Abstract
The role of chromatin-remodeling factors in transcription is well established, but the link between chromatin-remodeling complexes and DNA repair remains unexplored. Human Rvb1 and Rvb2 are highly conserved AAA+ ATP binding proteins that are part of various chromatin-remodeling complexes, such as Ino80, SNF2-related CBP activator protein (SRCAP), and Tip60/NuA4 complexes, but their molecular function is unclear. The depletion of Rvb1 increases the amount and persistence of phosphorylation on chromatin-associated H2AX after the exposure of cells to UV irradiation or to mitomycin C, cisplatin, camptothecin, or etoposide, without increasing the amount of DNA damage. Tip60 depletion, but not Ino80 or SRCAP depletion, mimics the effect of Rvb1 depletion on H2AX phosphorylation. Rvb1 is required for the histone acetyltransferase (HAT) activity of the Tip60 complex, and histone H4 acetylation is required prior to the dephosphorylation of phospho-H2AX. Thus, Rvb1 is critical for the dephosphorylation of phospho-H2AX due to the role of Rvb1 in maintaining the HAT activity of Tip60/NuA4, implicating the Rvb1-Tip60 complex in the chromatin-remodeling response of cells after DNA damage.
Rvb1 and Rvb2 (also known as Tip49 and Tip48, pontin52 and reptin52, and Tih1p and Tih2p) are two putative ATPases that are highly conserved in eukaryotes. Studies of different organisms have identified Rvb1 and Rvb2 in complexes involved in chromatin remodeling (16, 18, 21, 22, 25, 26, 29, 38) and suggested their importance in the related functions of transcriptional and developmental regulation (3, 5, 8, 13, 14, 36). The mammalian homologs have been implicated in at least two oncogenic pathways: Rvb1 associates with Myc, and dominant negative versions of Rvb1 inhibit cell transformation by Myc (12, 47). Rvb1 also functionally interacts with beta-catenin, an important mediator of the growth-suppressive signal from the adenomatous polyposis coli tumor suppressor protein (3, 4, 14). Despite the apparent involvement of Rvb1 and Rvb2 in important pathways, the molecular function of Rvb is still unclear.
Modulation in chromatin organization during transcription, replication, and DNA repair involves two classes of chromatin-remodeling factors (24, 41). The first class includes a group of ATP-dependent chromatin-remodeling factors that use the energy of ATP hydrolysis to mobilize nucleosomes and give access to the underlying DNA. The second includes acetylases, deacetylases, methylases, and demethylases that add or remove covalent modifications on histone tails and thus affect chromatin compaction. The chromatin-remodeling complexes containing metazoan Rvb include both the ATPase-type remodelers Ino80 and SNF2-related CBP activator protein (SRCAP, or Swr1) and the acetyltransferase remodeler human Tip60 (6, 11, 18, 20). The only molecular function of Rvb that is defined is in the recruitment of the ATPase Arp5 into the Saccharomyces cerevisiae Ino80 chromatin-remodeling complex, a function that is important for the remodeling activity (21, 22, 35). We therefore wondered whether metazoan Rvb had a similarly important role in the assembly or activity of an associated chromatin-remodeling complex.
DNA damage results in the phosphorylation of the histone variant, H2AX, on S139 by the ATM and ATR protein kinases. The phospho-H2AX serves as a marker for damaged DNA that recruits other proteins like MDC1, the Mre11-Rad50-Nbs1 (MRN) complex, 53BP1, and BRCA1 to the sites of DNA damage either to amplify the damage signal to the rest of the cell or to repair the DNA damage (37). Mice with a single copy of the H2AX gene are predisposed to genomic instability and cancers when engineered to have a mutation also in p53 (2, 7). Yeast with a mutation in H2A at S129, the site of damage-induced phosphorylation that is the equivalent of S139 in mammalian H2AX, show a defect in the activation of the checkpoint pathway in response to DNA damage in G1/S (17) and a defect in DNA double-strand break repair (10). The human and Drosophila Tip60 proteins remodel nucleosomes containing phospho-H2AV (the fly equivalent of phospho-H2AX) (19, 27). In this paper, we report that human Rvb1 and Tip60 are critical for the downregulation of phospho-H2AX in cells after DNA damage and that Rvb1 is required for the histone H4 acetyltransferase (HAT) activity of the human Tip60 complex. Additional results suggest that human Tip60 is required to remodel phospho-H2AX-containing nucleosomes at sites of DNA damage in vivo. Thus, we propose that metazoan Rvb is required for the proper assembly and activity of the Tip60 chromatin-remodeling complex and the response of a cell to DNA damage.
MATERIALS AND METHODS
Cell culture, transfection with siRNAs, and plasmids.
HeLa and 293T cells were cultured in Dulbecco's modified Eagle's medium containing 10% donor calf serum. For transfection with small interfering RNAs (siRNAs), cells were grown at ∼40% confluence in a 6-well plate and transfected at 24-h intervals with 100 nM annealed siRNA duplex (Invitrogen) by using Oligofectamine (Invitrogen). Four hours after the first transfection, 3× medium was added. Forty-eight hours after the first transfection, cells were split 1:2 to maintain the cells in log phase. Target sequences of the oligonucleotides used were as follows: control (GL2), CGTACGCGGAATACTTCGA; RVB1-A, TAAAGGAGACCAAGGAAGT; RVB1-B, TCTTCTCTCTCTCTCTCTA; INO80-A, TGACCTGCGTCTACACTTA; SRCAP-A, GGAAACGATTGAAGTTGAA; TIP60-A, TGATCGAGTTCAGCTATGA; and PP2A-A, CCTCGTGAATACAATTTAA. Human RVB1 and TIP60 were expressed in human cells from plasmids under the control of the cytomegalovirus promoter, whereas wild-type and mutant histone H4 proteins were expressed under the control of the EF1α promoter. Lysines 5, 8, 12, and 16 of histone H4 were mutated into alanine. Mutagenesis was performed by using QuikChange according to the instructions of the manufacturer (Stratagene). Short hairpin RNA (shRNA) expression vectors were purchased from Openbiosystems (39) and modified by expressing the shRNA cassette under the control of the RNA polymerase II promoter. Details about the shRNA construct can be provided upon request.
Antibodies.
Anti-Rvb1 antibody was generated as described before (35). Rabbit antibodies were raised against His-Ino80 (amino acids 230 to 486) and His-SRCAP (amino acids 1141 to 1395). His6-tagged proteins were expressed from the pET28a vector in Escherichia coli and purified on TALON metal affinity resin (Clontech) under denaturing conditions. Other antibodies used were as follows: anti-phospho-H2AX (Cell Signaling), anti-H2AX (Cell Signaling), anti-H2A (Cell Signaling), anti-Tip60 (a gift from Bruno Amati), anti-β-actin (Sigma), anti-Flag (Sigma), anti-acetyl-histone H4 (Upstate), anti-acetyl-lysine (Upstate), anti-histone H4 (Cell Signaling), and anti-cyclobutane pyrimidine dimer (anti-CPD; Medical and Biological Laboratories, Nagoya, Japan).
DNA damage and comet assay.
siRNA-transfected HeLa cells were either left untreated or treated with UV (20 J/m2) and harvested after 60 min of UV treatment. DNA damage was assayed by alkaline and neutral single-cell agarose gel electrophoresis essentially as described previously (33). Briefly, HeLa cells were transfected with siRNAs and 72 h later were either irradiated with UV or left unirradiated. Cells were collected after the indicated time, resuspended in ice-cold phosphate-buffered saline (PBS) at 3 × 105 cells/ml, and processed further according to the instructions of the manufacturer (Trevigen). Slides were subjected to electrophoresis (1 V/cm of distance between electrodes) either for 20 min (neutral) or for 40 min (alkaline), and cells were fixed with 70% ethanol and stained with Sybr green. Nuclei were visualized using epifluorescent illumination on a Zeiss microscope. The DNA damage in >100 cells for each experimental condition was quantified by determining the tail moment, a function of both the tail length and the intensity of the DNA in the tail relative to the total DNA, by using CometScore software (TriTek Corp.). Statistical analysis was by the Wilcoxon sum-of-rank test.
Measurement of CPD.
The amounts of CPD in UV-irradiated cells were measured by an enzyme-linked immunosorbent assay as described previously by using anti-CPD antibody (46). Briefly, cells were either left unirradiated or harvested post-UV irradiation at the times indicated in the figures. Genomic DNA was isolated, and the DNA concentration was calculated from the absorbance at 260 nm. DNA dilutions in PBS were prepared and added to the microtiter plates precoated with 0.003% protamine sulfate. The enzyme-linked immunosorbent assay was performed by following the instructions of the assay kit manufacturer (Medical and Biological Laboratories, Nagoya, Japan).
Western blotting, immunoprecipitation, and immunostaining.
For Western blotting, cells were lysed in IPH buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 0.5% [vol/vol] NP-40, 1 mM dithiothreitol, 20 mM NaF, and a protease inhibitor mix [Sigma]) and sonicated three times for 15 s each at 25% output by using a Branson microtip (3.2 mm) and a Fisher model 500 sonic dismembrator. Trichostatin A (1 μM) and sodium butyrate (5 mM) were added for 240 min before harvesting to prevent the deacetylation of histone H4 (see Fig. 7D). For immunoprecipitation, either anti-Flag M2 affinity gel (Sigma) or other test antibodies were prebound to protein A-Sepharose (GE Healthcare) for 2 h in lysis buffer and incubated with cell lysate for 4 h. Following extensive washing with lysis buffer, bound proteins were eluted by boiling in 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer.
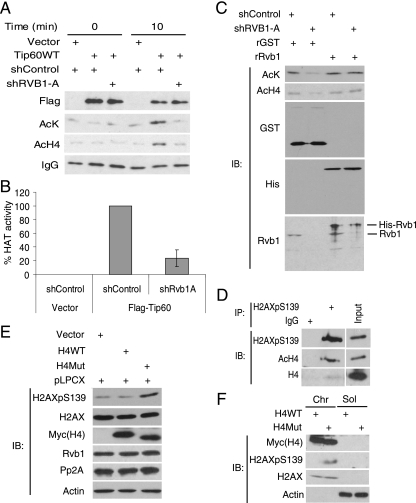
FIG. 7.
Rvb1 is required for the HAT activity of the Tip60 complex. (A) Rvb1 is required for the HAT activity of the Tip60 complex. 293T cells were transfected with a vector or a plasmid expressing FLAG-TIP60 together with a plasmid expressing an shRNA against RVB1 (shRVB1-A) or a control shRNA (shControl). The Tip60 complex was immunoprecipitated using anti-Flag antibody. Equal amounts of immune complexes were incubated for 0 or 10 min with core histones in the HAT assay. Acetylated histone H4 was detected by immunoblotting with anti-acetyl-lysine and anti-histone H4 antibodies. IgG, immunoglobulin G; AcK, acetyl-lysine; AcH4, acetyl histone H4; +, present. (B) Quantitation of HAT activity using anti-acetyl-lysine as measured in the analysis presented in panel A. Means ± SD of results from four experiments are shown. (C) Recombinant Rvb1 (rRvb1) restores the HAT activity of the Tip60 complex to cells depleted of Rvb1. The Flag-Tip60 complex was immunoprecipitated from cells transfected with the indicated plasmids and assayed for HAT activity as described for panel A. Either recombinant His6-Rvb1 (rRvb1) or nonspecific protein (recombinant glutathione S-transferase [rGST]) was added to the Tip60 complex as indicated. IB, immunoblot. (D) Phospho-H2AX coimmunoprecipitates with acetyl-histone H4. Anti-phospho-H2AX antibody was used for immunoprecipitation (IP) from lysates harvested 60 min post-UV irradiation. Immunoblotting was performed with the indicated antibodies. Ten percent inputs (input lane) are shown in parallel. H2AXpS139, H2AX phosphorylated on S139. (E) The expression of nonacetylable histone H4 increased total phospho-H2AX. 293T cells were transfected with the indicated plasmids, and cells were grown in the presence of puromycin for 48 h. Lysates were immunoblotted with anti-phospho-H2AX, anti-H2AX, anti-Myc (for wild-type and mutant histone H4 [H4WT and H4Mut]), anti-Rvb1, anti-PP2A, and anti-β-actin antibodies. pLPCX, plasmid containing the puromycin resistance gene. (F) Wild-type and nonacetylable histone H4 are incorporated equally into chromatin, but the nonacetylable H4 leads to an increase in the phosphorylation of chromatin-associated H2AX. The fractions were immunoblotted for the indicated proteins. The experiment was carried out as described for panel E. Chr, chromatin; Sol, soluble cellular extract separated from chromatin.
For immunostaining, cells cultured on cover glasses were fixed with 4% paraformaldehyde for 10 min and permeabilized with 0.2% Triton X-100 for 10 min. Following blocking with 3% bovine serum albumin (BSA) in PBS or Tris-buffered saline (TBS; for phospho-H2AX antibody) for 1 h, cells were incubated with antibody diluted in PBS or TBS containing 3% BSA for 1 h. After three washes with PBS or TBS, cells were incubated with either fluorescein isothiocyanate- or Texas Red-labeled anti-rabbit or anti-mouse immunoglobulin G antibody (DAKO) diluted in PBS or TBS containing 3% BSA for 30 min and washed three times with PBS or TBS. Cells were mounted onto a slide with Vectashield containing DAPI (4′,6′-diamidino-2-phenylindole; Vector Laboratory). The immunofluorescence analysis of chromatin-bound protein was done as described elsewhere (1). Briefly, cells were incubated in a Triton X-100 buffer (0.1% Triton X-100, 10 mM HEPES-KOH [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol) prior to formaldehyde fixation to remove detergent-sensitive proteins. Following extraction, immunostaining was performed as mentioned above. Chromatin fractions were isolated as described elsewhere (28).
HAT assay.
The Tip60 complex was immunoprecipitated as described above, washed twice in HAT assay buffer (50 mM Tris [pH 8.0], 10% glycerol, 0.1 mM EDTA, 1 mM dithiothreitol), and incubated in 25 μl of HAT assay buffer containing acetyl coenzyme A (100 μM) and core histone (2 μg) at 30°C for the times indicated in the figures before the addition of 25 μl of SDS sample buffer. Five microliters of the reaction mixture was separated by 15% SDS-PAGE, and acetylated protein was visualized by Western blotting using either anti-acetyl-lysine or anti-acetyl-histone H4 antibodies. Twenty microliters of the reaction mixture was resolved by 15% SDS-PAGE and stained with Coomassie blue to ensure the loading of equal amounts of histone H4. For the quantitation of HAT activity, the acetyl-lysine signal was quantitated by Scion imaging software and the background signal from vector-transfected cells was subtracted. The activity of the wild-type Tip60 complex was taken as 100%. Where indicated, recombinant Rvb1 was added to the Tip60 complex immunoprecipitate and incubated at 4°C for 4 h before the HAT assay.
RESULTS
The loss of Rvb1 results in an increase in phosphorylated H2AX.
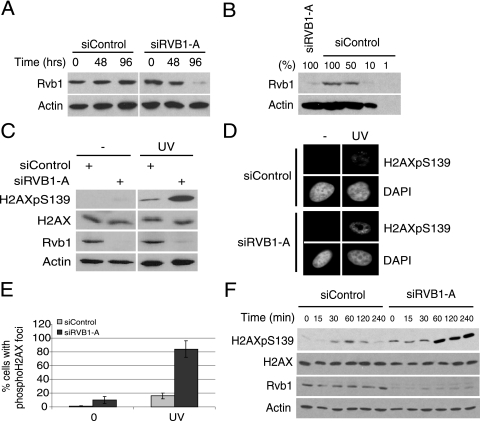
Members of the family of phosphatidylinositol 3-kinase-like kinases ATM, ATR, and DNA-PKcs are activated when cells are exposed to various kinds of DNA-damaging agents and phosphorylate H2AX at S139 as a marker for damaged DNA. Phospho-H2AX spans megabases flanking a DNA damage site and helps to recruit proteins involved in DNA damage repair. To understand the role of mammalian Rvb1 in the DNA damage response, we decreased the Rvb1 level by using siRNA. Transfection with siRVB1-A resulted in a 90% reduction in levels of Rvb1 after 72 h (Fig. 1A and B). Phosphorylation on H2AX was significantly increased without any change in H2AX levels in the absence of Rvb1 in UV-treated cells (Fig. 1C). An immunofluorescence analysis showed that both the intensity and the number of phospho-H2AX foci after UV damage were higher in the absence of Rvb1 than in the presence of Rvb1 (Fig. 1D and E). This increase in the phosphorylation of H2AX was also observed when Rvb1-depleted cells were treated with other kinds of DNA-damaging agents: mitomycin C, cisplatin, camptothecin, or etoposide (see Fig. S1 in the supplemental material). The loss of Rvb1 results in the continued accumulation of phosphorylation on H2AX without any of the downregulation seen in control cells after 60 min (Fig. 1F). These results suggest that the loss of Rvb1 increases the total level of phosphorylated H2AX without any change in H2AX levels.
FIG. 1.
The depletion of Rvb1 increases phospho-H2AX after DNA damage. (A) Depletion of Rvb1 protein using siRNA against RVB1. Lysates prepared from HeLa cells transfected with the indicated siRNAs at the indicated times were immunoblotted for Rvb1 protein and β-actin. (B) Semiquantitative Western blot analysis of Rvb1. The indicated amounts of HeLa cell lysates were loaded and blotted with anti-Rvb1 and anti-β-actin antibodies. One hundred percent equals 20 μg of protein loaded. (C) Increase in total phospho-H2AX after DNA damage. Lysates from HeLa cells were prepared after the cells were transfected with siRNA with or without UV. UV irradiation was carried out 72 h after the transfection of cells with siRNA, and cells were harvested 60 min post-UV irradiation. Results from immunoblotting with anti-phospho-H2AX, anti-H2AX, anti-Rvb1, and anti-β-actin antibodies are shown. +, present; −, absent; H2AXpS139, H2AX phosphorylated on S139. (D) Immunofluorescence analysis with anti-phospho-H2AX antibody. HeLa cells were transfected with the indicated siRNAs and processed for immunofluorescence by using anti-phospho-H2AX antibody. DNA damage was induced as mentioned in the legend to panel C. (E) Quantitation of phospho-H2AX foci. The percentages of cells positive for phospho-H2AX foci among cells transfected with the indicated siRNAs and those with (UV) and without (0) DNA damage in the analysis presented in panel D are shown. The experiment was repeated three times, and the means ± standard deviations (SD) are plotted. (F) The loss of Rvb1 results in the persistence of phospho-H2AX. HeLa cells were transfected with the indicated siRNAs. Cells were harvested at the indicated time points after UV irradiation, and lysates were immunoblotted with the indicated antibodies.
Reversal of Rvb1 knockdown phenotype.
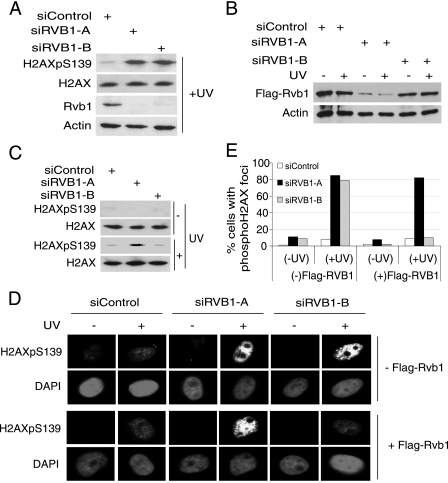
To rule out the possibility of an off-target activity of the siRNA, we rescued the phospho-H2AX accumulation with exogenous siRNA-resistant RVB1. A second RNA duplex (siRVB1-B) specific to a target site in the 3′ untranslated region of endogenous RVB1 was synthesized. Both duplexes (siRVB1-A and siRVB1-B) efficiently depleted cells of endogenous Rvb1 (Fig. 2A) and increased phospho-H2AX after UV irradiation (Fig. 2A). We established a cell line expressing exogenous FLAG-RVB1 without the 3′ untranslated region of RVB1. siRVB1-A and siRVB1-B knocked down endogenous and exogenous Rvb1 and only endogenous Rvb1, respectively (Fig. 2A and B). The increase in the phospho-H2AX level after knockdown by siRVB1-A (for endogenous and exogenous Rvb1) was not seen after knockdown by siRVB1-B, when exogenous Flag-Rvb1 persisted (Fig. 2C). The transfection of cells expressing Flag-Rvb1 with siRVB1-B did not increase the number of phospho-H2AX focus-positive cells or the intensity of phospho-H2AX foci (Fig. 2D and E). The restoration of the original phospho-H2AX phenotype by exogenous siRNA-resistant Rvb1 was also observed when other DNA-damaging agents were used (see Fig. S2 in the supplemental material). The increase in phospho-H2AX with two different siRNAs targeting different parts of RVB1 and the restoration of the phospho-H2AX downregulation by siRNA-resistant Flag-Rvb1 suggest that the increase in phospho-H2AX is due to the on-target depletion of Rvb1 and cannot be attributed to off-target activities of the siRNA duplexes.
FIG. 2.
The Rvb1 knockdown phenotype can be reversed by the expression of exogenous Rvb1 protein. (A) The depletion of endogenous Rvb1 by siRVB1-A and siRVB1-B increases phospho-H2AX. HeLa cell lysates were prepared 72 h after the transfection of cells with the indicated siRNAs. The decrease in total Rvb1 protein was detected using anti-Rvb1 antibody, and anti-β-actin was used as a loading control. +, present; H2AXpS139, H2AX phosphorylated on S139. (B) Exogenous Flag-Rvb1 was knocked down by siRVB1-A but not siRVB1-B. HeLa cells stably expressing Flag-Rvb1 were lysed after transfection with the indicated siRNAs with (+) or without (−) UV irradiation. The experiment was carried out as described for panel A. (C) Phospho-H2AX levels after Rvb1 are restored by expressing Flag-Rvb1 resistant to siRVB1-B. HeLa cells stably expressing Flag-Rvb1 were transfected for 72 h with the indicated siRNAs before UV irradiation and immunoblotting of lysates with the indicated antibodies. (D) Phospho-H2AX foci detected by immunofluorescence. HeLa cells with or without Flag-Rvb1 were transfected with the indicated siRNAs for 72 h and either left untreated or irradiated with UV. +, present; −, absent. (E) Quantitation of phospho-H2AX foci. The percentages of cells positive for phospho-H2AX foci among cells transfected with the indicated siRNAs and those with (+UV) and without (−UV) DNA damage in the analysis presented in panel D are shown.
Phosphatidylinositol 3-kinase-like kinases are not hyperactivated in the absence of Rvb1.
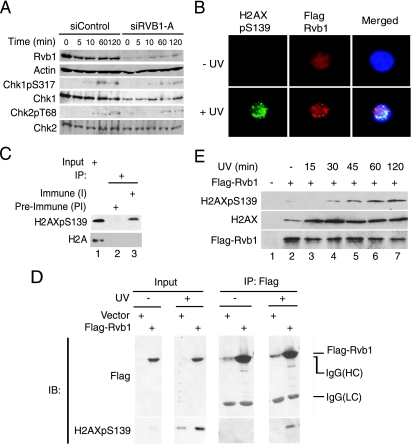
The increased phosphorylation of H2AX may be due to the hyperactivation of ATM and ATR kinases. To investigate this possibility, we examined the phosphorylation status of Chk1 and Chk2, checkpoint kinases that are known targets of ATR and ATM (37). The loss of Rvb1 did not increase the phosphorylation of either Chk1 or Chk2, suggesting that the increase in the total level of phospho-H2AX was not due to the hyperactivation of ATR or ATM (Fig. 3A).
FIG. 3.
Rvb1 colocalizes and interacts with phospho-H2AX after DNA damage. (A) The loss of Rvb1 does not hyperactivate ATM and ATR kinases. Lysates from HCT116 cells were prepared after the transfection of the cells with the indicated siRNAs and the harvesting of the cells at the indicated time points post-UV irradiation. The results of immunoblotting with antibodies to Chk1 phosphorylated on S317 (Chk1pS317) and Chk phosphorylated on T68 (Chk2pT68) and anti-Chk1, anti-Chk2, anti-Rvb1, and anti-β-actin antibodies are shown. (B) Colocalization of Rvb1 with phospho-H2AX after DNA damage. 293T cells were transfected with a Flag-Rvb1-expressing plasmid. Cells were either left untreated (− UV) or irradiated with UV (+ UV). Immunofluorescence analyses using anti-Flag (red) and anti-phospho-H2AX (green) antibodies were performed. The merged images show the colocalization of Rvb1 and phospho-H2AX in DAPI-stained nuclei (blue). H2AXpS139, H2AX phosphorylated on S139. (C) Endogenous Rvb1 interacts with phospho-H2AX. The results of immunoprecipitation (IP) using either preimmune (PI) or immune (I) antibody against Rvb1 from lysates prepared from HeLa cells after UV irradiation are shown. Western blotting was carried out with anti-phospho-H2AX and anti-H2A antibodies. Ten percent inputs (lane 1) are shown in parallel. +, present. (D) Exogenous Rvb1 interacts with phospho-H2AX. 293T cells were transfected with a Flag-Rvb1-expressing plasmid. Cells were either left untreated (−) or irradiated with UV. Cell lysates were used to immunoprecipitate Flag-Rvb1 and subjected to Western blotting using anti-Flag and anti-phospho-H2AX antibodies. Results obtained using inputs of 10% of wild-type levels (input lanes) are shown in parallel. Input lanes of the immunoblot for H2AXpS139 represent a lighter exposure to see the stimulation of phospho-H2AX signals after UV treatment. IgG, immunoglobulin G; HC, heavy chain; LC, light chain; IB, immunoblot. (E) Increased interaction of Rvb1 with H2AX after DNA damage. 293T cells were transfected with an empty vector or a plasmid expressing FLAG-RVB1. Cells were irradiated and harvested at the indicated times after UV treatment. Lysates were immunoprecipitated with anti-Flag antibody, and immunoprecipitates were subjected to Western blotting for H2AX, phospho-H2AX, and Rvb1. +, present; −, absent.
Rvb1 localizes to the DNA damage sites.
To understand the mechanism by which phospho-H2AX is increased after Rvb1 knockdown, we looked at the localization of Rvb1 after DNA damage. For this evaluation, Flag-Rvb1 was expressed in 293T cells and immunofluorescence analysis was performed using anti-Flag antibody. Rvb1 is nuclear and homogenously distributed in the nucleoplasm under normal conditions (Fig. 3B, upper panels). After irradiation with UV, the distribution of Rvb1 changes to form distinct nuclear foci within an hour (Fig. 3B, lower panels). Phospho-H2AX foci appeared and were colocalized with Rvb1 foci in cells irradiated with UV (Fig. 3B, lower panels). Of the cells with Rvb1 foci (n = 100), 65% had colocalized phospho-H2AX and Rvb1 foci, and 81% of Rvb1 foci (n = 106) were colocalized with phospho-H2AX foci. The interaction of Rvb1 with phospho-H2AX was confirmed by the immunoprecipitation of endogenous Rvb1 and Flag-Rvb1 (Fig. 3C and D). Rvb1 interacted with the phospho-H2AX induced after UV irradiation, and this interaction was seen within 30 min of UV irradiation (Fig. 3C, D, and E). Rvb1 also immunoprecipitated with unphosphorylated H2AX in unirradiated cells (Fig. 3E, lane 2). We are currently investigating the mechanism underlying the increase in association postirradiation, but collectively, these data show that Rvb1 interacts with phospho-H2AX and so may have a direct effect on phospho-H2AX-containing nucleosomes.
Rvb1 does not affect the DNA damage repair process.
The loss of Rvb1 may stimulate phospho-H2AX formation by repressing the repair of DNA damage. We therefore examined whether the knockdown of Rvb1 inhibits the DNA repair pathways in the short term when phospho-H2AX is increased. Single-strand and double-strand DNA breaks in the Rvb1-depleted cells were measured by the alkaline comet assay (33). The loss of Rvb1 produced a small increase of such breaks, even in the absence of DNA damage, but this increase was not remarkable by 60 min after UV irradiation (see Fig. S3 in the supplemental material). The neutral comet assay showed no increase in the levels of double-strand DNA breaks compared to those in control RNA interference-treated cells both before and after UV treatment (see Fig. S3 in the supplemental material). UV irradiation produces CPDs. The rate of CPD removal at 1 or 4 h post-UV irradiation was not decreased after Rvb1 depletion (see Fig. S4 in the supplemental material). Therefore, the loss of Rvb1 does not significantly repress the repair of DNA breaks or CPD within the 60-min time frame after which there is already a significant increase in phospho-H2AX accumulation.
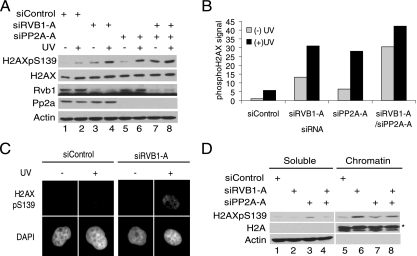
The loss of Rvb1 increases the phosphorylation of chromatin-bound H2AX.
The phospho-H2AX-associated Rvb1 may be required for the remodeling of chromatin prior to phosphatase action or for the activity of the phosphatase itself. Previously, it was suggested that protein phosphatase 1 (PP1) removes phospho-H2AX from repaired double-strand breaks (32). But recently, Chowdhury et al. carefully looked at the phosphatase activity of PP1 and PP2A both in vivo and in vitro and concluded that PP2A is the physiological phosphatase that dephosphorylates phospho-H2AX (9). The depletion of PP2A by siRNA (siPP2A-A) or the inhibition of the activity of PP2A by okadaic acid increased the phosphorylation of H2AX after radiation (Fig. 4A, lane 6, and B and data not shown). In unirradiated cells, the codepletion of Rvb1 and PP2A increased the phosphorylation of H2AX significantly compared to that in cells depleted of Rvb1 or PP2A individually (Fig. 4A, lanes 3, 5, and 7, and B), suggesting that Rvb1 and PP2A act at different steps during the removal of phospho-H2AX. The loss of PP2A increased phospho-H2AX in the soluble fraction (Fig. 4D, lane 3), while the loss of Rvb1 increased phospho-H2AX in the chromatin fraction (Fig. 4C and D, lane 6). The codepletion of Rvb1 and PP2A increased the phospho-H2AX in the chromatin fraction while decreasing it in the soluble fraction. These results suggest that Rvb1 is required to remodel chromatin before phospho-H2AX is accessible for dephosphorylation by PP2A.
FIG. 4.
Rvb1 is required for the removal of phospho-H2AX from chromatin. (A) Increase of phospho-H2AX after Rvb1 and PP2A depletion. Cell lysates prepared after the depletion of Rvb1 and PP2A alone or together were immunoblotted with the indicated antibodies. H2AXpS139, H2AX phosphorylated on S139; +, present; −, absent. (B) Quantitation of phospho-H2AX signals in panel A by using Scion imaging software. (C) Increased phospho-H2AX was present on chromatin after the knockdown of Rvb1. The phospho-H2AX foci seen in cells depleted of Rvb1 were bound to chromatin. HeLa cells transfected with the indicated siRNAs with (+) or without (−) UV irradiation were extracted in situ to remove non-chromatin-bound proteins prior to fixation and immunostaining for phospho-H2AX. (D) The loss of PP2A stabilizes phospho-H2AX in the soluble fraction, and this effect is dependent on Rvb1. HeLa cells transfected with the indicated siRNAs were fractionated 60 min after UV irradiation. The fractions were immunoblotted for the indicated proteins. *, cross-reacting band.
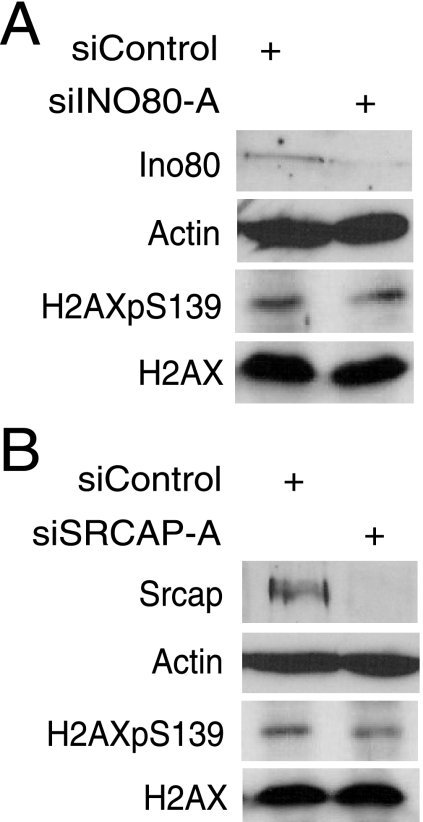
The knockdown of either Ino80 or SRCAP does not affect levels of phospho-H2AX after DNA damage.
Our earlier studies with yeast showed that Rvb1 and Rvb2 recruit Arp5 into the Ino80 complex and are required for the catalytic activity of the Ino80 complex (22), one of the chromatin-remodeling complexes involved in the DNA damage response (30, 34, 42, 44, 45). Human Ino80 (hIno80) is also associated with Rvb (20). The use of siINO80-A caused a 90% reduction in the Ino80 level (Fig. 5A and data not shown), but this reduction did not increase the level of phospho-H2AX after UV irradiation (Fig. 5A) either because we did not succeed in decreasing Ino80 below a threshold that can still sustain its activity or because Ino80 is not essential for the dephosphorylation of phospho-H2AX.
FIG. 5.
The depletion of either Ino80 or SRCAP does not affect levels of phospho-H2AX after DNA damage. (A) The depletion of Ino80 does not increase total phospho-H2AX levels after DNA damage. HeLa cells transfected with the indicated siRNAs were immunoblotted for total phospho-H2AX and H2AX protein. A decrease in Ino80 was detected by using anti-Ino80 antibody, whereas β-actin is shown as a loading control. Cells were treated with UV 72 h after transfection with siRNAs and harvested 60 min postirradiation. H2AXpS139, H2AX phosphorylated on S139; +, present. (B) The knockdown of SRCAP does not increase total phospho-H2AX levels after DNA damage. HeLa cells transfected with the indicated siRNAs were immunoblotted for total phospho-H2AX and H2AX. The depletion of SRCAP was detected by using anti-SRCAP antibody. The experiment was carried out as described for panel A.
Rvb1 and Rvb2 are also present in another complex, Swr1, that is involved in the exchange of H2A.Z (6, 22, 26, 29, 48). We therefore tested whether the SRCAP complex, the mammalian homolog of the Swr1 complex containing Rvb1 (6), is involved in the removal of phospho-H2AX from DNA damage sites. siSRCAP-A efficiently decreased the SRCAP protein, but the depletion of SRCAP did not increase phospho-H2AX relative to that in the control siRNA-treated cells (Fig. 5B). Therefore, Ino80 and SRCAP, two chromatin-remodeling factors associated with Rvb1, are not essential for dephosphorylating phospho-H2AX after DNA damage.
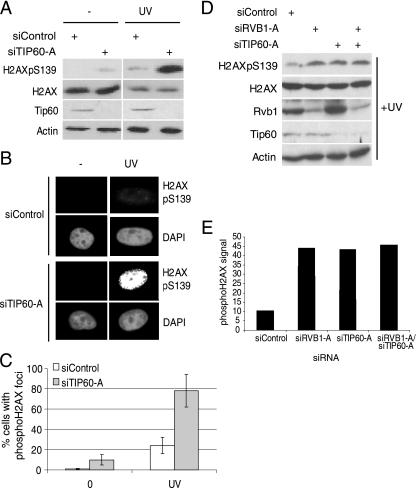
Tip60 is involved in the dephosphorylation of phospho-H2AX after DNA damage.
Tip60 is a histone acetyltransferase that is associated with Rvb1 (11, 18). Drosophila Tip60 downregulates phospho-H2AV (27), and human Tip60 acetylates H2AX and modulates the DNA damage response (18, 19, 31). To check whether the knockdown of human Tip60, like Rvb1, increases phospho-H2AX after DNA damage, an siRNA duplex targeting TIP60 (siTIP60-A) was used to decrease Tip60 (Fig. 6A). Tip60 depletion increased total phospho-H2AX after UV irradiation (Fig. 6A), and as in the case of Rvb1 depletion, the phospho-H2AX was chromatin bound (Fig. 6B and C). Although the individual knockdown of Rvb1 and Tip60 increased the phospho-H2AX, the codepletion of the two did not further increase the phospho-H2AX (Fig. 6D and E), suggesting that Rvb1 depletion and Tip60 depletion may inhibit the same step toward phospho-H2AX removal.
FIG. 6.
The depletion of Tip60 increases phospho-H2AX. (A) The depletion of Tip60 increases total phospho-H2AX levels after DNA damage. HeLa cells transfected with the indicated siRNAs with or without (−) UV irradiation were immunoblotted as shown. H2AXpS139, H2AX phosphorylated on S139; +, present. (B) Immunofluorescence after Tip60 depletion. HeLa cells transfected with the indicated siRNAs with or without UV irradiation were extracted in situ to remove non-chromatin-bound proteins prior to fixation and immunostaining for phospho-H2AX. (C) Percentages of cells positive for phospho-H2AX foci in the analysis presented in panel B. 0, no UV treatment. Means ± SD of results from three experiments are shown. (D) The codepletion of Rvb1 and Tip60 does not further increase phospho-H2AX levels after UV irradiation. UV-treated HeLa cells were depleted of Rvb1 and Tip60 either individually or together by using the indicated siRNAs. Western blotting analysis with anti-phospho-H2AX, anti-H2AX, anti-Rvb1, anti-Tip60, and anti-β-actin is shown. (E) Quantitation of phospho-H2AX signals in panel D (upper lane) using Scion imaging software.
Rvb1 is required for the histone acetyltransferase activity of the Tip60 complex.
To examine Rvb1's role in the biochemical activity of the Tip60 complex, we first established an assay for Tip60 complex-mediated HAT activity. For this assay, Flag immunoprecipitation was carried out with the cell lysates expressing vector alone, wild-type Flag-Tip60 (Tip60wt), or Flag-Tip60 mutated in the catalytic site (Tip60mut) (18). Equal amounts of Tip60wt and Tip60mut were immunoprecipitated, and Rvb1 specifically interacted with both Tip60wt and Tip60mut (see Fig. S5 in the supplemental material). In an in vitro acetyltransferase reaction, the Tip60wt complex transferred acetyl groups to lysines on histone H4 (see Fig. S6 in the supplemental material) as demonstrated by comparison to the background from either the Tip60mut complex or mock immunoprecipitates (see Fig. S7 in the supplemental material). Using this assay for the HAT activity of the Tip60 complex, we discovered that the Tip60 complex depleted of Rvb1 was inefficient in acetylating histone H4 in vitro (Fig. 7A and B). Therefore, Rvb1 is required for the histone acetyltransferase activity of the Tip60 complex.
Cellular Rvb1 depletion may result in the failure to synthesize or incorporate a critical component into the Tip60 complex or result in a malassembled complex that can be reactivated by adding Rvb1 in vitro. To distinguish between these possibilities, recombinant Rvb1 was purified from bacteria (see Fig. S8 in the supplemental material) and added to the Tip60 complex lacking endogenous Rvb1. Recombinant Rvb1 restored HAT activity to the Tip60 complex (Fig. 7C), suggesting that Rvb1 is required directly for the activity of the Tip60 complex. To distinguish whether Rvb1 is an essential cofactor for Tip60 activity or is required for the proper assembly or activity of the Tip60 complex, we added recombinant Rvb1 to recombinant Tip60 polypeptide (see Fig. S9 in the supplemental material). The HAT activity of the Tip60 polypeptide was not stimulated by Rvb1, suggesting that Rvb1 is required specifically for the correct assembly or activity of the Tip60 complex.
Phospho-H2AX downregulation requires histone H4 acetylation.
Histone H4 is known to be acetylated in vivo at DNA damage sites in a Tip60-dependent manner (31). When we immunoprecipitated about 20% of the phospho-H2AX from UV-treated cells, nearly 10% of the acetylated histone H4 coimmunoprecipitated with the phospho-H2AX (Fig. 7D), suggesting that almost 50% of the acetylated H4 may be associated with phospho-H2AX. We therefore hypothesized that nucleosomes at DNA damage sites may be acetylated on histone H4 before phospho-H2AX is remodeled and made available for dephosphorylation. To test this possibility, we overexpressed either wild-type histone H4 or histone H4 mutated in the lysines that are acetylated by Tip60. The expression of mutated H4 resulted in elevated phospho-H2AX without any change in the levels of Rvb1 or PP2A (Fig. 7E). Wild-type and mutant histone H4 were incorporated into the chromatin similarly (Fig. 7F), suggesting that the acetylation of histone H4 is required for the dephosphorylation of phospho-H2AX. Interestingly, the phospho-H2AX accumulated in the chromatin fraction, consistent with the model that the HAT activity of Tip60 is required to acetylate H4 before the phospho-H2AX can be remodeled and dephosphorylated.
DISCUSSION
In this study, we show that Rvb1 is required for the activity of the Tip60 complex and that the complex has a specific role in decreasing phospho-H2AX in vivo after DNA damage. The early detection of phospho-H2AX in Rvb1-depleted cells suggests that the latent period of the phospho-H2AX signal in wild-type cells is due to the constitutive activity of Rvb1 on phospho-H2AX-containing nucleosomes even early in the cell's response. At later time points, the Rvb1 deficit manifests itself as the continued accumulation of phospho-H2AX beyond a point when normal cells have started downregulating the phosphorylation. The identification of Rvb1 in various chromatin-remodeling complexes involved in the DNA damage response by us as well as others had suggested a role of the protein in the DNA damage response. The increase in phospho-H2AX levels in Rvb1-depleted cells subjected to DNA damage is in agreement with this possibility.
The overexpression of histone H4 that cannot be acetylated by Tip60 results in increased levels of chromatin-bound phospho-H2AX, consistent with a role of the histone H4 acetyltransferase, Tip60, in the dephosphorylation of phospho-H2AX. We cannot formally rule out that the increase in phospho-H2AX that we observed in cells was secondary to a transcriptional role of Tip60. Despite this caveat, we prefer the hypothesis that the Rvb1-Tip60 chromatin-remodeling factor is necessary at sites of DNA damage for the dephosphorylation of phospho-H2AX because the Tip60 complex associates with the sites of damaged DNA and is known to promote the opening of the chromatin by the posttranslational modification of the histone tails (this study and references 31 and 40).
S. cerevisiae phospho-H2AX is exchanged from the chromatin before dephosphorylation by PPH3 (23), and chromatin remodeling by the Drosophila Tip60 complex is important for the in vitro dephosphorylation of a related histone, phospho-H2AV (27). Although PP2A is known to remove the phosphate directly from phospho-H2AX in mammalian cells (9), a chromatin-remodeling step upstream of the dephosphorylation may be necessary. Our results show that the removal of Rvb1 or of Tip60 results in the accumulation of phospho-H2AX on the chromatin but that the removal of PP2A leads to the accumulation of the phospho-H2AX in the soluble fraction and the chromatin. The abundance of PP2A and the interaction of PP2A with H2AX are independent of a functional Rvb1-Tip60 complex (Fig. 4A and data not shown). These results are consistent with the possibility that chromatin remodeling by the Rvb1-Tip60 complex at sites of DNA damage is required before phospho-H2AX can be dephosphorylated by PP2A.
The hIno80 and SRCAP complexes are mammalian homologs of the Ino80 and Swr1 complexes, two chromatin-remodeling complexes in yeast known to contain Rvb1. Both the yeast proteins are recruited to double-strand breaks, and Ino80 is required for the eviction of phospho-H2A nucleosomes from the sites of DNA damage (34, 44). Decreasing hIno80 (or SRCAP) to 10% of the wild-type levels did not impair phospho-H2AX dephosphorylation in our experiments with mammalian cells, while similar decreases of Rvb1 or Tip60 impaired phospho-H2AX dephosphorylation. These results suggest that in mammalian cells, the Tip60 complex is of greater importance than the Ino80 complex in the dephosphorylation of phospho-H2AX.
Our results agree with those in a very recent paper saying that Tip60 associates with and is required for the mobilization of H2AX in the first 5 min after the induction of double-strand breaks with ionizing radiation (19, 27). Ikura et al., however, suggest a different mechanism, in which Tip60 acetylates H2AX on K5 and promotes UBC13-mediated polyubiquitination of H2AX on K119 prior to the release of H2AX from the chromatin (19). We have failed to see any acetylation of K5 on H2AX after UV irradiation (data not shown) and have instead noted acetylation on H4 associated with H2AX and have mimicked the phenotype of Tip60 depletion by overexpressing nonacetylable H4 (Fig. 7). The differences noted may be due to differences in the agents used to induce double-stranded DNA breaks: ionizing radiation breaks the DNA directly, while UV or chemicals induce breaks mostly after DNA replication across the sites of DNA damage. We prefer the explanation that both H4 and H2AX are important substrates of Tip60, with H2AX acetylation and ubiquitinylation being more important very early in the DNA damage response (within minutes) while H4 acetylation is more important on a longer time scale for curtailing the phospho-H2AX response.
The Tip60 complex has been implicated not only in DNA damage repair but also in the activation of checkpoint pathways (40). The Tip60 complex acetylates ATM after DNA damage, and this acetylation is required for the activation of ATM. As Rvb1 regulates the acetyltransferase activity of the Tip60 complex, a decrease in the specific activation of ATR/ATM may explain why phospho-Chk1 and phospho-Chk2 are not hyperactivated after Rvb1 depletion, despite the increase in phospho-H2AX. In addition, because other substrates of ATR or ATM are not hyperphosphorylated, we do not think that Rvb1 depletion increases the phosphorylation of H2AX by increasing the activity of ATR or ATM. Instead, we favor the hypothesis that the Tip60 complex is required for the dephosphorylation of phospho-H2AX and that it is the loss of this function that leads to the increased phosphorylation of H2AX after Tip60 or Rvb1 depletion. The chemical inhibition of Tip60 and Rvb1 is expected to prolong the phospho-H2AX response while maintaining the normal kinetics of activation and inactivation of the downstream Chk1 and Chk2 kinases that arrest the cell cycle. By delinking the kinetics of phospho-H2AX activation from Chk1 or Chk2 activation, inhibitors of the Rvb1-Tip60 complex may be useful as sensitizers for chemo- or radiotherapy of cancers.
A role for Rvb1 in the activity of the Tip60 complex links the activity of Rvb1 to that of critical tumor-suppressive and oncogenic transcription factors that are regulated by Tip60. Tip60 is required for the optimum function of p53 on specific p53-driven promoters (43), so that our results suggest that Rvb1 will also be required for p53-mediated pathways of apoptosis after DNA damage. Tip60 is also required for gene expression from Myc-driven promoters (15). Therefore, a role for Rvb1 in Tip60 activity may explain why Rvb1 is required for cell transformation by Myc (47). Similarly, a role for Rvb1 in the action of chromatin-remodeling complexes that interact with beta-catenin may explain the role of Rvb1 in the transmission of the growth-suppressive signal from the adenomatous polyposis coli tumor suppressor protein (4).
The only molecular function attributed to Rvb1 until now has been in the recruitment of Arp5 to the yeast Ino80 complex (22). The discovery reported here expands the role of Rvb1 in chromatin remodeling and highlights the importance of Rvb1 in the DNA damage response of mammalian cells. Future studies will investigate how the absence of Rvb1 impairs the HAT activity of the Tip60 complex and the consequences of Rvb1 depletion for the sensitivity of cells to DNA damage.
Supplementary Material
Acknowledgments
We thank the members of the Dutta laboratory for helpful discussions.
This work was supported partly by RO1 CA89406 to A.D.
Footnotes
Published ahead of print on 19 February 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alexandrow, M. G., and J. L. Hamlin. 2004. Cdc6 chromatin affinity is unaffected by serine-54 phosphorylation, S-phase progression, and overexpression of cyclin A. Mol. Cell. Biol. 241614-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassing, C. H., H. Suh, D. O. Ferguson, K. F. Chua, J. Manis, M. Eckersdorff, M. Gleason, R. Bronson, C. Lee, and F. W. Alt. 2003. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114359-370. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, A., S. Chauvet, O. Huber, F. Usseglio, U. Rothbacher, D. Aragnol, R. Kemler, and J. Pradel. 2000. Pontin52 and Reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J. 196121-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, A., O. Huber, and R. Kemler. 1998. Pontin52, an interaction partner of beta-catenin, binds to the TATA box binding protein. Proc. Natl. Acad. Sci. USA 9514787-14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellosta, P., T. Hulf, S. Balla Diop, F. Usseglio, J. Pradel, D. Aragnol, and P. Gallant. 2005. Myc interacts genetically with Tip48/Reptin and Tip49/Pontin to control growth and proliferation during Drosophila development. Proc. Natl. Acad. Sci. USA 10211799-11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, Y., J. Jin, L. Florens, S. K. Swanson, T. Kusch, B. Li, J. L. Workman, M. P. Washburn, R. C. Conaway, and J. W. Conaway. 2005. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 28013665-13670. [DOI] [PubMed] [Google Scholar]
- 7.Celeste, A., S. Difilippantonio, M. J. Difilippantonio, O. Fernandez-Capetillo, D. R. Pilch, O. A. Sedelnikova, M. Eckhaus, T. Ried, W. M. Bonner, and A. Nussenzweig. 2003. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114371-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, S. G., A. Bhoumik, L. Broday, V. Ivanov, B. Rosenstein, and Z. Ronai. 2001. TIP49b, a regulator of activating transcription factor 2 response to stress and DNA damage. Mol. Cell. Biol. 218398-8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhury, D., M. C. Keogh, H. Ishii, C. L. Peterson, S. Buratowski, and J. Lieberman. 2005. γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell 20801-809. [DOI] [PubMed] [Google Scholar]
- 10.Downs, J. A., N. F. Lowndes, and S. P. Jackson. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 4081001-1004. [DOI] [PubMed] [Google Scholar]
- 11.Doyon, Y., W. Selleck, W. S. Lane, S. Tan, and J. Cote. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 241884-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dugan, K. A., M. A. Wood, and M. D. Cole. 2002. TIP49, but not TRRAP, modulates c-Myc and E2F1 dependent apoptosis. Oncogene 215835-5843. [DOI] [PubMed] [Google Scholar]
- 13.Etard, C., D. Gradl, M. Kunz, M. Eilers, and D. Wedlich. 2005. Pontin and Reptin regulate cell proliferation in early Xenopus embryos in collaboration with c-Myc and Miz-1. Mech. Dev. 122545-556. [DOI] [PubMed] [Google Scholar]
- 14.Feng, Y., N. Lee, and E. R. Fearon. 2003. TIP49 regulates beta-catenin-mediated neoplastic transformation and T-cell factor target gene induction via effects on chromatin remodeling. Cancer Res. 638726-8734. [PubMed] [Google Scholar]
- 15.Frank, S. R., T. Parisi, S. Taubert, P. Fernandez, M. Fuchs, H. M. Chan, D. M. Livingston, and B. Amati. 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106297-307. [DOI] [PubMed] [Google Scholar]
- 17.Hammet, A., C. Magill, J. Heierhorst, and S. P. Jackson. 2007. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep. 8851-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102463-473. [DOI] [PubMed] [Google Scholar]
- 19.Ikura, T., S. Tashiro, A. Kakino, H. Shima, N. Jacob, R. Amunugama, K. Yoder, S. Izumi, I. Kuraoka, K. Tanaka, H. Kimura, M. Ikura, S. Nishikubo, T. Ito, A. Muto, K. Miyagawa, S. Takeda, R. Fishel, K. Igarashi, and K. Kamiya. 2007. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 277028-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin, J., Y. Cai, T. Yao, A. J. Gottschalk, L. Florens, S. K. Swanson, J. L. Gutierrez, M. K. Coleman, J. L. Workman, A. Mushegian, M. P. Washburn, R. C. Conaway, and J. W. Conaway. 2005. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J. Biol. Chem. 28041207-41212. [DOI] [PubMed] [Google Scholar]
- 21.Jonsson, Z. O., S. K. Dhar, G. J. Narlikar, R. Auty, N. Wagle, D. Pellman, R. E. Pratt, R. Kingston, and A. Dutta. 2001. Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J. Biol. Chem. 27616279-16288. [DOI] [PubMed] [Google Scholar]
- 22.Jonsson, Z. O., S. Jha, J. A. Wohlschlegel, and A. Dutta. 2004. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol. Cell 16465-477. [DOI] [PubMed] [Google Scholar]
- 23.Keogh, M. C., J. A. Kim, M. Downey, J. Fillingham, D. Chowdhury, J. C. Harrison, M. Onishi, N. Datta, S. Galicia, A. Emili, J. Lieberman, X. Shen, S. Buratowski, J. E. Haber, D. Durocher, J. F. Greenblatt, and N. J. Krogan. 2006. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature 439497-501. [DOI] [PubMed] [Google Scholar]
- 24.Khorasanizadeh, S. 2004. The nucleosome. From genomic organization to genomic regulation. Cell 116259-272. [DOI] [PubMed] [Google Scholar]
- 25.Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings, A. J. Link, H. D. Madhani, and J. Rine. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, D. P. Richards, X. Wu, A. Emili, T. R. Hughes, S. Buratowski, and J. F. Greenblatt. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 121565-1576. [DOI] [PubMed] [Google Scholar]
- 27.Kusch, T., L. Florens, W. H. Macdonald, S. K. Swanson, R. L. Glaser, J. R. Yates III, S. M. Abmayr, M. P. Washburn, and J. L. Workman. 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 3062084-2087. [DOI] [PubMed] [Google Scholar]
- 28.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 208602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303343-348. [DOI] [PubMed] [Google Scholar]
- 30.Morrison, A. J., J. Highland, N. J. Krogan, A. Arbel-Eden, J. F. Greenblatt, J. E. Haber, and X. Shen. 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119767-775. [DOI] [PubMed] [Google Scholar]
- 31.Murr, R., J. I. Loizou, Y. G. Yang, C. Cuenin, H. Li, Z. Q. Wang, and Z. Herceg. 2006. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 891-99. [DOI] [PubMed] [Google Scholar]
- 32.Nazarov, I. B., A. N. Smirnova, R. I. Krutilina, M. P. Svetlova, L. V. Solovjeva, A. A. Nikiforov, S. L. Oei, I. A. Zalenskaya, P. M. Yau, E. M. Bradbury, and N. V. Tomilin. 2003. Dephosphorylation of histone gamma-H2AX during repair of DNA double-strand breaks in mammalian cells and its inhibition by calyculin A. Radiat Res. 160309-317. [DOI] [PubMed] [Google Scholar]
- 33.Olive, P. L., J. P. Banath, and R. E. Durand. 1990. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the “comet” assay. Radiat. Res. 12286-94. [PubMed] [Google Scholar]
- 34.Papamichos-Chronakis, M., J. E. Krebs, and C. L. Peterson. 2006. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 202437-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu, X. B., Y. L. Lin, K. C. Thome, P. Pian, B. P. Schlegel, S. Weremowicz, J. D. Parvin, and A. Dutta. 1998. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J. Biol. Chem. 27327786-27793. [DOI] [PubMed] [Google Scholar]
- 36.Rottbauer, W., A. J. Saurin, H. Lickert, X. Shen, C. G. Burns, Z. G. Wo, R. Kemler, R. Kingston, C. Wu, and M. Fishman. 2002. Reptin and pontin antagonistically regulate heart growth in zebrafish embryos. Cell 111661-672. [DOI] [PubMed] [Google Scholar]
- 37.Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 7339-85. [DOI] [PubMed] [Google Scholar]
- 38.Shen, X., G. Mizuguchi, A. Hamiche, and C. Wu. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406541-544. [DOI] [PubMed] [Google Scholar]
- 39.Silva, J. M., M. Z. Li, K. Chang, W. Ge, M. C. Golding, R. J. Rickles, D. Siolas, G. Hu, P. J. Paddison, M. R. Schlabach, N. Sheth, J. Bradshaw, J. Burchard, A. Kulkarni, G. Cavet, R. Sachidanandam, W. R. McCombie, M. A. Cleary, S. J. Elledge, and G. J. Hannon. 2005. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 371281-1288. [DOI] [PubMed] [Google Scholar]
- 40.Sun, Y., X. Jiang, S. Chen, N. Fernandes, and B. D. Price. 2005. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. USA 10213182-13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teixeira, M. T., B. Dujon, and E. Fabre. 2002. Genome-wide nuclear morphology screen identifies novel genes involved in nuclear architecture and gene-silencing in Saccharomyces cerevisiae. J. Mol. Biol. 321551-561. [DOI] [PubMed] [Google Scholar]
- 42.Tsukuda, T., A. B. Fleming, J. A. Nickoloff, and M. A. Osley. 2005. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438379-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyteca, S., M. Vandromme, G. Legube, M. Chevillard-Briet, and D. Trouche. 2006. Tip60 and p400 are both required for UV-induced apoptosis but play antagonistic roles in cell cycle progression. EMBO J. 251680-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Attikum, H., O. Fritsch, and S. M. Gasser. 2007. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 264113-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Attikum, H., O. Fritsch, B. Hohn, and S. M. Gasser. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119777-788. [DOI] [PubMed] [Google Scholar]
- 46.Wang, H., L. Zhai, J. Xu, H. Y. Joo, S. Jackson, H. Erdjument-Bromage, P. Tempst, Y. Xiong, and Y. Zhang. 2006. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 22383-394. [DOI] [PubMed] [Google Scholar]
- 47.Wood, M. A., S. B. McMahon, and M. D. Cole. 2000. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 5321-330. [DOI] [PubMed] [Google Scholar]
- 48.Wu, W. H., S. Alami, E. Luk, C. H. Wu, S. Sen, G. Mizuguchi, D. Wei, and C. Wu. 2005. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 121064-1071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.