Abstract
Transcription factor C/EBP-β regulates a number of physiological responses. During an investigation of the growth-suppressive effects of interferons (IFNs), we noticed that cebpb−/− cells fail to undergo apoptosis upon gamma IFN (IFN-γ) treatment, compared to wild-type controls. To examine the basis for this response, we have performed gene expression profiling of isogenic wild-type and cebpb−/− bone marrow macrophages and identified a number of IFN-γ-regulated genes that are dependent on C/EBP-β for their expression. These genes are distinct from those regulated by the JAK-STAT pathways. Genes identified in this screen appear to participate in various cellular pathways. Thus, we identify a new pathway through which the IFNs exert their effects on cellular genes through C/EBP-β. One of these genes is death-associated protein kinase 1 (dapk1). DAPK1 is critical for regulating the cell cycle, apoptosis, and metastasis. Using site-directed mutagenesis, RNA interference, and chromatin immunoprecipitation assays, we show that C/EBP-β binds to the promoter of dapk1 and is required for the regulation of dapk1. Both mouse dapk1 and human dapk1 exhibited similar dependences on C/EBP-β for their expression. The expression of the other members of the DAPK family occurred independently of C/EBP-β. Members of the C/EBP family of transcription factors other than C/EBP-β did not significantly affect dapk1 expression. We identified two elements in this promoter that respond to C/EBP-β. One of these is a consensus C/EBP-β-binding site that constitutively binds to C/EBP-β. The other element exhibits homology to the cyclic AMP response element/activating transcription factor binding sites. C/EBP-β binds to this site in an IFN-γ-dependent manner. Inhibition of ERK1/2 or mutation of an ERK1/2 site in the C/EBP-β protein suppressed the IFN-γ-induced response of this promoter. Together, our data show a critical role for C/EBP-β in a novel IFN-induced cell growth-suppressive pathway via DAPK1.
The interferon (IFN) family of cytokines regulates a number of physiological responses including innate defenses against viral, bacterial, and parasitic pathogens; the development of neoplastic growth; and specific immunity (6, 21, 28, 31, 74, 78). IFN-induced activities are driven largely through an induction of cellular IFN-stimulated genes (ISGs). IFN-induced signals regulate the expression of several immediate-early genes involved in innate immunity against infectious pathogens through a relatively well-defined signaling pathway known as the JAK-STAT pathway (41). IFN-γ-induced activation of the tyrosine kinases JAK1 and Tyk2 leads to tyrosyl phosphorylation of the receptor and the STAT1 and STAT2 proteins, which causes the nuclear migration of STATs and the formation of a transcriptionally active DNA-binding complex, ISGF3, in association with a non-STAT protein, IRF9 (9). In the IFN-γ-initiated pathways, tyrosine kinases JAK1 and JAK2 cause the tyrosyl phosphorylation of the STAT1 protein, which migrates to the nucleus and binds the gene promoters that possess an IFN-γ-activated site (GAS) or GAS-like sites and stimulates gene expression (77). STATs are rapidly activated following the engagement of IFNs with their receptors, and their initial activation is terminated within an hour through the recruitment of nuclear export mechanisms (46), synthesis of inhibitors of JAKs (2, 12), and dephosphorylation of activated STATs (86) despite the presence of IFN in the extracellular environment. It is also known that IFNs, particularly gamma IFN (IFN-γ), continue to induce the expression of a number of cellular genes and their biological responses even after the cessation of the JAK-STAT pathways (10) and even in stat1−/− cells (26, 63). Gene regulatory pathways mediated by the transcription factors IRF1 and IRF8 are some examples of STAT-independent IFN-induced regulation (81, 84). Previously, we showed that the transcription factor C/EBP-β participates in an IFN-γ response pathway that involves its binding to a novel IFN-γ-responsive element, GATE, identified in the irf9 promoter (70, 90). GATE-driven transcription, although IFN dependent, does not involve a direct binding of STATs to the irf9 promoter and occurs through C/EBP-β in a kinetically delayed manner in the apparent absence of activated STAT1 and JAK1 (70, 90).
The C/EBPs are a group of transcription factors that belong to a superfamily constituted of CREB, Fos, Jun/AP-1, activating transcription factor (ATF), and Maf/Nrf. The C/EBP subfamily includes C/EBP-α, C/EBP-β, C/EBP-γ, C/EBP-δ, C/EBP-ɛ, and C/EBP-ζ (37, 39). These proteins participate in a number of biological responses including energy metabolism (17), fat storage, tissue differentiation (18, 73), hematopoiesis (59), the immune response (1), antibacterial defense (82), and female fertility (79). Among these proteins, C/EBP-β uniquely responds to a variety of extracellular and intracellular signals to mediate a number of responses (37, 39). Although the effect of C/EBP-β on irf9 is well characterized (70, 93), it is not clear whether C/EBP-β has any other gene targets in the IFN-signaling pathways. Recently, we noticed that IFN-γ-induced cell death is suppressed significantly in cebpb−/− cells, indicating the existence of other potential IFN-induced pathways driven through C/EBP-β. In order to examine the importance of C/EBP-β in IFN-signaling pathways, we have conducted microarray-based gene expression profiling in this investigation. Our data uncovered that a number of genes involved in various responses are dependent on C/EBP-β for their expression. Surprisingly, we have found that the basal and IFN-γ-induced expression of DAPK1, a cell death-activating serine/threonine protein kinase, is critically dependent on C/EBP-β. The loss of C/EBP-β completely suppressed the basal and IFN-induced transcription from the dapk1 promoter. The expression of dapk1 is not significantly modulated by other members of C/EBP family. Using RNA interference, promoter mutational analyses, and chromatin immunoprecipitation (IP) (ChIP) assays, we show that C/EBP-β directly binds to the dapk1 promoter and regulates its basal and IFN-γ-induced expression. Two elements, a distal consensus C/EBP-β-binding site (CBS) and a promoter-proximal cyclic AMP response element (CRE)/ATF binding site, appear to mediate these responses. We also show that in response to IFN-γ, the ERK1/2 proteins phosphorylate a critical threonine residue of C/EBP-β for inducing dapk1 expression. Thus, C/EBP-β links IFN signal transduction pathways to the control of cell growth through DAPK1.
MATERIALS AND METHODS
Cell lines, antibodies, and plasmids.
Isogenic wild-type (cebpb+/+) and cebpb−/− mouse embryonic fibroblasts (MEFs) were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 8% fetal bovine serum and 1% antibiotic-antimycotic agents as described previously (69). IFN-γ (PBL Biomedical Labs, Piscataway, NJ) was used at 500 U/ml in these experiments unless indicated otherwise. hTERT-HME1, a nononcogenic human mammary epithelial cell line (Clontech), was grown in MCDB 170 medium supplemented with bovine pituitary extract, hydrocortisone, insulin, gentamicin, human epidermal growth factor, amphotericin B, and 10% fetal bovine serum (Clonetics, Palo Alto, CA). This cell line was a gift from George Stark, Cleveland Clinic Foundation, Cleveland, OH. Wild-type and cebpd−/− MEFs were provided by Esta Sterneck, NCI—Frederick Cancer Research Facility (NCIFCRF), Frederick, MD. cebpb−/−/cebpd−/− double-knockout MEFs were provided by Sarah Gaffen, SUNY—Buffalo, Buffalo, NY. Expression vectors coding for the wild-type and the kinase-deficient (K42A) mutant of DAPK1 (15) were provided by Adi Kimchi, Weizmann Institute of Science, Tel Aviv, Israel. The three different isoforms of C/EBP-β, LAP1, LAP2, and LIP, seen in some cells, have been suggested to be products of translational initiation at internal methionines within the same open reading frame (39). However, they were also shown to be produced during the extraction of cells (5). We have used full-length murine C/EBP-β (LAP1 isoform that migrates at ∼38 kDa on sodium dodecyl sulfate [SDS]-polyacrylamide gel electrophoresis gels) throughout this study. This is the major isoform seen in bone marrow (BM) cells and MEFs under our culture conditions. That said, there was no significant difference between the LAP1 and LAP2 isoforms in terms of their abilities to transactivate the dapk1 promoter in our experiments (data not shown). C/EBP-β mutants Mut1 and Mut2 were described previously (92), while T189A and T189D were generated using PCR-directed mutagenesis using the primers shown in Table S4 in the supplemental material. Plasmid-based primers (shown in the third row of Table S4 in the supplemental material) were used in combination with the mutant primers to generate the PCR products. All mutants were expressed from the pCDNA 3.1 Neo vector. Sequence-verified mutants were used in the experiments. Rabbit antibodies specific for C/EBP-α, C/EBP-β, C/EBP-δ, caspase-8, and caspase-9 (Santa Cruz Biotech) and mouse monoclonal antibodies specific for DAPK1 and actin (Sigma-Aldrich, Inc.) were used in these experiments. Total extracellular signal-regulated kinase (ERK) and diphosphorylated ERK (ppERK) antibodies were obtained from Cell Signaling Technology, Inc. Rabbit polyclonal antibodies against the phospho-T189 form of C/EBP-β were provided by Peter Johnson, NCIFCRF, Frederick, MD. The ERK pathway inhibitors PD98059 and U0126 (24) and the phosphatidylinositol (PI) 3-kinase inhibitor LY294002 were purchased from Calbiochem, Inc. Death-agonistic Fas antibody was purchased from BD Biosciences, Inc.
Generation of BM cell lines.
To better define the impact of IFN-γ-stimulated C/EBP-β on cellular gene expression, we generated BM macrophage cell lines from wild-type (cebpb+/+) and cebpb−/− mice (91). The mice used were a mixture of 129/Sv and C57BL/6 strains maintained by a continuous intercrossing of heterozygous animals. This is due to the facts that cebpb−/− female mice are infertile and the homozygous deletion of cebpb causes embryonic lethality, which yields an extremely low frequency of viable cebpb−/− pups (79). The macrophage cell lines were obtained by infecting freshly isolated plastic-adherent BM macrophage cells with a recombinant J2 retrovirus that expresses v-Myc and v-Raf (8). These cells were cultured continuously in the presence of recombinant human macrophage colony-stimulating factor (M-CSF) (100 ng/ml) and were analyzed for the expression of Mac-1 and F-480 markers. Each cell line was greater than 90% positive for these markers, and by this criterion, they are monocyte/macrophage-like cells (data not shown). These cells were genotyped first to demonstrate their genetic identities. We have used a combination of two PCR primer sets with genomic DNA as a template, one that detects a wild-type locus yields a 294-bp product, and the other that detects the disrupted locus, with the npt gene present, yields a 351-bp product (Peter Johnson, NCIFCRF, Frederick, MD, personal communication). The cebpb-specific primers 5′-AGCCCCTACCTGGAGCCGCTCGCG (forward) and 5′-GCGCAGGGCGAACGGGAAACCG (reverse) and the npt gene-specific primers 5′-GTGCTCGACGTTGTCACTGAAGCGG (forward) and 5′-GATATTCGGCAAGCAGGCATCG (reverse) were used for genotyping. Tail DNAs from homozygous (+/+ and −/−) and heterozygous (+/−) mice were used as controls in these experiments. Cells were grown continuously in the presence of M-CSF. However, during IFN-γ treatment, M-CSF levels were lowered to 1/10 of the original concentration to minimize excessive mitogenic stimulus. To avoid passage-dependent cellular changes (induced by the myc-raf oncogenes used for immortalization), we have used cells in early passages (passages 5 to 8) to perform these experiments.
Apoptosis assays.
Cells were grown in chamber slides, treated with the indicated reagents, and fixed with formaldehyde. They were then stained with DAPI (4′,6′-diamidino-2-phenylindole) (250 ng/ml), washed, and observed under a fluorescent microscope for nuclear condensation and fragmentation. The numbers of nuclei undergoing apoptosis from several different fields were counted and expressed as a percentage of total nuclei. This method of determination of apoptosis directly correlates with other methods such as annexin V staining and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (3). We have used annexin V staining as another correlate of apoptosis using a commercially available tetramethyl rhodamine isocyanate (TRITC)-labeled annexin V cell labeling kit (Trevigen, Inc.). In rescue experiments, expression vectors carrying the gene of interest or the corresponding empty vector was electroporated into cebpb−/− cells (107 cells) along with vector pEGFP (Clontech, Inc.) using Nucleofector technology (Amaxa Inc.), as recommended by the manufacturer. After electroporation, the cells were distributed into different plates and treated with the indicated agents for 16 h. They were stained with TRITC-annexin V and subjected to fluorescence-activated cell sorter (FACS) analysis. The percentage of green fluorescent protein (GFP)-positive cells that became TRITC positive was estimated for determining the magnitude of apoptosis. Expression of the transfected gene product was determined using Western blot analyses. Caspase activities were determined using a commercially available colorimetric assay kit as recommended by the manufacturer (Biosource International, Inc.).
Gene expression profiling.
To identify IFN-γ-regulated C/EBP-β-dependent genes, we screened a microarray consisting of 15,000 individual mouse cDNAs (the NIA mouse 15K collection). Poly(A)+ RNA isolated from cebpb+/+ and cebpb−/− BM macrophage cell lines was stimulated with IFN-γ (500 U/ml) for various lengths of time (1, 2, 4, 8, 12, 16, 20, and 24 h) and used for probe preparation. Although these cells were normally cultured with M-CSF, they were moved to a medium with a low M-CSF during IFN-γ treatment to avoid potential signaling conflicts with M-CSF. Greater than 90% of cells were viable under these conditions. Pooled poly(A)+ RNAs from three independent batches of cells were used for the microarray profiling. The IFN-treated samples were pooled. Control RNAs were prepared from the untreated samples. The resultant mRNAs were converted to cDNAs and labeled with Cy3 and Cy5 dyes. They were then hybridized to a mouse 15K cDNA array, washed, and scanned at the Walter Keck Biotechnology Resource Facility at Yale University, New Haven, CT, using established protocols (11), and the data were analyzed using the GENEPIX 3.0 program. For each cell type, untreated samples were compared to IFN-γ-treated samples. Genes induced in cebpb+/+ cells in response to IFN-γ were first identified. They were then compared to those expressed in cebpb−/− cells. Genes that were expressed in response to IFN-γ by more than twofold in cebpb+/+ but not in cebpb −/− cells were considered to be C/EBP-β-dependent IFN-γ-induced genes. Conversely, genes whose expression was suppressed in response to IFN-γ in cebpb+/+ but not in cebpb −/− cells were considered to be C/EBP-β-dependent IFN-γ-repressed genes.
Lentiviral shRNAs.
Lentiviral vectors carrying short hairpin RNAs (shRNAs) specific for human and mouse cebpb, mouse cebpa and cebpd, and mouse erk1 and erk2 were purchased from Open Biosystems, Inc. Virus stocks were prepared as recommended by the supplier (49). Mouse and human cebpb-specific shRNAs (nucleotides in boldface type) have the following sequences: 5′-CCGGGCCCTGAGTAATCACCTTAAACTCGAGTTTAAGGTGATTACTCAGGGCTTTTT and 5′-TGCTGTTGACAGTGAGCGCTGTGTACAGATGAATGATAAATAGTGAAGCCACAGATGTATTTATCATTCATCTGTACACATTGCCTACTGCCTCGGA, respectively. Both these shRNAs target the 3′ untranslated region of the cebpb mRNAs in their respective species. Because their target sequences are not identical, these shRNAs could not direct the degradation of the endogenous mRNAs when infected in cells of another species. Hence, we used them as scrambled shRNA controls in some experiments. All shRNAs were expressed using pLKO1-puro, a lentiviral expression vector in which shRNAs are generated under the control of the human U6 promoter. This vector also carries a puromycin resistance marker gene under the control of the human phosphoglycerate kinase gene promoter, which allows the selection of transfected cells. To produce infectious lentiviral particles, each shRNA expression plasmid (3 μg) was mixed with vectors pCMV-dR8.2dvpr (2.7 μg) and pCMV-VSVg (0.3 μg) and transfected into HEK-293T cells using Fugene 6 reagent (Roche) as described previously (49). Medium from these cultures was collected daily for 5 days, pooled, passed through a 0.45-μm filter, and used as a source for lentiviral shRNAs. Knockdown of expression of the target gene was assessed by performing Western blot analyses.
Reverse transcription (RT)-PCR analyses.
Cells (70% confluent) were treated with IFN-γ (500 U/ml). After washing with phosphate-buffered saline, they were lysed, and RNA was extracted using RNAzol reagent (Tel-Test Inc.). Total RNA was used for cDNA synthesis by using a kit (Invitrogen, Inc.). The resultant cDNA was used as a template in real-time PCR utilizing Sybr chemistry by using gene-specific primers (see Table S1 in the supplemental material). Ribosomal protein L32 (rpl32) transcript abundance was used as an internal control under these conditions using primers 5′-TTAAGCGTAACTGGCGGAAACC (forward) and 5′-CAGTAAGATTTGTTGCACATCAGC (reverse). The mean relative abundance of specific transcripts was calculated on the basis of threshold cycle values as described in our recent studies (36). All reactions were performed in triplicates. Each experiment was repeated with three separate batches of RNA.
Northern and Western blot analyses.
Northern blot analysis was performed as described previously (90). After SDS-PAGE, proteins were transferred from the gel onto a nylon membrane, blocked with 4% milk in Tris-buffered saline containing 1% Triton X-100 for 1 h, and incubated with primary antibodies (1:1,000 dilution) overnight at 4°C. The membranes were washed with Tris-buffered saline containing 1% Triton X-100 for 5 min twice and incubated with horseradish peroxidase-conjugated secondary antibody (Amersham) at a 1:2,000 dilution for 1 h. The membrane was washed, and the blot was developed using enhanced chemiluminescence reagents (Pierce, Inc.).
Reporter gene assays.
Cells were transfected with the indicated luciferase reporter constructs (300 ng) along with a β-actin-β-galactosidase reporter plasmid (100 ng). In some experiments, the pcDNA3.1 vector or the same vector coding for various C/EBPs (100 ng) was cotransfected. Luciferase activity was determined after various treatments and normalized to that of β-galactosidase (69). Each experiment was repeated at least three times, and triplicate samples were employed for each sample. Depending on which construct was used for the experiments, basal luciferase activities observed with vector pGL3-basic or pGL3-promoter were subtracted from the experimental values.
Isolation of dapk1 promoter and generation of its derivatives.
Genomic DNA prepared from C57BL/6 mouse tails was used as a template to amplify a 3-kb fragment upstream of the mouse dapk1 gene using primers 5′-CGACGCGTCGCTAGTTATATGTTTGTTCAC-3′ (forward) and 5′-CCGCTCGAGCGGTCCGCTGCGCTCGCGGCTCCT-3′ (reverse) (based on a mouse sequence in the GenBank database). The underlined sequences represent MluI and XhoI restriction sites, respectively. A PCR product was generated using Platinum Pfx (Invitrogen, Inc.) and cloned into the MluI and XhoI sites of the pGL3-basic vector (Promega, Inc.). Various deletion constructs carrying different lengths of the promoter region were generated using specific primers (see Table S2 in the supplemental material). To test the promoter independence of specific motifs, DNA fragments derived from the dapk1 promoter were cloned into the pGL3-promoter vector (Promega, Madison, WI).
The mDAPK 1.2-kb construct was used as a template for introducing point mutations. Primers used for site-directed mutagenesis are indicated in Table S3 in the supplemental material. Mutants were generated by two rounds of PCR. The first round of amplification involved a specific mutant primer and a flanking primer (vector-specific primer RV3 or GL2). The products of the first PCR were purified and mixed in an equimolar ratio, and a second round of PCR was performed using the flanking primers (RV3 and GL2). The second-round PCR product was digested with KpnI and BglII and cloned into the pGL3-basic vector utilizing the same sites. For the generation of double-mutant mDAPK, 1.2-kb CRE/ATFmut was used as a template in PCR with CBS-specific mutant primers. The CBS core sequence was mutated from TGGC to GTCT, and the CRE/ATF core sequence was mutated from GACG to GGTC. These wild-type or mutant constructs were also used as templates for generating a 649-bp promoter fragment, which included the CBS and CRE/ATF sites but lacked the native dapk1 promoter, and cloned into the pGL3-promoter vector, in which the simian virus 40 (SV40) early promoter drove the basal expression of the luciferase gene.
Wild-type 1.2-kb mDAPK was used as a template in PCR with specific primers (see Table S4 in the supplemental material) to generate dapk1 promoter fragments (200 bp) that contain either CBS or CRE/ATF sites. The promoter fragments were cloned into the KpnI and BglII sites of the pGL3-promoter vector. Sequence-verified constructs were used in the experiments. hDAPK1-Luc, where a fragment at positions −1750 to +280 derived from the human dapk1 promoter drives the expression of a luciferase gene (35), was provided by R. H. Chen, Institute of Molecular Medicine, National Taiwan University, Taiwan.
ChIP assay.
ChIP assays were performed using a commercially available kit from the Upstate Biotech, Inc., as recommended by the manufacturer, with minor modifications. After cross-linking the chromatin with 1% formaldehyde for 5 min, cells were scraped and collected. The cells were sonicated seven times for 15 s with 30-s intervals under ice by using a Bronson sonicator. The average fragment size was 500 bp under these conditions. An equal amount of chromatin was incubated with at least 5μg of either C/EBP-β-specific immunoglobulin G (IgG) or normal rabbit IgG (Santa Cruz Biotech) at 4°C overnight. IP products were collected after incubation with protein G-coated magnetic beads (Active Motif, Inc.). The beads were washed, protein-DNA cross-links were reversed, and DNA was purified by phenol-chloroform extraction and ethanol precipitation. Purified DNA from the input and IP samples were subjected to real-time PCR with specific primers (see Table S5 in the supplemental material).
In vitro phosphorylation of C/EBP-β protein.
Mouse C/EBP-β cloned in pQE-31 (Qiagen, Inc.) was expressed as a hexahistidine-tagged protein in Escherichia coli. The recombinant protein was purified to homogeneity after two rounds of chromatography on Ni-nitrilotriacetic acid agarose columns and was used as a substrate for in vitro phosphorylation. The ERK1 and ERK2 proteins were immunoprecipitated using specific antibodies. Normal rabbit IgG was used as a negative control for these reactions. The immunocomplexes were bound to protein G/protein A agarose beads (Santa Cruz Biotech) and washed with IP buffer. The beads were washed twice and incubated in 20 μl of protein kinase assay buffer (50 mM HEPES, 0.1 mM EDTA, 0.01% Brij 35, 0.1 mg/ml bovine serum albumin, 0.1% β-mercaptoethanol, 150 mM NaCl) and 2 μg of recombinant C/EBP-β protein. ATP mix (10 μl) was added and incubated for 20 min at 30°C. ATP mix consisted of 930 μl protein kinase assay buffer, 6 μl 50 mM ATP (pH 7.0), 20 μl 2.0 M MgCl2, and 44 μl of [γ-32P]ATP (10 mCi/ml). Reactions were terminated by boiling the samples, and the contents were loaded onto a 10% SDS-PAGE gel. The gels were Western blotted onto nylon membranes. The membranes were washed and autoradiographed. In some experiments, labeled bands were identified using a Molecular Dynamics PhosphorImager, and band intensities (arbitrary units) were quantified.
RESULTS
C/EBP-β-defective macrophage cells fail to undergo apoptosis.
Since IFNs cause cell growth arrest and death to exert their tumor-suppressive actions, we first examined if the loss of C/EBP-β affected cellular sensitivity to IFN-γ treatment. We used two isogenic BM macrophage cell lines generated from cebpb+/+ and cebpb−/− mice. These genotypes of cells were ensured first using PCR (Fig. 1A). As expected, wild-type and cebpb−/− loci yielded 294- and 351-bp PCR products, respectively. Tail snip DNAs prepared from mice of different cebpb−/− genotypes were used as positive controls in these experiments. Protein extracts from cebpb+/+ and cebpb−/− cells were also subjected to Western blot analysis with C/EBP-α- and C/EBP-β-specific antibodies (Fig. 1B). As anticipated, only C/EBP-β was absent in the mutant cells. Importantly, the loss of C/EBP-β did not affect the expression of C/EBP-α. Levels of actin protein were comparable between these cells, indicating equal loading of the protein.
FIG. 1.
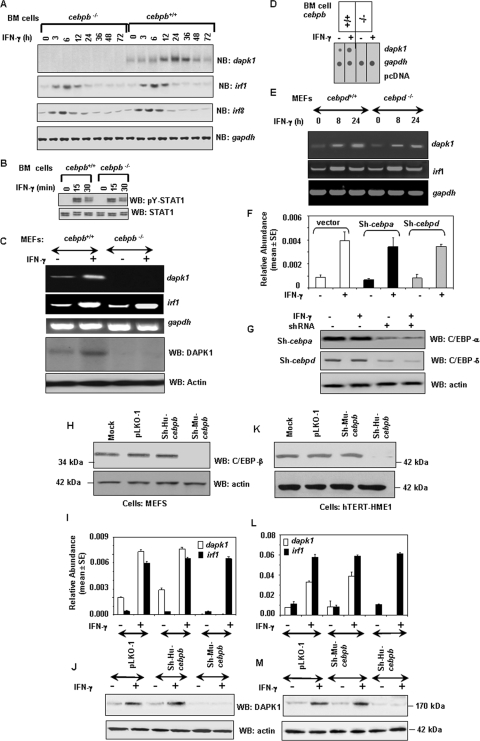
Loss of cebpb down-regulates IFN-γ-induced apoptosis. (A) Genotyping of macrophage cells was performed using PCR. MT, mouse tail DNA; KO, knockout allele; WT, wild-type allele. (B) Western blot (WB) analysis of C/EBP-β protein expression in BM cells. About 80 μg of total protein was used for Western blot analysis. Blots were probed with the indicated antibodies. (C) Photomicrograph showing IFN-γ-induced nuclear damage in wild-type BM cells after staining with DAPI. Magnification, ×60. White bar, 5 μm. Note the condensation of chromatin after IFN treatment. (D) Quantification of cell death as monitored by nuclear damage. Baseline cell death values of 1.7% and 1.9% were subtracted from the experimental values in the cebpb+/+ and cebpb−/− cells, respectively. γ, IFN-γ (500 U/ml); T, TNF-α (100 ng/ml); γ/T, IFN-γ/TNF-α combination. Treatments were performed for 18 h. Each bar represents the mean ± standard error (SE) for three separate experiments. (E) IFN-γ-induced cell death as measured by TRITC-annexin V staining. Annexin V staining was performed using a commercially available kit (Trevigen, Inc.) as recommended by the supplier. FACS analysis was performed to quantify positively stained cells. Baseline values of 1.9% and 2.1% were subtracted from the experimental values in the cebpb+/+ and cebpb−/− cells, respectively. Each bar represents the mean ± SE for three separate experiments. (F) Western blot analysis of the expression of apoptotic caspases in cebpb+/+ and cebpb−/− cells. Sixty micrograms of total protein was used for Western blot analysis with the indicated antibodies. (G) Activation of caspase-8 (after ligation with Fas antibody [3 μg/ml] for 8 h) and caspase-9 (after UV irradiation [254 nm and 40 mJ/cm2] for 10 min) in cebpb+/+ and cebpb−/− BM cells. Caspase activity was assayed using a commercially available colorimetric assay kit that employs enzyme-specific substrates (Biosource International, Inc.). Filled bars, BM-cebpb+/+; open bars, BM-cebpb−/−. (H) Induction of apoptosis by adriamycin (A) (100 μg/ml for 8 h) and UV radiation (254 nm and 40 mJ/cm2 for 10 min) and with no treatment (N). Apoptosis was quantified as described above (E). Each bar represents the mean ± SE of five separate samples. (I) Restoration of IFN-γ-induced apoptosis following transfection of C/EBP-β. cebpb−/− BM cells were electroporated with 4 μg each of either the pcDNA3.1 vector or the same vector coding for the mouse C/EBP-β cDNA along with 2 μg of the pEGFP expression vector using Amaxa Nucleofector technology. Cells were grown for 36 h prior to stimulation with IFN-γ, TNF-α, and their combination, as described above, and stained with TRITC-annexin V. GFP-positive and TRITC-positive/GFP-positive cells were quantified after FACS analysis. TRITC-positive/GFP-positive cells were expressed as a percentage of total GFP-positive cells. Each bar represents the mean ± SE (n = 5/sample). The blots below this graph show a Western blot analysis of the cells used in I with the indicated antibodies. Total protein (68 μg) was used for the analysis.
We next tested the effect of IFN-γ on these cells. IFN-γ is known to cause growth suppression via apoptosis. We have also used tumor necrosis factor alpha (TNF-α) as a control in these experiments because it is known to induce apoptosis in many cell types. Apoptosis was monitored using two methods: (i) DAPI staining for nuclear damage and (ii) annexin V binding for cell surface changes. A typical nuclear damage pattern observed with DAPI staining after IFN-γ treatment of cebpb+/+ cells is shown in Fig. 1C. Diffused chromatin was found in the nuclei of untreated cells. IFN-γ treatment caused nuclear DNA condensation and fragmentation in these cells. The number of nuclei with condensed/fragmented chromatin (dead cells) was quantified from multiple fields (n = 30) in each case and expressed as a percentage of total nuclei (Fig. 1D). Treatment of cebpb+/+ cells with IFN-γ caused a significant rise in apoptosis (15-fold) as measured by nuclear damage. Although TNF-α alone did not cause any significant cell death in these cells, it significantly (P > 0.001) augmented IFN-γ-induced cell death. In contrast, neither IFN-γ nor IFN-γ/TNF-α caused such robust apoptosis in cebpb−/− cells. A similar profile was observed upon annexin V staining of the cells (Fig. 1E). IFN-γ caused a sharp increase in the number of annexin V-positive cells (dead cells) in cebpb+/+ but not in cebpb−/− cells. TNF-α treatment augmented it further. The lack of apoptosis in cebpb−/− cells was not due to a reduction in the levels of caspase-8 and caspase-9 in these cells, as cebpb+/+ and cebpb−/− cells had comparable levels of these proteins (Fig. 1F). More importantly, caspase-8 activity was stimulated equivalently upon ligation with a death-agonistic Fas antibody in both cell types. Similarly, caspase-9 activity was induced comparably following UV irradiation of cebpb+/+ and cebpb−/− cells (Fig. 1G). Thus, IFN-induced cell death is defective in cebpb−/− cells. Consistent with these observations, both these cell types were equally sensitive to adriamycin- and UV-induced apoptosis (Fig. 1H), indicating that there was no generalized loss of apoptotic response in cebpb−/− cells. We next examined if the restoration of C/EBP-β would promote IFN-γ-induced cell death in cebpb−/− cells. Cells were electroporated with empty pCDNA3.1 expression vector or the same vector carrying C/EBP-β along with a pEGFP expression vector (for tracking the transfected cells). After 24 to 36 h of transfection, cells were treated with IFN-γ for 24 h and then stained with TRITC-labeled annexin V. GFP-TRITC double-positive cells (dead) were expressed as a percentage of the total GFP-positive cells after FACS analysis (Fig. 1I). Indeed, the restoration of C/EBP-β, but not the control vector, reestablished the IFN-γ-induced apoptotic program. Expression of C/EBP-β was confirmed by performing a Western blot analysis of the transfected cell extracts with C/EBP-β- and GFP-specific antibodies (Fig. 1I, bottom). As expected, C/EBP-β was detected only in cells transfected with the expression vector carrying it. The GFP blot shows comparable loading in these lanes. No such augmentation of IFN-γ-induced cell death occurred following the transfection of expression vectors coding for either C/EBP-α or C/EBP-δ into cebpb−/− cells (data not shown). Thus, C/EBP-β is critical for the IFN-γ-induced cell death response(s). Although not shown here, similar apoptotic effects were observed with IFN-α in these cells (data not shown).
Identification of C/EBP-β-dependent IFN-γ-stimulated genes.
Since IFN-γ failed to induce apoptosis, we next examined a molecular basis for it by profiling the differences in gene expression between wild-type and cebpb−/− cells. The NIA mouse 15K cDNA microarray was screened with RNA isolated from wild-type and cebpb−/− cells. In these experiments, three independent batches of RNAs from cells treated with IFN-γ for various times (2, 4, 6, 12, 16, and 24 h) were pooled. Similarly, three independent batches of RNAs from untreated cells were prepared and pooled. Poly(A)+ RNA from each sample (cebpb+/+ untreated and IFN-γ treated and cebpb −/− untreated and IFN-γ treated) was prepared. Labeled cDNA probes generated from these mRNAs were hybridized to the cDNA microarrays, and changes in gene expression were determined. Genes differentially expressed (induced or repressed) in response to IFN-γ by about twofold were identified by comparing the gene expression patterns observed with untreated and IFN-γ-treated cebpb+/+ cells. A similar analysis was performed with cebpb−/− cells. The differences in IFN-γ-regulated expression patterns were then compared between wild-type and cebpb−/− cells to finally identify IFN-regulated genes whose expression was dependent on C/EBP-β. Genes whose expressions were induced and repressed by IFN-γ in a C/EBP-β-dependent manner are listed in Tables 1 and 2, respectively. About 50 genes were dependent on C/EBP-β for their IFN-γ-induced expression. Twelve genes were suppressed by IFN-γ in the presence of C/EBP-β.
TABLE 1.
C/EBP-β-dependent IFN-γ-induced genes
| Transcript | Fold change |
|---|---|
| ADP ribosylation-like factor 6-interacting protein | 2.973 |
| Alkaline phosphatase | 2.471 |
| ATP-dependent DNA helicase II | 2.724 |
| Cold-induced RNA-binding protein | 2.437 |
| Collagen α2(v) chain precursor | 2.654 |
| Cyclic AMP phosphoprotein 19 kDa | 2.086 |
| Cytochrome B5 | 2.667 |
| Dead box polypeptide 5 | 2.250 |
| Death-associated protein kinase 1 | 4.258 |
| Degenerative spermatocyte homologue | 2.606 |
| Dystrophia myotonica protein containing WD repeat | 2.601 |
| Estradiol-17-β dehydrogenase 4 | 2.636 |
| Eukaryotic translation initiation factor 4A | 2.573 |
| Extracellular protease inhibitor | 2.662 |
| Ferritin light chain 1 | 2.716 |
| Fibulin 1 | 2.416 |
| FK506-binding protein 6 | 2.410 |
| Heterogenous nuclear ribonucleoprotein A3 | 2.478 |
| HSP86 | 3.234 |
| IRF9/ISGF3g/p48 | 3.276 |
| Limb-expressing 1 homologue | 4.505 |
| Lymphocyte antigen 57 | 3.170 |
| Lymphocyte cytosolic protein 2 | 2.651 |
| Membrane protein C21 orf4 | 2.831 |
| Microsomal signal peptidase 18 kDa | 2.867 |
| Na-K-Cl cotransporter | 3.466 |
| NDRG4 protein | 2.630 |
| Nuclear protein 220 | 2.616 |
| p130 | 4.578 |
| Phosphatidyl-choline transfer protein | 2.443 |
| Phosphoprotein enriched in astrocytes 15 | 2.870 |
| Proliferation-associated protein 2G4 | 3.099 |
| Proline-rich protein MP-2 | 3.175 |
| Protein p8 | 4.336 |
| Proteasome psMA6 | 3.482 |
| RAC-2 | 6.984 |
| RAD-18 homologue | 2.710 |
| Retinoblastoma-binding protein 7 | 2.564 |
| RNA-dependent helicase p68 | 2.493 |
| Septin-3 | 2.907 |
| Serine protease inhibitor | 2.403 |
| Serine protease stubble | 2.721 |
| Signal recognition particle 54 kDa | 2.305 |
| Skeletal muscle LIM protein 3 | 3.036 |
| Tumor-related protein | 3.300 |
| Tyrosine protein kinase Lyn | 2.170 |
| Ubiquitin-activating enzyme II | 2.589 |
| Zinc finger protein 36 | 3.702 |
| SERP1 | 8.035 |
TABLE 2.
C/EBP-β-dependent IFN-γ-repressed genes
| Transcript | Fold change |
|---|---|
| Cathepsin C | −1.532 |
| Nuclear factor erythroid derivative 2a | −2.527 |
| Urokinase plasminogen activator receptor | −1.509 |
| Expressed sequence c77180 | −0.509 |
| Fc receptor IgG | −0.500 |
| 5-Lipooxygenase-activating protein | −2.491 |
| Cytochrome 8-245 | −1.433 |
| Ectonucleoside triphosphate diphosphohydrolase | −2.424 |
| Cyclin D2 | −3.315 |
| X-1-specific transcript | −1.313 |
| Zyxin, binds alpha actinin and CRP | −1.305 |
| Hypothetical protein spx19 | −1.228 |
Some of these genes are known to be regulated by IFNs. The inducible genes appear to have many different functions such as signal transduction (e.g., the Lyn tyrosine kinase, RAC-2, and fibulin-1). Several protease inhibitors such as extracellular proteinase inhibitor and serine protease inhibitor were also induced in response to IFN treatment. Ubiquitylating enzymes, proteasome subunits, were also found in these genes. In most cases, the critical transcription factor mediating the IFN effects had not been identified. Some of these gene products, such as RAC-2 and p130 (Rb related), are modulated by type I IFNs (60, 87). The p8 protein codes for a homologue of a bovine chemokine (85). Some genes like estradiol-17β-dehydrogenase 4 are involved in estrogen biosynthesis and may control cell growth (89). Interestingly, none of the previously defined STAT1-dependent genes were identified in this screen. Thus, it appears that very little overlap exists between IFN-regulated STAT1-driven genes and IFN-induced C/EBP-β-driven ISGs.
We have confirmed the IFN-γ-induced C/EBP-β-driven expression of some of these genes using real-time PCR (Fig. 2). Transcripts of the RNA-dependent helicase, RAC-2, and septin-3 were induced by 4.25-, 3-, and 4-fold, respectively, in wild-type cells in the presence of IFN-γ. These genes are involved in cell division, signaling, and transcription. Consistent with gene expression profiling data, IFN-γ did not induce the expression of these transcripts in cebpb−/− cells. We showed that the expression of the IRF9/ISGF3γ/p48 gene was C/EBP-β and IFN dependent in previous studies (70). A similar observation was made with another gene, dapk1 (see below for details). We also tested the effect of IFN-γ on the expression of cyclin D2, given its potential role in cell growth control. Its expression in cebpb+/+ cells was repressed in the presence of IFN-γ but not in cebpb−/− cells. Since the biological significance of many of these genes in IFN-γ-induced actions is unclear at this stage, we focused the rest of the study on dapk1, which is known to participate in IFN-γ-induced cell death pathways (19). A similar profile of IFN-γ-induced C/EBP-β-dependent expression of these genes (data not shown) was observed in MEFs. Therefore, we used MEFs in further studies.
FIG. 2.
Quantitative RT-PCR analysis of the expression of some transcripts identified in gene expression profiling experiments. Transcript-specific primers listed in Table S1 in the supplemental material were used with cDNA prepared from cebpb+/+ and cebpb−/− BM cells after 12 h of IFN-γ treatment. The transcripts coding for ribosomal protein L32 were used as an internal control. Relative transcript abundance was calculated based on the threshold cycle values. Each bar shows the mean ± SE of five samples. RNA dep helicase, RNA-dependent helicase.
Expression of dapk1 is dependent on C/EBP-β.
Since dapk1 was identified as an IFN-γ-regulated gene in gene expression profiling, we next examined if its expression is affected by the loss of C/EBP-β. In cebpb+/+ macrophage cells, IFN-γ caused a time-dependent induction of dapk1 (Fig. 3A). Notably, by 12 h, a significant rise in dapk1 mRNA occurred, which remained high until 36 h and declined thereafter. By 24 h, there was a fourfold-higher expression of dapk1 mRNA in cebpb+/+ macrophage cells. In contrast, neither basal nor IFN-γ-induced expression of dapk1 mRNA was observed in cebpb−/− cells. All lanes had equal amounts of mRNA as determined by a reprobing of these blots with gapdh. We next investigated if the lack of dapk1 expression in cebpb−/− cells was due to a generalized loss of the IFN-γ response in these cells by taking two approaches: (i) measuring the expression patterns of two other IFN-induced mRNAs corresponding to irf1 and irf8 genes and (ii) monitoring IFN-γ-induced tyrosyl phosphorylation of STAT1, a critical regulator of these mRNAs. The expression levels of irf1 and irf8 mRNAs were induced equivalently by IFN-γ in cebpb+/+ and cebpb−/− cells (Fig. 3A, middle). There was no significant difference between cebpb+/+ and cebpb−/− cells with respect to the kinetics of induction and the down-regulation of these mRNAs. As reported previously in other studies, both these mRNAs are induced by IFN-γ treatment within 3 h and began to decline by 12 h. In a similar manner, IFN-γ caused a rapid activation and a decline of the tyrosyl phosphorylation of STAT1α and STAT1β in both cell types (Fig. 3B). There was no difference in the levels of total STAT1 in these cells. Thus, the STAT1-dependent arm of IFN signaling was unaffected by the loss of C/EBP-β. Lastly, we examined if dapk1 was induced at the transcriptional level by C/EBP-β after IFN treatment in these cells by using a nuclear runoff transcription assay (Fig. 3D). IFN-γ induced the transcription of dapk1 mRNA robustly in cebpb+/+ but not in cebpb−/− cells. Even the basal transcription of this was absent in cebpb−/− cells.
FIG. 3.
Basal and IFN-γ-induced expression of dapk1 is C/EBP-β dependent. (A) cebpb+/+ and cebpb−/− cells were stimulated with IFN-γ for various hours, and poly(A)+ RNA was prepared. About 3 μg of RNA was used for Northern blot (NB) analysis with the indicated gene probes labeled with 32P. The blots were washed and exposed to X-ray films to detect the bands. Blot regions corresponding to the specific bands are shown. (B) Activation of STAT1 via phosphorylation at tyrosine 701 by IFN-γ treatment. The upper and lower bands correspond to the STAT1α and STAT1β proteins, respectively. (C) Expression of endogenous dapk1 mRNA and protein in cebpb+/+ and cebpb−/− MEFs. RT-PCR with gene-specific primers was used to detect the specific transcripts indicated on the right. RT-PCR for irf1 and dapk1 was performed with RNA isolated after 6 and 12 h of IFN treatment, respectively. Western blot (WB) analysis was performed with specific antibodies after separating 85 μg of total protein by SDS-PAGE and Western transfer. (D) Nuclear runoff transcription. Nuclei were isolated after stimulating the BM cells with IFN-γ for 8 h and incubated with 32P-lableled UTP for 45 min. The labeled RNAs were extracted and hybridized to specific DNAs affixed on a nylon filter. pcDNA3.1 and gapdh were used as negative and positive controls, respectively, in this experiment. (E) RT-PCR analysis of the expression of dapk1, irf1, and gapdh transcripts using RNA extracted form IFN-γ-treated cebpd+/+ and cebpd−/− MEFs. (F and G) Effect of shRNA-mediated knocking down of cebpa and cebpd on the IFN-induced expression of dapk1. Wild-type BM cells were infected with lentiviral vectors coding for cebpa- and cebpd-specific shRNAs or the control empty vector (pLKO-1). Real-time PCR was carried out using dapk1-specific primers. (G) Western blot analysis of cell extracts (80 μg from each sample). (H and K) Effect of cebpb-specific shRNAs on the expression of endogenous C/EBP-β in MEFs (H) and hTERT-HME1 (K) cells. Lentiviral vector (pLKO-1)-encoded species-specific shRNAs against C/EBP-β transcript were used for knocking down the expression of endogenous C/EBP-β. (I and L) Real-time PCR analysis of IFN-induced expression of dapk1 and irf1 transcripts in MEFs (I) and hTERT-HME1 (L) cells. (J and M) Western blot analyses of the DAPK1 and actin proteins in MEFs (J) and hTERT-HME1 (M) cells.
To rule out a possibility that it is a cell type-specific effect, we employed isogenic MEFs derived from cebpb+/+ and cebpb−/− mice. Based on the information from Fig. 3A, we chose 12 h of IFN treatment for examining these aspects. RT-PCR analyses showed a clear induction of dapk1 mRNA (about three- to fourfold based on various experiments) in cebpb+/+ MEFs (Fig. 3C). The basal and IFN-induced expressions of dapk1 were undetectable in cebpb−/− MEFs, while irf1 mRNA was readily induced equivalently in both cell types, as noted with macrophages. The expression of the irf1 transcript was monitored at 6 h post-IFN treatment, given its early inductive pattern. Consistent with these data, Western blot analyses revealed basal and IFN-γ-induced expression of the DAPK1 protein in cebpb+/+ but not in cebpb−/− MEFs. Together, these experiments establish an important role for C/EBP-β in regulating the expression of dapk1. Since experiments with MEFs and BM cells yielded very similar results, we used MEFs for the rest of the promoter analyses.
Lastly, we also examined if dapk1 expression was affected in MEFs lacking cebpd, a member of the C/EBP family of transcription factors (Fig. 3E). No significant difference in the magnitudes of the basal and IFN-γ-induced expression of dapk1 mRNA was observed between cebpd+/+ and cebpd−/− MEFs. The expression of irf1 mRNA (an internal control for IFN-γ-induced activity) was unaffected by the lack of C/EBP-δ in the cells. To verify these observations further, we knocked down the expression of cebpa and cebpd mRNAs using lentiviral shRNA vectors (Fig. 3F and G). shRNA-mediated knockdown of expression of these transcription factors did not significantly affect the basal and IFN-induced expression of the dapk1 transcript as measured by real-time PCR. These results are consistent with those obtained with cebpd−/− MEFs. The scrambled shRNA controls for these experiments yielded results similar to those for the vector control (data not shown).
To further validate the data observed with cebpb−/− cells, we used shRNAs capable of targeting cebpb mRNA to degradation via RNA interference. Human and mouse shRNAs capable of targeting endogenous mRNA for destruction in a species-specific manner (because of sequence differences) were used in these experiments. Based on this specificity, human shRNA, which targets endogenous cebpb mRNA in human cells, serves as a negative control (scramble) in mouse cells. A similar relationship exists between mouse cebpb-specific shRNA and human cebpb mRNA. We used a nononcogenic mammary epithelial cell line and MEFs for these studies. We first tested the abilities of these shRNAs to target C/EBP-β expression. Both shRNAs suppressed the expression of the endogenous C/EBP-β protein in cells of their respective species (Fig. 3H and K). They failed to act as specific shRNAs when cells of another species were infected. As expected, the empty lentiviral vectors and mock infection did not affect the expression of C/EBP-β in both species. Western blot analysis with actin served to demonstrate equal loading in these cells. The effect of these shRNAs on the expression of endogenous dapk1 mRNA was tested. Real-time PCR analysis of dapk1 transcript showed that human and mouse cebpb-specific shRNAs suppressed the basal and IFN-induced expression of dapk1 mRNAs in cells of their respective species (Fig. 3I and L). They failed to inhibit dapk1 expression upon transfection into cells derived from a heterologous species. In contrast, the IFN-γ-induced expression of the irf1 transcript was unaffected by cebpb-specific shRNAs. These data show a specific requirement of C/EBP-β for the expression of dapk1 in an IFN-regulated pathway. The knockdown of C/EBP-β also resulted in a loss of basal and IFN-γ-induced expression of the DAPK1 protein (Fig. 3J and M).
Other members of the DAPK family do not require C/EBP-β for their expression.
There are four additional members of the DAPK family, each of which is involved in various biological processes including apoptosis. These are DAPK2, DRK2, DAPK3, and ZIP kinase. We next examined if the expression of other members of the DAPK family is also similarly regulated by IFN-γ by using real-time PCR (see Fig. S1 in the supplemental material). All these mRNAs were constitutively expressed in cells, and IFN-γ did not significantly up-regulate them. More importantly, a loss of C/EBP-β did not affect the expression of any of these genes. Thus, the IFN-γ-induced expression of dapk1 is exclusively dependent on C/EBP-β.
C/EBP-β promotes IFN-γ-dependent gene expression from the dapk1 promoter.
Since the above-described studies provide indirect evidence for the role of C/EBP-β in regulating dapk1, we wanted to examine if C/EBP-β directly regulated transcription from the dapk1 promoter. We isolated a 3-kb DNA fragment corresponding to the promoter of dapk1 from the C57BL/6 mouse background. This fragment was cloned upstream of the firefly luciferase gene in the promoterless pGL3-basic vector and used for transient transfection assays in cebpb+/+ and cebpb−/− MEFs. Basal and IFN-γ-induced expressions of the reporter gene were seen only in wild-type and not in cebpb−/− MEFs (Fig. 4A). Approximately 3.5-fold-greater luciferase activity was seen after IFN-γ treatment. These observations suggest that the transcriptional induction of dapk1 by IFN-γ was dependent on C/EBP-β. We next tested if similar effects could be seen in presence of lentiviral shRNAs in wild-type MEFs (Fig. 4B). Mu-cebpb-shRNA but not Hu-cebpb-shRNA or/and the empty expression vector suppressed the basal and IFN-γ-induced expression of the reporter gene.
FIG. 4.
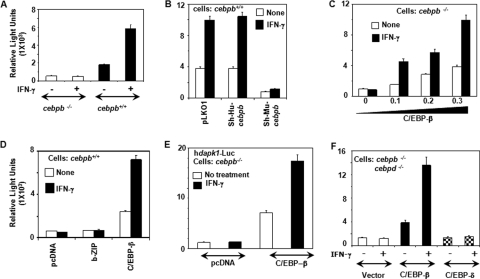
Basal and IFN-γ-induced gene expression from the dapk1 promoter is dependent on C/EBP-β. MEFs were transfected with the mdapk1-3K-Luc construct (300 ng) along with a β-actin-β-galactosidase reporter (100 ng) (A to C and F). (C) Increasing amounts of C/EBP-β (0 to 0.3 μg) were cotransfected into the cells. In the case of the control, 0.3 μg of empty vector was used. Total transfected DNA was kept constant (usually less than 700 ng) by adding pCDNA3.1 vector DNA as required. Lentiviral shRNA vectors are described in the legend of Fig. 3. (D) Mutant C/EBP-β containing only the b-ZIP domain fails to support the IFN-induced transcription of dapk1-luc. (E) A luciferase construct driven by the human dapk1 promoter (region at positions −1.75 to +280) was used for the analysis. The cell types used in each case are indicated. (F) Effects of C/EBP-β and C/EBP-δ on the expression of mDAPK1-Luc in cebpb−/−/cebpd−/− double-knockout cells. Luciferase activity was normalized to that of β-galactosidase in each experiment. Normalized luciferase activity was plotted as relative light units. Each bar represents the mean ± SE of triplicates. Each experiment was repeated three times.
In the next experiment, we examined if a restoration of C/EBP-β could reinstate the basal and IFN-γ-induced response of the promoter. cebpb−/− MEFs were transfected with mDAPK1-Luc along with increasing amounts of a C/EBP-β expression vector, and luciferase reporter activity was measured. Empty expression vector was used as a control. The C/EBP-β expression vector, but not the empty expression vector, rescued the basal and the IFN-γ-induced expression of the reporter (Fig. 4C). The lack of dapk1 expression in cebpb−/− cells was not due to differential transfectability of these cells. To demonstrate this aspect, we transfected cebpb+/+ and cebpb−/− cells with an IFN-γ-inducible STAT1-dependent luciferase vector, pIRE-Luc, and a constitutively active vector, CMV-Luc (see Fig. S2A and S2B in the supplemental material). Both these reporters expressed equivalently in cebpb+/+ and cebpb−/− cells. To further support the role of C/EBP-β in regulating the dapk1 promoter, we expressed only the DNA binding domain (b-ZIP) of the C/EBP-β protein. Only the wild-type C/EBP-β protein, but not b-ZIP, promoted the basal and IFN-γ-induced expression of dapk1 (Fig. 4D). A similar effect of C/EBP-β was observed with hDAPK1-Luc, driven by the promoter of human dapk1 (Fig. 4E). In cebpb−/− cells, the coexpression of wild-type C/EBP-β, but not an empty expression vector, significantly up-regulated the basal and IFN-γ-induced expression of luciferase. Thus, both the human and mouse dapk1 promoters are similarly responsive to C/EBP-β and IFN-γ. Since both human and mouse promoters responded similarly to C/EBP-β and IFN-γ, we used the mouse promoter for the rest of the study.
Other members of the C/EBP family do not significantly induce dapk1 expression.
Next, we also checked the expression of DAPK1-Luc in cebpb and cebpd double-knockout cells (Fig. 4F). The expression of only C/EBP-β, but not C/EBP-δ, promoted basal and IFN-γ-induced expression from the dapk1 promoter. Additionally, in cebpb−/− BM cells, the expression of C/EBP-α or C/EBP-δ did not restore basal and IFN-induced expression from the dapk1 promoter (data not shown). The coexpression of only C/EBP-β significantly induced the basal and IFN-γ-induced expression of luciferase. Lastly, consistent with these observations, the IFN-induced expression of the dapk1-luc reporter was not significantly augmented by other members of this family (see Fig. S2C in the supplemental material). Thus, C/EBP-β appears to be the major transcriptional inducer of dapk1, and other members of the C/EBP-family do not appear to substitute for C/EBP-β in regulating dapk1 vis-à-vis promoter-binding activity. Like IFN-γ, IFN-α/β-induced expression of dapk1-luc was also dependent on C/EBP-β (data not shown).
Two independent C/EBP-β-responsive elements mediate the IFN response.
Given the C/EBP-β responsiveness of the dapk1 promoter, we sought to identify these elements. Therefore, we first performed a serial deletion analysis of the promoter. These studies revealed that progressive deletions leading from the 5′ end of the promoter to the −1.2-kb region sustained maximal transcription of the basal and IFN-γ-induced expression of the luciferase reporter (Fig. 5A). Further deletions caused a significant decline (P > 0.001) in basal and IFN-γ-induced expression from the dapk1 promoter. For example, the mDAPK1 -0.5-kb construct, which contained only the upstream 500 bp of the dapk1 promoter, was not as efficient as the mDAPK1 1.2-kb construct. Further deletions caused a total loss of promoter activity. Thus, a 1-kb fragment located between −0.2 and −1.2 kb of the dapk1 promoter contains elements required for basal and IFN-γ-induced expression.
FIG. 5.
Identification and analysis of the critical elements required for C/EBP-β-dependent IFN-γ-driven responses of the dapk1 promoter. Open bars, no treatment; filled bars, IFN-γ. (A) Deletion analysis of the mouse dapk1 promoter. Deletion constructs were generated using specific primers listed in Table S2 in the supplemental material. (B) The potential response elements were subjected to site-directed mutagenesis using the primers described in Table S4 in the supplemental material. (C) Mutant and wild-type (WT) constructs within a 649-bp fragment of the dapk1 promoter were PCR amplified using the primers listed in row 3 of Table S3 in the supplemental material and cloned upstream of the SV40 early promoter in the pGL3-promoter vector. Mutants and wild-type constructs shown in B were used as templates for generating these mutants. (D) Promoter subfragments corresponding to the distal CBS and proximal CRE/ATF sites were amplified separately and cloned into the pGL3-promoter vector using the primers shown in Table S3 in the supplemental material. Transfection and luciferase activities were determined and quantified as described in the legend of Fig. 4.
Since the mDAPK1 1.2-kb construct provided maximal induction of the promoter, we analyzed the promoter fragment for potential C/EBP-β-responsive and/or transcription factor binding sites. Sequence analysis with the MatInspector program (http://www.genomatix.de), which not only analyzes the transcription factor binding motifs but also provides statistical probabilities for the likelihood of a potential binding site in a given DNA sequence, predicted several theoretical transcription factor binding sites. Among these was a potential CBS in the distal region of the promoter (Fig. 5B). Henceforth, this site will be referred to as the “distal CBS.” We introduced point mutations into the core sequence that will disrupt its ability to respond to C/EBP-β. A comparison of the IFN responsiveness of this mutant to that of the wild-type dapk1 promoter revealed no significant differences in terms of their basal and IFN-γ-induced responses. In light of this observation, we searched for other C/EBP-β-responsive elements. In the promoter-proximal region, there was a CREB/ATF-like binding site (proximal CRE/ATF). We mutated this element and tested its sensitivity to C/EBP-β and IFN-γ. Although the basal transcription driven by this mutant promoter was weakly affected, the IFN-γ-induced response was significantly blunted. Since mutations in both elements did not completely suppress the promoter response, we mutated both elements and tested the promoter response to C/EBP-β and IFN-γ. The double mutant completely lost its response to both C/EBP-β and IFN-γ, compared to the wild-type and single mutants. Thus, two independent response elements mediate the basal and IFN-γ-induced responses from the dapk1 promoter. The CRE/ATF site appears to be the most critical for the IFN-γ-induced response.
A 649-bp region of the dapk1 promoter is sufficient for driving the IFN-γ response mediated by C/EBP-β on a heterologous promoter.
In order to define if the promoter fragment containing the distal CBS and proximal CRE/ATF site was sufficient to confer C/EBP-β and IFN-γ responsiveness to a heterologous promoter, we generated the following constructs. A 649-bp promoter fragment containing wild-type and mutant binding sites was PCR amplified using the primers shown in row 3 of Table S3 in the supplemental material. Wild-type and mutant promoters shown in Fig. 5B were used as templates to generate the PCR products. These products lack the native dapk1 promoter. They were placed upstream of the SV40 promoter in the pGL3-promoter vector. The wild-type and individual mutants were transfected into cebpb−/− cells along with a C/EBP-β expression vector, and luciferase activity was measured (Fig. 5C). The pGL3-promoter vector itself did not respond to IFN-γ treatment. The wild type and CBS mutants responded to C/EBP-β and IFN-γ as observed in the case of the native promoter. The CRE/ATF mutant significantly lost its IFN response and was still inducible by C/EBP-β at the basal level. However, inactivation of both CBS and CRE/ATF elements completely suppressed the basal and IFN-γ-induced response, dependent on C/EBP-β. This mutant behaved like the parental pGL3-promoter vector. Thus, although the CBS mutation does not affect the promoter responses, in the absence of the CRE/ATF site, it seems to provide a weak IFN response.
The individual response elements derived from the dapk1 promoter respond to C/EBP-β in the absence of the other but are insufficient for generating the full promoter response.
To determine if either of these response elements is sufficient for inducing transcription, we have used ∼200-bp fragments derived from the dapk1 promoter bearing the distal CBS or the proximal CRE/ATF sites. These fragments were cloned upstream of the SV40 early promoter and used for transient transfection analyses in the presence and absence of C/EBP-β in cebpb−/− cells (Fig. 5D). The magnitude of gene expression that occurred with these constructs was compared to that of a control construct in which a 649-bp element (containing both elements) drove the expression of the luciferase reporter under the control of the SV40 early promoter.
The CBS and CRE/ATF reporters are expressed poorly in the absence of C/EBP-β. The construct carrying the distal CBS responded to C/EBP-β but failed to promote an IFN-γ response on its own. The construct bearing the proximal CRE/ATF was induced by C/EBP-β and responded to IFN-γ treatment. Its basal response was weaker than that observed with a construct bearing the distal CBS. Although the proximal CRE/ATF construct responded significantly to IFN-γ, it was not as efficient as the 649-bp construct, which contained both these elements. The 649-bp construct responded to IFN-γ and C/EBP-β a significantly (P > 0.001) higher levels than both the distal CBS and the proximal CRE/ATF constructs. Thus, the proximal CRE/ATF site can act as an IFN-γ-inducible element on its own but is not sufficient to generate the maximal basal and IFN-γ response in the absence of the distal CBS. This result is consistent with observations made with the mDAPK1 0.5-kb construct (which lacked the distal CBS), whose basal and IFN-γ-induced expression was lower than that of the mDAPK 1.2-kb construct, which harbors both distal CBS and proximal CRE/ATF elements (Fig. 5D).
IFN-dependent recruitment of C/EBP-β to the dapk1 promoter.
We next investigated whether C/EBP-β bound to the elements defined in the mutational analyses using ChIP assays with promoter region-specific primers (see Table S5, row 1, in the supplemental material). cebpb+/+ and cebpb−/− MEFs were first stimulated with IFN-γ for 12 h, chromatin was cross-linked, and soluble chromatin was prepared. ChIP assays were performed using dapk1 distal promoter-specific primers. A typical ChIP assay is shown in Fig. 6A. As expected, no ChIP products were observed with cebpb−/− MEFs. In cebpb+/+ MEFs, C/EBP-β was recruited to this site constitutively, and IFN-γ treatment did not significantly affect it. The normal IgG and no-IgG control reactions did not yield any PCR products, showing the specificity of these ChIP reactions. The input chromatin levels were comparable between the cell lines and treatments. A similar profile of C/EBP-β binding was noted in a real-time PCR analysis of ChIP products (Fig. 6B). No significant increase in C/EBP-β binding to the distal CBS of the dapk1 promoter occurred following IFN-γ treatment for several hours.
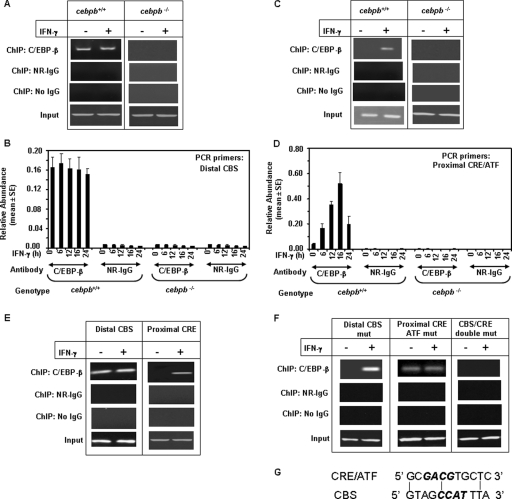
FIG. 6.
Effect of IFN-γ on the binding of C/EBP-β to the distal CBS and proximal CRE/ATF sites of the dapk1 promoter in MEFs. ChIP assays were performed using a commercially available kit (Upstate Biotech, Inc.). (A and C) Typical PCR patterns obtained in ChIP assays with distal CBS-specific (A) and proximal CRE/ATF-specific (C) primers (see Table S5 in the supplemental material). For input control reactions, one-fifth of the soluble chromatin used for the ChIP analysis was employed. Twenty-six cycles of PCR were performed in each case. NR IgG, normal rabbit IgG. C/EBP and IgG were used at 10 μg each/reaction. (B and D) Real-time PCR analysis of dapk1 promoter fragments recovered in ChIP assays performed with the indicated antibodies using chromatin prepared from wild-type and cebpb−/− MEFs. Each bar represents the mean abundance of dapk1 promoter fragments ± SE of six separate reactions from two independent experiments. (E) Wild-type MEFs were transfected with constructs carrying only the distal CBS or the proximal CRE/ATF (shown at the bottom of D) for 36 h. After stimulation with IFN-γ, chromatin was prepared and used for ChIP assays. For the distal CBS construct, primers 180mDAPK-R and RV3 were used for PCR. For the proximal CRE, primers 200mDAPK-F and GL2 were used for PCR. The rest of the controls for this experiment were essentially similar to those described above (A). (F) ChIP analysis of cells transfected with the single- and double-mutant constructs shown at bottom of panel C. The plasmid-specific primers RV3 and GL2 were used for PCR analysis of the ChIP products. (G) Comparison of the sequences of CBS and CRE/ATF elements of the dapk1 promoter. Core sequences are shown in boldface, italicized type. Vertical lines show the homologous nucleotides.
We next examined if an IFN-γ-inducible recruitment of C/EBP-β to the proximal CRE/ATF site occurred using ChIP assays with the primers listed in row 2 of Table S5 in the supplemental material. A typical PCR profile is shown in Fig. 6C. In cebpb−/− MEFs, no PCR products were detected, although the control reactions indicated sufficient DNA input. In cebpb+/+ MEFs, no detectable binding of C/EBP-β occurred in the unstimulated sate. However, IFN-γ treatment induced C/EBP-β recruitment to the promoter in a time-dependent manner, which persisted up to 16 h (Fig. 6D). No PCR products were observed in the control ChIP reactions performed with no IgG and normal IgG. Thus, the recruitment of C/EBP-β to the proximal CRE/ATF site of the dapk1 promoter was induced in an IFN-γ-dependent manner.
The differential recruitment of C/EBP-β to the promoter-proximal CRE/ATF and distal CBS was studied using another approach. The luciferase plasmids bearing the individual ∼200-bp promoter fragments (shown in Fig. 5D) were transfected into cebpb+/+ cells. Cells were treated with IFN-γ for 12 h, DNA-protein complexes were fixed in vivo with formaldehyde treatment, and soluble chromatin was prepared and immunoprecipitated with the indicated antibodies. PCR was performed with plasmid (RV3 or GL2) and promoter fragment-specific primer pairs (see Table S3 in the supplemental material). These primers pairs distinguish the endogenous promoter from the plasmid-based one. For the distal CBS construct, we used primers 180mDAPK-R and RV3 for PCR. In the case of the proximal CRE/ATF construct, primers 200mDAPKF and GL2 were used for PCR. As shown in Fig. 6E, the binding of C/EBP-β to the distal promoter fragment bearing the CBS was constitutive, and IFN-γ did not change such binding. In contrast, the binding of C/EBP-β to the proximal promoter fragment bearing the CRE/ATF site was induced following IFN-γ treatment. Its binding was barely detected in the absence of IFN-γ treatment. The control ChIP reactions performed with no IgG and normal rabbit IgG did not yield any products.
Lastly, we used promoter constructs bearing mutations in individual CBS and CRE/ATF sites (shown in Fig. 5C) and the double mutant for ChIP assays for demonstrating a differential recruitment of C/EBP-β to the promoter. Thirty-six hours after transfection of the plasmids into wild-type MEFs, cells were treated with IFN-γ for 12 h, and soluble chromatin was prepared after cross-linking with formaldehyde. Chromatin was immunoprecipitated with the indicated antibodies, and PCR was performed using plasmid-specific GL2 and RV3 primer pairs (Fig. 6F). As expected, PCR products were not detected in the control ChIP reactions with control IgG or no IgG. ChIP assays with the distal CBS mutant, which contained a normal CRE/ATF site, showed IFN-γ-inducible binding of C/EBP-β to the promoter. In contrast, ChIP assays with the CRE/ATF mutant, which contained a normal distal CBS, showed basal binding of C/EBP-β to the promoter, which was not further stimulated by IFN-γ. The ChIP assay with the double mutant, consistent with a loss of its IFN-γ response, did not produce any PCR products. The chromatin inputs were comparable between treated and untreated conditions. Thus, C/EBP-β recruitment to proximal CRE/ATF, but not to the distal CBS, is IFN-γ induced. Importantly, C/EBP-β binding to the CBS and CRE/ATF occurred independently of each other. Notably, despite its ability to bind C/EBP-β, the CRE/ATF site exhibited a weak homology to distal CBS (Fig. 6G). Only 3 of the 11 nucleotides were conserved between these motifs. Electrophoretic mobility shift assays with CBS and CRE/ATF oligonucleotides also showed a similar pattern of binding of C/EBP-β in response to IFN-γ (see Fig. S3 in the supplemental material).
IFN-γ-induced recruitment of C/EBP-β to the CRE/ATF site is dependent on the ERK signaling pathway.
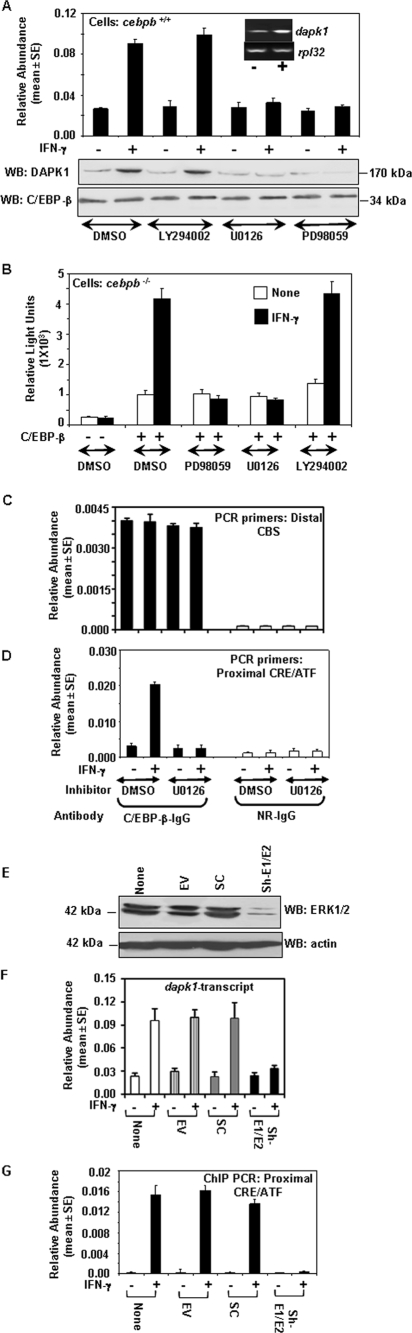
Since IFN-γ treatment causes an inducible recruitment of C/EBP-β to the dapk1 promoter, we examined if this action requires previously reported IFN-induced ERK signaling pathways (32). We have used two inhibitors, PD98059 and U0126, that specifically block ERK1/2 activation via MEK1/2. As a control, we used an unrelated inhibitor, LY294002, which blocks the PI3-kinase pathways. We first tested the effects of these inhibitors on the expression of endogenous dapk1 mRNA in wild-type MEFs using real-time PCR (Fig. 7A). Results from a Western blot analysis of the DAPK1 protein also correlated with the RNA expression profile (Fig. 7A, bottom). Only the ERK pathway inhibitors, but not the control inhibitor LY294002 or dimethyl sulfoxide (DMSO), blocked the IFN-γ-induced expression of dapk1 mRNA and protein. These inhibitors did not affect the basal expression of dapk1. In the next experiment, cebpb−/− MEFs were transfected with the mDAPK-1.2K-Luc construct in the absence and presence of C/EBP-β and then stimulated with IFN-γ in the absence and presence of these inhibitors (Fig. 7B). PD98059 and U0126, but not LY294002, blocked the IFN-γ-induced C/EBP-β-dependent expression of luciferase. We next examined if this effect could be related to the IFN-induced binding of C/EBP-β to the distal CBS and proximal CRE/ATF sites of the dapk1 promoter using quantitative ChIP assays. Since the LY294002 inhibitor was no different from DMSO and PD98059 was not so different from U0126 in terms of their effects on the dapk1 promoter, we compared the effects of DMSO to those of U0126 on the IFN-induced recruitment of C/EBP-β to the dapk1 promoter in MEFs. C/EBP-β binding to the distal CBS was unaffected by IFN-γ, and U0126 did not affect it (Fig. 7C). However, the IFN-γ-inducible binding of C/EBP-β to the CRE/ATF site that was noted in the controls was blocked in the U0126-treated cells (Fig. 7D).
FIG. 7.
Mitogen-activated protein kinases, ERK1 and ERK2, are required for IFN-induced expression of dapk1 and DNA binding of C/EBP-β to the dapk1 promoter. (A) Effect of various inhibitors (10 μM) on the IFN-γ-induced expression of dapk1. Wild-type MEFs were treated with IFN-γ in the absence and presence of various inhibitors. The inhibitors were added to the cells 30 min prior to IFN treatment for 12 h. Real-time PCR was carried out. The inset images show typical induction profiles of dapk1 and rpl32 (control) transcripts as analyzed by RT-PCR. The Western blot (WB) at the bottom shows effects of inhibitors on DAPK1 protein expression. (B) Effects of ERK pathway inhibitors on the C/EBP-β-dependent induction of the dapk1 promoter. Luciferase assays were performed in cebpb−/− MEFs as described in the legend of Fig. 5. (C and D) Quantification of the IFN-induced binding of C/EBP-β to the dapk1 promoter. ChIP products obtained with the indicated antibodies were subjected to real-time analysis with promoter-region-specific primers. IFN-γ and U0126 treatments of wild-type MEFs were performed as described above. (E) ERK1/2 knockdown in normal MEFs. Cells were infected with a mixture of lentiviral vectors coding for shRNAs that can target murine ERK1/2. Lysates were prepared, and a Western blot analysis of ERK1/2 was performed using specific antibodies. EV, empty vector (pLKO1); SC, pLKO1 vector carrying scrambled shRNA sequences; sh-E1+E2, shRNAs that can target ERK1 and ERK2 expressed from the pLKO1 vector. (F) Effect of ERK1/2b knockdown on the expression of dapk1 mRNA in normal MEFs. After infecting cells with lentiviral vectors carrying the indicated shRNAs (as in panel E) for 3 days, cells were stimulated with IFN-γ for 12 h. RNA was prepared, and a real-time PCR analysis of dapk1 transcripts was performed. (G) Quantitative ChIP assay for the binding of C/EBP-β to the CRE/ATF of dapk1 in MEFs after the knockdown of erk1/erk2.
The importance of ERK1/2 in the IFN-γ-induced regulation of the dapk1 promoter was further assessed using the following corroborative experiments. In the first experiment, wild-type MEFs were infected with lentiviral vectors coding for control RNAs or shRNAs coding for erk1/2. The knockdown of endogenous ERK1/2 was ensured with a Western blot analysis of cellular lysates. Only the erk1/2 shRNAs, but not the scrambled control shRNA, knocked down greater than 90% of protein expression (Fig. 7E). These cells were then treated with IFN-γ for 16 h, and the expression of endogenous dapk1 mRNA was monitored. dapk1 mRNA was not significantly induced by IFN-γ in the presence of erk1/2-specific shRNAs (Fig. 7F). As expected, its induction was unaffected by empty vector and scrambled shRNA. Similarly, these erk1/2 shRNAs, but not the controls, knocked down the IFN-γ-induced expression of endogenous dapk1 expression in wild-type BM cells (data not shown). Thus, the ERK1/2 requirement for the IFN-induced expression of dapk1 was not a cell type-specific effect. Lastly, we examined the recruitment of C/EBP-β to the CRE/ATF site of the dapk1 promoter using quantitative ChIP assays. Normal IFN-γ-induced recruitment of C/EBP-β to the CRE/ATF site of dapk1 occurred in the control cells. However, it was blunted in cells expressing the erk1/2 shRNAs (Fig. 7G). The control ChIP reactions with normal rabbit antibody were negative for PCR products (data not shown).
Since these two kinases function redundantly, we used MEFs from erk1−/− mice for the next experiment. The use of these cells also permitted a rescue of ERK1 to confirm the specificity of the ERKs in regulating dapk1 (see below). We first ensured the IFN-dependent activation of ERK1/2 using phospho-specific antibodies (Fig. 8A). Since erk1−/− MEFs expressed ERK2, in contrast to the wild-type cells, a single ppERK band was seen in these extracts. We next measured the endogenous dapk1 mRNA in these cells following IFN treatment. Although only ERK2 was expressed in erk1−/− cells, it was able to sustain a level of IFN-induced expression of dapk1 comparable to that found in wild-type cells (Fig. 8B).
FIG. 8.
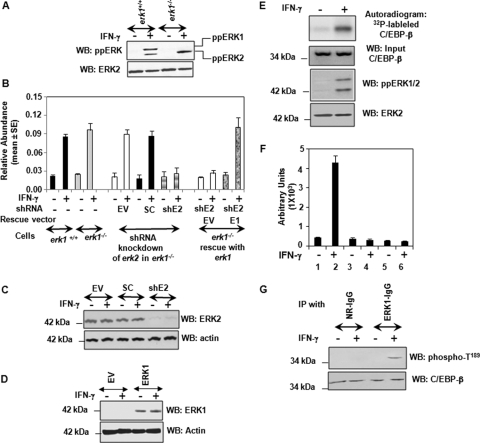
ERK-dependent regulation and phosphorylation of C/EBP-β in response to IFN-γ. (A) Activation of ERKs in wild-type and erk1−/− MEFs. Cells were stimulated with IFN-γ for 1 h, and lysates (30 μg) were subjected to Western blot (WB) analysis with phospho-ERK-specific (activated form) and native ERK2-specific antibodies. (B) ERK1 and ERK2 are critical for the IFN-γ-induced expression of dapk1 mRNA. Real-time PCR analysis of the dapk1 transcript is shown. The middle part of this graph shows the effect of ERK2 knockdown in erk1−/− MEFs on DAPK1 expression. The last four bars show the results of a rescue experiment. Following the knockdown of erk2 expression in erk1−/− MEFs with shRNAs, cells were transfected with an expression vector coding for murine ERK1. shE2, erk2-specific shRNA; SC, scrambled shRNA; EV, empty vector. (C) Effects of lentivirus-encoded erk2 shRNAs on ERK2 expression in erk1−/− MEFs. Western blot analysis of the cell lysates (45 μg) is shown. (D) Cells infected with lentiviral vector erk2-specific shRNAs (see panel C) were electroporated with empty vector (pCDNA3.1) or the same vector coding for ERK1. Western blot analysis of the lysates was performed with the indicated antibodies. (E) Phosphorylation of recombinant C/EBP-β protein by ERK1/2. Wild-type MEFs were stimulated with IFN-γ for 2 h, and ERK1 and ERK2 were immunoprecipitated using protein G/protein A agarose beads (Santa Cruz Biotech). The bead-bound IP products were washed twice with the reaction buffer prior to incubation with recombinant C/EBP-β (2 μg) in the presence of [32P]ATP. The beads were spun down, and supernatants were collected, separated by SDS-PAGE, and transferred onto a nylon membrane prior to autoradiography. A typical profile is shown. These reactions were also probed with ERK2-, ppERK1-, and C/EBP-β-specific antibodies. (F) Quantification of ERK1/2-induced C/EBP-β phosphorylation. Lysates were immunoprecipitated with ERK1/2 antibodies (lanes 1 and 2), normal rabbit antibody (lanes 3 and 4), and no antibody (lanes 5 and 6), and in vitro phosphorylation was performed as described below (G). The reaction products were separated by SDS-PAGE and transferred onto a nylon membrane prior to phosphorimaging. Radioactive signals (arbitrary units) in the C/EBP-β band were quantified and presented. Each bar shows the mean ± SE (n = 3). (G) In vitro phosphorylation of C/EBP-β was performed as described above (E), except that cold ATP was used for phosphorylation. Following this, the reaction products were subjected to a Western blot analysis with native and C/EBP-β-phospho-T189-specific antibodies.
Next, we knocked down the expression of ERK2 using lentiviral expression vectors and measured the IFN-γ-induced expression of dapk1 using real-time PCR (Fig. 8B). As anticipated, only erk2-specific shRNA, but not the control shRNAs, suppressed the IFN-induced expression of dapk1 mRNA. The steady-state expression of dapk1 mRNA was unaffected under these conditions. The suppression of ERK2 expression was confirmed by Western blot analysis. Only the erk2-specific shRNA but not the scrambled shRNA or the empty vector knocked down >95% of ERK2 expression (Fig. 8C). In a follow-up experiment, we verified the specificity of ERK signaling. Since erk2 shRNAs target the mRNA within the coding region, it was difficult to generate mutant versions of the ERK2 protein that would restore the ERK activity in these cells. Since these cells lacked erk1, we chose to restore ERK1 in cells already expressing erk2 shRNAs. Indeed, upon the restoration of ERK1, but not empty vector, the IFN-γ-induced expression of dapk1 was observed (Fig. 8B). The restoration of ERK1 in the cells was confirmed with a Western blot analysis of the cellular lysates with an ERK1-specific antibody (Fig. 8D).
We next determined if activated ERK1/2 could directly phosphorylate purified recombinant C/EBP-β protein. Wild-type MEFs were stimulated with IFN-γ for 2 h, and activated ERK1/2 was immunoprecipitated using ERK1/2-specific antibodies. The IP products were incubated with bacterially expressed, purified C/EBP-β protein in the presence of [γ-32P]ATP. The reaction products were separated by SDS-PAGE and then transferred onto a nylon membrane. The membranes were washed and exposed to X-ray films to detect the phosphorylated bands. Figure 8E shows a typical profile. ERK1/2 from IFN-γ-treated cells strongly induced the phosphorylation of C/EBP-β protein compared to the control. A basal level of ERK1/2 activity was seen in the uninduced lysates (probably because of serum in the medium). The activation of ERK1/2 in the IFN-stimulated cell lysates was confirmed by Western blot analysis using ppERK-specific antibodies. Total ERK levels and C/EBP input into the reactions were comparable between these samples. Figure 8F shows a quantitative view of the reactions. In these experiments, the incorporation of 32P was quantified after PhosphorImager analysis of the blots. Only the IP products of ERK1/2-specific IgG, but not those of control IgG, were able to induce the phosphorylation of the C/EBP-β protein. A ninefold increase in the ERK-induced phosphorylation of C/EBP-β occurred following IFN-γ treatment.
Since the above-described experiment did not indicate whether phosphorylation occurred at a specific residue on the C/EBP-β protein, we performed a Western blot analysis of the in vitro reaction products with specific antibodies that could detect phosphorylation at the T189 residue of the C/EBP-β protein. We have used these antibodies in the past to demonstrate phosphorylation at the T189 residue of the endogenous C/EBP-β protein. Consistent with our previous observations, phospho-T189 C/EBP-β protein was detected by these antibodies upon incubation of the in vitro reaction mixtures with ERK1/2 (Fig. 8G). No phosphorylated C/EBP-β protein was detected when recombinant C/EBP-β protein was incubated with the IP products of a control antibody. Thus, ERK1/2 directly phosphorylated C/EBP-β at the T189 residue.
To further prove an important role for the T189 residue of C/EBP-β in regulating dapk1 expression, we transfected cebpb−/− MEFs with expression vectors coding for wild-type C/EBP-β and its mutant lacking the ERK1/2-targeted sites (Fig. 9A), along with the mDAPK-1.2K-Luc reporter (Fig. 9B). A control mutant, Mut1, in which the adjacent serine residues were mutated to alanines, was also used in these studies. The ERK-activated site in Mut1 was kept intact. Mut2 lacked the critical TPSP motif of the C/EBP-β protein. In Mut T189A, the critical threonine was converted to an alanine. The use of cebpb−/− MEFs for this study permitted an unequivocal demonstration of the effect of the mutations on gene expression. In the absence of any C/EBP-β, the reporter gene was not induced by IFN-γ. Both the wild-type and Mut1 constructs supported IFN-γ-inducible expression of the reporter equivalently. The Mut2 and Mut T189A constructs did not support IFN-induced transcription from the promoter. All mutants equivalently supported the basal transcription of the dapk1 promoter. To further demonstrate the importance of phosphorylation at the T189 residue, we engineered a phosphomimetic mutation at this site by replacing the T189 with an aspartic acid (Mut T189D). This mutant robustly stimulated transcription without IFN-γ treatment. A very similar effect of the mutants on the endogenous dapk1 mRNA was noted (Fig. 9C). The differences in the expression levels of the reporter genes could not be attributed to different levels of expression of wild-type and mutant constructs. All mutants expressed equivalently (Fig. 9D). These blots were also probed with antibodies specific for the C/EBP-β-phospho-T189 form. As expected, these antibodies detected only the wild-type and Mut 1 proteins from the IFN-γ-stimulated lysates. Although it functioned normally, the T189D mutant was not detected by these antibodies because it lacked the pT189 residue. Lastly, we checked the ability of the mutant forms of C/EBP-β to be recruited to the CRE/ATF site of dapk1 using ChIP assays (Fig. 9E). Although C/EBP-β proteins coded by the wild-type and Mut1 constructs bound to the CRE/ATF element in an IFN-γ-dependent manner, the proteins encoded by Mut 2 and Mut T189A failed to do so. The T189D mutant was recruited to the CRE-ATF site independently of IFN-γ treatment. The basal bindings of these mutants to the distal CBS were comparable and were unaffected by IFN-γ treatment (data not shown). Thus, an ERK1-controlled pathway through the GTPS motif of regulatory domain 2 (RD2) of C/EBP-β is critical for the inducible binding and promotion of IFN-regulated transcription through the CRE/ATF site of the dapk1 promoter.
FIG. 9.
The T189 residue within RD2 of C/EBP-β is required for the IFN-γ-inducible expression of DAPK1. (A) Mutations introduced into C/EBP-β cDNA. TAD, transcription activation domain; Basic, domain rich in basic amino acids; LZ, leucine zipper domain. The mutated amino acids are indicated in each case. (B and C) Effect of cebpb mutations on the IFN-induced expression of dapk1. cebpb−/− MEFs were electroporated with the indicated mutants along with mdapk1.2K-Luc. A portion of the transfected cells was used for the luciferase assay, and the remainder was used for real-time PCR analysis of the dapk1 transcript. (D) Expression of mutant and wild-type (Wt) versions of C/EBP-β in the lysates of transfected cells. Western blot (WB) analysis with indicated antibodies was conducted using 100 μg of total protein. (E) Quantitative ChIP assay for the recruitment of C/EBP-β to the CRE/ATF site. IFN-γ treatment was done for 12 h.
Reexpression of dapk1 into cebpb−/− BM cells restores IFN-γ-induced apoptosis.
Since our studies showed a dependence of dapk1 on C/EBP-β for its expression, and cebpb−/− macrophages behaved like dapk1-deficient cells, we examined if the restoration of dapk1 expression into these cells would reestablish the IFN-γ-induced apoptotic program (Fig. 10A). Cells were electroporated with expression vectors coding for wild-type and kinase-deficient mutants along with the pEGFP vector. They were then treated with IFN-γ for 24 h and subjected to FACS analysis after staining with TRITC-labeled annexin V. Neither the empty expression vector nor the kinase-deficient mutant supported IFN-γ-induced apoptosis in these cells. However, wild-type DAPK1 not only promoted a higher basal level of apoptosis than the controls but also augmented it further in the presence of IFN-γ. The differential effects of mutant and wild-type DAPK1 on these cells could not be attributed to differences in the levels of their expression (Fig. 10B).
FIG. 10.

Restoration of dapk1 expression reestablishes IFN-γ-induced apoptosis in cebpb−/− BM cells. (A) Cells were electroporated with 4 μg of an empty pcDNA3.1 vector or the same vector expressing either the wild-type or the kinase-deficient (KD) mutant (K42A) of DAPK1 along with the pEGFP expression vector (2 μg) for tracking the transfected cells. The cells were then treated with IFN-γ (500 U/ml) for 24 h and stained with TRITC-annexin V (red fluorescence). Cells were subjected to FACS analysis. Green-positive (GFP-positive) and green/red-positive (GFP-positive/TRITC-positive) cells were estimated. The double-positive cells were estimated as a percentage of total GFP-positive cells to determine the magnitude of cell death. (B) Western blot (WB) analysis of the DAPK1 expression. A portion of transfected and IFN-treated cells was lysed, and the expression levels of DAPK1 and GFP were monitored with specific antibodies.
DISCUSSION
The transcription factor C/EBP-β is a major regulator of metabolic homeostasis (17). Apart from this, C/EBP-β also participates in disparate processes such as cell differentiation, growth, immune responses, neoplastic growth, development of the reproductive system, and pro- and antigrowth pathways (38, 50, 72, 73, 79, 82). For example, it is necessary for mediating ras-induced carcinogenesis in skin keratinocytes (96). Other studies showed that C/EBP-β autoregulates through IGF-1 expression for providing survival signals (91) in macrophage tumor cells. In the stellate cells of the liver, the phosphorylation of C/EBP-β at T217 by CCl4-induced ribosomal S6 kinase (RSK) creates an inhibitory site that prevents the activation of caspase-1 and caspase-8 to protect cells (13). C/EBP-β has also been identified as a survival factor in certain anaplastic lymphomas (57) and in relapsing Wilms’ tumors (42). In contrast, the loss of C/EBP-β results in the development of a lymphoproliferative disorder in mice and a loss of Th1 immune responses (72). The cebpb−/− macrophages fail to inhibit intracellular bacterial and fungal infections and lack the ability to kill heterologous tumors (82). It is unclear exactly how these apparently diverse effects are mediated by a single transcription factor. The regulatory spectrum of this protein has been further expanded with the availability of gene knockout mice and expression profiling technologies.
In this report, we have uncovered a new IFN-γ-induced growth regulatory pathway that is dependent on C/EBP-β. The loss of cebpb resulted in an inhibition of IFN-γ-induced cell death. The differential IFN sensitivity of the cebpb−/− cells could not be due to a loss of the primary response from the IFN-γ receptor, given the normal activation of STAT1 and the STAT1-dependent genes irf1 and irf8 in cebpb−/− cells (Fig. 3) and the expression and activation of a number of apoptosis-associated factors such as caspase-8 and caspase-9. Since the apoptotic response can be rescued following the transfection of C/EBP-β into cebpb−/− cells, it cannot be attributed to some secondary changes resulting from the loss of C/EBP-β. Thus, some C/EBP-β-regulated gene product(s) is critical for inducing IFN-γ-stimulated cell death.
Indeed, the expression of a number of IFN-γ-stimulated genes was affected by the loss of C/EBP-β. These C/EBP-β-dependent ISGs participate in a wide variety of biological activities. Although we have not exhaustively examined every gene, we have confirmed a select few to be IFN-induced C/EBP-β-regulated genes (Fig. 2 and 3). These include the signaling molecule RAC-2, the transcriptional regulator RNA-dependent helicase, and the cytoskeletal assembly protein septin-3. We have also shown that some genes are repressed by C/EBP-β in the presence of IFN-γ, e.g., cyclin D2. Thus, C/EBP-β may play a broader role in IFN-γ-signaling pathways, which were not appreciated previously. More importantly, the absence of previously described STAT-inducible ISGs in the group of C/EBP-β-regulated ISGs reported here shows the specificity of this regulatory pathway. We recognize that the pooling strategy employed for array analysis may result in some differentially expressed genes being undetected (e.g., if gene induction in one of the pooled samples is offset by repression at another), and we do not suggest that our analysis represents a comprehensive description of all C/EBP-β-dependent IFN-inducible genes. Because six samples were pooled, if a change in gene expression is observed at only one of the time points, the detection of that change is predicted to be reduced by sixfold based on the dilution of the target mRNA. This assumes that such genes are expressed at only one time point but not at others. However, most IFN-induced genes are induced across a window of 4 to 6 h (from our past experiences). Accordingly, during a 24-h period of treatment, there would be at least six overlapping points. When this factor is taken into consideration, it would lower the chances of dilution of transcripts by sixfold. This would still allow for a sufficient representation of most transcripts in the IFN-induced sample above the basal level. Our experimental values showed that about 0.33% of the total genes screened are C/EBP-β-dependent IFN-inducible genes. These numbers agree with most array studies with single-agent inducers, which show an induction of anywhere from 0.1 to 2% of the genes depending on the stimulus. Thus, we believe that if a dilution of transcripts occurred, because of pooling, it would have a moderate effect on the detection of some genes in this array.
Interestingly, the cell death-associated gene dapk1 and the previously reported irf9 gene were also present in this set of IFN-γ-induced C/EBP-β-dependent genes. DAPK1 is a calcium-calmodulin-dependent serine/threonine protein kinase, which suppresses tumor growth and metastasis via the induction of apoptosis (15, 19, 34). It was originally isolated as a regulator of IFN-γ-activated cell death using a genetic screen (19), whose down-regulation inhibited IFN-γ-induced cell death in several cell types. The catalytic activity of DAPK1 is stimulated following the dephosphorylation of a critical serine residue located at position 308 and by association with calmodulin (75, 76). Although DAPK1 was originally described as being an IFN-γ-dependent cell death regulator, the overexpression of catalytically inactive mutants of DAPK1 interfered with TNF-α- and Fas-induced cell death in some human cell lines (16). However, the BM cells used in our study were sensitive to Fas-induced apoptosis (Fig. 1F) despite the lack of DAPK1 expression. Thus, the requirement of DAPK1 for inducing apoptosis may be cell type dependent. One recent study reported that TNF induces cell death in MEFs lacking C/EBP-β (64). In our hands, cebpb+/+ and cebpb−/− BM cells and MEFs (data not shown) were resistant to TNF-induced cell death. It is likely that these cells harbor some unknown inhibitors of TNF-induced cell death. Lastly, death induction by TNF-α is extremely slow in cebpb−/− MEFs (64), which suggests the involvement of some secondary factors in that process. Additionally, there could be differences in the passage and immortalization status of MEFs used between these two studies. In our report, all studies were conducted with primary MEFs or BM cells in early passages. Since our primary aim is to understand IFN-induced cell death, we have not investigated the differences in TNF action further. Other death stimuli such as C6-ceramide (56), inhibition of mitochondrial respiration, unliganded UNC5H2 (44), transforming growth factor β (35), and DNA damage also require DAPK for inducing cell death (7). DAPKs modulate cytoskeletal remodeling, membrane blebbing, and autophagy (7, 33). Indeed, a loss of dapk1 expression was reported previously for a number of human cancers (7). Although methylation appears to be one major mechanism, in several cases, dapk1 expression is silenced without apparent methylation (23, 27, 30, 52, 67, 97). Mutational inactivation of dapk1 is almost rare in most cancers (7), except in some familial chronic lymphocytic leukemias, where its expression down-regulated by a unique polymorphism (65). dapk1−/− MEFs are immortalized at a higher rate than their wild-type counterparts, indicating a major role for it in cellular senescence (7). Lastly, the loss of dapk1 results in a reduced expression of p53 and p19ARF (66). Given the central role of DAPK1 in multiple growth regulatory processes, it is conceivable that its expression and activity are tightly regulated.
We have shown a cell-independent requirement of C/EBP-β for dapk1 expression. A direct role for C/EBP-β in IFN-induced dapk1 expression was supported by the following: (i) the restoration of its expression following the expression of C/EBP-β in cebpb−/− cells (Fig. 1 and 4), (ii) a loss of its induction in the presence of C/EBP-β-specific shRNAs (Fig. 3), and (iii) the binding of C/EBP-β to specific elements in the dapk1 promoter (Fig. 5 and 6). Remarkably, among the members of the dapk family, only the expression of dapk1 required C/EBP-β (see Fig. S1 in the supplemental material). On the other hand, none of the other members of C/EBP family, except C/EBP-β, appear to induce its expression even when overexpressed (see Fig. S2 in the supplemental material). Consistent with this, the shRNA-induced loss of either C/EBP-α or C/EBP-δ expression did not affect the IFN-induced expression of dapk1 in wild-type BM cells (Fig. 3F). Neither C/EBP-α nor C/EBP-δ up-regulated dapk1 expression in cebpb−/− BM cells and MEFs (data not shown). Similarly, dapk1 expression was unaffected in cebpd−/− cells, and the restoration of C/EBP-β alone was sufficient for inducing it in cebpb−/−/cebpd−/− double-knockout cells (Fig. 4). Thus, C/EBP-β exclusively regulates dapk1 in response to IFN-γ.
Mutational analysis revealed two motifs, a promoter-proximal CRE/ATF-binding site and a distal CBS, through which C/EBP-β may regulate dapk1. The inactivation of both these elements completely suppressed basal and IFN-induced gene expression from the dapk1 promoter. Surprisingly, mutation in the CRE/ATF but not the CBS significantly reduced both basal and IFN-induced expression of DAPK1. This mutant was still weakly induced by IFN-γ. This result indicates that in the absence of a functional CRE/ATF site, the distal CBS may substitute for it either on its own or in association with other elements in the promoter for promoting a weaker IFN-induced response. Thus, CRE/ATF can now be added to the list of IFN-responsive elements. Although CRE/ATF was able to respond to IFN-γ, it was not as efficient as the 649-bp promoter fragment in terms of its magnitude. The requirement of two sites of engagement with the promoter suggests that contacts between proteins binding at these locations collaborate in the formation of the enhanceosome during gene induction. ChIP assays demonstrated an IFN-γ-stimulated binding of C/EBP-β to the CRE/ATF site but not to the CBS. One explanation for this differential binding is the lack of homology between CRE/ATF and the CBS (Fig. 8C). It is also possible that in addition to C/EBP-β, other proteins bind at the CRE/ATF in an IFN-dependent manner. This issue is being investigated. Previous studies reported that CREs may act as C/EBP-binding sites in some instances (25, 55, 80, 94). Remarkably, the recruitment of C/EBP-β to the CRE/ATF site of the dapk1 promoter is kinetically slower. These observations suggest that other events such as the removal of an inhibitory factor and/or the induction or activation of the other positive-acting factors that cooperate with C/EBP-β may be necessary for IFN-induced dapk1 expression to occur. One recent study showed that an increased binding of HoxB7 to a variant version of human dapk1 found in certain familial chronic lymphocytic leukemias suppressed its basal expression (65). However, this mutant site (exon 16) is outside the dapk1 promoter reported here. Transforming growth factor β regulates the expression of dapk1 through SMADs (35). The mouse and human dapk1 promoters have SMAD binding sites. It is unclear whether the SMAD binding sites have a role in regulating dapk1 transcription in response to IFN-γ. We are currently investigating the roles of these and other transcription factor binding sites in regulating dapk1.
Lastly, we have examined a mechanism by which IFN-γ-induced C/EBP-β stimulates the expression of the dapk1 promoter. The inhibition of ERK1/2 using shRNAs and chemical inhibitors suppressed IFN-γ-driven dapk1 expression because they interfered with the IFN-γ-induced binding of C/EBP-β to CRE/ATF (Fig. 7 and 8). The transfection of ERK1 back into cells lacking ERK1/2 rescued the IFN-induced expression of dapk1, indicating the specificity of ERKs in regulating dapk1. Activated ERK1/2 proteins directly phosphorylated recombinant C/EBP-β protein at the T189 site. Consistent with these observations, the T189A mutant failed to drive IFN-induced transcription from the dapk1 promoter. In contrast, the T189D mutant, bearing a phosphomimetic amino acid, activated dapk1 transcription independently of IFN treatment. The T189D mutant bound to the dapk1 promoter without requiring IFN treatment. Thus, IFN-γ-induced ERK-dependent phosphorylation of C/EBP-β may permit its association with CRE/ATF. Indeed, other studies have shown an important role for the phosphorylation of this site in regulating C/EBP-β-mediated transcriptional regulation in the context of other genes in different cell types (54, 58, 83). ERKs readily phosphorylated a synthetic peptide containing the ERK phosphorylation site of human C/EBP-β in vitro (51). It appears that a conformational change in the C/EBP-β protein caused by cytokine-induced phosphorylation at T189 may allow its recruitment to some promoters and its association with other proteins (47). A recent report showed that the phosphorylation of the DAPK1 protein by RSK, a downstream enzyme of the ERK pathway, leads to an inhibition of its apoptotic activity (4). This phosphorylation occurs independently of the PI3-kinase-AKT/PKB pathway. The S289 of the DAPK1 protein was phosphorylated by RSK. Thus, the ERK signaling pathways may control the DAPK1 at two levels: (i) at the level of transcription through the activation of C/EBP-β in the current study and (ii) at a posttranslational level by acting as an attenuator of apoptosis (4). Conversely, DAPK1 in turn inactivates ERK activity (14). Thus, DAPK1 may act as an off switch to terminate the ERK-C/EBP-β pathway.
The biological connection between DAPK1, C/EBP-β, and IFNs can be reconciled in the light of several recent and past observations. Autophagy is a major intracellular catabolic process that conserves energy resources under conditions of stress by digesting damaged cellular organelles (45, 48). It is also crucial for limiting the spread of intracellular pathogens (40). Although viewed as separate processes until recently, apoptosis and autophagy have significant overlaps (45, 88). In addition, autophagy appears to play a central role in antigen presentation, survival and death of T cells, and the development of adaptive immunity (71). Macrophages play central role in many of these processes.
Among the several mechanisms of innate immunity is the targeting of intracellular pathogens to lysosome-mediated destruction (20). Some bacteria escape the phagosome and enter the cytosol to multiply or in some cases modify the phagosome to prevent fusion with the lysosome, allowing them to replicate within the phagosomal compartment (53). However, in response to these escapees, the immune system activates autophagy, which targets intracellular bacteria for sequestration within autophagosomes, ultimately delivering them to the lysosomes, where they are destroyed. IFN-γ induces autophagy (22, 61), which inhibits the growth of intracellular bacterial pathogens and some viruses (29, 43, 62, 68). Interestingly, one of the major defects in cebpb−/− mice is the inability of macrophages to kill intracellular bacterial and fungal pathogens (72, 82, 95) and heterologous tumors due to defective phagocytic activity. As mentioned above, DAPK1 participates in both apoptosis and autophagic vesicle formation (7, 33). These events may suppress tumor and bacterial proliferation in vivo. In the event of excessive bacterial loads, the default mechanism would be suicide of infected macrophages. Since DAPK1 controls both autophagy and apoptosis, we hypothesize that the IFN-γ-regulated C/EBP-β-dependent expression of dapk1 may control host defenses against bacterial infection and tumor growth in vivo. Another long-term effect of the loss of C/EBP-β in mice is the development of a lymphoproliferative disorder (72), which may also be relevant to DAPK1, given the loss of its expression in several human lymphoid and myeloid leukemias (7). Thus, the IFN-γ-ERK-C/EBP-β-DAPK axis may play a major role in regulating pathogens and tumor growth.
Supplementary Material
Acknowledgments
These studies are supported by NIH grant CA78282 to D.V.K.
We thank Peter Johnson for providing cebpb−/− mice, Esta Sterneck for cebpd−/− MEFs, Sarah Gaffen for cebpb−/−/cebpd−/− double-knockout MEFs, Howard Young for J2 retrovirus-producing cell lines, and Adi Kimchi for dapk1 expression vectors.
Footnotes
Published ahead of print on 4 February 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akira, S., and T. Kishimoto. 1997. NF-IL6 and NF-kappa B in cytokine gene regulation. Adv. Immunol. 651-46. [PubMed] [Google Scholar]
- 2.Alexander, W. S., and D. J. Hilton. 2004. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu. Rev. Immunol. 22503-529. [DOI] [PubMed] [Google Scholar]
- 3.Angell, J. E., D. J. Lindner, P. S. Shapiro, E. R. Hofmann, and D. V. Kalvakolanu. 2000. Identification of GRIM-19, a novel cell death-regulatory gene induced by the interferon-beta and retinoic acid combination, using a genetic approach. J. Biol. Chem. 27533416-33426. [DOI] [PubMed] [Google Scholar]
- 4.Anjum, R., P. P. Roux, B. A. Ballif, S. P. Gygi, and J. Blenis. 2005. The tumor suppressor DAP kinase is a target of RSK-mediated survival signaling. Curr. Biol. 151762-1767. [DOI] [PubMed] [Google Scholar]
- 5.Baer, M., and P. F. Johnson. 2000. Generation of truncated C/EBPbeta isoforms by in vitro proteolysis. J. Biol. Chem. 27526582-26590. [DOI] [PubMed] [Google Scholar]
- 6.Belardelli, F., and M. Ferrantini. 2002. Cytokines as a link between innate and adaptive antitumor immunity. Trends Immunol. 23201-208. [DOI] [PubMed] [Google Scholar]
- 7.Bialik, S., and A. Kimchi. 2006. The death-associated protein kinases: structure, function, and beyond. Annu. Rev. Biochem. 75189-210. [DOI] [PubMed] [Google Scholar]
- 8.Blasi, E., D. Radzioch, L. Merletti, and L. Varesio. 1989. Generation of macrophage cell line from fresh bone marrow cells with a myc/raf recombinant retrovirus. Cancer Biochem. Biophys. 10303-317. [PubMed] [Google Scholar]
- 9.Bluyssen, H. A., R. Muzaffar, R. J. Vlieststra, A. C. van der Made, S. Leung, G. R. Stark, I. M. Kerr, J. Trapman, and D. E. Levy. 1995. Combinatorial association and abundance of components of interferon-stimulated gene factor 3 dictate the selectivity of interferon responses. Proc. Natl. Acad. Sci. USA 925645-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boehm, U., T. Klamp, M. Groot, and J. C. Howard. 1997. Cellular responses to interferon-γ. Annu. Rev. Immunol. 15749-795. [DOI] [PubMed] [Google Scholar]
- 11.Botwell, D., and J. Sambrook (ed.). 2003. DNA microarrays, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Brysha, M., J. G. Zhang, P. Bertolino, J. E. Corbin, W. S. Alexander, N. A. Nicola, D. J. Hilton, and R. Starr. 2001. Suppressor of cytokine signaling-1 attenuates the duration of interferon gamma signal transduction in vitro and in vivo. J. Biol. Chem. 27622086-22089. [DOI] [PubMed] [Google Scholar]
- 13.Buck, M., V. Poli, T. Hunter, and M. Chojkier. 2001. C/EBPbeta phosphorylation by RSK creates a functional XEXD caspase inhibitory box critical for cell survival. Mol. Cell 8807-816. [DOI] [PubMed] [Google Scholar]
- 14.Chen, C. H., W. J. Wang, J. C. Kuo, H. C. Tsai, J. R. Lin, Z. F. Chang, and R. H. Chen. 2005. Bidirectional signals transduced by DAPK-ERK interaction promote the apoptotic effect of DAPK. EMBO J. 24294-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen, O., E. Feinstein, and A. Kimchi. 1997. DAP-kinase is a Ca2+/calmodulin-dependent, cytoskeletal-associated protein kinase, with cell death-inducing functions that depend on its catalytic activity. EMBO J. 16998-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen, O., B. Inbal, J. L. Kissil, T. Raveh, H. Berissi, T. Spivak-Kroizaman, E. Feinstein, and A. Kimchi. 1999. DAP-kinase participates in TNF-alpha- and Fas-induced apoptosis and its function requires the death domain. J. Cell Biol. 146141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Croniger, C., M. Trus, K. Lysek-Stupp, H. Cohen, Y. Liu, G. J. Darlington, V. Poli, R. W. Hanson, and L. Reshef. 1997. Role of the isoforms of CCAAT/enhancer-binding protein in the initiation of phosphoenolpyruvate carboxykinase (GTP) gene transcription at birth. J. Biol. Chem. 27226306-26312. [DOI] [PubMed] [Google Scholar]
- 18.Darlington, G. J., S. E. Ross, and O. A. MacDougald. 1998. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 27330057-30060. [DOI] [PubMed] [Google Scholar]
- 19.Deiss, L. P., E. Feinstein, H. Berissi, O. Cohen, and A. Kimchi. 1995. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 915-30. [DOI] [PubMed] [Google Scholar]
- 20.Dorn, B. R., W. A. Dunn, Jr., and A. Progulske-Fox. 2002. Bacterial interactions with the autophagic pathway. Cell. Microbiol. 41-10. [DOI] [PubMed] [Google Scholar]
- 21.Dunn, G. P., C. M. Koebel, and R. D. Schreiber. 2006. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 6836-848. [DOI] [PubMed] [Google Scholar]
- 22.Espert, L., P. Codogno, and M. Biard-Piechaczyk. 2007. Involvement of autophagy in viral infections: antiviral function and subversion by viruses. J. Mol. Med. 85811-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esteller, M., P. G. Corn, S. B. Baylin, and J. G. Herman. 2001. A gene hypermethylation profile of human cancer. Cancer Res. 613225-3229. [PubMed] [Google Scholar]
- 24.Favata, M. F., K. Y. Horiuchi, E. J. Manos, A. J. Daulerio, D. A. Stradley, W. S. Feeser, D. E. Van Dyk, W. J. Pitts, R. A. Earl, F. Hobbs, R. A. Copeland, R. L. Magolda, P. A. Scherle, and J. M. Trzaskos. 1998. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 27318623-18632. [DOI] [PubMed] [Google Scholar]
- 25.Fawcett, T. W., J. L. Martindale, K. Z. Guyton, T. Hai, and N. J. Holbrook. 1999. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem. J. 339135-141. [PMC free article] [PubMed] [Google Scholar]
- 26.Gil, M. P., E. Bohn, A. K. O'Guin, C. V. Ramana, B. Levine, G. R. Stark, H. W. Virgin, and R. D. Schreiber. 2001. Biologic consequences of Stat1-independent IFN signaling. Proc. Natl. Acad. Sci. USA 986680-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez-Gomez, P., M. J. Bello, D. Arjona, J. Lomas, M. E. Alonso, J. M. De Campos, J. Vaquero, A. Isla, M. Gutierrez, and J. A. Rey. 2003. Promoter hypermethylation of multiple genes in astrocytic gliomas. Int. J. Oncol. 22601-608. [PubMed] [Google Scholar]
- 28.Gresser, I., and F. Belardelli. 2002. Endogenous type I interferons as a defense against tumors. Cytok. Growth Factor Rev. 13111-118. [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez, M. G., S. S. Master, S. B. Singh, G. A. Taylor, M. I. Colombo, and V. Deretic. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119753-766. [DOI] [PubMed] [Google Scholar]
- 30.Harden, S. V., Y. Tokumaru, W. H. Westra, S. Goodman, S. A. Ahrendt, S. C. Yang, and D. Sidransky. 2003. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin. Cancer Res. 91370-1375. [PubMed] [Google Scholar]
- 31.Hertzog, P. J., L. A. O'Neill, and J. A. Hamilton. 2003. The interferon in TLR signaling: more than just antiviral. Trends Immunol. 24534-539. [DOI] [PubMed] [Google Scholar]
- 32.Hu, J., S. K. Roy, P. S. Shapiro, S. R. Rodig, S. P. Reddy, L. C. Platanias, R. D. Schreiber, and D. V. Kalvakolanu. 2001. ERK1 and ERK2 activate CCAAAT/enhancer-binding protein-beta-dependent gene transcription in response to interferon-gamma. J. Biol. Chem. 276287-297. [DOI] [PubMed] [Google Scholar]
- 33.Inbal, B., S. Bialik, I. Sabanay, G. Shani, and A. Kimchi. 2002. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J. Cell Biol. 157455-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inbal, B., O. Cohen, S. Polak-Charcon, J. Kopolovic, E. Vadai, L. Eisenbach, and A. Kimchi. 1997. DAP kinase links the control of apoptosis to metastasis. Nature 390180-184. [DOI] [PubMed] [Google Scholar]
- 35.Jang, C. W., C. H. Chen, C. C. Chen, J. Y. Chen, Y. H. Su, and R. H. Chen. 2002. TGF-beta induces apoptosis through Smad-mediated expression of DAP-kinase. Nat. Cell Biol. 451-58. [DOI] [PubMed] [Google Scholar]
- 36.Kalakonda, S., S. C. Nallar, D. J. Lindner, J. Hu, S. P. Reddy, and D. V. Kalvakolanu. 2007. Tumor-suppressive activity of the cell death activator GRIM-19 on a constitutively active signal transducer and activator of transcription 3. Cancer Res. 676212-6220. [DOI] [PubMed] [Google Scholar]
- 37.Kalvakolanu, D. V. 2003. Alternate interferon signaling pathways. Pharmacol. Ther. 1001-29. [DOI] [PubMed] [Google Scholar]
- 38.Katz, S., E. Kowenz-Leutz, C. Muller, K. Meese, S. A. Ness, and A. Leutz. 1993. The NF-M transcription factor is related to C/EBP beta and plays a role in signal transduction, differentiation and leukemogenesis of avian myelomonocytic cells. EMBO J. 121321-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lekstrom-Himes, J., and K. G. Xanthopoulos. 1998. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 27328545-28548. [DOI] [PubMed] [Google Scholar]
- 40.Levine, B., and V. Deretic. 2007. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 7767-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levy, D. E., and J. E. Darnell. 2002. STATs: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3651-662. [DOI] [PubMed] [Google Scholar]
- 42.Li, W., P. Kessler, H. Yeger, J. Alami, A. E. Reeve, R. Heathcott, J. Skeen, and B. R. Williams. 2005. A gene expression signature for relapse of primary Wilms tumors. Cancer Res. 652592-2601. [DOI] [PubMed] [Google Scholar]
- 43.Liang, X. H., L. K. Kleeman, H. H. Jiang, G. Gordon, J. E. Goldman, G. Berry, B. Herman, and B. Levine. 1998. Protection against fatal Sindbis virus encephalitis by Beclin, a novel Bcl-2-interacting protein. J. Virol. 728586-8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Llambi, F., F. C. Lourenco, D. Gozuacik, C. Guix, L. Pays, G. Del Rio, A. Kimchi, and P. Mehlen. 2005. The dependence receptor UNC5H2 mediates apoptosis through DAP-kinase. EMBO J. 241192-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maiuri, M. C., E. Zalckvar, A. Kimchi, and G. Kroemer. 2007. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 8741-752. [DOI] [PubMed] [Google Scholar]
- 46.McBride, K. M., C. McDonald, and N. C. Reich. 2000. Nuclear export signal located within the DNA-binding domain of the STAT1 transcription factor. EMBO J. 196196-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng, Q., A. Raha, S. Roy, J. Hu, and D. V. Kalvakolanu. 2005. IFN-gamma-stimulated transcriptional activation by IFN-gamma-activated transcriptional element-binding factor 1 occurs via an inducible interaction with CAAAT/enhancer-binding protein-beta. J. Immunol. 1746203-6211. [DOI] [PubMed] [Google Scholar]
- 48.Mizushima, N. 2007. Autophagy: process and function. Genes Dev. 212861-2873. [DOI] [PubMed] [Google Scholar]
- 49.Moffat, J., D. A. Grueneberg, X. Yang, S. Y. Kim, A. M. Kloepfer, G. Hinkle, B. Piqani, T. M. Eisenhaure, B. Luo, J. K. Grenier, A. E. Carpenter, S. Y. Foo, S. A. Stewart, B. R. Stockwell, N. Hacohen, W. C. Hahn, E. S. Lander, D. M. Sabatini, and D. E. Root. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 1241283-1298. [DOI] [PubMed] [Google Scholar]
- 50.Muller, C., E. Kowenz-Leutz, S. Grieser-Ade, T. Graf, and A. Leutz. 1995. NF-M (chicken C/EBP beta) induces eosinophilic differentiation and apoptosis in a hematopoietic progenitor cell line. EMBO J. 146127-6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakajima, T., S. Kinoshita, T. Sasagawa, K. Sasaki, M. Naruto, T. Kishimoto, and S. Akira. 1993. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc. Natl. Acad. Sci. USA 902207-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narayan, G., H. Arias-Pulido, S. Koul, H. Vargas, F. F. Zhang, J. Villella, A. Schneider, M. B. Terry, M. Mansukhani, and V. V. Murty. 2003. Frequent promoter methylation of CDH1, DAPK, RARB, and HIC1 genes in carcinoma of cervix uteri: its relationship to clinical outcome. Mol. Cancer 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogawa, M., T. Yoshimori, T. Suzuki, H. Sagara, N. Mizushima, and C. Sasakawa. 2005. Escape of intracellular Shigella from autophagy. Science 307727-731. [DOI] [PubMed] [Google Scholar]
- 54.Park, B. H., L. Qiang, and S. R. Farmer. 2004. Phosphorylation of C/EBPbeta at a consensus extracellular signal-regulated kinase/glycogen synthase kinase 3 site is required for the induction of adiponectin gene expression during the differentiation of mouse fibroblasts into adipocytes. Mol. Cell. Biol. 248671-8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park, E. A., A. L. Gurney, S. E. Nizielski, P. Hakimi, Z. Cao, A. Moorman, and R. W. Hanson. 1993. Relative roles of CCAAT/enhancer-binding protein beta and cAMP regulatory element-binding protein in controlling transcription of the gene for phosphoenolpyruvate carboxykinase (GTP). J. Biol. Chem. 268613-619. [PubMed] [Google Scholar]
- 56.Pelled, D., T. Raveh, C. Riebeling, M. Fridkin, H. Berissi, A. H. Futerman, and A. Kimchi. 2002. Death-associated protein (DAP) kinase plays a central role in ceramide-induced apoptosis in cultured hippocampal neurons. J. Biol. Chem. 2771957-1961. [DOI] [PubMed] [Google Scholar]
- 57.Piva, R., E. Pellegrino, M. Mattioli, L. Agnelli, L. Lombardi, F. Boccalatte, G. Costa, B. A. Ruggeri, M. Cheng, R. Chiarle, G. Palestro, A. Neri, and G. Inghirami. 2006. Functional validation of the anaplastic lymphoma kinase signature identifies CEBPB and BCL2A1 as critical target genes. J. Clin. Investig. 1163171-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piwien-Pilipuk, G., O. MacDougald, and J. Schwartz. 2002. Dual regulation of phosphorylation and dephosphorylation of C/EBPbeta modulate its transcriptional activation and DNA binding in response to growth hormone. J. Biol. Chem. 27744557-44565. [DOI] [PubMed] [Google Scholar]
- 59.Poli, V. 1998. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J. Biol. Chem. 27329279-29282. [DOI] [PubMed] [Google Scholar]
- 60.Prada-Delgado, A., E. Carrasco-Marin, G. M. Bokoch, and C. Alvarez-Dominguez. 2001. Interferon-gamma listericidal action is mediated by novel Rab5a functions at the phagosomal environment. J. Biol. Chem. 27619059-19065. [DOI] [PubMed] [Google Scholar]
- 61.Pyo, J. O., M. H. Jang, Y. K. Kwon, H. J. Lee, J. I. Jun, H. N. Woo, D. H. Cho, B. Choi, H. Lee, J. H. Kim, N. Mizushima, Y. Oshumi, and Y. K. Jung. 2005. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J. Biol. Chem. 28020722-20729. [DOI] [PubMed] [Google Scholar]
- 62.Radtke, A. L., and M. X. O'Riordan. 2006. Intracellular innate resistance to bacterial pathogens. Cell. Microbiol. 81720-1729. [DOI] [PubMed] [Google Scholar]
- 63.Ramana, C. V., M. P. Gil, Y. Han, R. M. Ransohoff, R. D. Schreiber, and G. R. Stark. 2001. Stat1-independent regulation of gene expression in response to IFN-gamma. Proc. Natl. Acad. Sci. USA 986674-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ranjan, P., and J. M. Boss. 2006. C/EBPbeta regulates TNF induced MnSOD expression and protection against apoptosis. Apoptosis 111837-1849. [DOI] [PubMed] [Google Scholar]
- 65.Raval, A., S. M. Tanner, J. C. Byrd, E. B. Angerman, J. D. Perko, S. S. Chen, B. Hackanson, M. R. Grever, D. M. Lucas, J. J. Matkovic, T. S. Lin, T. J. Kipps, F. Murray, D. Weisenburger, W. Sanger, J. Lynch, P. Watson, M. Jansen, Y. Yoshinaga, R. Rosenquist, P. J. de Jong, P. Coggill, S. Beck, H. Lynch, A. de la Chapelle, and C. Plass. 2007. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell 129879-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raveh, T., G. Droguett, M. S. Horwitz, R. A. DePinho, and A. Kimchi. 2001. DAP kinase activates a p19ARF/p53-mediated apoptotic checkpoint to suppress oncogenic transformation. Nat. Cell Biol. 31-7. [DOI] [PubMed] [Google Scholar]
- 67.Reddy, A. N., W. W. Jiang, M. Kim, N. Benoit, R. Taylor, J. Clinger, D. Sidransky, and J. A. Califano. 2003. Death-associated protein kinase promoter hypermethylation in normal human lymphocytes. Cancer Res. 637694-7698. [PubMed] [Google Scholar]
- 68.Rich, K. A., C. Burkett, and P. Webster. 2003. Cytoplasmic bacteria can be targets for autophagy. Cell. Microbiol. 5455-468. [DOI] [PubMed] [Google Scholar]
- 69.Roy, S. K., J. Hu, Q. Meng, Y. Xia, P. S. Shapiro, S. P. Reddy, L. C. Platanias, D. J. Lindner, P. F. Johnson, C. Pritchard, G. Pages, J. Pouyssegur, and D. V. Kalvakolanu. 2002. MEKK1 plays a critical role in activating the transcription factor C/EBP-beta-dependent gene expression in response to IFN-gamma. Proc. Natl. Acad. Sci. USA 997945-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roy, S. K., S. J. Wachira, X. Weihua, J. Hu, and D. V. Kalvakolanu. 2000. CCAAT/enhancer-binding protein-beta regulates interferon-induced transcription through a novel element. J. Biol. Chem. 27512626-12632. [DOI] [PubMed] [Google Scholar]
- 71.Schmid, D., and C. Munz. 2007. Innate and adaptive immunity through autophagy. Immunity 2711-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Screpanti, I., L. Romani, P. Musiani, A. Modesti, E. Fattori, D. Lazzaro, C. Sellitto, S. Scarpa, D. Bellavia, G. Lattanzio, F. Bistoni, L. Frati, R. Cortese, A. Gulino, A. Ciliberto, F. Constantini, and V. Poli. 1995. Lymphoproliferative disorder and imbalanced T-helper response in C/EBP beta-deficient mice. EMBO J. 141932-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seagroves, T. N., S. Krnacik, B. Raught, J. Gay, B. Burgess-Beusse, G. J. Darlington, and J. M. Rosen. 1998. C/EBPbeta, but not C/EBPalpha, is essential for ductal morphogenesis, lobuloalveolar proliferation, and functional differentiation in the mouse mammary gland. Genes Dev. 121917-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55255-281. [DOI] [PubMed] [Google Scholar]
- 75.Shani, G., S. Henis-Korenblit, G. Jona, O. Gileadi, M. Eisenstein, T. Ziv, A. Admon, and A. Kimchi. 2001. Autophosphorylation restrains the apoptotic activity of DRP-1 kinase by controlling dimerization and calmodulin binding. EMBO J. 201099-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shohat, G., T. Spivak-Kroizman, O. Cohen, S. Bialik, G. Shani, H. Berrisi, M. Eisenstein, and A. Kimchi. 2001. The pro-apoptotic function of death-associated protein kinase is controlled by a unique inhibitory autophosphorylation-based mechanism. J. Biol. Chem. 27647460-47467. [DOI] [PubMed] [Google Scholar]
- 77.Shuai, K., G. R. Stark, I. M. Kerr, and J. E. Darnell, Jr. 1993. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science 2611744-1746. [DOI] [PubMed] [Google Scholar]
- 78.Stark, G. R., I. M. Kerr, B. R. G. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67227-264. [DOI] [PubMed] [Google Scholar]
- 79.Sterneck, E., L. Tessarollo, and P. F. Johnson. 1997. An essential role for C/EBPbeta in female reproduction. Genes Dev. 112153-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tae, H. J., S. Zhang, and K. H. Kim. 1995. cAMP activation of CAAT enhancer-binding protein-beta gene expression and promoter I of acetyl-CoA carboxylase. J. Biol. Chem. 27021487-21494. [DOI] [PubMed] [Google Scholar]
- 81.Tamura, T., and K. Ozato. 2002. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J. Interf. Cytok. Res. 22145-152. [DOI] [PubMed] [Google Scholar]
- 82.Tanaka, T., S. Akira, K. Yoshida, M. Umemoto, Y. Yoneda, N. Shirafuji, H. Fujiwara, S. Suematsu, N. Yoshida, and T. Kishimoto. 1995. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell 80353-361. [DOI] [PubMed] [Google Scholar]
- 83.Tang, Q. Q., M. Gronborg, H. Huang, J. W. Kim, T. C. Otto, A. Pandey, and M. D. Lane. 2005. Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc. Natl. Acad. Sci. USA 1029766-9771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. Irf family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19623-655. [DOI] [PubMed] [Google Scholar]
- 85.Teixeira, M. G., K. J. Austin, D. J. Perry, V. D. Dooley, G. A. Johnson, B. R. Francis, and T. R. Hansen. 1997. Bovine granulocyte chemotactic protein-2 is secreted by the endometrium in response to interferon-tau (IFN-tau). Endocrine 631-37. [PubMed] [Google Scholar]
- 86.ten Hoeve, J., M. de Jesus Ibarra-Sanchez, Y. Fu, W. Zhu, M. Tremblay, M. David, and K. Shuai. 2002. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol. Cell. Biol. 225662-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thomas, N. S., A. R. Pizzey, S. Tiwari, C. D. Williams, and J. Yang. 1998. p130, p107, and pRb are differentially regulated in proliferating cells and during cell cycle arrest by alpha-interferon. J. Biol. Chem. 27323659-23667. [DOI] [PubMed] [Google Scholar]
- 88.Thorburn, A. 2007. Apoptosis and autophagy: regulatory connections between two supposedly different processes. Apoptosis 131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vihko, P., P. Harkonen, P. Soronen, S. Torn, A. Herrala, R. Kurkela, A. Pulkka, O. Oduwole, and V. Isomaa. 2004. 17 beta-hydroxysteroid dehydrogenases—their role in pathophysiology. Mol. Cell. Endocrinol. 21583-88. [DOI] [PubMed] [Google Scholar]
- 90.Weihua, X., V. Kolla, and D. V. Kalvakolanu. 1997. Interferon-g induced transcription of the murine ISGF3g (p48) gene is mediated by novel factors. Proc. Natl. Acad. Sci. USA 94103-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wessells, J., S. Yakar, and P. F. Johnson. 2004. Critical prosurvival roles for C/EBPβ and insulin-like growth factor I in macrophage tumor cells. Mol. Cell. Biol. 243238-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Williams, S. C., M. Baer, A. J. Dillner, and P. F. Johnson. 1995. CRP2 (C/EBP beta) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. EMBO J. 143170-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xiao, W., L. Wang, X. Yang, T. Chen, D. Hodge, P. F. Johnson, and W. Farrar. 2001. CCAAT/enhancer-binding protein beta mediates interferon-gamma-induced p48 (ISGF3-gamma) gene transcription in human monocytic cells. J. Biol. Chem. 27623275-23281. [DOI] [PubMed] [Google Scholar]
- 94.Yamada, K., D. T. Duong, D. K. Scott, J. C. Wang, and D. K. Granner. 1999. CCAAT/enhancer-binding protein beta is an accessory factor for the glucocorticoid response from the cAMP response element in the rat phosphoenolpyruvate carboxykinase gene promoter. J. Biol. Chem. 2745880-5887. [DOI] [PubMed] [Google Scholar]
- 95.Yogi, Y., M. Endoh, T. Tanaka, S. Akira, H. Okamura, and H. Nomaguchi. 1999. Bacteria killing by macrophages via NF-IL6 gene dependent mechanism: the susceptibility to Mycobacterium leprae in NF-IL6 knockout mice. Nihon Hansenbyo Gakkai Zasshi 6897-108. [DOI] [PubMed] [Google Scholar]
- 96.Zhu, S., K. Yoon, E. Sterneck, P. F. Johnson, and R. C. Smart. 2002. CCAAT/enhancer binding protein-beta is a mediator of keratinocyte survival and skin tumorigenesis involving oncogenic Ras signaling. Proc. Natl. Acad. Sci. USA 99207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zochbauer-Muller, S., K. M. Fong, A. K. Virmani, J. Geradts, A. F. Gazdar, and J. D. Minna. 2001. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 61249-255. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.