibody against FLAG epitope (α-FLAG). Proteins were resolved on a 16% gel. (Right) Anti-FLAG Western blot of input WCE and anti-FLAG IP. Proteins were resolved on a 4 to 20% gel. (C) Purified, glutathione Sepharose-immobilized GST-Bmh2 resolved on 4 to 12% gel and stained with colloidal blue. Lane M, molecular size standards. (D) (Top) Western blot analysis using anti-FLAG antibody (α-FLAG) to detect FLAG-tagged H3 in input and proteins bound to GST-Bmh2 after incubation in the presence of WCE derived from strains containing wild-type and K14R H3, washing at 350 mM NaCl, and resolution by SDS-PAGE (4 to 12% gel). (Bottom) The same blot was probed with anti-GST antibody (α-GST) to detect GST-Bmh2 as a loading control. The numbers to the sides of the gels are the sizes (in kilodaltons) of the proteins.
To investigate whether Bmh1 was bound to the phosphoacetylated peptide, we incubated peptide-linked resin in the presence of yeast WCE containing C-terminally three-FLAG-tagged Bmh1 (Fig. 2B, right panel) and extensively washed the resin in buffer containing 350 mM NaCl as before. Western blot analysis with antibody against the FLAG epitope revealed that Bmh1 was indeed specifically interacting with the H3S10phK14ac peptide (Fig. 2B, left panel).
Given that 14-3-3 proteins are known to interact with phosphorylated proteins, it is not surprising that H3S10ph facilitates interaction with the histone. Intriguingly, K14ac appeared to be equally important for this interaction to survive the stringent washes used in this technique. We wondered whether this could be an artifact of using immobilized peptides and thus examined whether this interaction could still occur in the reverse situation where the 14-3-3 protein was immobilized and the histones were free in solution. The advantage of using yeast as a model system to study the role of histone modifications is that strains can be generated in which the only copy of a specific histone gene is mutated at one or more defined residues. This allows us to substitute certain amino acids which are modified with residues that cannot be modified but maintain a charge similar to the original amino acid. Thus, we can rigorously investigate the importance of this posttranslational modification in a specific process. We took advantage of this technique to determine whether H3K14ac was truly important for binding of the yeast 14-3-3 proteins. We engineered a strain in which H3K14 was mutated to arginine (H3K14R). A single N-terminal FLAG epitope tag was also incorporated into the H3 gene. We then performed an affinity assay using purified GST-Bmh2 immobilized on glutathione resin (Fig. 2C) and yeast WCE from strains containing wild-type H3 or H3K14R. We focused on the K14R substitution, since this was the key difference from the previous study, and the clear novel prediction is that the single substitution should lower binding of 14-3-3 proteins (24). The resin was washed extensively with buffer containing 350 mM NaCl, and the binding proteins were resolved by SDS-PAGE and transferred to nitrocellulose. Western blot analysis with anti-GST antibody confirmed that equal amounts of GST-Bmh2 were used for the assay (Fig. 2D, bottom panel), and then the same blot was probed with anti-FLAG antibody to detect H3 (Fig. 2D, top right panel). We found that Bmh2 had an increased affinity for wild-type H3, which contained intact K14, compared to H3K14R where K14 could not be acetylated. The amounts of H3 in these two samples were similar (Fig. 2D, input, top left panel), indicating that acetylation of K14 is indeed required for the interaction of 14-3-3 proteins with H3.
14-3-3 binding is direct and increased by acetylation.
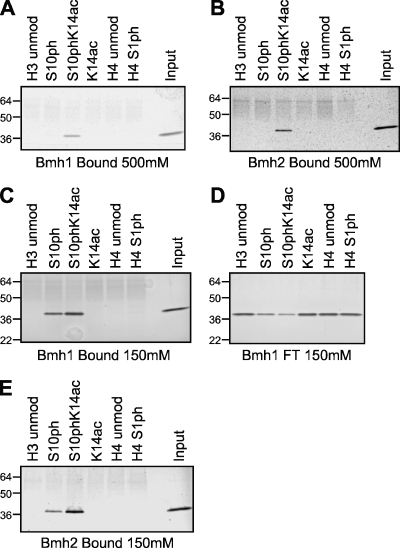
We then sought to determine whether binding of 14-3-3 family proteins to modified H3 was direct or whether other accessory proteins were necessary for this interaction. For this, we performed binding reactions with peptide-linked resin and six-His-tagged Bmh1 or Bmh2 purified from bacteria followed by extensive washing with high-ionic-strength buffer containing 500 mM NaCl. After SDS-PAGE and silver staining, it was determined that both yeast 14-3-3 isoforms bind directly to the phosphoacetylated H3 tail peptide (Fig. 3A and B). Binding was not detected for the singly modified H3 peptide S10ph or K14ac, the unmodified H3 peptide, and control unmodified H4 or H4S1ph peptide.
FIG. 3.
Recombinant Bmh1 and Bmh2 directly bind to H3S10 phK14ac peptides and also bind to H3S10ph peptides at lower ionic strength. (A) Bmh1 bound to H3 and H4 peptide resin after washing in the presence of 500 mM NaCl, resolution by SDS-PAGE (4 to 12% gel), and silver staining. unmod, unmodified. (B) Bmh2 bound to resin as described above for panel A. (C) Bmh1 bound to H3 and H4 peptide resin after binding and washing in the presence of 150 mM NaCl, resolution by SDS-PAGE (4 to 12% gel), and silver staining. (D) Flowthrough (FT) fractions of unbound Bmh1 from the binding experiment in panel C. (E) Bmh2 bound to resin after washing at 150 mM NaCl as described above for panel A. The numbers to the left of the gels are the sizes (in kilodaltons) of the proteins.
Recently, it was reported that 14-3-3 interacts with H3 when S10 is phosphorylated and that acetylation of K9 and K14 on the same histone tail does not interfere with this interaction (24). Our results do lend support to this observation, except that we could not detect binding of 14-3-3 to H3S10ph in the absence of K14ac. Our binding experiments were performed using very stringent conditions with binding and/or washing steps performed in buffers containing 300 mM (or more) NaCl. Note that the direct binding experiments with Bmh1 and Bmh2 were analyzed at 500 mM NaCl. This suggested that the interaction between the yeast 14-3-3 proteins and H3S10phK14ac was stable enough to survive extensive washing at higher ionic strength, whereas the potential interaction with H3S10ph may not survive under these conditions. Thus, we tested this hypothesis by decreasing the ionic strength of the binding and washing buffers to enable this potentially weaker interaction to persist. Indeed, when binding of purified Bmh1 or Bmh2 to the immobilized peptides was examined at 150 mM NaCl, we could detect interaction of the 14-3-3 proteins with the singly modified H3S10ph peptide (Fig. 3C and E, respectively). Notably, though, the interaction with the doubly modified H3S10phK14ac peptide still appeared to be stronger. This is not only obvious in the bound material (Fig. 3C and 4D) but also evident in the flowthrough fractions (Fig. 3D for Bmh1, data not shown for Bmh2, and Fig. 4E for 14-3-3).
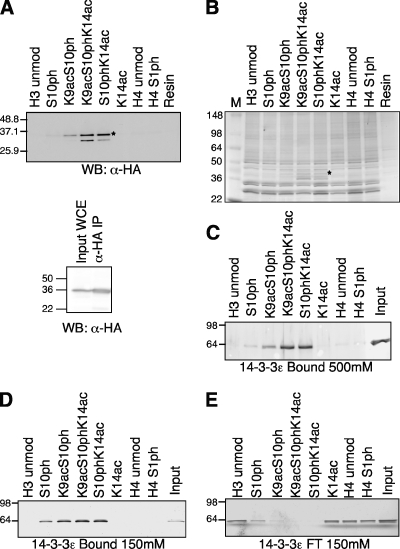
FIG. 4.
The influence of H3K9ac on Bmh1 binding is less than that of H3K14ac. (A) (Top) Western blot (WB) of proteins bound to modified or unmodified (unmod) H3 and H4 peptides after incubation with yeast WCE containing three-HA-tagged Bmh1, washing in the presence of 350 mM NaCl, resolution by SDS-PAGE (4 to 20% gel), transferring to nitrocellulose, and probing with antibody against HA epitope (α-HA). (Bottom) Anti-HA Western blot of input WCE and anti-HA IP (4 to 12% gel). (B) Silver-stained gel of bound proteins from the same samples as in the experiment in panel A (4 to 20% gel). Lane M, molecular size standards. (C) Recombinant GST-tagged human 14-3-3ɛ bound to peptide resin after washing in the presence of 500 mM NaCl, resolution by SDS-PAGE (4 to 12% gel), and silver staining. (D) 14-3-3ɛ bound to peptide resin after washing in the presence of 150 mM NaCl, resolution by SDS-PAGE (4 to 12% gel), and silver staining. (E) Flowthrough (FT) fractions of unbound 14-3-3ɛ from the binding experiment in panel D (4 to 12% gel). The numbers to the left of the gels are the molecular sizes (in kilodaltons) of the proteins.
Given that acetylation of H3K14 was enhancing the relative interaction of the 14-3-3 proteins with H3S10ph, we decided to examine the influence of another known site of acetylation, H3K9. Here, we incubated yeast WCE containing C-terminally HA-tagged Bmh1 with peptide-linked resin (Fig. 4A, bottom panel), and this time we included the H3K9acS10ph peptide modified twice and the H3K9acS10phK14ac peptide modified three times in the analysis. The binding of Bmh1 to the H3 tail peptides in the presence or absence of modifications was monitored by Western blotting with antibody against the HA epitope (Fig. 4A). A silver-stained gel of the bound material is included here as a loading control (Fig. 4B [note that the Bmh1 binding can be detected in the stained gel]). Once again, the Bmh1 interaction with H3 showed a strong preference for the linked S10phK14ac modifications over S10ph alone (Fig. 4A). Interestingly, K9ac did lead to some increase in Bmh1 binding in combination with S10ph, but this interaction was not as strong as that of S10phK14ac, suggesting that the K14ac plays a more important role in regulating the interaction of 14-3-3 proteins with H3 in the presence of S10ph.
We further investigated the direct binding of purified GST-tagged human 14-3-3ɛ to H3S10ph in the presence of K14ac or K9ac (Fig. 4C). As expected, at 500 mM NaCl, 14-3-3 bound strongly to H3S10phK14ac, while the binding to S10ph alone was minimal. Similar to the result with yeast WCE, the interaction of 14-3-3ɛ was increased in the presence of K9ac, but the strength of this binding was not as great as with the double modification S10phK14ac. Even at lower ionic strength, the influence of K9ac on the increase of this interaction is not as prominent as that of K14ac. This is seen in both the bound protein (Fig. 4D) and is also evident in the flowthrough fractions (Fig. 4E). Again, this indicated that K14 is likely to be the main acetylation site involved in recruiting 14-3-3 to the histone tail in vivo.
As mentioned earlier, a previous report indicated that acetylation did not enhance binding of 14-3-3 to H3S10ph (24). Surface plasmon resonance and ITC were employed to show that the binding constants for the H3S10ph and phosphoacetylated (H3K9acS10phK14ac) peptides with 14-3-3 were similar (24). Indeed, as described above, at 150 mM NaCl, we could detect 14-3-3 binding to H3S10ph in our peptide affinity assays, but in our assay, the binding did not appear to be as strong as that seen for H3S10phK14ac. Moreover, at 500 mM NaCl, the interaction with the single modified H3S10ph peptide was undetectable.
We decided to investigate this further using ITC to more quantitatively measure the affinity of Bmh1 for the H3S10ph and H3S10phK14ac peptides (Fig. 5). We carried out these studies with Bmh1 protein in the reaction vessel and titrated with the respective peptide. We initially carried out these studies in PBS buffer (150 mM NaCl) and observed similar dissociation constants of Bmh1 for both peptides (24 μM for H3S10ph and 26 μM for H3S10phK14ac, respectively). These dissociation constraints are slightly lower than those that had been reported in the earlier study for human 14-3-3 proteins binding to the same peptide substrates with 250 mM salt (78 μM and 92 μM, respectively) (24). We next carried out analogous ITC studies at an increased NaCl concentration of 500 mM and found that at this higher salt concentration that there was a clear Bmh1 binding preference for the dually modified peptide substrate. Specifically, while the dual-modified H3S10phK14ac peptide still retains a well measured dissociation constant with Bmh1 protein (281 μM), the interaction of the single modified H3S10ph peptide is not detectable under the same conditions. Taken together, the ITC studies presented here demonstrates that under certain conditions Bmh1 has a clear preference for the dual H3S10phK14ac mark over the single H3S10ph mark.
FIG. 5.
Isothermal titration calorimetry of Bmh1 binding to histone H3 peptides with different modifications. For each panel (A to D), the top panel shows the raw data for injections of the peptide into the Bmh1 solution as described in Materials and Methods; the lower section shows the integrated heats of injections, where panels A and B depict S10p binding at 150 mM and 500 mM NaCl, respectively, and panels C and D depict S10pK14ac binding at 150 mM and 500 mM NaCl, respectively. At the bottom of the figure, thermodynamic parameters involved in interaction of Bmh1 with modified histone H3 peptides are shown. The single binding site constant Kd and the heat of binding ΔH were used as fitting parameters in analysis of these data with the MicroCal software. The free energy and entropy changes for binding were then calculated by using the following relationships: ΔG = −RT ln Kd and ΔG = ΔH − TΔS, respectively, where R is the gas constant, T is temperature, and ΔS is the change in substrate concentration. In each case, parameters are reported with associated errors of the fit. The large error rate in S10Pho (500 mM NaCl) titration (indicated by an asterisk) shows that the binding is too weak to be detected.
H3K14ac is crucial in recruitment of Bmh1 to the GAL1 promoter.
Our finding that H3K14ac leads to increased binding of 14-3-3 proteins with H3 compared to H3S10ph alone under stringent high-salt conditions in vitro prompted us to investigate the effect of H3K14ac on Bmh1 recruitment in vivo. A previous report suggested that 14-3-3 is recruited to the S10ph-regulated promoters c-fos and c-jun during transcriptional activation in mammalian cells (24). We previously reported that both H3S10ph and H3K14ac were required for full transcriptional activation of GAL1 in yeast (22). Thus, we decided to examine whether Bmh1 and Bmh2 played a role in transcriptional activation of this well-characterized gene.
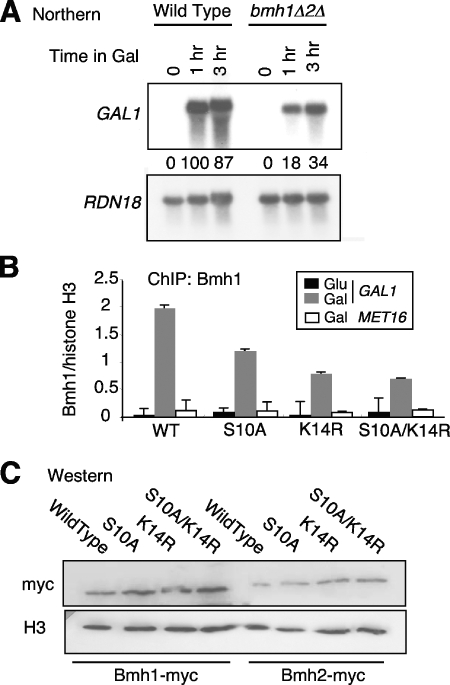
We first analyzed whether Bmh1 and Bmh2 are required to achieve optimal transcriptional activation of the GAL1 gene. While single BMH gene deletions are viable, double deletions are generally lethal (15, 40, 43). However, the double bmh1 bmh2 deletion is viable in the Σ1278b strain background (31). Thus, we used this double deletion strain, RRY1217, to analyze the influence of Bmh1 and Bmh2 on GAL1 transcriptional activation. Northern blot analysis revealed that, in the presence of galactose, the level of GAL1 transcription in the double deletion strain is only about one-third to one-fifth that of the wild-type strain (Fig. 6A). 18S RNA levels serve as a loading control. These levels are similar to the defects we previously observed in GAL1 transcription in the histone substitution strains (22). Thus, Bmh1 and Bmh2 play a role in transcriptional activation of GAL1.
FIG. 6.
Yeast 14-3-3 proteins are required for optimal GAL1 transcriptional activation in vivo, and H3K14ac plays a major role in regulating Bmh1 recruitment to GAL1 during transcription. (A) Northern blot analysis of GAL1 and control 18S RNA levels in wild-type and bmh1 bmh2 double deletion strains after growth in the presence of galactose to induce GAL1 transcription. (B) ChIP of Bmh1 in wild-type (WT) H3 and H3 mutant strains S10A, S10AK14R, and K14R. Recruitment of Bmh1 to the GAL1 and MET16 promoters were analyzed by real-time PCR with primers specific for the 5′ regions of the genes. Signals are normalized for immunoprecipitation recovery and calculated as a percentage of total IP material. The Bmh1 signal is shown normalized to H3 levels in the same chromatin sample at the GAL1 promoter. (C) Western blot analysis of Bmh1 and Bmh2 myc epitope-tagged proteins in the H3 substitution strains (wild type, S10A, S10AK14R, and K14R), normalized to histone H3 levels.
We then investigated whether Bmh1 is recruited to the GAL1 promoter during galactose induction. In our previous studies, we determined that both H3S10ph and H3K14ac exist at the GAL1 promoter during transcriptional activation (22). Thus, if Bmh1 is important for transcriptional activation of this gene and if it interacts with the dual modification (H3S10phK14ac), it would be predicted to be recruited to this promoter upon galactose induction. To test this hypothesis, we again took advantage of histone mutation strains, reasoning that, if each modification in the dual pattern S10phK14ac is involved, then recruitment should be lowered by each single substitution mutant and by the double substitution mutant—indeed, that the single substitutions should lower recruitment is the important prediction of a dual modification pattern. Thus, we looked for the presence of Bmh1 at the GAL1 promoter in wild-type H3, the double H3 mutant S10AK14R, and single H3 mutant S10A and K14R strains. ChIP as performed using antibodies against Bmh1, and PCR primers specific to the 5′ region of the GAL1 open reading frame were used to analyze Bmh1 recruitment to this location of the gene (Fig. 6B). The Bmh1 signal was normalized against H3 levels. The levels of Bmh1 and Bmh2 are not lowered in the histone substitution strains (Fig. 6C). We found that Bmh1 recruitment increases more than 10-fold going from repressing conditions (glucose) to activating conditions (galactose). Importantly, Bmh1 recruitment is decreased by about 75% in the H3S10AK14R strain compared to the wild-type H3 strain (Fig. 6B). Remarkably, the majority of this decreased recruitment was caused by the lack of K14ac rather than S10ph, as judged by recruitment levels in the single H3 K14R and S10A mutants (Fig. 6B). Bmh1 levels were reduced by about 75% in the H3K14R strain and less than 50% in the H3S10A strain compared to the wild type. MET16 was used as a negative-control locus, since it is not induced in galactose-containing media; we found no recruitment of Bmh1 to MET16 and no change in the histone substitution strains (Fig. 6B). These results suggest that, in vivo, K14ac is more important than S10ph for Bmh1 recruitment to GAL1 during transcriptional activation.
DISCUSSION
This study was designed to identify proteins that specifically recognize the histone modification pattern H3S10phK14ac. Histone peptide affinity assays were performed under very stringent conditions (washing the peptide resin in buffer containing 300 to 500 mM NaCl) with the intention of distinguishing stably interacting proteins. Here, we identified known phospho-binding 14-3-3 proteins as H3 tail-binding molecules. By this technique, it was clear that 14-3-3 preferentially binds to H3 peptides that are not only phosphorylated on S10 but are also acetylated on K14 (Fig. 1 and 2). Both of these modifications are necessary for the interaction to survive higher-ionic-strength washes, as no binding is detected for the singly modified peptides, H3S10ph or H3K14ac, under these conditions. This is true for both mammalian 14-3-3 proteins as well as the yeast homologues Bmh1 and Bmh2. Moreover, in the reverse scenario where full-length histones were employed to analyze binding to immobilized Bmh2, the K14R mutant H3, which lacks acetylation at K14, is unable to bind to Bmh2 (Fig. 2D). This suggests that, indeed, H3K14ac is required for the interaction between 14-3-3 and H3, and not just S10ph alone.
This result was somewhat surprising because during the course of this study, another group reported that 14-3-3 bound to H3S10ph alone (24). Careful analysis of this work, however, revealed that a singly modified H3S10ph peptide was not employed in this group's peptide affinity assay to directly compare the single modification to the dual modification pattern with K9ac or K14ac. Nevertheless, the previous analysis did include the singly modified S10ph peptide in surface plasmon resonance and ITC analyses (techniques performed at less-stringent, lower-ionic-strength conditions than those used in our experiments) and found that 14-3-3 had similar affinities for H3 peptides which were modified once by S10ph or modified three times by K9acS10phK14ac. We felt that direct comparison of the binding of 14-3-3 proteins with single and dual modifications under various salt conditions in vitro, combined with in vivo analyses possible in yeast, would help resolve the different findings.
We employed salt titration studies to our direct binding experiments (Fig. 3 and 4C to E) and found that 14-3-3 proteins could bind to the singly modified H3S10ph at lower ionic strength (150 mM NaCl). While the peptide affinity assays suggested that the 14-3-3 proteins still slightly preferred the interaction with the phosphoacetylated peptide over the phosphorylated peptide, our ITC results argued that Bmh1 had equal affinity for the two peptides at lower ionic strength (Fig. 5). This latter result could be partially explained, however, by nonspecific interactions between the 14-3-3 protein and the peptide which are removed by extensive washing in the peptide affinity assays. Indeed, at higher ionic strength, while the overall levels of binding decreased, the ITC results showed a clear relative preference for the interaction with the doubly modified H3S10phK14ac tail peptide over the S10ph peptide. It might also be argued that high-ionic-strength buffers in these analyses are not physiologically relevant to test the true nature of an interaction that occurs in vivo. Thus, it is possible that under these in vitro conditions, the higher concentration of salt ions would favor hydrophobic interactions (K14ac) and destabilize hydrophilic interactions (S10ph) in an artificial manner. Since the “true” ionic strength or microenvironment within the cell where this interaction occurs is unknown, we reasoned that the physiological relevance of interactions between specific histone modifications and their interacting proteins should be investigated in vivo.
We used yeast as a model system to study the role of this combinatorial H3S10phK14ac histone modification pattern on Bmh1 recruitment to GAL1 during transcriptional activation where both of these modifications are known to exist (22). We generated histone H3 mutation strains in which the only copy of the H3 gene was altered such that S10ph could not occur (S10A), K14ac could not occur (K14R), or neither modification could occur (S10AK14R). ChIP analysis revealed that both modifications are important for bringing Bmh1 to the GAL1 gene (Fig. 6B), i.e., there is a reduction in Bmh1 recruitment in both the S10A and K14R mutant strains compared to the wild-type strain where both modifications could occur. In fact, H3K14ac appears to be more important in the recruitment of Bmh1 than S10ph, as the K14R mutant displayed a 75% reduction in Bmh1 recruitment compared to less than 50% decrease for the S10A strain. Thus, in vivo, both K14ac and S10ph on H3 appear to be important for interaction of 14-3-3 proteins with the histone tail.
Our study provides evidence for the role of a histone modification pattern in recruitment of a specific effector protein during transcriptional activation. We note that, while our manuscript was under revision, another study was published which arrives at essentially the same conclusion, i.e., that 14-3-3 recognizes a dual modification pattern of phosphoacetylation on histone H3 (46). Previous investigations of tandem bromodomains or studies with multiple chromodomains have shown that they can recognize two identical modifications. For example, interaction of Brd4 with mitotic chromatin requires multiple sites of acetylation and both of its bromodomains (11). Moreover, in Arabidopsis, ITC studies indicated that two chromodomains of the CHROMOMETHYLASE3 (CMT3) DNA methyltransferase directly associated with the doubly methylated histone tail of H3. This interaction occurred only when H3 was simultaneously methylated at both the H3 lysine 9 and 27 positions, suggesting that productive binding resulted from cooperation between subunits (19). However, to our knowledge, our finding that a combinatorial pattern of two dissimilar modifications is involved in recruitment of one protein is unique. The existence of other combinatorial modification patterns within histone proteins suggests that this may be of general importance in genomic regulation.
Acknowledgments
The anti-Bmh1 antibody was a kind gift from G. P. H. van Heusden. We thank Viji Shridhar for help with cloning of Bmh1 and Bmh2, Kristin Ingvarsdottir for the FLAG-H3 strain, David Bungard for the FLAG-H3K14R strain, J. Govin for help in preparing figures, and W. Alston for help in preparing the text for publications.
An NRSA Training Program in Basic Cancer Research training grant (5T32 CA09171-28) and an American Cancer Society postdoctoral fellowship made possible by the Aramark Corporation supported the work of W.W. Research was supported by grants from NIH (GM55360) and NSF (MCB-9604208) to S.L.B.
Footnotes
Published ahead of print on 11 February 2008.
REFERENCES
- 1.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111381-392. [DOI] [PubMed] [Google Scholar]
- 2.Aitken, A. 2006. 14-3-3 proteins: a historic overview. Semin. Cancer Biol. 16162-172. [DOI] [PubMed] [Google Scholar]
- 3.Anest, V., J. L. Hanson, P. C. Cogswell, K. A. Steinbrecher, B. D. Strahl, and A. S. Baldwin. 2003. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature 423659-663. [DOI] [PubMed] [Google Scholar]
- 4.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12142-148. [DOI] [PubMed] [Google Scholar]
- 5.Bertos, N. R., A. H. Wang, and X. J. Yang. 2001. Class II histone deacetylases: structure, function, and regulation. Biochem. Cell Biol. 79243-252. [PubMed] [Google Scholar]
- 6.Chen, F., and P. D. Wagner. 1994. 14-3-3 proteins bind to histone and affect both histone phosphorylation and dephosphorylation. FEBS Lett. 347128-132. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, P., C. D. Allis, and P. Sassone-Corsi. 2000. Signaling to chromatin through histone modifications. Cell 103263-271. [DOI] [PubMed] [Google Scholar]
- 8.Cheung, P., K. G. Tanner, W. L. Cheung, P. Sassone-Corsi, J. M. Denu, and C. D. Allis. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5905-915. [DOI] [PubMed] [Google Scholar]
- 9.Clements, A., A. N. Poux, W. S. Lo, L. Pillus, S. L. Berger, and R. Marmorstein. 2003. Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol. Cell 12461-473. [DOI] [PubMed] [Google Scholar]
- 10.DeManno, D. A., J. E. Cottom, M. P. Kline, C. A. Peters, E. T. Maizels, and M. Hunzicker-Dunn. 1999. Follicle-stimulating hormone promotes histone H3 phosphorylation on serine-10. Mol. Endocrinol. 1391-105. [DOI] [PubMed] [Google Scholar]
- 11.Dey, A., F. Chitsaz, A. Abbasi, T. Misteli, and K. Ozato. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 1008758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 111475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischle, W., Y. Wang, and C. D. Allis. 2003. Binary switches and modification cassettes in histone biology and beyond. Nature 425475-479. [DOI] [PubMed] [Google Scholar]
- 14.Fischle, W., Y. Wang, and C. D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15172-183. [DOI] [PubMed] [Google Scholar]
- 15.Gelperin, D., J. Weigle, K. Nelson, P. Roseboom, K. Irie, K. Matsumoto, and S. Lemmon. 1995. 14-3-3 proteins: potential roles in vesicular transport and Ras signaling in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 9211539-11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imhof, A., and A. P. Wolffe. 1999. Purification and properties of the Xenopus Hat1 acetyltransferase: association with the 14-3-3 proteins in the oocyte nucleus. Biochemistry 3813085-13093. [DOI] [PubMed] [Google Scholar]
- 17.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 18.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128693-705. [DOI] [PubMed] [Google Scholar]
- 19.Lindroth, A. M., D. Shultis, Z. Jasencakova, J. Fuchs, L. Johnson, D. Schubert, D. Patnaik, S. Pradhan, J. Goodrich, I. Schubert, T. Jenuwein, S. Khorasanizadeh, and S. E. Jacobsen. 2004. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 234286-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo, W.-S., R. Trievel, J. Rojas, L. Duggan, D. Allis, R. Marmorstein, and S. Berger. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5917-926. [DOI] [PubMed] [Google Scholar]
- 21.Lo, W. S., L. Duggan, N. C. Emre, R. Belotserkovskya, W. S. Lane, R. Shiekhattar, and S. L. Berger. 2001. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 2931142-1146. [DOI] [PubMed] [Google Scholar]
- 22.Lo, W. S., E. R. Gamache, K. W. Henry, D. Yang, L. Pillus, and S. L. Berger. 2005. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J. 24997-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 24.Macdonald, N., J. P. Welburn, M. E. Noble, A. Nguyen, M. B. Yaffe, D. Clynes, J. G. Moggs, G. Orphanides, S. Thomson, J. W. Edmunds, A. L. Clayton, J. A. Endicott, and L. C. Mahadevan. 2005. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3. Mol. Cell 20199-211. [DOI] [PubMed] [Google Scholar]
- 25.Meek, S. E., W. S. Lane, and H. Piwnica-Worms. 2004. Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J. Biol. Chem. 27932046-32054. [DOI] [PubMed] [Google Scholar]
- 26.Millar, C. B., and M. Grunstein. 2006. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell Biol. 7657-666. [DOI] [PubMed] [Google Scholar]
- 27.Morillon, A., J. O'Sullivan, A. Azad, N. Proudfoot, and J. Mellor. 2003. Regulation of elongating RNA polymerase II by forkhead transcription factors in yeast. Science 300492-495. [DOI] [PubMed] [Google Scholar]
- 28.Nowak, S. J., and V. G. Corces. 2004. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 20214-220. [DOI] [PubMed] [Google Scholar]
- 29.Pan, S., P. C. Sehnke, R. J. Ferl, and W. B. Gurley. 1999. Specific interactions with TBP and TFIIB in vitro suggest that 14-3-3 proteins may participate in the regulation of transcription when part of a DNA binding complex. Plant Cell 111591-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prigent, C., and S. Dimitrov. 2003. Phosphorylation of serine 10 in histone H3, what for? J. Cell Sci. 1163677-3685. [DOI] [PubMed] [Google Scholar]
- 31.Roberts, R. L., H. U. Mosch, and G. R. Fink. 1997. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell 891055-1065. [DOI] [PubMed] [Google Scholar]
- 32.Rosenquist, M., P. Sehnke, R. J. Ferl, M. Sommarin, and C. Larsson. 2000. Evolution of the 14-3-3 protein family: does the large number of isoforms in multicellular organisms reflect functional specificity? J. Mol. Evol. 51446-458. [DOI] [PubMed] [Google Scholar]
- 33.Sassone-Corsi, P., C. A. Mizzen, P. Cheung, C. Crosio, L. Monaco, S. Jacquot, A. Hanauer, and C. D. Allis. 1999. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science 285886-891. [DOI] [PubMed] [Google Scholar]
- 34.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 40341-45. [DOI] [PubMed] [Google Scholar]
- 35.Tang, Y., M. V. Poustovoitov, K. Zhao, M. Garfinkel, A. Canutescu, R. Dunbrack, P. D. Adams, and R. Marmorstein. 2006. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat. Struct. Mol. Biol. 13921-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomson, S., A. L. Clayton, C. A. Hazzalin, S. Rose, M. J. Barratt, and L. C. Mahadevan. 1999. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 184779-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomson, S., A. L. Clayton, and L. C. Mahadevan. 2001. Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol. Cell 81231-1241. [DOI] [PubMed] [Google Scholar]
- 38.Tzivion, G., Y. H. Shen, and J. Zhu. 2001. 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene 206331-6338. [DOI] [PubMed] [Google Scholar]
- 39.van Hemert, M. J., G. P. van Heusden, and H. Y. Steensma. 2001. Yeast 14-3-3 proteins. Yeast 18889-895. [DOI] [PubMed] [Google Scholar]
- 40.van Heusden, G. P., D. J. Griffiths, J. C. Ford, T. F. C. Chin-A-Woeng, P. A. Schrader, A. M. Carr, and H. Y. Steensma. 1995. The 14-3-3 proteins encoded by the BMH1 and BMH2 genes are essential in the yeast Saccharomyces cerevisiae and can be replaced by a plant homologue. Eur. J. Biochem. 22945-53. [PubMed] [Google Scholar]
- 41.van Heusden, G. P., and H. Y. Steensma. 2006. Yeast 14-3-3 proteins. Yeast 23159-171. [DOI] [PubMed] [Google Scholar]
- 42.van Heusden, G. P., A. L. van der Zanden, R. J. Ferl, and H. Y. Steensma. 1996. Four Arabidopsis thaliana 14-3-3 protein isoforms can complement the lethal yeast bmh1 bmh2 double disruption. FEBS Lett. 391252-256. [DOI] [PubMed] [Google Scholar]
- 43.van Heusden, G. P., T. J. Wenzel, E. L. Lagendijk, H. Y. de Steensma, and J. A. van den Berg. 1992. Characterization of the yeast BMH1 gene encoding a putative protein homologous to mammalian protein kinase II activators and protein kinase C inhibitors. FEBS Lett. 302145-150. [DOI] [PubMed] [Google Scholar]
- 44.Wang, W., and D. C. Shakes. 1996. Molecular evolution of the 14-3-3 protein family. J. Mol. Evol. 43384-398. [DOI] [PubMed] [Google Scholar]
- 45.Waterman, M. J., E. S. Stavridi, J. L. Waterman, and T. D. Halazonetis. 1998. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat. Genet. 19175-178. [DOI] [PubMed] [Google Scholar]
- 46.Winter, S., E. Simboeck, W. Fischle, G. Zupkovitz, I. Dohnal, K. Mechtler, G. Ammerer, and C. Seiser. 2008. 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J. 2788-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yaffe, M. B. 2002. How do 14-3-3 proteins work?—Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 51353-57. [DOI] [PubMed] [Google Scholar]
- 48.Yaffe, M. B., and A. E. Elia. 2001. Phosphoserine/threonine-binding domains. Curr. Opin. Cell Biol. 13131-138. [DOI] [PubMed] [Google Scholar]
- 49.Yaffe, M. B., and S. J. Smerdon. 2001. PhosphoSerine/threonine binding domains: you can't pSERious? Structure 9R33-R38. [DOI] [PubMed] [Google Scholar]
- 50.Yaffe, M. B., and S. J. Smerdon. 2004. The use of in vitro peptide-library screens in the analysis of phosphoserine/threonine-binding domain structure and function. Annu. Rev. Biophys. Biomol. Struct. 33225-244. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto, Y., U. N. Verma, S. Prajapati, Y. T. Kwak, and R. B. Gaynor. 2003. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature 423655-659. [DOI] [PubMed] [Google Scholar]
- 52.Zegerman, P., B. Canas, D. Pappin, and T. Kouzarides. 2002. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J. Biol. Chem. 27711621-11624. [DOI] [PubMed] [Google Scholar]