Abstract
The histone H3 lysine 79 methyltransferase DOT1L/KMT4 can promote an oncogenic pattern of gene expression through binding with several MLL fusion partners found in acute leukemia. However, the normal function of DOT1L in mammalian gene regulation is poorly understood. Here we report that DOT1L recruitment is ubiquitously coupled with active transcription in diverse mammalian cell types. DOT1L preferentially occupies the proximal transcribed region of active genes, correlating with enrichment of H3K79 di- and trimethylation. Furthermore, Dot1l mutant fibroblasts lacked H3K79 di- and trimethylation at all sites examined, indicating that DOT1L is the sole enzyme responsible for these marks. Importantly, we identified chromatin immunoprecipitation (ChIP) assay conditions necessary for reliable H3K79 methylation detection. ChIP-chip tiling arrays revealed that levels of all degrees of genic H3K79 methylation correlate with mRNA abundance and dynamically respond to changes in gene activity. Conversion of H3K79 monomethylation into di- and trimethylation correlated with the transition from low- to high-level gene transcription. We also observed enrichment of H3K79 monomethylation at intergenic regions occupied by DNA-binding transcriptional activators. Our findings highlight several similarities between the patterning of H3K4 methylation and that of H3K79 methylation in mammalian chromatin, suggesting a widespread mechanism for parallel or sequential recruitment of DOT1L and MLL to genes in their normal “on” state.
Histone lysine methylation encodes genomic functions into the chemical state of nucleosomes (38). The collective actions of lysine methyltransferase and lysine demethylase enzymes maintain a landscape of steady-state methylation of histones around which eukaryotic DNA is packaged. Histone methylation can facilitate or abrogate a variety of protein-protein interactions occurring along the chromatin fiber, thus permitting stable regulation over localized regions of the genome. Several recent high-throughput descriptions of histone lysine methylation across mammalian genomes have documented the pervasiveness of this form of epigenetic organization (2, 15, 23). However, the full biological significance of most histone lysine methylation pathways in mammals has yet to be revealed.
Methylation of histone H3 at lysine 79 (H3K79) is conserved among most eukaryotic species. In budding yeast, nearly 90% of histone H3 bears monomethylation (H3K79me1), dimethylation (H3K79me2), or trimethylation (H3K79me3) at lysine 79, all catalyzed exclusively by the histone methyltransferase Dot1 (27, 46). H3K79 methylation is widely distributed across the euchromatic yeast genome but markedly depleted at heterochromatic mating-type, ribosomal DNA, and telomeric loci (26, 30). Genes in these regions are controlled by silent information regulator (SIR) proteins, which can bind nucleosomes and silence transcription (reviewed in reference 33). Genetic, as well as biochemical, evidence suggests a mutual antagonism between H3K79 methylation by Dot1 and the association of SIR proteins with chromatin (1, 10, 26, 46). Therefore, the action of Dot1 in yeast serves to impose a boundary that confines SIR proteins to silent telomeric heterochromatin. In Drosophila, H3K79 methylation is also a prominent modification within euchromatin, with levels in transcribed regions correlating with gene activity (35). Mutations in the Drosophila Dot1 ortholog grappa lead to Polycomb and Tri-thorax-group phenotypes (37), suggesting that H3K79 methylation may influence developmentally regulated gene expression in multicellular eukaryotes.
DOT1L, the mammalian ortholog, displays enzymatic properties similar to those of its counterpart in yeast (9). Accordingly, H3K79 methylation can be detected on mammalian histones by mass spectrometry, with monomethylation being the most abundant species and correlating with the fraction of histone H3 modified by acetylation, suggesting enrichment at active genes (53). Several reports of individual genes in mammalian cells have correlated H3K79 methylation with transcriptional activation but also with gene repression (15, 17, 26, 54). All three degrees of H3K79 methylation were recently examined across the human genome by Solexa sequencing of DNA obtained by chromatin immunoprecipitation (ChIP) (2). It was reported that H3K79me3 is enriched at both silent and active genes, with silent regions having overall higher levels of this modification. H3K79me1 and H3K79me2 were not found to show a significant preference for either active or silent genes. These seemingly contradictory descriptions of H3K79 methylation within mammalian chromatin highlight the need to better understand the mechanisms that recruit DOT1L to genomic sites in vivo and the relationship of H3K79 methylation with gene transcription.
The MLL1 gene, which encodes a histone H3K4 methyltransferase, frequently undergoes chromosomal translocations in acute leukemia. The resulting oncogenic fusion protein encodes the N terminus of MLL1 (which lacks methyltransferase activity) fused to the C terminus of a heterogeneous group of partner molecules (19). Interestingly, several fusion partners of MLL1 encode proteins that bind directly or indirectly to DOT1L (5, 25, 28). The inappropriate recruitment of DOT1L to MLL1-regulated genes in the Hox cluster leads to H3K79 hypermethylation, increased transcription, and a block in hematopoietic cell differentiation (28). Thus, DOT1L has the potential to be an important regulator of gene expression in mammalian cells and represents a potential therapeutic target in this disease.
In this study, we examined the recruitment of DOT1L and the patterning of mono-, di-, and trimethylation of H3K79 under dynamic conditions in several mammalian cell lineages. Our findings revealed the ubiquitous nature of DOT1L recruitment and H3K79 methylation at actively transcribed chromatin. We also observed strong similarities between the patterning of H3K79 methylation and that of H3K4 methylation within mammalian chromatin that may reflect parallel pathways specifying gene activity or antagonizing gene silencing. Our study also revealed a novel pattern of H3K79 methylation that correlates with binding of DNA-binding activators at regulatory elements.
MATERIALS AND METHODS
Dot1l mutant MEFs.
Murine embryonic stem cell line RRR032, obtained from BayGenomics, contains a gene trap integration within intron 12 of the Dot1l gene. RRR032 embryonic stem cells were injected into C57BL/6 blastocysts to generate chimeric mice, which were then bred to obtain heterozygous Dot1l mutant animals. Mouse embryonic fibroblast (MEF) cultures were derived from embryonic day 13.5 embryos obtained from matings between heterozygous mice. The genotype of homozygous Dot1l mutant MEFs was determined by PCR and reverse transcription (RT)-PCR, confirming integration of the gene trap between exons 12 and 13. Cells were immortalized by serial passaging.
ChIP.
Sonicated chromatin was prepared essentially as previously described (11). Cells were cross-linked either with formaldehyde alone (1% for 10 min) or with sequential ethylene glycol bis(succinimidyl succinate) (Z1565; Pierce) (1.5 mM for 30 min), followed by formaldehyde (1% for 10 min), as described previously (52). Sequential cross-linking was used for DOT1L, MLL1, and ASH1L ChIP experiments. Antibodies for immunoprecipitation were prebound to protein G-agarose beads prior to incubation with chromatin. Washing, elution, and extraction conditions were as previously described (11). A detailed description of the ChIP protocol used may be found at http://stokes.chop.edu/web/blobel/.
Quantitative PCR (qPCR) analysis was performed on an ABI 7900HT with Sybr green. Each immunoprecipitated DNA sample was quantified by using the average of duplicate PCRs and referenced to a standard curve generated from serially diluted input material, thereby normalizing for primer efficiency and differences in the starting cell numbers. Two to five biological replicates were performed. Error bars represent standard deviations. All ChIP-qPCR signals were normalized to the input (labeled as IP/input on the y axis), which should be considered arbitrary units on a relative enrichment scale. While the units on the y axis are arbitrary, the immunoprecipitation (IP) efficiencies of most of the antibodies giving signals ranged from 0.01% to 10%, depending on the antibody used. Importantly, occupancy was interpreted by comparing IP/input values to negative control regions analyzed in parallel (e.g., CD4, Hbb-IVR16), which are displayed on each graph. IP/input signals have been normalized to total histone H3 where indicated.
DNA microarray analysis.
Input and immunoprecipitated DNAs were amplified by in vitro transcription as previously described (22). The microarray was custom designed with 105,000 features containing overlapping 60-mer oligonucleotides centered every 25 bp across the chromosomal regions of interest and printed by Agilent. The array contained 74 genes that were densely tiled through the transcribed region and up to 50 kb upstream of the transcription start site (TSS) and 10 kb downstream of the poly(A) sequence without tiling into neighboring genes or through a repetitive sequence. Full experimental details may be found at http://stokes.chop.edu/web/blobel/.
Antibodies.
The antibodies used in this study were anti-FLAG M2 (F1804; Sigma), anti-DOT1L (A300-954A or 953A; Bethyl), anti-H3K4me3 (07-473; Upstate), anti-H3K4me2 (07-030; Upstate), anti-H3K4me1 (ab8895; Abcam), anti-H3K79me2/me3 (ab2621; Abcam), anti-H3K79me2 (ab3594; Abcam), anti-H3K79me1 (ab2886; Abcam), anti-H3K36me3 (ab9050; Abcam), anti-total RNA polymerase II (pol II) (sc-899; Santa Cruz), anti-total histone H3 (ab1791; Abcam), anti-GATA-1 (sc-265; Santa Cruz), and anti-PPARγ (sc7196; Santa Cruz).
Supplemental materials and methods.
Additional information regarding cell culture conditions, ChIP protocols, and ChIP-chip analysis and the sequences of all of the primers used may be found at http://stokes.chop.edu/web/blobel/.
RESULTS
DOT1L recruitment is coupled with active gene transcription in mammalian cells.
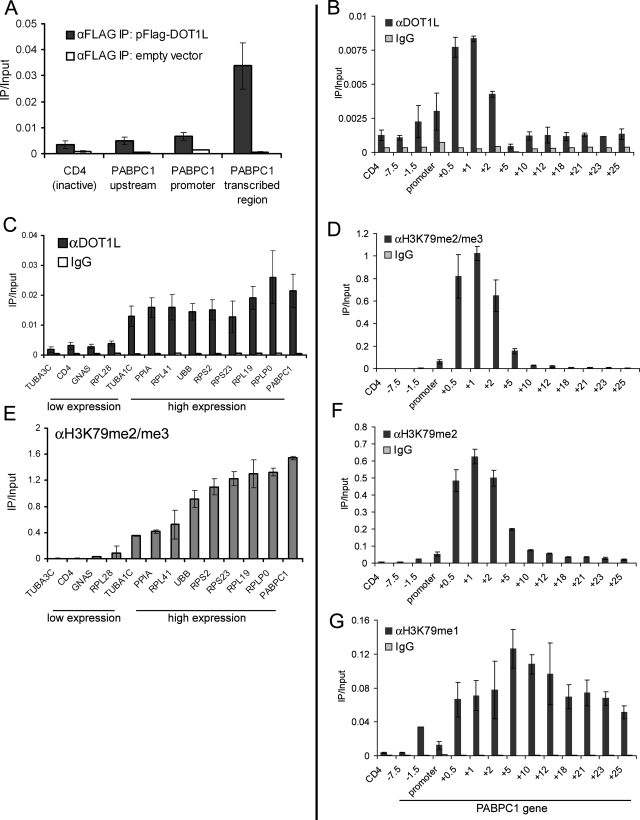
In Saccharomyces cerevisiae, Dot1 and H3K79 trimethylation have been detected ubiquitously in the transcribed region of active genes (26, 30, 36), suggesting that Dot1 is a general factor coupled with transcription elongation. In mammalian cells, however, several reports have linked H3K79 methylation with the active state of transcription (15, 17, 45) but also with the repressed state (2, 54). Thus, it is unclear if mammalian DOT1L recruitment is linked with transcription elongation in a general fashion, as is observed in yeast. To begin addressing this issue, we examined the occupancy of FLAG-DOT1L expressed transiently in HEK293T cells. Following the verification of protein expression by Western blotting, ChIP was performed with anti-FLAG antibodies, examining for occupancy at active and inactive sites. FLAG-DOT1L displayed pronounced occupancy in the transcribed region of the highly active PABPC1 gene (+2 kb) relative to the inactive CD4 gene (Fig. 1A). Occupancy was also low upstream (−7.5 kb) and at the promoter region of PABPC1 (−0.5 kb), suggesting a link with transcription elongation and correlating with our previously described distribution of H3K79 methylation at this gene (45). Similar results were obtained with HA-DOT1L (see Fig. 1A at http://stokes.chop.edu/web/blobel/). As a positive control, we were able to measure the occupancy of FLAG-MLL1 in the transcribed region of PABPC1 but not at the inactive CD4 gene (see Fig. 1B at http://stokes.chop.edu/web/blobel/), validating that occupancy detected with overexpressed molecules can mimic that of endogenous protein (14).
FIG. 1.
Human DOT1L occupies transcribed regions in a ubiquitous fashion. (A) ChIP in HEK293T cells transfected with pFLAG-DOT1L or empty vector. (B, D, F, and G) ChIP with the indicated antibodies in HEK293T cells across the PABPC1 gene with PCR primers at the indicated locations relative to the TSS. CD4 is a negative control region. (C and E) ChIP-qPCR analysis of HeLa cells at genes with high or low expression with the indicated antibodies. The DOT1L antibody is A300-954A. Primers amplify within the +0.5-kb region relative to the TSS. IgG, immunoglobulin G.
To validate the results obtained with tagged DOT1L and to explore the generality of its association with active chromatin, we measured by ChIP the profile of endogenous DOT1L at a panel of genes with either low or high expression in HeLa cells. Using previously published expression array data (32), we randomly chose four genes with low mRNA levels and nine genes with highly abundant mRNAs in HeLa cells. The expression pattern of these genes was confirmed by ChIP with anti-pol II antibodies (see Fig. 2 at http://stokes.chop.edu/web/blobel/). DOT1L occupancy was examined with two independently raised antibodies, which gave similar results. DOT1L occupancy was consistently detected in the transcribed region of the nine highly active genes at levels significantly above those observed at the four genes with low expression (Fig. 1C; see Fig. 3 at http://stokes.chop.edu/web/blobel/). A similar pattern of endogenous DOT1L was also observed in HEK293T cells (see Fig. 4 at http://stokes.chop.edu/web/blobel/). Occupancy of DOT1L also increased upon induction of Hbb-b1 transcription in differentiating murine erythroid cells (see Fig. 5 at http://stokes.chop.edu/web/blobel/). Taken together, these findings suggest that DOT1L associates with actively transcribed regions in a general fashion in diverse mammalian cell lineages.
FIG. 2.
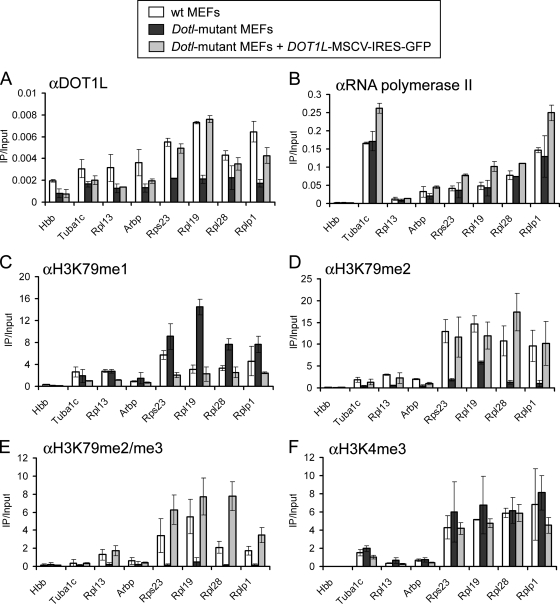
DOT1L is required for H3K79 di- and trimethylation at active genes. (A to F) ChIP analysis of wild-type (wt) MEFs, homozygous Dot1l mutant MEFs, or Dot1l mutant MEFs infected with DOT1L-murine stem cell virus-IRES-GFP. Levels of histone modification were normalized to total histone H3. Primers amplify within the +0.5-kb region relative to the TSS, except for Hbb, which amplifies a noncoding segment of the β-globin locus (IVR16 primers) and serves as a negative control.
FIG. 3.
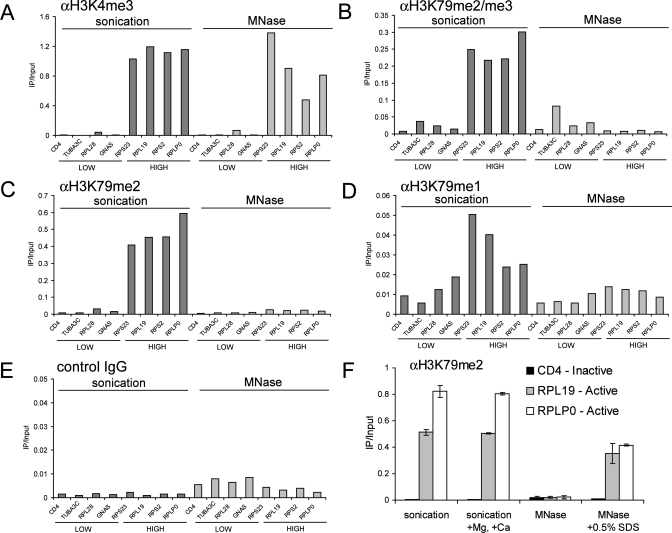
SDS treatment of cross-linked chromatin samples facilitates detection of H3K79 methylation by ChIP. (A to E) ChIP performed with HeLa chromatin prepared either by sonication or by MNase digestion with the indicated antibodies. (F) ChIP for H3K79me2 including sonicated chromatin treated with 3 mM MgCl2 and 1.5 mM CaCl2 or MNase-digested chromatin treated with 0.5% SDS. ChIP primers amplify within the +0.5-kb region relative to the TSS. IgG, immunoglobulin G.
FIG. 4.
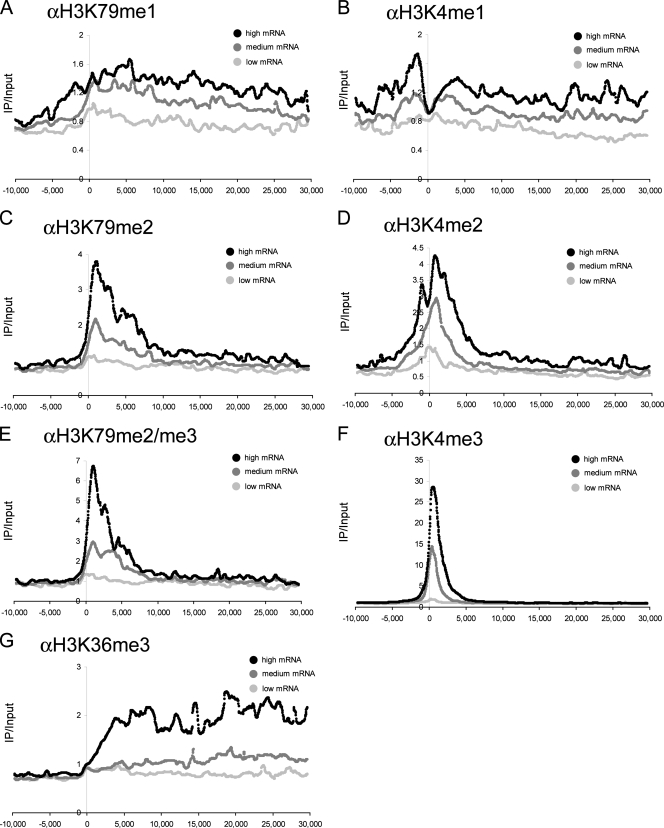
Levels of H3K79 methylation correlate with gene expression in mammalian cells. (A to G) ChIP-chip analysis of methylation with the indicated antibodies. The graphs shown are composite representations of similarly expressed genes with a moving median of 25 adjacent data points (spanning 660 bp). Each location is relative to the TSS, measured in base pairs. All methylation signals were normalized to the input and total histone H3.
FIG. 5.
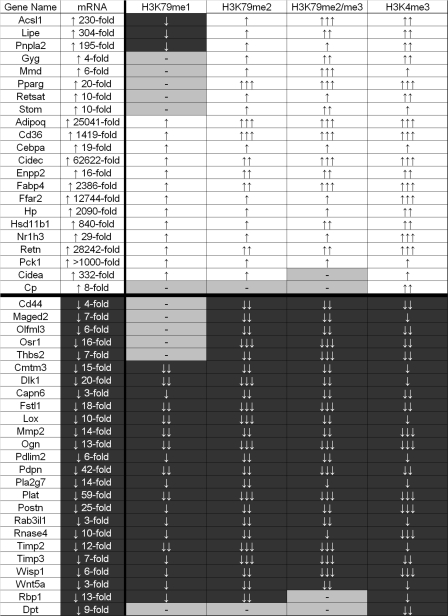
Dynamic acquisition or removal of H3K79 and H3K4 methylation reflects the transcriptional activity of genes regulated by adipogenesis. A summary of ChIP-chip data from 47 dynamic genes is shown. ↑ represents a 1.5- to 2.5-fold change. ↑↑ represents a 2.5- to 7-fold change. ↑↑↑ represents a >7-fold change. A minus sign represents no significant change. All methylation changes were normalized to the input and total histone H3.
Next, we sought to compare the occupancy of DOT1L to the patterning of H3K79me1, -me2, and -me3 by ChIP. Commercially available anti-H3K79-methyl antibodies were obtained and evaluated for specificity in ChIP assays by two approaches. First, we used wild-type and DOT1 deletion S. cerevisiae strains to assess the possible cross-reactivity of our antibodies with other methyl marks. We found that signals obtained by ChIP with H3K79me1, -me2, and -me3 antibodies were all abolished in the strain lacking DOT1, suggesting minimal cross-reactivity with other transcription-associated sites of methylation in this organism (e.g., H3K4 and H3K36) (see Fig. 6 at http://stokes.chop.edu/web/blobel/). Second, we evaluated antibody specificity toward the mono-, di-, or trimethyl state of H3K79 by using methylated peptides as competitors in ChIP assays performed with HeLa cell chromatin (see Fig. 6 at http://stokes.chop.edu/web/blobel/). All three antibodies displayed maximum reactivity toward the specific degree of methylation present in the antigen. H3K79 monomethyl-specific antibodies displayed negligible cross-reactivity with the other degrees of methylation. The anti-dimethyl H3K79 antibody displayed a slight reactivity toward monomethyl H3K79 but greatly preferred its bona fide antigen. However, the trimethyl antibody displayed significant cross-reactivity with dimethylated H3K79 peptide. Although the trimethylated peptide competed 10 times more effectively than the dimethylated peptide, the considerable degree of cross-reactivity precludes reliable distinction between H3K79me3 and H3K79me2. Therefore, we refer to signals obtained with this antibody as H3K79me2/me3.
FIG. 6.
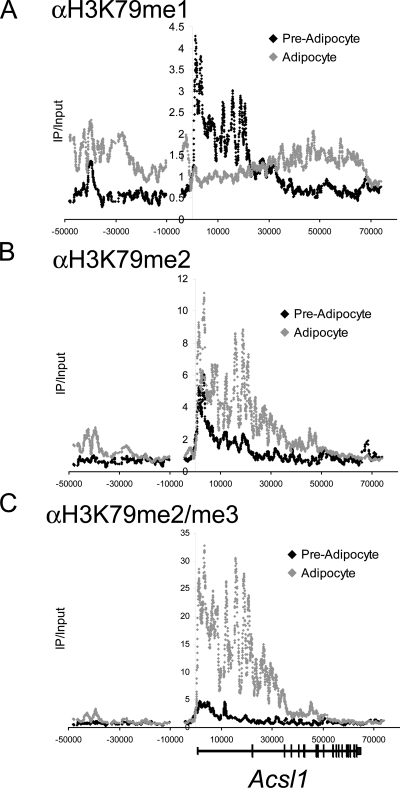
H3K79 monomethylation is converted into di- and trimethylation upon induction of Acsl1 transcription. ChIP-chip analysis of H3K79 methylation at Acsl1 in either undifferentiated (preadipocytes) or differentiated (adipocytes) 3T3-L1 cells is shown. All methylation signals were normalized to the input and total histone H3.
Using these tested antibodies, we compared the methylation of H3K79 to the occupancy of DOT1L at 13 amplicons spaced across a 33-kb region encompassing the active PABPC1 gene in HEK293T cells. Importantly, H3K79me2, H3K79me2/3, and DOT1L were similarly distributed across PABPC1, with maximum enrichment in the first 2 kb of the transcribed region and lower levels observed both upstream and further downstream (Fig. 1B, D, and F). In contrast, H3K79me1 displayed a broader localization pattern, extending upstream, as well as downstream, of the region enriched for DOT1L (Fig. 1G). The pattern of DOT1L recruitment along PABPC1 is distinct from total pol II, with the latter displaying a bias toward the promoter region (see Fig. 7 at http://stokes.chop.edu/web/blobel/). Instead, the peak density of DOT1L occupancy along PABPC1 is at the proximal transcribed region, correlating with the site where transcription elongation is initiated. In addition, DOT1L occupancy is well correlated with the presence of H3K79 di- and trimethylation, suggesting that DOT1L might be essential for maintaining these modifications.
FIG. 7.
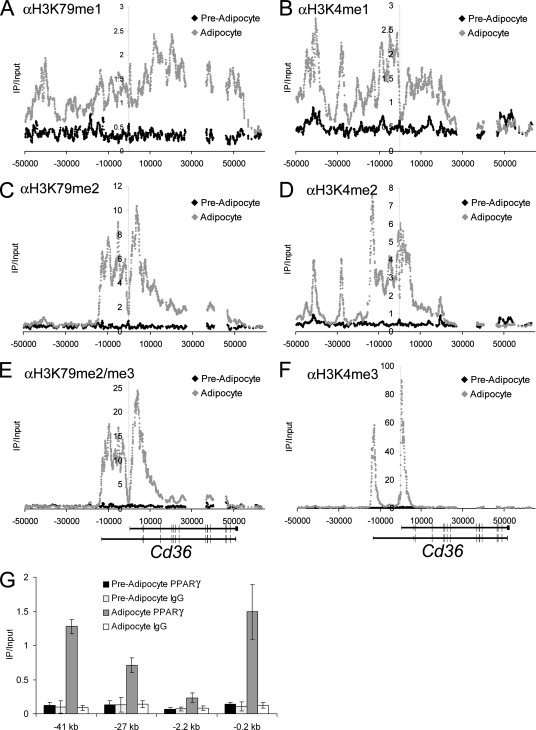
Distal upstream locations of H3K79 monomethylation at Cd36 correlate with occupancy of PPARγ. (A to F) ChIP-chip mapping of H3K4 methylation and H3K79 methylation at the Cd36 locus. All methylation signals were normalized to the input and total histone H3. (G) PPARγ ChIP-qPCR performed at the indicated regions of Cd36 relative to the TSS. The positive control for PPARγ occupancy is −0.2 kb. −2.2 kb is a negative control site. IgG, immunoglobulin G.
ASH1L and MLL1, both H3K4 methyltransferases, are general factors coupled with the “on” state of transcription (13, 14). Notably, both enzymes displayed an occupancy pattern similar to that of DOT1L, preferring active over inactive genes (see Fig. 8A and B at http://stokes.chop.edu/web/blobel/). In addition, levels of H3K4me3 and H3K79me2/me3 were also correlated among these sites (Fig. 1E; see Fig. 8C at http://stokes.chop.edu/web/blobel/). These findings suggest that recruitment of these three methyltransferases might be mechanistically coordinated by a subset of elongating pol II that preferentially occupies proximal transcribed regions.
FIG. 8.
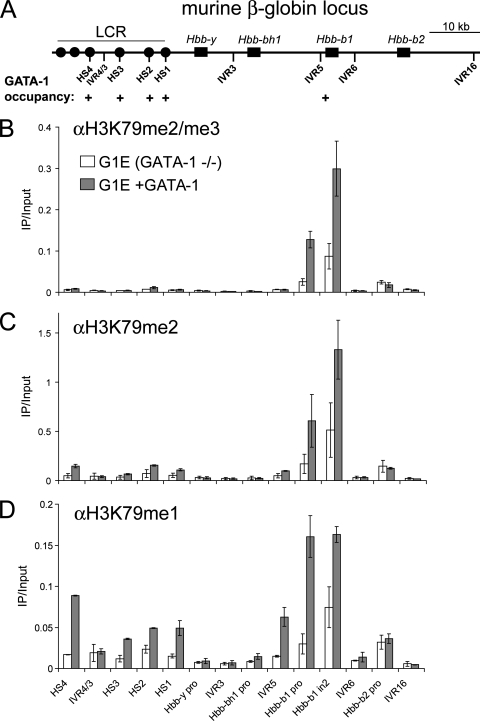
GATA-1 is required for H3K79 monomethylation at the LCR of the β-globin locus. (A) Schematic of the murine β-globin locus with known GATA-1 binding sites indicated (18). (B to D) ChIP-qPCR with the indicated antibodies detecting occupancy at various regions of the murine β-globin locus in G1E cells expressing GATA-1-ER before or after 24 h of estradiol treatment. Signals were normalized to the total histone H3 and the input. IVR, intervening region; pro, promoter; in2, intron 2.
Mammalian DOT1L is required for H3K79 di- and trimethylation at transcribed regions.
While 10 different mammalian enzymes are known to methylate H3K4 (34), DOT1L is the only known methyltransferase with specificity for H3K79. To test whether DOT1L is indeed the sole enzyme that establishes H3K79 methylation at transcribed chromatin, we examined H3K79 methylation in MEFs carrying a homozygous gene trap insertion in the Dot1l gene (6, 42). The site of gene trap integration (intron 12) is expected to truncate Dot1l expression at amino acid 335, eliminating a critical nucleosome binding region at amino acids 390 to 407 required for full DOT1L methyltransferase activity in vitro (24). ChIP analysis with anti-DOT1L antibodies recognizing an epitope C terminal to the site of gene trap integration confirmed loss of full-length DOT1L in the transcribed region of several highly active genes (Fig. 2A). These genes are transcribed normally in the mutant MEFs, as determined by RT-PCR (data not shown) and pol II ChIPs (Fig. 2B), indicating that the lack of DOT1L is due to the gene trap mutation and is not a result of reduced transcription. Importantly, di- and trimethylation of H3K79 were severely reduced in Dot1l mutant cells at all of the active sites examined (Fig. 2D and E). In contrast, monomethylation of H3K79 was largely intact and even increased at several sites (Fig. 2C). We observed normal levels of H3K4me3 at active genes in the mutant MEFs, indicating that the methylation changes were specific to H3K79 (Fig. 2F). To verify that alterations in H3K79 methylation were attributed to the Dot1l mutation and were not a cell culture artifact, we expressed wild-type human DOT1L in these cells with murine stem cell virus-IRES-GFP. Importantly, levels of H3K79 methylation in Dot1l mutant cells were restored to the levels found in wild-type MEFs (Fig. 2A to F). These results support DOT1L as the principal mammalian enzyme responsible for di- and trimethylation of H3K79 in transcribed regions.
The maintenance of H3K79me1 in Dot1 mutant cells was unexpected, as DOT1L is the only known mammalian methyltransferase with specificity for H3K79. It is possible that the Dot1l gene trap mutation does not completely abolish methyltransferase activity in vivo, as some residual in vitro activity of the DOT1L protein was retained when amino acids 390 to 407 were deleted (24). However, we cannot rule out the existence of another H3K79 methyltransferase in mammalian cells that may partially compensate for DOT1L loss.
Sodium dodecyl sulfate (SDS) treatment of cross-linked chromatin facilitates H3K79 methylation detection by ChIP.
Our findings presented thus far support the presence of DOT1L and H3K79 methylation as a ubiquitous feature in the transcribed region of active genes. However, a recent genome-wide methylation study with human CD4+ T cells described a pattern of H3K79 methylation that did not correlate with gene transcription (2). In that study, H3K79me1 and H3K79me2 displayed a minimal preference for active or repressed sites and H3K79me3 was detected at higher levels at repressed genomic regions (2). While the antibodies used in this prior study are identical to those used here, the prior study relied exclusively on micrococcal nuclease (MNase) digestion of chromatin whereas we used sonication. To test whether the method of preparing chromatin influences the detection of H3K79 methylation, we compared the two techniques directly (for detailed protocols, see http://stokes.chop.edu/web/blobel/). ChIP with MNase-digested chromatin did not yield significant enrichment at active genes compared to inactive sites, whereas robust enrichments were observed when sonicated chromatin was used (Fig. 3B to D; see Fig. 9 at http://stokes.chop.edu/web/blobel/). Antibodies recognizing other modifications on histone H3 (anti-H3K9acetyl and anti-H3K4me3) produced similar results with both methods (Fig. 3A; see Fig. 9 at http://stokes.chop.edu/web/blobel/). We next exchanged buffer components between the two protocols to evaluate whether a specific reagent was responsible for inhibiting or facilitating H3K79 detection. Strikingly, exposing MNase-digested chromatin to 0.5% SDS (which is used in the sonication protocol) restored the detection of H3K79 dimethylation (Fig. 3F). This suggests that partial denaturation by SDS facilitates the exposure of methylated H3K79 within formaldehyde-cross-linked chromatin. We believe that the results obtained with SDS-treated chromatin likely represent the true pattern of H3K79 methylation since the latter closely resembles the occupancy of DOT1L (Fig. 1). In addition, H3K79 methylation signals obtained with SDS-treated chromatin were strongly reduced in Dot1l mutant cells (Fig. 2). These results emphasize the need for optimization of buffer conditions for the proper detection of different epitopes by immunoprecipitation.
FIG. 9.
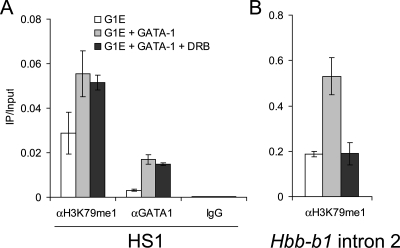
Intergenic transcription is not required to establish H3K79me1 at the LCR. ChIP was performed with G1E(Bcl-XL) cells expressing GATA-1-ER with the indicated antibodies before or after 18 h of treatment with estradiol with or without DRB. (A) HS1 primers. (B) Hbb-b1 intron 2 primers. IgG, immunoglobulin G.
ChIP-chip tiling array analysis of H3K79 methylation during adipocyte differentiation.
With this technical consideration in mind, we sought to reevaluate the general relationship between H3K79 methylation and gene transcription in mammalian chromatin by using ChIP-chip tiling arrays. Specifically, we examined to what extent mRNA levels correlate with methylation of H3K79. As a control, we also analyzed H3K4me3, H3K4me2, H3K4me1, and H3K36me3, which have all been profiled extensively in mammalian chromatin and found to correlate with gene activity (2-4), although in some instances, H3K4me3 can be present prior to productive transcription elongation (15). To explore the dynamics of these modifications, we used an established tissue culture model of adipogenesis whereby 3T3-L1 preadipocytes can be induced to differentiate into adipocytes (12). Cell maturation is accompanied by robust activation and repression of gene transcription and thus represents an ideal system for studying potential changes in histone methylation. Using Affymetrix microarray data generated with cDNA prepared from 3T3-L1 cells before and after differentiation (data not shown), we chose 47 genes that were induced or repressed by adipogenesis. As controls, we included several transcriptionally silent genes, as well as several constitutively active genes (72 genes in total). Upon confirming mRNA changes by RT-PCR (see Fig. 10 at http://stokes.chop.edu/web/blobel/), we designed an oligonucleotide tiling array covering the nonrepetitive locus of each gene, including the entire transcribed region, 50 kb upstream, and 10 kb downstream without tiling into neighboring genes.
First, we evaluated the relationship between steady-state levels of histone methylation and transcription by generating composite methylation profiles of genes with similar levels of expression (2, 15, 30). Each gene in the array was grouped into low, medium, or high expression on the basis of percentile rank (see Fig. 11 at http://stokes.chop.edu/web/blobel/). Methylation profiles of all of the genes within each category were aligned at the TSS, and a moving-median composite curve was generated. This analysis revealed that all degrees of H3K79 methylation were enriched at genes in a manner correlating with their levels of expression (Fig. 4A, C, and E; see Fig. 12 at http://stokes.chop.edu/web/blobel/). Levels of H3K4me1, -me2, and -me3 and H3K36me3 also correlated positively with mRNA abundance, consistent with prior genome-wide analyses (2, 15, 23). The relationship between H3K79 methylation and transcription differs demonstrably from a previous report (2), which we attribute to the aforementioned differences in preparing chromatin.
The spatial patterning of H3K79 methylation became more focally enriched with each successive degree of methylation. H3K79me2/me3 tended to be limited (but with greater overall enrichment) to the proximal 7 kb of the transcribed region, whereas H3K79me1 tended to extend further along the transcribed region to distances greater than 30 kb. H3K79me1 could also be found at intergenic regions positioned upstream of TSSs (Fig. 4A). Notably, the spatial relationship for H3K79me1/me2/me3 was highly similar to that observed for H3K4me1/me2/me3. Both H3K79me2/me3 and H3K4me2/me3 displayed a highly overlapping distribution at the 5′ end of active genes, whereas H3K4me1 and H3K79me1 more closely resembled the 3′ distribution of H3K36me3 in distal transcribed regions. The close similarity between H3K79 methylation and H3K4 methylation at active chromatin further supports a shared mechanism that coordinates DOT1L and MLL-like methyltransferase activities in vivo.
We also evaluated the dynamic relationship between H3K79 methylation and changes in gene expression linked with 3T3-L1 cell differentiation (Fig. 5). More than 90% of the genes whose expression is altered during adipogenesis showed changes in the levels of H3K79me2/me3 in their transcribed regions paralleling the observed changes in mRNA level. Together with the results above (Fig. 4), these findings further strengthen the correlation between transcription and H3K79 methylation. Methylation changes observed by ChIP-chip were confirmed at all genes by an independently replicated ChIP experiment analyzed by qPCR (data not shown). Four of the 47 dynamically regulated genes did not display the expected correlation between levels of H3K79me2/me3 methylation and changes in mRNA (Cidea, Cp, Rbp1, and Dpt). This could reflect the posttranscriptional regulation of these genes. An alternative explanation is that the specific activity of DOT1L might be subject to regulation at select genes, perhaps through interaction with other transcription factors. Nevertheless, our data strongly support parallel H3K79 and H3K4 methylation pathways at most mammalian genes when actively transcribed.
ChIP-chip analysis revealed two basic categories of genic H3K79 methylation dynamics during adipogenesis. Thirty genes displayed a correlation of all degrees of H3K79 methylation in the transcribed region with gene activity, as represented by the profiles of the downregulated Wisp1 and upregulated Cidec genes (see Fig. 13 at http://stokes.chop.edu/web/blobel/). Interestingly, a second category of methylation dynamics was observed at 13 genes that displayed changes in di- and trimethylation of H3K79 that correlated with gene transcription but without the expected change in monomethylation (Fig. 5). In fact, several upregulated genes became depleted of H3K79me1 precisely over regions that became maximally enriched for H3K79me2/me3, as shown for Acsl1 (Fig. 6). By RT-PCR, Acsl1 displayed detectable expression in preadipocytes prior to 230-fold upregulation in adipocytes (see Fig. 10 at http://stokes.chop.edu/web/blobel/). Thus, the transition from low- to high-level transcription of several genes can be correlated with the conversion of H3K79me1 into H3K79me2/me3.
One possibility is that H3K79me1 (without H3K79me2/me3) represents a “poised” chromatin state. To pursue this question further, we examined H3K79 methylation in murine embryonic stem cells, where a subset of genes destined for imminent activation display a bivalent pattern of H3K4 and H3K27 trimethylation (4). Notably, five out of eight bivalent genes were also enriched for H3K79me1 but not for H3K79me2/me3 (see Fig. 14 at http://stokes.chop.edu/web/blobel/), consistent with H3K79me1 marking certain genes in their poised state.
Intergenic enrichment of H3K79 monomethylation upstream of the Cd36 gene correlates with PPARγ occupancy.
Unexpectedly, our ChIP-chip methylation profiles revealed intergenic locations for H3K79 monomethylation. As an example, Cd36 displayed a prominent peak of H3K79me1 centered at 41 kb upstream of the TSS in addition to a broad area of enrichment in the transcribed region (Cd36 also has a second TSS 13 kb upstream) (Fig. 7A). A second area of lower H3K79me1 enrichment was also seen at around 27 kb upstream (Fig. 7A). The presence of H3K79me1 at these sites was confirmed in a biological replicate analyzed by ChIP-qPCR (see Fig. 15 at http://stokes.chop.edu/web/blobel/). Of note, while the H3K79me2 antibody reproducibly gives weak signals at intergenic sites when analyzed by qPCR (see Fig. 15 at http://stokes.chop.edu/web/blobel/), this may be due to a low-level cross-reactivity with H3K79me1 (see Fig. 6 at http://stokes.chop.edu/web/blobel/). H3K4me1 and H3K4me2 were also localized to these two upstream locations (Fig. 7B and D), which previously has been shown to be a methylation pattern indicative of enhancer regions (2, 3, 16). Importantly, intergenic monomethylation of H3K79 and H3K4 was significantly increased in adipocytes relative to preadipocytes, in accordance with Cd36 transcription. Together with the failure to detect intergenic transcription in these regions (data not shown), this led us to examine whether foci enriched for H3K79me1 represent enhancer regions bound by transcriptional activators.
The nuclear receptor PPARγ activates Cd36 transcription through binding sites in the proximal promoter region (−0.2 kb) (43). Given the importance of PPARγ to Cd36 expression, we investigated whether it might additionally function at distal regulatory elements and contribute to establishing H3K79me1. Using ChIP, we found significant enrichment for PPARγ in adipocytes but not preadipocytes at both the −41- and −27-kb regions enriched for H3K79me1. The signals were comparable to the proximal promoter (−0.2 kb) and significantly above an intervening control region (−2.2 kb) (Fig. 7G). Thus, occupancy of PPARγ at distal regulatory elements of Cd36 correlates with the acquisition of H3K79me1 and H3K4me1/me2. This methylation pattern is distinct from that observed near the TSSs, where H3K79me2/me3 and H3K4me3 were strongly enriched (Fig. 7C, E, and F). These findings suggest that intergenic H3K79 monomethylation demarcates gene-regulatory elements.
Seventy-five locations of intergenic H3K79me1 were identified across the panel of 47 tiled genes (see Fig. 16 at http://stokes.chop.edu/web/blobel/). Importantly, 97% of these sites were also marked by enrichment of H3K4me1, which has previously been shown to be a methyl mark predictive of enhancer activity (16). This highlights an additional similarity between H3K4 methylation and H3K79 methylation, suggesting that both modifications can demarcate enhancer regions. However, it should be pointed out that intergenic H3K4me1 generally displayed greater overall enrichment and was seen more frequently across tiled genes (110 sites) than H3K79me1 (75 sites). Thus, H3K4me1 may display greater sensitivity in annotating potential regulatory regions, while H3K79me1 might be more selective for certain classes of regulatory DNA elements. Future work will examine the broader range of elements that possess H3K79me1 and whether a consistent relationship exists with specific sequence features. Our findings suggest that patterning of the degree of H3K79 methylation across mammalian genomes may have utility in distinguishing potential regulatory elements (H3K79me1 alone) from the transcribed region of active genes (H3K79me1/me2/me3).
GATA-1 is required for H3K79 monomethylation at the LCR of the murine β-globin locus.
To investigate further the relationship between H3K79me1 and transcription factor occupancy, we examined H3K79 methylation at the well-characterized regulatory elements of the murine β-globin locus. High-level transcription of Hbb-b1 requires occupancy of the erythroid transcription factor GATA-1 at the proximal promoter and at the upstream locus control region (LCR). While H3K79 dimethylation has previously been observed at a few sites across the β-globin locus, a relationship with transcription factor binding or enhancer activity was not determined (17). To evaluate a potential relationship between GATA-1 binding to its target enhancer elements and H3K79 methylation, we performed ChIP-qPCR studies with the GATA-1 null erythroid cell line G1E. Conditional activation of a stably expressed fusion of GATA-1 and the ligand binding domain of the estrogen receptor (GATA-1-ER) leads to direct activation of Hbb-b1 transcription (48). We measured the profile of H3K79 methylation at 14 regions across the β-globin locus in GATA-1-ER-expressing G1E cells treated for 24 h with estradiol. Similar to observations at many adipogenesis-regulated genes, all degrees of H3K79 methylation were acquired at the activated Hbb-b1 transcribed region upon GATA-1 activation, with H3K79me1 bearing a somewhat broader localization pattern than H3K79me2/me3 (Fig. 8B to D). However, GATA-1 binding to each site within the LCR correlated with significant increases in H3K79me1 but not at an intervening region (IVR4/3) (Fig. 8D). H3K79me2/me3 signals were limited to the Hbb-b1 transcribed region (Fig. 8B and C). These results suggest that GATA-1 may establish a pattern of intergenic H3K79me1 at bound elements within the LCR in parallel with the pattern of H3K79me1/me2/me3 established at Hbb-b1 upon gene activation.
To evaluate a possible contribution of intergenic transcription to the establishment of H3K79me1 at the LCR, we activated GATA-1 in the presence of the pTEFb kinase inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), which potently inhibits Hbb-b1, as well as intergenic transcription at the β-globin locus (18). To permit survival of G1E cells during DRB treatment, a G1E cell line overexpressing the antiapoptotic molecule Bcl-XL was used (49). In addition, the duration of estradiol treatment was reduced to 18 h. RT-PCR revealed that intergenic transcription of DNase-hypersensitive site 1 (HS1) of the LCR was not significantly increased following GATA-1 activation (see Fig. 17 at http://stokes.chop.edu/web/blobel/). In addition, DRB treatment strongly reduced the level of detectable HS1, as well as Hbb-b1 transcripts, consistent with prior findings (18). Notably, we found that DRB treatment does not reduce GATA-1-induced accumulation of H3K79me1 at HS1 (Fig. 9A). In contrast, H3K79me1 in the transcribed region of Hbb-b1 failed to accumulate in the presence of DRB (Fig. 9B). As a control, we observed that GATA-1 occupied the LCR normally under these conditions (Fig. 9A). This result suggests that intergenic H3K79 monomethylation can be initiated by DNA-binding activators independently of transcription elongation.
The observations that GATA-1 and PPARγ can be correlated with the acquisition of H3K79me1 at bound elements suggest that these transcription factors might be able to recruit DOT1L to modify chromatin at bound sites. Several attempts to detect a physical association between GATA-1 and DOT1L were unsuccessful. In addition, occupancy of DOT1L at the LCR was low compared to the Hbb-b1 transcribed region and did not change significantly following the activation of GATA-1-ER (see Fig. 5 at http://stokes.chop.edu/web/blobel/). When considered together with the observation that H3K79me1 levels are maintained in DOT1L-deficient fibroblasts, this leaves open the question of whether DOT1L is, in fact, the enzyme generating H3K79 monomethylation at intergenic locations.
DISCUSSION
H3K79 di- and trimethylation: a cause or a consequence of gene transcription?
The results presented here support DOT1L as a general factor linked with recently initiated transcription elongation complexes. First, endogenous DOT1L occupancy was consistently observed in the proximal transcribed regions of all of the active genes examined, including tissue-specific as well as housekeeping genes. Second, ChIP-chip studies demonstrated that H3K79me2/me3, modifications performed solely by DOT1L, displayed a strong correlation with gene activity. Therefore, our findings support the DOT1L pathway as a pervasive feature within active mammalian euchromatin, with the majority of active genes being marked with H3K79me2/me3.
Despite the strong correlation between H3K79 di- and trimethylation and transcriptional activity, our findings with Dot1l mutant fibroblasts demonstrated that many active genes can be transcribed at normal levels despite significant depletion of H3K79me2/me3 in the transcribed region. This raises the possibility that levels of H3K79me2/me3 might reflect gene activity without being truly required for the active state. However, while Dot1l mutant MEFs are viable, mice failed to progress through fetal development (6). The same is true for the DOT1 ortholog (grappa) in Drosophila, which is an essential gene required for normal development (37). Dot1 deletion strains of yeast are viable, but a subset of genes under the regulation of SIR proteins are affected (41, 46). On the basis of these observations across different species, we hypothesize that a subset of developmentally regulated mammalian genes require DOT1L for proper expression. Preliminary RT-PCR data suggest gene-specific expression alterations in Dot1l mutant fibroblasts (data not shown). However, the ubiquitous nature of DOT1L occupancy at most active genes precludes us from distinguishing direct from indirect requirements. Future genome-wide expression analysis in wild-type and Dot1l mutant cells will examine the broader spectrum of DOT1L-requiring genes. It also remains a possibility that H3K79 methylation participates in additional cellular processes distinct from transcriptional regulation, such as in maintaining genomic stability (51).
Why might only a subset of genes require H3K79me2/me3 for transcription? Genetic experiments with yeast suggest that Dot1 might functionally oppose epigenetic silencing (41). We speculate that a requirement for mammalian DOT1L might be imposed by the presence of certain mammalian gene-silencing complexes (e.g., HP1 or Polycomb) over proximal transcribed regions. H3K79me2/me3 might alter critical protein-protein interactions that enable promoter escape by elongation complexes through potentially repressive chromatin. To this end, it is noteworthy that repressive H3K9me3 and HP1 have been observed in the transcribed body of active mammalian genes (23, 31, 44). In addition, Polycomb complexes have been shown to selectively inhibit the initiation kinetics of transcription elongation (7, 8) rather than access of general transcription machinery to promoter regions. Therefore, placement of H3K79me2/me3 within proximal transcribed chromatin would be ideally located to promote the passage of elongating RNA pol II under such conditions.
Intergenic H3K79 monomethylation: a mark of enhancer regions.
Our observation of H3K79 monomethylation at intergenic locations expands the potential role for this modification in processes beyond transcription elongation. Transcription factors (e.g., GATA-1 and PPARγ) or their cofactors may recruit an H3K79 monomethyltransferase to label surrounding chromatin at bound elements. Long-range enhancer function is likely to require a sophisticated network of molecular interactions to communicate with target gene promoters. Histone H3 lysine 79 monomethylation may provide unique binding surfaces along the chromatin fiber to stabilize interactions between enhancers and their target promoters, thereby rendering high-level gene transcription. It will be interesting in future work to identify recognition modules for the H3K79me1 mark and how they might affect transcription factor action over large distances.
It should be emphasized that our findings leave open the question whether DOT1L is, in fact, responsible for H3K79 monomethylation in mammalian cells, as stable DOT1L occupancy does not precisely correlate with locations of H3K79me1 and levels of this modification were normal in Dot1l mutant MEFs. This issue could be resolved by testing additional Dot1l mutant alleles for a defect in H3K79me1, as well as a more detailed biochemical evaluation of DOT1L substrate preferences in vitro.
Skewing the normal balance of H3K4 methylation and H3K79 methylation in acute leukemia.
One theme that emerged from this work is the parallel nature of H3K4 methylation and H3K79 methylation in mammalian chromatin. DOT1L and MLL-like molecules may share a requirement for the PAF elongation complex (55), H2B ubiquitylation (56), and a Cps35 homolog for recruitment and/or activity, similar to their orthologs in yeast (20, 21, 36). Interestingly, the gene for MLL1 can undergo translocations in acute leukemia with partner genes that encode proteins (AF10, AF4, ENL, and AF9) that bind directly or indirectly to DOT1L. The resulting MLL1 fusion molecule has lost its native H3K4 methyltransferase domain and instead recruits the H3K79 methyltransferase activity of DOT1L. Thus, the physiologic coupling of H3K4 methylation and H3K79 methylation described here would be expected to skew toward more extensive H3K79 methylation at the expense of H3K4. This is thought to specifically lead to upregulation of Hox gene expression (28); however, our findings here raise the possibility that such fusion proteins might also upset the global balance among these modifications, on the basis of the generalized occupancy patterns of both MLL1 and DOT1L in the normal cellular state.
The interplay between MLL1 and DOT1L in leukemia suggests that, despite their similarities, these two methylation pathways have differing potencies at select target gene loci. This might relate to different effector molecules being recruited or displaced by these two modifications. H3K4 methylation may recruit nucleosome-remodeling complexes (39, 50), RNA-splicing machinery (40), TFIID (47), or possibly prevent de novo DNA CpG methylation (29). A biochemical mechanism by which H3K79 methylation alters gene expression in mammalian cells awaits the identification of methylation-sensitive effector molecules with specificity for this modification that can influence transcriptional outcomes.
Acknowledgments
We thank members of the Blobel and Lazar laboratories for discussion and comments on the manuscript; Klaus Kaestner, Peter White, and the Penn Genomics and Gene Targeting Core for help with the processing and analysis of microarrays; Ken Zaret and Mitch Weiss for helpful advice; Ali Shilatifard for comments on the manuscript; Brian Brunk and the Penn Center for Bioinformatics for help with the design of microarray oligonucleotides; Yi Zhang for providing the pFLAG-hDOT1L PCDNA3 plasmid; and Eric Rappaport for technical support of real-time PCR equipment.
This work was supported by NIH grants DK58044 and DK54937 to G.A.B. and DK49780 to M.A.L. C.R.V. was supported by NIH training grant T32 HL007150. M.S. was supported by a Mentored Fellowship award from the American Diabetes Association.
Footnotes
Published ahead of print on 19 February 2008.
REFERENCES
- 1.Altaf, M., R. T. Utley, N. Lacoste, S. Tan, S. D. Briggs, and J. Cote. 2007. Interplay of chromatin modifiers on a short basic patch of histone h4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 281002-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120169-181. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, B. E., T. S. Mikkelsen, X. Xie, M. Kamal, D. J. Huebert, J. Cuff, B. Fry, A. Meissner, M. Wernig, K. Plath, R. Jaenisch, A. Wagschal, R. Feil, S. L. Schreiber, and E. S. Lander. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125315-326. [DOI] [PubMed] [Google Scholar]
- 5.Bitoun, E., P. L. Oliver, and K. E. Davies. 2007. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum. Mol. Genet. 1692-106. [DOI] [PubMed] [Google Scholar]
- 6.Botuyan, M. V., J. Lee, I. M. Ward, J. E. Kim, J. R. Thompson, J. Chen, and G. Mer. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 1271361-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breiling, A., B. M. Turner, M. E. Bianchi, and V. Orlando. 2001. General transcription factors bind promoters repressed by Polycomb group proteins. Nature 412651-655. [DOI] [PubMed] [Google Scholar]
- 8.Dellino, G. I., Y. B. Schwartz, G. Farkas, D. McCabe, S. C. Elgin, and V. Pirrotta. 2004. Polycomb silencing blocks transcription initiation. Mol. Cell 13887-893. [DOI] [PubMed] [Google Scholar]
- 9.Feng, Q., H. Wang, H. H. Ng, H. Erdjument-Bromage, P. Tempst, K. Struhl, and Y. Zhang. 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 121052-1058. [DOI] [PubMed] [Google Scholar]
- 10.Fingerman, I. M., H. C. Li, and S. D. Briggs. 2007. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev. 212018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forsberg, E. C., K. M. Downs, and E. H. Bresnick. 2000. Direct interaction of NF-E2 with hypersensitive site 2 of the beta-globin locus control region in living cells. Blood 96334-339. [PubMed] [Google Scholar]
- 12.Green, H., and O. Kehinde. 1975. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell 519-27. [DOI] [PubMed] [Google Scholar]
- 13.Gregory, G. D., C. R. Vakoc, T. Rosovskia, X. Zheng, S. Patel, T. Nakamura, E. Canaani, and G. A. Blobel. 2007. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol. Cell. Biol. 278466-8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guenther, M. G., R. G. Jenner, B. Chevalier, T. Nakamura, C. M. Croce, E. Canaani, and R. A. Young. 2005. Global and Hox-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. USA 1028603-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guenther, M. G., S. S. Levine, L. A. Boyer, R. Jaenisch, and R. A. Young. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 13077-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heintzman, N. D., R. K. Stuart, G. Hon, Y. Fu, C. W. Ching, R. D. Hawkins, L. O. Barrera, S. Van Calcar, C. Qu, K. A. Ching, W. Wang, Z. Weng, R. D. Green, G. E. Crawford, and B. Ren. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39311-318. [DOI] [PubMed] [Google Scholar]
- 17.Im, H., C. Park, Q. Feng, K. D. Johnson, C. M. Kiekhaefer, K. Choi, Y. Zhang, and E. H. Bresnick. 2003. Dynamic regulation of histone H3 methylated at lysine 79 within a tissue-specific chromatin domain. J. Biol. Chem. 27818346-18352. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, K. D., J. A. Grass, C. Park, H. Im, K. Choi, and E. H. Bresnick. 2003. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol. Cell. Biol. 236484-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krivtsov, A. V., and S. A. Armstrong. 2007. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 7823-833. [DOI] [PubMed] [Google Scholar]
- 20.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11721-729. [DOI] [PubMed] [Google Scholar]
- 21.Lee, J. S., A. Shukla, J. Schneider, S. K. Swanson, M. P. Washburn, L. Florens, S. R. Bhaumik, and A. Shilatifard. 2007. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 1311084-1096. [DOI] [PubMed] [Google Scholar]
- 22.Liu, C. L., S. L. Schreiber, and B. E. Bernstein. 2003. Development and validation of a T7 based linear amplification for genomic DNA. BMC Genomics 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikkelsen, T. S., M. Ku, D. B. Jaffe, B. Issac, E. Lieberman, G. Giannoukos, P. Alvarez, W. Brockman, T. K. Kim, R. P. Koche, W. Lee, E. Mendenhall, A. O'Donovan, A. Presser, C. Russ, X. Xie, A. Meissner, M. Wernig, R. Jaenisch, C. Nusbaum, E. S. Lander, and B. E. Bernstein. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Min, J., Q. Feng, Z. Li, Y. Zhang, and R. M. Xu. 2003. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell 112711-723. [DOI] [PubMed] [Google Scholar]
- 25.Mueller, D., C. Bach, D. Zeisig, M. P. Garcia-Cuellar, S. Monroe, A. Sreekumar, R. Zhou, A. Nesvizhskii, A. Chinnaiyan, J. L. Hess, and R. K. Slany. 2007. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 1104445-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng, H. H., D. N. Ciccone, K. B. Morshead, M. A. Oettinger, and K. Struhl. 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA 1001820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng, H. H., Q. Feng, H. Wang, H. Erdjument-Bromage, P. Tempst, Y. Zhang, and K. Struhl. 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 161518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okada, Y., Q. Feng, Y. Lin, Q. Jiang, Y. Li, V. M. Coffield, L. Su, G. Xu, and Y. Zhang. 2005. hDOT1L links histone methylation to leukemogenesis. Cell 121167-178. [DOI] [PubMed] [Google Scholar]
- 29.Ooi, S. K., C. Qiu, E. Bernstein, K. Li, D. Jia, Z. Yang, H. Erdjument-Bromage, P. Tempst, S. P. Lin, C. D. Allis, X. Cheng, and T. H. Bestor. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448714-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122517-527. [DOI] [PubMed] [Google Scholar]
- 31.Regha, K., M. A. Sloane, R. Huang, F. M. Pauler, K. E. Warczok, B. Melikant, M. Radolf, J. H. Martens, G. Schotta, T. Jenuwein, and D. P. Barlow. 2007. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol. Cell 27353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross, D. T., U. Scherf, M. B. Eisen, C. M. Perou, C. Rees, P. Spellman, V. Iyer, S. S. Jeffrey, M. Van de Rijn, M. Waltham, A. Pergamenschikov, J. C. Lee, D. Lashkari, D. Shalon, T. G. Myers, J. N. Weinstein, D. Botstein, and P. O. Brown. 2000. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 24227-235. [DOI] [PubMed] [Google Scholar]
- 33.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72481-516. [DOI] [PubMed] [Google Scholar]
- 34.Ruthenburg, A. J., C. D. Allis, and J. Wysocka. 2007. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 2515-30. [DOI] [PubMed] [Google Scholar]
- 35.Schübeler, D., D. M. MacAlpine, D. Scalzo, C. Wirbelauer, C. Kooperberg, F. van Leeuwen, D. E. Gottschling, L. P. O'Neill, B. M. Turner, J. Delrow, S. P. Bell, and M. Groudine. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 181263-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shahbazian, M. D., K. Zhang, and M. Grunstein. 2005. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell 19271-277. [DOI] [PubMed] [Google Scholar]
- 37.Shanower, G. A., M. Muller, J. L. Blanton, V. Honti, H. Gyurkovics, and P. Schedl. 2005. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics 169173-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shilatifard, A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75243-269. [DOI] [PubMed] [Google Scholar]
- 39.Sims, R. J., III, C. F. Chen, H. Santos-Rosa, T. Kouzarides, S. S. Patel, and D. Reinberg. 2005. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 28041789-41792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sims, R. J., III, S. Millhouse, C. F. Chen, B. A. Lewis, H. Erdjument-Bromage, P. Tempst, J. L. Manley, and D. Reinberg. 2007. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 28665-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson, C. Goggin, M. Mahowald, and D. E. Gottschling. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150613-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stanford, W. L., J. B. Cohn, and S. P. Cordes. 2001. Gene-trap mutagenesis: past, present and beyond. Nat. Rev. Genet. 2756-768. [DOI] [PubMed] [Google Scholar]
- 43.Teboul, L., M. Febbraio, D. Gaillard, E. Z. Amri, R. Silverstein, and P. A. Grimaldi. 2001. Structural and functional characterization of the mouse fatty acid translocase promoter: activation during adipose differentiation. Biochem. J. 360305-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vakoc, C. R., S. A. Mandat, B. A. Olenchock, and G. A. Blobel. 2005. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell 19381-391. [DOI] [PubMed] [Google Scholar]
- 45.Vakoc, C. R., M. M. Sachdeva, H. Wang, and G. A. Blobel. 2006. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell. Biol. 269185-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Leeuwen, F., P. R. Gafken, and D. E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109745-756. [DOI] [PubMed] [Google Scholar]
- 47.Vermeulen, M., K. W. Mulder, S. Denissov, W. W. Pijnappel, F. M. van Schaik, R. A. Varier, M. P. Baltissen, H. G. Stunnenberg, M. Mann, and H. T. Timmers. 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 13158-69. [DOI] [PubMed] [Google Scholar]
- 48.Weiss, M. J., C. Yu, and S. H. Orkin. 1997. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell. Biol. 171642-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welch, J. J., J. A. Watts, C. R. Vakoc, Y. Yao, H. Wang, R. C. Hardison, G. A. Blobel, L. A. Chodosh, and M. J. Weiss. 2004. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 1043136-3147. [DOI] [PubMed] [Google Scholar]
- 50.Wysocka, J., T. Swigut, H. Xiao, T. A. Milne, S. Y. Kwon, J. Landry, M. Kauer, A. J. Tackett, B. T. Chait, P. Badenhorst, C. Wu, and C. D. Allis. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 44286-90. [DOI] [PubMed] [Google Scholar]
- 51.Wysocki, R., A. Javaheri, S. Allard, F. Sha, J. Cote, and S. J. Kron. 2005. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell. Biol. 258430-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng, P. Y., C. R. Vakoc, Z. C. Chen, G. A. Blobel, and S. L. Berger. 2006. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. BioTechniques 41694, 696, 698. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, K., J. S. Siino, P. R. Jones, P. M. Yau, and E. M. Bradbury. 2004. A mass spectrometric “Western blot” to evaluate the correlations between histone methylation and histone acetylation. Proteomics 43765-3775. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, W., X. Xia, D. I. Jalal, T. Kuncewicz, W. Xu, G. D. Lesage, and B. C. Kone. 2006. Aldosterone-sensitive repression of ENaCα transcription by a histone H3 lysine-79 methyltransferase. Am. J. Physiol. Cell Physiol. 290C936-C946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu, B., S. S. Mandal, A. D. Pham, Y. Zheng, H. Erdjument-Bromage, S. K. Batra, P. Tempst, and D. Reinberg. 2005. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 191668-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, B., Y. Zheng, A. D. Pham, S. S. Mandal, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2005. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell 20601-611. [DOI] [PubMed] [Google Scholar]