Abstract
ONYX-015 (dl1520) is an E1B 55-kilodalton protein-deficient replicating adenovirus that is currently in clinical trials as an antitumor agent. On the basis of the observation that the E1B 55kD gene product is able to bind to and inactivate p53, ONYX-015's mechanism of action is proposed to involve selective replication in and killing of p53-deficient cells. While its efficacy as a therapeutic agent appears evident, the virus's mechanism of cellular selectivity, including a possible role of p53 in this regard, is less clear. Indeed, there have been a number of recent reports suggesting that the p53 status of target cells does not reliably predict ONYX-015 replication or cell killing. To address the role of p53 in ONYX-015 selectivity, we have undertaken a rigorous analysis of the behavior of this virus in small airway-derived primary human epithelial cells expressing either dominant-negative or gain-of-function mutant p53 genes. Examination of small airway epithelial cells expressing a variety of p53 mutant alleles revealed that while all were able to inhibit endogenous p53 activity, only one allele examined, 248W, demonstrated a markedly increased ability to facilitate ONYX-015 replication. This allele is a member of a group of p53 mutants (know as class I mutants) characterized by retention of global structural conformation but loss of DNA-binding activity. These observations indicate that the nature of the p53 mutation affects ONYX-015 replication, help reconcile disparate published findings, and may provide criteria by which to direct clinical application of ONYX-015.
The p53 gene is mutated or lacking in at least 50% of human cancers, including multiple tumor types (24, 37). p53 inactivation is typically caused by somatic point mutations, most frequently in the region of the gene corresponding to the central, DNA-binding domain of the protein. Several of these hotspot mutations result in disruption of the sequence-specific DNA-binding ability of p53 (4, 18). On the basis of crystal structure and conformation-specific antibody information, p53 mutants have been divided into two classes: class I mutants are those that contain aberrant residues at the sites of direct DNA-protein contact (thereby inhibiting efficient DNA binding) but otherwise retain structural integrity, whereas class II mutants are those in which the global protein structure (tertiary or quaternary) has been disrupted in a manner that prevents DNA binding (4, 39). In addition, a third category of mutant p53 has been described on the basis of its ability to inhibit endogenous p53 and corresponds to the C terminus of p53, including the tetramerization domain (38, 44). All three p53 mutant types are known to act as dominant-negative inhibitors of p53, presumably by interacting with wild-type p53 as partners in a nonfunctional tetrameric complex.
Some dominant-negative alleles of p53 demonstrate gain-of-function properties. For example, expression of certain alleles of mutant p53 in p53 null cells yields increased tumors in animal models (49) and promotes enhanced transformation of established cell lines (7, 9) compared with expression of the p53 null controls alone. In another report, some mutant p53 alleles, but not others, were shown to inhibit a G2-M spindle checkpoint, resulting in endoreduplication (19). Further evidence of gain of function was seen in a murine model in which in vivo expression of a mutant p53 (172H, equivalent to human 175H) yielded an altered tumor spectrum and a dramatically higher rate of tumor metastasis than that of the null allele (30). Finally, although the mechanism is unknown, recent evidence suggests that the ability of certain p53 mutants to interact with and inhibit the p53-related protein p73 may contribute to a gain-of-function phenotype (6, 11, 31).
p53 is activated in response to a variety of cellular stress signals, including DNA damage, aberrant proliferation, nucleotide deprivation, and hypoxia (40, 43), of which one or more are predicted to occur following viral infection. Activation of p53 results in cell growth arrest or apoptosis, depending on the stimulus and cell type (2, 29, 48). Interestingly, the p53 response in epithelial cells (the physiological target of adenovirus) appears to be qualitatively different from that in fibroblasts or transformed cells, with which it has been more extensively analyzed. Specifically, a variety of primary epithelial cells, including prostate, breast, bronchial, and keratinocyte cells, show an attenuated or absent G1 checkpoint response following IR exposure (5, 10, 13, 32, 36), the basis of which is not clear. Of note, the steady-state levels of p53 are often higher in epithelial cells, due seemingly to a longer protein half-life (5). The effect of these differences on the epithelial cell's response to adenoviral infection is not known.
Given the prevalence of p53 mutations in human cancers, p53 deficiency represents an attractive criterion upon which to base a tumor-specific cancer therapeutic. To this end, McCormick and colleagues have pioneered the clinical application of a genetically modified adenovirus, dl1520 (termed ONYX-015), that is proposed to selectively replicate in, and thereby kill, p53-deficient tumor cells (3, 22). Initial clinical studies show promise, particularly when combined with conventional chemotherapeutic compounds, and the agent is currently being tested in phase III clinical trials (26, 27, 34). Originally constructed and described by Barker and Berk (1), ONYX-015 lacks the virally encoded 55-kDa E1B gene (E1B 55kD). In addition to its known late-phase functions, the E1B 55kD protein is known to bind cellular p53, repress its transcriptional activity, and promote its degradation, thereby leading to an inactivation of p53-mediated checkpoints. These latter functions of E1B 55kD are proposed to provide ONYX-015's mechanism of selectivity. Cells containing an intact p53 pathway are thus predicted to inhibit replication of an E1B 55kD-deficient virus. In contrast, p53-deficient cells, such as those of a tumor, would be expected to allow efficient viral replication and subsequent cell killing.
The proposed basis for ONYX-015's oncolytic specificity has been challenged. While a number of studies support a role for p53 and p53 pathway components in determining viral selectivity (3, 12, 22, 28, 41, 50), others have failed to do so. Indeed, a number of groups have argued that ONYX-015 replication is independent of the host cell's p53 status (17, 20, 21, 42, 46, 47). The reasons for the apparent disparity are not entirely clear but are likely to arise in part from the different experimental models used. Typically, the adenoviral responses in cells of similar p53 status (but of various cellular backgrounds) have been compared with those in cells of differing p53 status. Uncontrolled differences in tissue of origin, genetic background, and infectibility all confound such comparisons. Furthermore, various readouts have been used as assay endpoints, including cell viability (17, 20), cytopathic effect (CPE) (42), viral burst measured in PFU/cell (17, 21, 46) and PFU/ml (42), hexon positivity (46), and CPE index (20), thereby further complicating interpretation and cross-comparison. A number of studies have presented models in which internally controlled matched pairs of cells have been compared. Typically, a p53 wild-type tumor cell line has been stably transfected with a vector expressing dominant-negative p53 or vector-only control to generate a matched pair that can be assayed for response following viral infection with either wild-type or E1B 55kD-deficient adenoviruses (12, 21, 42, 46). While this model represents an improved approach, it, too, suffers from experimental limitations. First, numerous studies now indicate that few, if any, tumor cell lines are truly wild type for p53 function; in most cell lines bearing the sequence of wild-type p53, p14ARF is either lacking or silenced (45). This is significant in light of a recent report suggesting that p14ARF is required for selective inhibition of ONYX-015 replication (41). Second, stably transfected cell lines are derived from clonal isolates that may have acquired other genetic changes in the selection process. Finally, in all cases only one p53 mutant allele was examined, resulting in limited analysis of potential allele-specific differences. Furthermore, certain mutant alleles are only weakly dominantly negative, thereby necessitating a clear demonstration of their loss of endogenous p53 function. Matched cell pairs have also been described in reports of studies using a temperature-sensitive allele of p53, and phenotypic comparisons were analyzed at restrictive and permissive temperatures (17, 21). This approach is further confounded by the fact that the E1B 55kD-deficient viruses are themselves cold sensitive (17, 21, 23).
In this paper, we describe the results of experiments designed to test the role of p53 in modulating E1B 55kD-deficient adenoviral replication by comparing wild-type control and p53 function-deficient primary airway epithelial cells, a physiologic target of adenovirus. We used early-passage primary cells to ensure that both efferent and afferent p53 pathways were intact. A variety of p53 mutant alleles were then utilized to render the cells p53 deficient. Nonclonal cell populations were examined, and loss of p53 function was demonstrated in cells expressing the various mutant alleles. Our results show that loss of p53 function per se is not sufficient to enable efficient replication of ONYX-015; however, one particular allele did demonstrate a gain of function in rescuing viral replication.
MATERIALS AND METHODS
Materials.
Small airway epithelial cells (SAEC) and cell culture growth media were obtained from Clonetics (BioWhittaker, Inc., Walkersville, Md.). Viruses (ONYX-015 and WT-D) were obtained from Onyx Pharmaceuticals, Richmond, Calif. Mutant p53 143A (pBabe-Puro) was obtained from T. Tlsty (University of California San Francisco). The genetic suppressor element (GSE) clone was obtained from V. Ossovskaya (University of California San Francisco). The ΦNX-ampho retroviral packaging cell line was obtained from G. Nolan, Stanford, Calif. Anti-p53 antibodies (DO-1, CM-1) were kindly provided by D. Lane, Dundee, Scotland.
Methods. Plasmid construction.
p53 mutants 175H and 248W were generated by site-specific mutagenesis, and full-length human cDNAs or the GSE fragment, corresponding to a 94-amino-acid peptide derived from the C terminus of rat p53 (38), were subcloned into either LXSN (Clontech Laboratories, Inc.) or pBabePuro (33) vectors.
Cell culture.
Passage 2 SAEC were derived from multiple donors and grown (using predefined medium) in cultures according to the manufacturer's recommendations to approximately 40% confluence prior to infection with amphotropic retrovirus with either a vector control or a p53 mutant, as described below. Cells containing the transduced gene were enriched by 48 h of drug selection with either neomycin (G418) (500 μg/ml) or puromycin (1 μg/ml) (Table 1). SAEC proved equally sensitive to either drug, with most noninfected cells dying within 24 h. Equal cell numbers were transferred to 6-well plates, grown to confluence, and allowed to arrest via contact growth inhibition (as measured by propidium iodide-fluorescence-activated cell sorter analysis) prior to infection with either ONYX-015 or WT-D adenovirus at a multiplicity of infection (MOI) of 10.
TABLE 1.
Description of p53 mutants useda
| p53 mutation | Mutation type | Species | Selection/vector |
|---|---|---|---|
| 143A | Class II (conformation) | Human | Puromycin/ pBabe-puro |
| 175H | Class II (conformation) | Human | G418/LXSN |
| 248W | Class I (DNA contact) | Human | G418/LXSN |
| Residues 275-368 | GSE | Rat | G418/LXSN |
All p53 genes used were full-length human cDNAs, with the exception of GSE, which corresponds to a 94-amino-acid peptide derived from rat p53 (38). This collection of p53 alleles includes mutants of all three classes of mutation (see text). Drug selections were as indicated.
Retroviral expression and infection.
ΦNX-ampho cells were transfected at roughly 60% confluence with Lipofectamine Plus (Invitrogen), and viral supernatant was collected at 48 and 72 h posttransfection, treated with Polybrene (4 μg/ml), filtered through 0.22-um-pore-size Steriflip filters (Millipore Corp.), and used directly for SAEC infection. The ΦNX-ampho cell medium was replaced and subsequently collected in a sequential viral harvest. SAEC cells were incubated with viral supernatant (10 ml in a T150 flask) for 6 h, washed twice, and then returned to growth in predefined medium. Following a second, repeated round of retroviral infection 24 h later, 50 to 80% of the cells were transduced as determined by parallel infection with a green fluorescent protein-expressing virus and by fluorescence-activated cell sorter analysis (data not shown).
Characterization of mutant p53-containing SAEC cells.
Subconfluent SAEC transduced with either the vector control or p53 mutants were measured for their ability to incorporate bromodeoxyuridine (BrdU) following etoposide treatment. Cells in duplicate wells were incubated with etoposide (Sigma) (500 ng/ml) for 24 h, washed, and grown in fresh medium for an additional 24 h. BrdU (Amersham-Life Science) (10 uM) was added during the last 6 h of growth, the cells were fixed with MeOH-acetone (1:1), and the incorporated BrdU was detected using a biotinylated anti-BrdU antibody (Alexis Biochemicals) and fluorescently labeled streptavidin (Rockland Inc., Gilbertsville, Pa.). Fluorescent nuclei were counted using a fluorescence microscope (Olympus BX60) and compared with total nuclei (DAPI [4′,6′diamidino-2-phenylindole] stained) to determine the percentage of BrdU-positive cells. Four fields were scored from each sample and averaged, and standard deviations were calculated.
Ionizing radiation and Western blot analysis.
Samples for protein analysis were prepared at various times postinfection. Protein lysates were prepared from subconfluent SAEC transduced with either the vector control or p53 mutants at 15 h following UV exposure (5 J/m2). Cells were washed three times in ice-cold phosphate-buffered saline, scraped in phosphate-buffered saline, and pelleted for 5 min at 2,000 × g. Cell pellets were resuspended in ELB (250 mM NaCl, 50 mM HEPES [pH 7.4], 0.1%NP-40) containing 2 mM dithiothreitol, 5 mM MgCl2, 1 mM Pefabloc, 1 mM EDTA, 25 mM NaF, 1 mM vanadate, and Complete protease inhibitor cocktail (Roche Molecular Biochemicals) and incubated on ice for 30 min. Following centrifugation at 21,000 × g for 10 min, the supernatant was removed and the protein concentration was determined (Bio-Rad protein detection kit). Lysate (30 μg per lane) was loaded on Novex sodium dodecyl sulfate-polyacrylamide electrophoresis gels and, following electrophoresis, the protein was electrophoretically transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore). The membranes were cut into separate molecular-weight ranges and probed as indicated with either anti-Mdm2 (SMP19; NeoMarkers), anti-p53 (DO-1 and CM-1; gifts from David Lane), or anti-p21 (Transduction Laboratories) antibodies.
Adenoviral infection and viral yield analysis.
Contact-inhibited SAEC transduced with either the vector control or p53 mutants were infected at an MOI of 10 with either WT-D or ONYX-015 virus. At 48 and 72 h postinfection, cells were scraped in their media (2 ml) and 1 ml of the suspension was transferred to a 1.5-ml Eppendorf tube. Following three cycles of freezing and thawing (in a sequence consisting of a dry-ice-EtOH bath followed by a 37°C bath and 10 s of vortex treatment), the sample was centrifuged at 5,000 rpm for 5 min and the supernatant was collected. The virus concentration of the supernatant was determined in duplicate using HEK293 cells in an enzyme-linked immunosorbent assay (ELISA)-based viral quantification assay as previously described (25). Briefly, the viral standard (ONYX-015) and unknown samples were serially diluted and used to infect HEK293 cells in a 96-well plate. At various times postinfection, the medium was removed, the cells and virus were fixed, and viral particles were detected using anti-adenoviral antibodies and an ELISA-based detection system. The viral yields shown represent the peak yields of triplicate averages from either the 48- or the 72-h time point. Typically, this represents the 48-h time point for WT-D-infected and the 72-h time point for ONYX-015-infected cells.
RESULTS
To evaluate the role of p53 function in determining ONYX-015 replication, we used early-passage primary SAEC as a model system. These cells were chosen for a number of reasons. First, they represent a physiologically relevant target cell of human adenovirus. Second, they are highly infectible, with more than 95% of cells infected at an MOI of 10 PFU/cell (data not shown). Third, SAEC are isolated from normal human donors and are genetically wild type and therefore presumed to contain intact afferent and efferent p53 pathways. Last, as described below, the use of SAEC offers the ability to compare multiple different p53 mutant alleles in a genetically matched, nonclonal cell population.
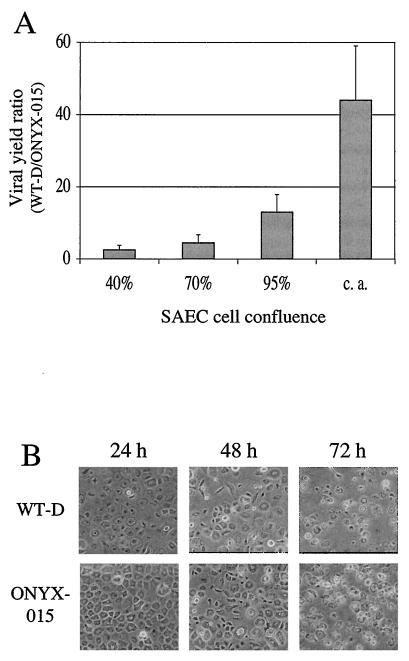
As determined by viral yield assay, SAEC enable selective replication of WT-D (wild-type control adenovirus) compared to ONYX-015 virus, particularly under conditions of contact inhibition and growth arrest (Fig. 1A). This result resembles a previously reported observation by Goodrum and Ornelles (16), who showed that the yield of E1B-55K-deficient virus was inversely related to the plating density at which HeLa cells were infected. On the basis of this observation, we performed all subsequent infections and viral yield analyses under conditions of contact arrest (S phase < 1%; data not shown). Interestingly, despite the virus's attenuated level of replication, ONYX-015 induced a morphological change similar to CPE in contact-arrested SAEC comparable to that induced by WT-D (Fig. 1B). Thus, CPE is not simply a function of viral replication and cannot be used as a readout for productive viral infection.
FIG. 1.
Characterization of SAEC. (A) SAEC were infected at a range of cell plating densities with either WT-D or ONYX-015 virus, and viral yields were determined. Results of viral yield levels determined by ELISA assay (see Materials and Methods) at 72 postinfection are shown as the ratio of WT-D viral yield to that of ONYX-015. Error bars indicate standard deviations. (B) Assessment of CPE in contact-arrested SAEC at various times following infection with either WT-D or ONYX-015.
To abolish p53 function in SAEC, we exogenously expressed a variety of mutant p53 genes. The mutants chosen represented all three p53 mutant categories: mutants 143A and 175H are examples of class II or conformational mutants in which p53 activity has been disrupted by a global change in tertiary structure; 248W is representative of a class I or contact site mutant in which p53 residues directly involved in protein-DNA contact sites have been altered and yet protein conformation remains intact; and, finally, GSE is a 94-amino-acid polypeptide derived from a region of p53 that includes the C-terminal oligomerization domain (38). Genes expressing the various p53 mutants or the appropriate vector controls (Table 1) were introduced into SAEC by means of amphotropic retrovirus delivery (Fig. 2; also see Materials and Methods), and drug-resistant cells were obtained. High levels of p53 were detected in all cells expressing mutant versions of p53 but not in vector-only control cells (Fig. 3b, −UV lanes). The elevated levels of full-length p53 observed in GSE-expressing cells represent endogenous, wild-type p53, whose expression is presumably increased in response to loss of the Mdm2-dependent negative-feedback loop (29).
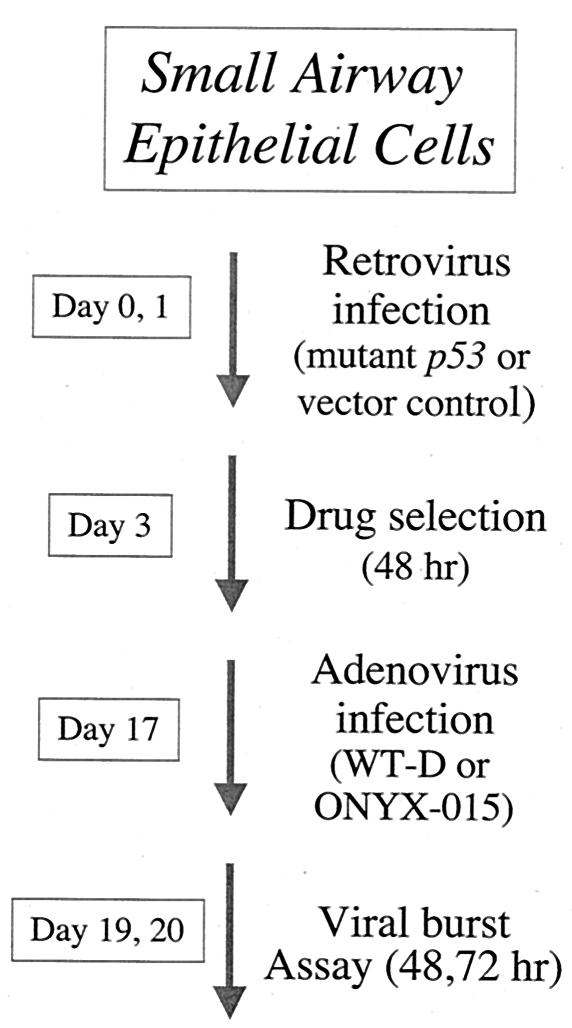
FIG. 2.
Outline of the experimental approach in early-passage (2) SAECs (Clonetics Corp.). See Materials and Methods for details.
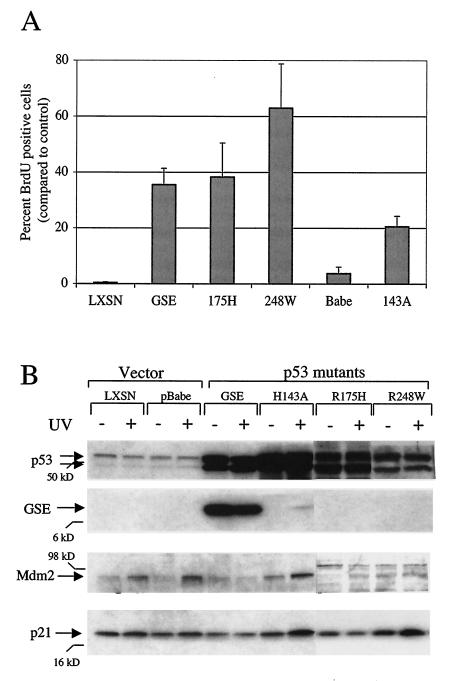
FIG. 3.
Characterization of SAEC containing mutant p53 or vector control. (A) Percentages of BrdU-positive cells following etoposide treatment compared to percentages of control (untreated) cells. Error bars indicate standard deviations. (B) Response of cells to UV irradiation. The results of immunoblot analysis of p53, GSE, Mdm2, and p21 expression at various times following exposure to UV at 5 J/m2 (+) or mock treatment (−) are indicated. Molecular mass markers are indicated.
Following selection, the cells were characterized to assess their p53 function. We chose two well-characterized p53 assays: cell cycle arrest in response to double-stranded DNA breaks and induction of p53 target genes in response to UV irradiation (Fig. 3). Subconfluent SAEC transduced with either the vector control or p53 mutants were treated with the topoisomerase II inhibitor etoposide and assayed for their ability to arrest cell cycle entry, as determined by BrdU incorporation. All of the mutant p53 constructs examined abolished this checkpoint, albeit to various degrees (Fig. 3A), indicating that p53 function was indeed compromised in these cells. In contrast, the etoposide checkpoint was intact in both nontransduced cells (data not shown) and vector-only control cells (Fig. 3A, lanes LXSN and pBabe), as evidenced by the presence of little or no BrdU incorporation.
To further characterize p53 function, cells transduced with either the vector control or p53 mutants were UV irradiated and cell lysates were then examined by Western blot analysis for the expression of p53 target genes (Fig. 3B). Interestingly, the increased level of p53 protein seen in some cell types in response to UV irradiation was not seen in SAEC. This is consistent with recent reports showing attenuated increases in p53 levels following DNA damage in primary epithelial cells (5, 10, 32) for reasons that are presently unclear. Increases were noted, however, in the level of the p53 target genes Mdm2 and, to a modest extent, p21 following UV irradiation (Fig. 3B; compare + versus − lanes and LXSN versus pBabe lanes). Notably, these increases were attenuated to various degrees in SAEC expressing the mutant p53 genes, thereby indicating elimination of p53 function in certain cases. The GSE mutant appeared to be most effective in blunting p53's transcriptional response to UV (Fig. 3B and data not shown), and 143A appeared to have the weakest effect in both assays. Thus, by each of these two criteria, SAEC expressing the various mutant p53 alleles were demonstrated to have altered and attenuated p53 function and, as such, were suitable for use in evaluating the role of p53 in replication of a ΔE1B 55kD adenovirus.
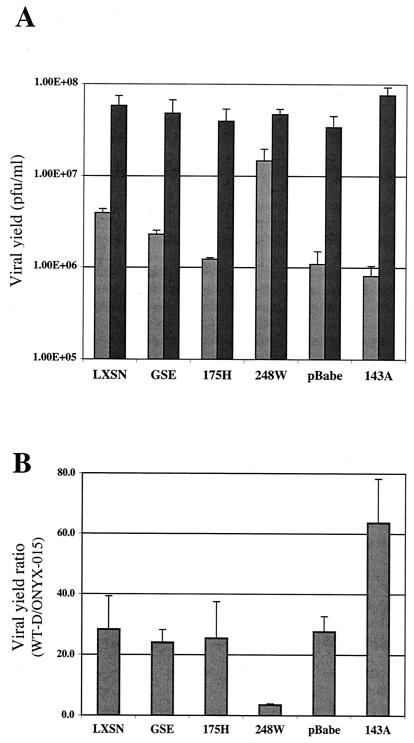
To measure adenoviral replication in these cells, pooled, drug-resistant populations of retrovirally infected SAECs were plated on six-well dishes, grown to confluence, and allowed to arrest due to prolonged contact inhibition. As previously shown (Fig. 1A), contact-dependent growth arrest was required to obtain strong, selective decrease in ONYX-015 virus replication compared to that seen with WT-D virus in human primary SAECs. Prior to adenoviral infection, a sample of the cells was harvested and assessed by propidium iodide staining for cell cycle analysis. Both the vector control and mutant p53-containing cells were growth arrested with at least 90% of cells in G0/G1 (data not shown). Cells in duplicate wells were infected with either WT-D or ONYX-015 adenovirus at an MOI of 10 (see Materials and Methods), and viral yields were determined at either 48 or 72 h postinfection (Fig. 4A). The ratio of the peak WT-D viral yield to that of ONYX-015 was compared to provide an estimate of the relative degree of viral replication (Fig. 4B). As expected, both vector control samples showed an approximate 30-fold-enhanced replication of WT-D relative to ONYX-015 (Fig. 4B). Interestingly, expression of mutant p53 affected ONYX-015 replication to various degrees and depended upon the specific p53 mutant that was examined. Cells that expressed GSE and 175H (p53 mutants that each showed strong evidence of blocking p53 activity in functional assays) (Fig. 3), showed only a moderate affect in rescuing ONYX-015 replication (Fig. 4). This suggests that loss of p53 function alone is not sufficient to restore the level of replication of E1B 55kD-deficient adenovirus to that of WT-D. In contrast, cells expressing the 248W p53 mutant were unique in showing a marked increase in ONYX-015 replication compared with vector-only control cells, yielding an average viral yield ratio of 3.5 (Fig. 4B). The fact that this mutant was unique in its ability to rescue E1B 55kD-deficient viral replication suggests that some other activity of the mutant beyond simple elimination of endogenous wild-type p53 function was required to restore viral replication. Surprisingly, cells expressing the p53 mutant 143A showed increased discrimination between WT-D and ONYX-015; replication of the E1B 55kD-deficient adenovirus was more attenuated in these cells than in vector-only control cells. We postulate that this result may be due to partial wild-type activity of this temperature-sensitive p53 mutant (see Discussion).
FIG. 4.
Viral yield in SAEC containing mutant p53 or vector control. (A) Viral yield for either ONYX-015 or WT-D virus measured at 72 h from a representative experiment. The light and dark bars represent ONYX-015 and WT-D, respectively. The values represent average viral yield measurements determined in triplicate from duplicate experiments. Units indicated are the equivalent of PFU/ml (see Materials and Methods). Error bars indicate standard deviation. (B) Ratio of WT-D to ONYX-015 average peak viral yields. The values shown represent the peak yields from either the 48- or 72-h time point. In some cases this represents the 48-h time point of WT-D-infected and the 72-h time point of ONYX-015-infected cells. Results represent data from two independent experiments, and standard deviations are indicated.
DISCUSSION
In understanding human cancer, one theme that is now clear is that disabling a cell's p53 function is a critical and nearly universal event in tumorigenesis. In this study, a genetic means was used to selectively abolish p53 function. This approach provides the ability to create multiple matched experimental and control cell populations to investigate the contribution of a single gene product in an otherwise wild-type cell. In this case, p53 function was altered in cells representing the physiologic target for a novel therapeutic adenovirus, leading to several interesting and important findings.
We determined that loss of p53 function alone was not sufficient to allow ONYX-015 virus replication. This is consistent with several recent reports and is in contradiction to ONYX-015's purported mechanism of action. In cell populations expressing either of two strong dominant-negative p53 mutants, GSE and 175H, almost no effect was seen on ONYX-015 replication in comparison to that of control cells. Although loss of p53 function was determined by a variety of assays, it is possible that such mutants did not inhibit all aspects of p53 function and that the component of p53 action required for viral attenuation remained intact. Since we do not currently understand p53's exact mechanism in this regard, we cannot rule this out as a possibility. A number of observations, however, suggest that this is unlikely. Structural characterization of p53 and the use of conformation-specific immunological probes indicate that mutations at residue 175 alter protein conformation and disrupt sequence-specific DNA binding (39), generally considered the cardinal function of p53. Similarly, GSE assembles with p53 to form a hetero-oligomeric complex that is unable to bind DNA. Furthermore, one of these mutants, 175H, not only arises as one of the most frequent p53 mutations in human tumors (18) but has now been shown to predispose mice to malignancies when expressed in germ line cells (30). Mutation at residue 175, thus, appears sufficient to abolish critical p53 functions required for tumor suppression.
In this study, one p53 allele, 248W, acted to enhance ONYX-015 virus replication. In this regard, we propose the existence of a gain-of-function role for this allele. There is precedence for gain of function in p53 mutants. Specifically, in assays such as growth in soft agar and oncogenic transformation, expression of certain p53 mutant alleles in a p53 null background show evidence of higher levels of tumorigenicity than cells of the null background alone (7, 9, 49). In an observation that may account for some of these findings, it has been shown that certain p53 mutants that disrupt conformation (class II) demonstrate gain of function in eliminating a cell cycle spindle checkpoint (19). Moreover, certain p53 mutants have the ability to bind to and disrupt the function of p73, a p53-related protein (6, 11, 31), thereby providing another possible gain-of-function mechanism. Further evidence comes from recent work that suggests a gain-of-function role for mutant p53 in promoting androgen-independent growth of prostate cancer cells in culture (35). In the case of 248W, the mechanism of action in enhancing E1B 55kD-deficient viral replication is not clear. Importantly, residue 248 is the most frequent site of p53 mutation in human cancers. Among the p53 mutants assayed in this report, 248W is unique in that it is a contact site (class I) mutant. Mutants of this class are believed to retain an overall wild-type structural conformation but harbor point mutations at specific residues required for site-specific DNA binding. In the case of adenoviral replication, a mutant of this type may act to override a cellular checkpoint that inhibits viral replication by binding and sequestration of p53 conformation-dependent protein partners that negatively regulate viral replication. Specifically, another class I mutant (273H) has been shown to constitutively interact with topoisomerase I (15) and promote gene amplification in Saos-2 cells (8). Alternatively, a mutant allele such as this may provide more favorable conditions for viral replication by relocalizing p53 to specific subcellular compartments or molecular complexes that enhance viral replication or by inducing additional cellular components, such as molecular chaperones, that facilitate viral biogenesis (14).
Even 248W, the mutant p53 allele with the greatest ability to rescue viral replication, did not increase the level of replication of ONYX-015 virus to that of wild-type adenovirus. Given the known requirement for the E1B 55-kDa protein in other viral processes, such as the regulation of RNA export and translation, it is not surprising that complete rescue was not observed. Surprisingly, 143A expression led to an apparent increased discrimination between WT-D virus and ONYX-015. This effect was due to an increase in WT-D viral yield as well as a decrease in that of ONYX-015 (Fig. 4A). One possible explanation may be the partial wild-type phenotype that this particular p53 allele is known to demonstrate, since it can alternate between mutant and wild-type conformations in a temperature-dependent manner. We speculate that high levels of p53 with partial wild-type function may result in a superinduction of p53 activity.
ONYX-015 shows clinical promise as a novel agent in human tumor therapy and is now being evaluated in phase III clinical trials of head and neck cancer. As such, its molecular mechanism of action is of obvious interest. Similarly, an understanding of specific genetic characteristics that promote ONYX-015 replication would provide a valuable means to guide clinical intervention. Within the limited p53 sequence data available from phase II ONYX-015 trials, there are a few (3 of 22) examples of p53 mutations of the class I type. These are currently being evaluated for correlation with clinical outcome.
Acknowledgments
We are grateful to Valeria Ossovskaya and Martin Oft for helpful suggestions and advice and to Leisa Johnson and Michael Korn for valuable comments on the manuscript. We thank members of the Lane (University of Dundee) and Tlsty (UCSF) laboratories for p53 antibodies and plasmid constructs.
REFERENCES
- 1.Barker, D. D., and A. J. Berk. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156:107-121. [DOI] [PubMed] [Google Scholar]
- 2.Bates, S., and K. H. Vousden. 1996. p53 in signaling checkpoint arrest or apoptosis. Curr. Opin. Genet. Dev. 6:12-18. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff, J. R., D. H. Kirn, A. Williams, C. Heise, S. Horn, M. Muna, L. Ng, J. A. Nye, A. Sampson-Johannes, A. Fattaey, and F. McCormick. 1996. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science 274:373-376. [DOI] [PubMed] [Google Scholar]
- 4.Cho, Y., S. Gorina, P. D. Jeffrey, and N. P. Pavletich. 1994. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 265:346-355. [DOI] [PubMed] [Google Scholar]
- 5.Delmolino, L., H. Band, and V. Band. 1993. Expression and stability of p53 protein in normal human mammary epithelial cells. Carcinogenesis 14:827-832. [DOI] [PubMed] [Google Scholar]
- 6.Di Como, C. J., C. Gaiddon, and C. Prives. 1999. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol. Cell. Biol. 19:1438-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dittmer, D., S. Pati, G. Zambetti, S. Chu, A. K. Teresky, M. Moore, C. Finlay, and A. J. Levine. 1993. Gain of function mutations in p53. Nat. Genet. 4:42-46. [DOI] [PubMed] [Google Scholar]
- 8.El-Hizawi, S., J. P. Lagowski, M. Kulesz-Martin, and A. Albor. 2002. Induction of gene amplification as a gain-of-function phenotype of mutant p53 proteins. Cancer Res. 62:3264-3270. [PubMed] [Google Scholar]
- 9.Eliyahu, D., D. Michalovitz, and M. Oren. 1985. Overproduction of p53 antigen makes established cells highly tumorigenic. Nature 316:158-160. [DOI] [PubMed] [Google Scholar]
- 10.Gadbois, D. M., and B. E. Lehnert. 1997. Cell cycle response to DNA damage differs in bronchial epithelial cells and lung fibroblasts. Cancer Res. 57:3174-3179. [PubMed] [Google Scholar]
- 11.Gaiddon, C., M. Lokshin, J. Ahn, T. Zhang, and C. Prives. 2001. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 21:1874-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganly, I., Y. T. Kim, B. Hann, A. Balmain, and R. Brown. 2001. Replication and cytolysis of an E1B-attenuated adenovirus in drug-resistant ovarian tumour cells is associated with reduced apoptosis. Gene Ther. 8:369-375. [DOI] [PubMed] [Google Scholar]
- 13.Girinsky, T., C. Koumenis, T. G. Graeber, D. M. Peehl, and A. J. Giaccia. 1995. Attenuated response of p53 and p21 in primary cultures of human prostatic epithelial cells exposed to DNA-damaging agents. Cancer Res. 55:3726-3731. [PubMed] [Google Scholar]
- 14.Glotzer, J. B., M. Saltik, S. Chiocca, A. I. Michou, P. Moseley, and M. Cotten. 2000. Activation of heat-shock response by an adenovirus is essential for virus replication. Nature 407:207-211. [DOI] [PubMed] [Google Scholar]
- 15.Gobert, C., A. Skladanowski, and A. K. Larsen. 1999. The interaction between p53 and DNA topoisomerase I is regulated differently in cells with wild-type and mutant p53. Proc. Natl. Acad. Sci. USA 96:10355-10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodrum, F. D., and D. A. Ornelles. 1997. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J. Virol. 71:548-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodrum, F. D., and D. A. Ornelles. 1998. p53 status does not determine outcome of E1B 55-kilodalton mutant adenovirus lytic infection. J. Virol. 72:9479-9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenblatt, M. S., W. P. Bennett, M. Hollstein, and C. C. Harris. 1994. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 54:4855-4878. [PubMed] [Google Scholar]
- 19.Gualberto, A., K. Aldape, K. Kozakiewicz, and T. D. Tlsty. 1998. An oncogenic form of p53 confers a dominant, gain-of-function phenotype that disrupts spindle checkpoint control. Proc. Natl. Acad. Sci. USA 95:5166-5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall, A. R., B. R. Dix, S. J. O'Carroll, and A. W. Braithwaite. 1998. p53-dependent cell death/apoptosis is required for a productive adenovirus infection. Nat. Med. 4:1068-1072. [DOI] [PubMed] [Google Scholar]
- 21.Harada, J. N., and A. J. Berk. 1999. p53-independent and -dependent requirements for E1B-55K in adenovirus type 5 replication. J. Virol. 73:5333-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heise, C., A. Sampson-Johannes, A. Williams, F. McCormick, D. D. Von Hoff, and D. H. Kirn. 1997. ONYX-015, an E1B gene-attenuated adenovirus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat. Med. 3:639-645. [DOI] [PubMed] [Google Scholar]
- 23.Ho, Y. S., R. Galos, and J. Williams. 1982. Isolation of type 5 adenovirus mutants with a cold-sensitive host range phenotype: genetic evidence of an adenovirus transformation maintenance function. Virology 122:109-124. [DOI] [PubMed] [Google Scholar]
- 24.Hollstein, M., B. Shomer, M. Greenblatt, T. Soussi, E. Hovig, R. Montesano, and C. C. Harris. 1996. Somatic point mutations in the p53 gene of human tumors and cell lines: updated compilation. Nucleic Acids Res. 24:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, L., A. Shen, L. Boyle, J. Kunich, K. Pandey, M. Lemmon, T. Hermiston, M. Giedlin, F. McCormick, and A. Fattaey. 2002. Selectively replicating adenoviruses targeting deregulated E2F activity are potent, systemic antitumor agents. Cancer Cell 1:325-337. [DOI] [PubMed] [Google Scholar]
- 26.Khuri, F. R., J. Nemunaitis, I. Ganly, J. Arseneau, I. F. Tannock, L. Romel, M. Gore, J. Ironside, R. H. MacDougall, C. Heise, B. Randlev, A. M. Gillenwater, P. Bruso, S. B. Kaye, W. K. Hong, and D. H. Kirn. 2000. A controlled trial of intratumoral ONYX-015, a selectively-replicating adenovirus, in combination with cisplatin and 5-fluorouracil in patients with recurrent head and neck cancer. Nat. Med. 6:879-885. [DOI] [PubMed] [Google Scholar]
- 27.Kirn, D., T. Hermiston, and F. McCormick. 1998. ONYX-015: clinical data are encouraging. Nat. Med. 4:1341-1342. [DOI] [PubMed] [Google Scholar]
- 28.Lee, H., J. Kim, B. Lee, J. W. Chang, J. Ahn, J. O. Park, J. Choi, C. O. Yun, B. S. Kim, and J. H. Kim. 2000. Oncolytic potential of E1B 55 kDa-deleted YKL-1 recombinant adenovirus: correlation with p53 functional status. Int. J. Cancer 88:454-463. [DOI] [PubMed] [Google Scholar]
- 29.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 30.Liu, G., T. J. McDonnell, R. Montes de Oca Luna, M. Kapoor, B. Mims, A. K. El-Naggar, and G. Lozano. 2000. High metastatic potential in mice inheriting a targeted p53 missense mutation. Proc. Natl. Acad. Sci. USA 97:4174-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marin, M. C., C. A. Jost, L. A. Brooks, M. S. Irwin, J. O'Nions, J. A. Tidy, N. James, J. M. McGregor, C. A. Harwood, I. G. Yulug, K. H. Vousden, M. J. Allday, B. Gusterson, S. Ikawa, P. W. Hinds, T. Crook, and W. G. Kaelin, Jr. 2000. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat. Genet. 25:47-54. [DOI] [PubMed] [Google Scholar]
- 32.Meyer, K. M., S. M. Hess, T. D. Tlsty, and S. A. Leadon. 1999. Hum. mammary epithelial cells exhibit a differential p53-mediated response following exposure to ionizing radiation or UV light. Oncogene 18:5795-5805. [DOI] [PubMed] [Google Scholar]
- 33.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nemunaitis, J., I. Ganly, F. Khuri, J. Arseneau, J. Kuhn, T. McCarty, S. Landers, P. Maples, L. Romel, B. Randlev, T. Reid, S. Kaye, and D. Kirn. 2000. Selective replication and oncolysis in p53 mutant tumors with ONYX-015, an E1B-55kD gene-deleted adenovirus, in patients with advanced head and neck cancer: a phase II trial. Cancer Res. 60:6359-6366. [PubMed] [Google Scholar]
- 35.Nesslinger, N. J., X. B. Shi, and R. W. DeVere White. 2003. Androgen-independent growth of LNCaP prostate cancer cells is mediated by gain-of-function mutant p53. Cancer Res. 63:2228-2233. [PubMed] [Google Scholar]
- 36.Nigro, J. M., K. D. Aldape, S. M. Hess, and T. D. Tlsty. 1997. Cellular adhesion regulates p53 protein levels in primary human keratinocytes. Cancer Res. 57:3635-3639. [PubMed] [Google Scholar]
- 37.Nigro, J. M., S. J. Baker, A. C. Preisinger, J. M. Jessup, R. Hostetter, K. Cleary, S. H. Bigner, N. Davidson, S. Baylin, P. Devilee, et al. 1989. Mutations in the p53 gene occur in diverse human tumour types. Nature 342:705-708. [DOI] [PubMed] [Google Scholar]
- 38.Ossovskaya, V. S., I. A. Mazo, M. V. Chernov, O. B. Chernova, Z. Strezoska, R. Kondratov, G. R. Stark, P. M. Chumakov, and A. V. Gudkov. 1996. Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc. Natl. Acad. Sci. USA 93:10309-10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prives, C. 1994. How loops, beta sheets, and alpha helices help us to understand p53. Cell 78:543-546. [DOI] [PubMed] [Google Scholar]
- 40.Prives, C., and P. A. Hall. 1999. The p53 pathway. J. Pathol. 187:112-126. [DOI] [PubMed] [Google Scholar]
- 41.Ries, S. J., C. H. Brandts, A. S. Chung, C. H. Biederer, B. C. Hann, E. M. Lipner, F. McCormick, and W. M. Korn. 2000. Loss of p14ARF in tumor cells facilitates replication of the adenovirus mutant dl1520 (ONYX-015). Nat. Med. 6:1128-1133. [DOI] [PubMed] [Google Scholar]
- 42.Rothmann, T., A. Hengstermann, N. J. Whitaker, M. Scheffner, and H. zur Hausen. 1998. Replication of ONYX-015, a potential anticancer adenovirus, is independent of p53 status in tumor cells. J. Virol. 72:9470-9478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz, D., and V. Rotter. 1998. p53-dependent cell cycle control: response to genotoxic stress. Semin. Cancer Biol. 8:325-336. [DOI] [PubMed] [Google Scholar]
- 44.Shaulian, E., I. Haviv, Y. Shaul, and M. Oren. 1995. Transcriptional repression by the C-terminal domain of p53. Oncogene 10:671-680. [PubMed] [Google Scholar]
- 45.Sherr, C. J. 2001. The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol. 2:731-737. [DOI] [PubMed] [Google Scholar]
- 46.Steegenga, W. T., N. Riteco, and J. L. Bos. 1999. Infectivity and expression of the early adenovirus proteins are important regulators of wild-type and ΔE1B adenovirus replication in human cells. Oncogene 18:5032-5043. [DOI] [PubMed] [Google Scholar]
- 47.Turnell, A. S., R. J. Grand, and P. H. Gallimore. 1999. The replicative capacities of large E1B-null group A and group C adenoviruses are independent of host cell p53 status. J. Virol. 73:2074-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vousden, K. H. 2000. p53: death star. Cell 103:691-694. [DOI] [PubMed] [Google Scholar]
- 49.Wolf, D., N. Harris, and V. Rotter. 1984. Reconstitution of p53 expression in a nonproducer Ab-MuLV-transformed cell line by transfection of a functional p53 gene. Cell 38:119-126. [DOI] [PubMed] [Google Scholar]
- 50.Yang, C. T., L. You, C. C. Yeh, J. W. Chang, F. Zhang, F. McCormick, and D. M. Jablons. 2000. Adenovirus-mediated p14(ARF) gene transfer in human mesothelioma cells. J. Natl. Cancer Inst. 92:636-641. [DOI] [PubMed] [Google Scholar]