Abstract
The transcription factor Adr1 activates numerous genes in nonfermentable carbon source metabolism. An unknown mechanism prevents Adr1 from stably binding to the promoters of these genes in glucose-grown cells. Glucose depletion leads to Snf1-dependent binding. Chromatin immunoprecipitation showed that the Adr1 DNA-binding domain could not be detected at the ADH2 promoter under conditions in which the binding of the full-length protein occurred. This suggested that an activation domain is required for stable binding, and coactivators may stabilize the interaction with the promoter. Artificial recruitment of Mediator tail subunits by fusion to the Adr1 DNA-binding domain overcame both the inhibition of promoter binding and glucose repression of ADH2 expression. In contrast, an Adr1 DNA-binding domain-Tbp fusion did not overcome glucose repression, although it was an efficient activator of ADH2 expression under derepressing conditions. When Mediator was artificially recruited, ADH2 expression was independent of SNF1, SAGA, and Swi/Snf, whereas ADH2 expression was dependent on these factors with wild-type Adr1. These results suggest that in the presence of glucose, the ADH2 promoter is accessible to Adr1 but that other interactions that occur when glucose is depleted do not take place. Artificial recruitment of Mediator appears to overcome this requirement and to allow stable binding and transcription under normally inhibitory conditions.
The glucose-repressed genes of Saccharomyces cerevisiae are excellent models for studying regulated promoters (8, 39). Glucose depletion can increase transcription of these genes several hundredfold, and much is known about the activators and binding sites involved. For example, the ADH2 (alcohol dehydrogenase) gene is regulated by the zinc finger activator Adr1, which binds to the 22-bp, palindromic UAS1 (2, 5, 22, 41, 50), and Cat8, a zinc knuckle transcription factor that binds to UAS2, a carbon source response element adjacent to UAS1 (20, 24, 25, 39, 47, 59). Adr1 and Cat8 directly activate numerous other genes in nonfermentative metabolism (24, 47). Unlike Cat8, whose levels are low in glucose-repressed cells, Adr1 is present in the nucleus under these conditions (5, 42). Although the UAS1 sequence is in a nucleosome-free region (51), Adr1 appears to be regulated at the level of promoter binding, since chromatin immunoprecipitation (ChIP) assays fail to detect binding under repressing conditions (47, 58). When glucose is depleted, Adr1 binds to its cognate promoters in a Snf1-dependent fashion (58). If Snf1 is activated in the presence of glucose by inactivating Reg1, the regulatory subunit of the PP1-type protein phosphatase, Adr1 binding and transactivation can be detected at a low level (18, 58).
In the presence of glucose, Adr1 appears to be competent to bind DNA. Adr1 purified from repressed cells binds the UAS1 sequence in vitro (49). A recombinant “mini-Adr1” with the DNA-binding domain (DBD) and one of its four activation domains forms preinitiation complexes on an immobilized DNA template and activates transcription using nuclear extracts from either repressed or derepressed cultures (58). Altering chromatin structure in vivo by deleting histone H3 N-terminal tails or histone deacetylase genes leads to promoter binding in the presence of glucose by Adr1 (46, 52). Overexpression of ADR1 from a strong promoter, high-copy-number plasmid, or multiple integrated copies of the gene leads to a weak constitutive expression of ADH2, which suggests that mass action can force DNA binding of Adr1 at sufficiently high concentrations (6, 15, 19, 27).
The mechanism that permits Adr1 promoter binding is unknown. Three possibilities are low-glucose-induced changes in chromatin structure, posttranslational modification of Adr1, or stabilizing interaction with coactivators. The chromatin structure of Adr1-dependent promoters undergoes a dramatic change in derepression (1, 51), but since these large changes require ADR1, they are presumed to occur after Adr1 interacts with the promoter. The region around the phosphorylated Ser230 of Adr1 appears to have an inhibitory influence on Adr1 activity (14, 16), but this is not part of the DBD, and an S230A mutation that enhances ADH2 expression does not affect DNA binding assayed in vitro (49). The third possibility, not mutually exclusive of the others, is stabilization by coactivators that are recruited to the promoter upon derepression. Tanaka (48) found that activation domains influence transcription factor DNA binding, with increased numbers of activation domains corresponding to an increase in stable binding, suggesting the possibility that interactions with the recruited initiation complex contribute to binding.
To study the requirements for stable binding by Adr1, we analyzed Adr1 binding by ChIP and assessed ADH2 expression by quantitative real-time PCR (qRT-PCR). To test the hypothesis that Adr1 binding is stabilized through coactivator interaction, we fused the Adr1 DBD to coactivator subunits and tested for the binding and activation of ADH2. This approach, known as activator bypass or artificial recruitment, has been used to study coactivator functions at the CUP1 (38) and GAL (34) promoters and, more generally, to test the recruitment model of preinitiation complex formation (36). In some cases, recruiting Tbp, Mediator, Tafs, SAGA, or even Snf1 to a promoter is sufficient to transcribe a reporter in the absence of an activation domain (9-11, 21, 26, 33, 37, 56). Several features of Adr1 make it attractive for this analysis. Its domains and recognized promoters are extensively characterized, so instead of an engineered reporter system, we can use the chromosomal loci of activated genes to assay binding and gene expression. The DBD alone is transcriptionally inactive even when expressed from the strong ADH1 promoter on a multicopy plasmid (31). When fused to the VP16 herpesvirus transcription activation domain, the DBD confers regulated expression upon a UAS1-containing reporter gene (42). UAS1 is nucleosome free in several Adr1-dependent promoters, including ADH2 (1, 51), providing access to the chromatin. In addition, ADH2 expression is Snf1 dependent like many glucose-repressed genes but is not repressed by Mig1 or other DNA-bound repressors (18, 31, 39), which makes its activation by Adr1 easier to study.
We found that the Adr1 DBD alone did not stably bind to the ADH2 promoter, but when fused to a tail subunit of Mediator, it bound to the ADH2 promoter even under repressing conditions. Moreover, the Adr1 DBD-Mediator fusion protein was able to activate the transcription of ADH2 in the absence of Snf1 or subunits of SAGA or Swi/Snf. In contrast, ADH2 expression activated by the artificial recruitment of Tbp was strongly glucose repressed. Thus, Mediator recruitment to the ADH2 promoter may play an important role in overcoming glucose repression.
MATERIALS AND METHODS
Strains and primers.
The Saccharomyces cerevisiae strains used are shown in Table 1. TYY309 and TYY317 are based on PJ69-4a (28). Epitope tags were introduced by the method of Knop et al. (32). Sequences of the oligonucleotides are available upon request. Yeast strains were grown as described previously (40). Repressing medium contained 5% glucose; derepressing medium contained 0.05% glucose.
TABLE 1.
Yeast strains
| Strain | Relevant characteristics | Reference or source |
|---|---|---|
| TYY201 (W303-1a) | MATaade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 57 |
| TYY202 | TYY201 adr1Δ1::LEU2 | 57 |
| TYY203 | TYY201 ADH2::YIpADH2/lacZ | 57 |
| TYY204 | TYY202 ADH2::YIpADH2/lacZ | 57 |
| TYY309 | PJ69-4a (MATatrp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ) Δadr1Δ1::LEU2 | This study |
| TYY317 | TYY309 gcn5Δ | This study |
| TYY497 | TYY204 (W303-1a) adr1Δ1::LEU2 ADH2::YIpADH2/lacZ(trp1::HIS3) | This study |
| TYY498 | TYY497 snf1Δ::URA3 | This study |
| TYY540 | TYY497 med15Δ::NAT1 | This study |
| TYY541 | TYY497 med2Δ::kanMX | This study |
| TYY542 | TYY497 med3Δ::kanMX | This study |
| TYY543 | TYY497 med16(sin4)Δ::kanMX | This study |
| TYY804 | TYY497 med17(srb4)Δ::NAT1 plus YCpsrb4-138(leu2::URA3) | This study |
| ECY1 | MATα his3-Δ200 leu2-3,112 trp1 ura3-52 YIp-ADH2 promoter-lacZ::URA3 Δadr1::NAT1 | This study |
| ECY4 | ECY1 with SNF1-ADR1(1-172 DBD)-myc::HIS3 | This study |
| ECY5 | ECY1 with MED15-ADR1(1-172 DBD)-myc::HIS3 | This study |
| ECY6 | ECY1 with GCN5-ADR1(1-172 DBD)-myc::HIS3 | This study |
| ECY10 | ECY1 with MED3-ADR1(1-172 DBD)-myc::HIS3 | This study |
| ECY11 | ECY1 with MED4-ADR1(1-172 DBD)-myc::HIS3 | This study |
| ECY12 | ECY1 with MED18-ADR1(1-172 DBD)-myc::HIS3 | This study |
| ECY13 | ECY10 with MED14-HA::kanMX | This study |
| ECY14 | ECY11 with MED15-HA::kanMX | This study |
| ECY15 | ECY12 with MED14-HA::kanMX | This study |
| ECY16 | ECY12 with MED14-HA::kanMX | This study |
| CTYTY61 | MATα his3-Δ200 leu2-3,112 trp1 ura3-52 YIp-ADH2 promoter-lacZ::URA3 | This study |
| CTYTY66 | MATα his3-Δ200 leu2-3,112 trp1 ura3-52 YIp-ADH2 promoter-lacZ::TRP1 Δsnf1::URA3 | This study |
| CTYTY75 | ECY10 with Δsnf1::kanMX | This study |
ChIP and real-time qPCR.
ChIP and gene-specific PCR with gel electrophoresis were performed as described previously (47). Real-time qPCR data from the ChIP experiments were generated with an MJResearch Chromo4 system, using ABI SYBRMaster mix. Data were analyzed using the method of Steger et al. (44) or of Bryant and Ptashne (7). Briefly, the amounts of DNA in the ChIP and total DNA samples were quantified relative to a standard curve for an Adr1-bound promoter and for a nonbound telomeric control region. The ratio of the DNAs was determined using the formula (ChIP DNAbound promoter/total DNAbound promoter)/ (ChIP DNAtelomeric control/total DNAtelomeric control). The data are presented as the ratios of specific to nonspecific binding, expressed as percentages or increases over background measured with primers to the telomeric control.
RNA isolation from 10 to 20 ml of cells was performed by acid phenol extraction at 65°C for 1 h (13). Residual DNA in the RNA preparation was reduced by treatment with DNase (Ambion) by following the manufacturer's recommendations. cDNA synthesis was performed with SuperscriptIII (Invitrogen) by following the manufacturer's protocol. qRT-PCR for measuring mRNA levels was performed as described above, in duplicate, using a 1:300 dilution of the cDNA. A standard curve was generated from ACT1 primers and used to quantitate all of the RNA levels.
Immunoprecipitations and Western blots.
All antibodies were obtained from Santa Cruz Biochemicals (Santa Cruz, CA). Immunoprecipitations to concentrate samples for Western blots and coimmunoprecipitations were carried out as described by Strahl-Bolsinger et al. (45), without DNase I treatment and using 2 μg monoclonal antihemagglutinin (anti-HA) (F-7) or 6 μg monoclonal anti-myc (9E10). Western blot analyses were performed according to the manufacturer's instructions for the Odyssey infrared imaging system (Licor Biosciences, Lincoln, NE), using a dilution of 1:500 to 1:1,000 of polyclonal anti-HA (Y-11) or monoclonal anti-myc (9E10) as the primary antibody.
Artificial recruitment strains.
Plasmids encoding the 172 N-terminal amino acids of Adr1 fused to Med15 (Gal11) or Tbp were constructed by generating PCR fragments of the 284 C-terminal amino acids of Med 15 or the entire open reading frame (ORF) product of TBP (minus the first three amino acids). The primers generated PstI restriction sites for cloning into a pRS314-based plasmid containing a portion of the ADR1 gene with a His6 tag. Digestion with PstI and ligation of the PCR fragment allowed the insertion of the C terminus of Med15 or the Tbp ORF product at Adr1 amino acid 172.
Mediator-Adr1 protein fusion strains were created by integrating the portion of ADR1 that encodes the DBD (amino acids 1 to 172), in frame, to the 3′ end of candidate genes, using a PCR-based epitope-tagging method (32). The integrating fragments also added a 3-myc tag and the HIS3 marker. They were generated by PCR using the plasmid pEC2 as a template. To generate pEC2, a PCR fragment was made using the Roche Expand PCR kit with primers CTO ADR1 1-172 S2 and CTO ADR1 1-172 S3 and plasmid pYM4 (32) as a template. Yeast strain BY4741 was cotransformed with the resulting PCR fragment and the ADR1-containing plasmid pKD16 (19). In vivo recombination between pKD16 and the ADR1 1-172 PCR fragment truncated the ADR1 ORF in pKD16 with a myc tag and kanMX6 marker. The kanMX6 marker was switched to HIS3 (53) to generate pEC2. When used as the template in a PCR with primers that had 40 to 60 homologous nucleotides on either side of the stop codon of a target gene, a fragment was generated that would add ADR1 encoding amino acids 1 to 172 and a 3-myc tag, all marked with HIS3, to the 3′ end of a target gene. Mediator-ADR1 fusions were confirmed by colony PCR and Western blot analyses.
β-Galactosidase assays.
β-Galactosidase assays (23) were performed using three cultures or transformants.
RESULTS
Adr1-DBD is not stably bound to the ADH2 promoter.
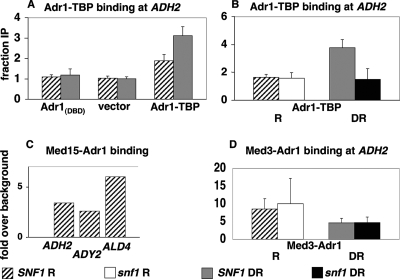
By several criteria, Adr1 has access to its binding site in the ADH2 promoter even under repressing conditions. We hypothesized that one reason Adr1-promoter binding cannot be detected under these conditions is because stable binding requires the recruitment of coactivators. To test this hypothesis, we assayed for the binding of the Adr1 DBD to the ADH2 promoter. The Adr1 DBD alone cannot activate transcription but can effect a slight remodeling of chromatin (17). To measure promoter binding directly, ChIP analysis was performed with the epitope-tagged Adr1 DBD. No occupation of the ADH2 promoter could be detected (Fig. 1A). The same low levels of promoter DNA were detected in the ChIP samples by qPCR when samples from either repressing or derepressing conditions were assayed, levels that were comparable to the level of nonspecific binding to a telomeric control sequence. Since full-length Adr1 exhibits regulated binding and activation (58), and since fusion to an activation domain, either VP16 or TADIII of Adr1, allows the regulated transcription of ADH2 (42), adding an activation domain to the Adr1 DBD is sufficient for stable binding and transcriptional activation.
FIG. 1.
Binding of Adr1 DBD fusions to Adr1-dependent promoters. ChIP was performed and the results were quantitated as described in Materials and Methods with strains with the indicated Adr1 DBD fusions or controls. (A) ChIP for the Adr1 DBD only (plasmid pAdr1Δ172 [17]), negative control vector (pKD8 [17]), or Adr1-TBP (Materials and Methods), all in ECY1 and grown under repressing conditions (hatched bars) or after 4 h of derepression (gray bars). Data are expressed as the ratios described in Materials and Methods. (B) ChIP of Adr1-TBP in a SNF1 strain (TYY497), repressed (R) (hatched bar) or 4-h derepressed (DR) (gray bar), and a snf1Δ strain (TYY498), repressed (white bar) or 4-h derepressed (black bar). Data are expressed as described for panel A. (C) ChIP of Med15-Adr1 under repressing conditions using strain ECY5. Data are expressed as increases over background from an untagged, repressed adr1Δ strain (ECY1). (D) ChIP of Med3-Adr1 in repressed SNF1 (ECY10) (hatched bar) and snf1Δ (CTYTY75) (white bar) strains and in the same strains after 4 h of derepression (gray and black bars, respectively). Data are expressed as described for panel C. Error bars in panels A, B, and D indicate standard errors of the means for two or three biological replicates.
Artificial recruitment of Mediator by Adr1 relieves glucose repression of ADH2.
Since the primary known role of an activation domain is to bring coactivators to the promoter (36), we tested the effects of artificial recruitment of coactivators by fusing them directly to the Adr1 DBD. Fusion of the Adr1 DBD to Med15 (Gal11) (see the review by Biddick and Young [3] for nomenclature and the arrangement of subunits) and Med3 (Pgd1), two subunits of the tail module of Mediator, created fusion proteins that activated ADH2 transcription in the presence and absence of glucose. Table 2 shows the activation of an ADH2-lacZ reporter gene by various fusion proteins and by wild-type Adr1. The first Adr1 DBD fusion protein tested, Adr1-Med15, contained the 280 C-terminal amino acids of Med15 fused to the C terminus of the Adr1 DBD. This fusion protein was 50 times more active under repressing conditions than wild-type Adr1, and its activation increased a further fivefold under derepressing conditions, reaching the same high level of activity as that promoted by wild-type Adr1 (Table 2). When the entire Med15 ORF product was fused in frame to the N terminus of the Adr1 DBD by an integrative targeting method, the fusion protein (designated Med15-Adr1 to distinguish it from the other Med15 fusion protein) was also active under repressing conditions (Table 2).
TABLE 2.
Activation of an integrated ADH2 promoter-lacZ reporter by coactivator-Adr1 DBD fusions
| Straina | Activator (on a plasmid or integrated)b | β-Gal activityc
|
|
|---|---|---|---|
| Repressed | Derepressed | ||
| TYY204 | None | 5.4 (2) | 55 (9) |
| Wild-type Adr1 | 5 (2) | 810 (100) | |
| Adr1-Med15 (Gal11) | 260 (90) | 1,400 (100) | |
| ECY10 | Med3 (Pgd1)-Adr1 | 1,000 (87) | 980 (120) |
| ECY11 | Med4-Adr1 | 18 (1.8) | 56 (6.1) |
| ECY12 | Med18 (Srb5)-Adr1 | 90 (1.8) | 150 (8.8) |
| ECY5 | Med15-Adr1 | 160 (31) | 1,600 (540) |
| ECY4 | Snf1-Adr1 | 14 (6.2) | 68 (13) |
| ECY6 | Gcn5-Adr1 | 44 (5.1) | 110 (19) |
| TYY497 | None | 5 (1) | 25 (5) |
| ADR1-TBP | 5 (2) | 250 (4) | |
| Wild type | 2 (1) | 230 (10) | |
Strains are described in Table 1. All strains are adriΔ.
Activators for strains TYY204 and TYY497 were on plasmids, and the activators for ECY strains were integrated at the coactivator locus as described in Materials and Methods. Adr1-Med15 (Gal11) is the Adr1 DBD fused to the 280 C-terminal amino acids of Gal11 as described in Materials and Methods. It is carried on a TRP1-CEN3-ARS1 plasmid. Adr1 DBD-Tbp fusion contains amino acids 1 to 172 of Adr1 fused at the C terminus to the entire SPT15 ORF, as described in Materials and Methods, and is carried on a TRP1-CEN3-ARS1 plasmid.
β-Galactosidase activity is expressed in Miller units with standard deviations in parentheses. Derepression, 4 h in 0.05% glucose.
Fusion of the entire ORF product of another Mediator tail component, Med3, to the N terminus of the Adr1 DBD created a fusion protein that was even more active under repressing conditions than that created by the fusion of Med15 to the Adr 1 DBD (Table 2), and its activity was comparable to that of wild-type Adr1 under derepressing conditions. The high constitutive activation is not the result of overexpression of the fusion protein genes. Expression of the MED3-ADR1 gene was 3.5-fold lower than that of the ADR1 gene, as determined by qPCR of mRNA. Expression of ADR1-MED15 was 2.3-fold higher than that of ADR1, yet this fusion is a weaker activator than Med3-Adr1. Also, ADR1-MED15 was expressed from the same promoter on the same plasmid as an ADR1-TBP ORF fusion, whose phenotype was very different (see below), even though its expression level should have been comparable.
Fusion of the Adr1 DBD to the C terminus of two other Mediator components, Med4 and Med18, subunits of the middle and head modules of Mediator, respectively, produced fusion proteins that were less active under both repressing and derepressing growth conditions than fusions to the tail subunits of Mediator (Table 2). Fusion of the entire ORF product of GCN5, a component of the coactivator SAGA, and the entire ORF product of SNF1 to the Adr1 DBD produced fusion proteins that were weakly active, as assayed by reporter gene expression (Table 2). Wishing to concentrate on the strongest phenotypes, we used the Med3 and Med15 fusions for further expression analyses.
Fusion of the entire ORF product of TBP to the Adr1 DBD produced a fusion protein that was comparable in its regulation and activity to wild-type Adr1 (Table 2), activating ADH2-lacZ expression only in the absence of glucose. Thus, Tbp behaved like a typical activation domain when fused to the Adr1 DBD: it activated expression, and the activation of ADH2-lacZ expression was strongly glucose repressed.
To analyze the expression of Adr1-dependent genes from endogenous chromosomal loci, the activity of the Mediator and Tbp fusions to the Adr1 DBD was confirmed by qPCR analysis of ADH2 mRNA. Consistent with the reporter assay data in Table 2, Adr1-Med15 activated ADH2 under repressing conditions to nearly the wild-type derepressed level ( Table 3). Fusion to the entire ORF product of the Mediator tail component MED3 produced the most active fusion protein under both repressing and derepressing conditions (Table 3). Adr1-TBP stimulated a small amount of expression of the endogenous ADH2 locus under repressing conditions, and activation under derepressing conditions was comparable to that of wild-type Adr1 (Table 3). In summary, while fusion of the Adr1 DBD to Gcn5, Tbp, or Snf1 produced weak constitutive activators, fusion to Mediator tail subunits could completely overcome the glucose repression of ADH2 expression.
TABLE 3.
Adr1-coactivator activation of ADH2 expression measured by real-time qRT-PCR
| Strain | Activator |
ADH2 mRNA level/ACT1 mRNA levela
|
|
|---|---|---|---|
| Repressedb | Derepressedc | ||
| TYY497 | None | 0.025 (0.0015) | NAd |
| Wild-type Adr1 | 0.012 (0.01) | 1.45 (0.08) | |
| Adr1-Med15 | 0.92 (0.09) | NA | |
| CTYTY61 | Wild-type Adr1 | NA | 1.3 |
| ECY10 | Med3-Adr1 | 1.9 | 15.5 |
| ECY1 | None | 0.002 | 0.012 |
| Adr1-TBP | 0.030 | 2.0 | |
Standard deviations, when measured, are given in parentheses.
5% glucose.
For 4 to 6 h (0.05% glucose).
NA, not assayed.
Artificial recruitment of Mediator by Adr1 causes constitutive DNA binding.
ChIP assays were performed to see if the fusion proteins were affecting Adr1-dependent genes indirectly or directly. Med15-Adr1, Med3-Adr1, and Adr1-Med15 were detected at the ADH2, ADY2, and ALD4 promoters under repressing conditions, suggesting that they activate ADH2 expression directly (Fig. 1 and data not shown). In contrast, the Adr1 DBD-TBP fusion, which activated a low level of ADH2 expression under repressing conditions (Table 2), showed only slightly higher than background levels of repressed ADH2 binding, and binding increased approximately twofold under derepressing conditions (Fig. 1A and B). The low level of Adr1-TBP binding and the lack of binding seen for the Adr1 DBD only (Fig. 1A) indicate that constitutive binding and expression are neither general phenomena of all Adr1 DBD fusions nor properties of the Adr1 DBD itself, when liberated from its transactivation domains.
Adr1-Mediator fusions incorporate into Mediator complexes.
Mutations in some Mediator components allow activator-independent gene expression (29, 54). Thus, an aberrant form of Mediator might act at ADH2 outside the context of the normal Mediator complex. We used two assays to test for the possibility of anomalous Mediator activation. First, an ADH2-lacZ reporter gene was assayed in strains carrying an Adr1-Med15 plasmid and deleted for each one of the Mediator tail subunit genes. Expression was reduced under both repressing and derepressing conditions when MED2, MED3, and MED16 were deleted but not when MED15 was deleted, presumably because the fusion protein could functionally replace wild-type Med15 (Table 4). The requirement for other tail subunits suggests that Adr1-Med15 functions within the context of an intact Mediator tail.
TABLE 4.
Adr1-Med15 activation depends on the Mediator tail
| Strain | Activator | Mediator mutation | β-Gal activitya
|
ADH2 mRNA level/ACT1 mRNA levelb
|
||
|---|---|---|---|---|---|---|
| Repressed conditions | Derepressed conditions | Repressed conditions | Derepressed conditions | |||
| TYY204 | Adr1-Med15 | None (WT) | 260 (100) | 1,400 (100) | ||
| TYY540 | Adr1-Med15 | med15 (gal11)Δ | 400 (150) | 1,200 (86) | ||
| TYY541 | Adr1-Med15 | med2Δ | 36 (14) | 250 (18) | ||
| TYY542 | Adr1-Med15 | med3 (pgd1)Δ | 18 (6.9) | 410 (29) | ||
| TYY543 | Adr1-Med15 | med16 (sin4)Δ | 120 (46) | 210 (15) | ||
| TYY309 | Adr1 | None (WT) | 0.002 | 7.1 | ||
| TYY309 | Adr1-Med15 | None (WT) | 0.92 | 13 | ||
| TYY317 | Adr1-Med15 | gcn5Δ | 0.90 | 1.5 | ||
Values are expressed as Miller units, with percentages of β-galactosidase activity of the wild-type Mediator shown in parentheses. The standard deviation of triplicate assays was about 20%. Derepression, overnight in 0.05% glucose.
Derepression, 14 h in 0.05% glucose.
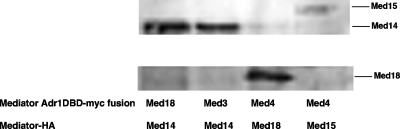
Second, strains were constructed in which both a Mediator component and a Mediator-Adr1 DBD fusion were epitope tagged. Coimmunoprecipitations were performed to assay for in vivo interactions. Figure 2 shows that Med14 (Rgr1) coimmunoprecipitated with Med18-Adr1 and Med3-Adr1 and that Med18 and Med15 coimmunoprecipitated with Med4-Adr1. Interaction between the fusions and subunits from different Mediator modules (30) suggested that the Mediator-Adr1 DBD fusions could be incorporated into Mediator complexes.
FIG. 2.
Mediator subunits coimmunoprecipitate with Mediator-Adr1 DBD fusion proteins. Coimmunoprecipitation was performed with strains with Myc-tagged Mediator-Adr1 DBD fusions and HA-tagged Mediator components (ECY13, ECY14, ECY15, and ECY16). Immunoprecipitations with anti-Myc monoclonal antibodies were Western blotted with anti-HA polyclonal antibodies as described in Materials and Methods.
Artificial recruitment of Mediator overcomes the requirement for SAGA and Swi/Snf at the ADH2 promoter.
Most promoters require several coactivators for efficient transcription. ADH2 expression, for example, requires Mediator, SAGA, NuA4, and Swi/Snf for the efficient recruitment of polymerase II, chromatin remodeling, and transcription (4, 12, 52). To assess the role of SAGA in ADH2 expression when it is activated by artificial recruitment of Mediator, qRT-PCR analysis was used to measure transcript levels in the absence of the histone deacetylase component, Gcn5, of SAGA. As shown in Table 4, Adr1-Med15 activated ADH2 expression to a high level in the absence of Gcn5, whereas wild-type Adr1 has a strong dependence on this coactivator subunit (12). ADH2 activation by Adr1-Med15 was also uncompromised by the deletion of another component of SAGA (ADA1) or of an essential subunit of Swi/Snf (SNF2; data not shown). This suggested that direct recruitment of Mediator could overcome the requirement for additional coactivators and that coactivators may be redundant with regard to Adr1 stabilization.
Adr1 can bind in the absence of individual subunits of Mediator.
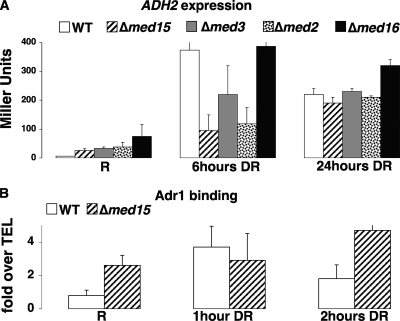
To test the hypothesis that Adr1 binding can be stabilized by any of several coactivators, we tested for the binding of wild-type Adr1 and the activation of ADH2 in Med15 and other Mediator mutants. When MED15, MED3 (PGD1), or MED2 was deleted from a strain with wild-type Adr1, ADH2 derepression was significantly slowed, although not abolished. Deletion of MED16 (SIN4) did not reduce ADH2 expression. In each of the mutants, there was a low level of constitutive ADH2 expression detected that was Adr1 dependent (Fig. 3A and data not shown). Since the deletion of MED15 had the strongest effect on ADH2 expression, the binding of Adr1 in the strain with this deletion was measured by ChIP analysis. There was no effect on Adr1 binding in the absence of MED15, and significant binding was detected under repressing conditions (Fig. 3B). Thus, the absence of Med15 did not significantly reduce Adr1 binding, although the early kinetics of expression could be significantly affected.
FIG. 3.
Adr1 binding and activation in Mediator mutants. (A) Beta-galactosidase assays of strains TYY497 (wild type [WT]), TYY540 (med15Δ), TYY541 (med2Δ), TYY542 (med3Δ), and TYY543 (med16Δ), all with CEN-TRP1 plasmid pKD16, which contains the ADR1 gene with a C-terminal HA tag. Cultures were assayed under repressing conditions (R) or after 6 or 24 h of derepression (DR). (B) ChIP for Adr1 in the wild-type and med15Δ strains used for panel A under repressing conditions (R) or after 1 or 2 h of derepression (DR). Data are expressed as increases over background from an unbound telomeric (TEL) region. Error bars show standard deviations from triplicate measurements of each sample.
Transcription by artificial recruitment of Mediator does not require Snf1.
Since Snf1 is normally required for promoter binding by Adr1 (58), we determined whether DNA binding and transcriptional activation by Mediator-Adr1 DBD fusions also require Snf1. Snf1 is inactive in in vitro kinase assays when isolated from glucose-grown cells (55), so it seemed likely that the expression caused by Adr1-Mediator fusions under repressed conditions would be Snf1 independent, even though the expression of most Adr1-dependent genes is Snf1 dependent (57). qPCR of RNA isolated from strains containing either wild-type Adr1 or Med3-Adr1 DBD fusions grown under repressing and derepressing conditions showed that the activation of several Adr1-dependent genes by Med3-Adr1 in the presence of glucose was independent of Snf1 (Table 5). As observed previously for ADH2 (Table 3), the derepression of several other genes was elevated relative to their expression in the presence of wild-type Adr1 when Med3-Adr1 was the activator (compare the values in Table 5 for Med3-Adr1 and wild-type Adr1 for SNF1). The levels of ADH2 and ATO3 expression were comparable in the presence and absence of Snf1, and repressed expression was similar to activation in derepressed SNF1 with wild-type Adr1 as the activator. The constitutive activation of ALD4 and ACS1 by Med3-Adr1 was lower than the activation of the derepressed wild type but still had a high degree of Snf1 independence. Many of the genes (ADH2, ATO3, ALD4, ACS1, the FDH genes, and ADY2) that were strongly Snf1 dependent when wild-type Adr1 was the activator showed enhanced derepression in the presence of Adr1-Med3 in the snf1 mutant ADH2 expression when Adr1-Med15 was the activator was also Snf1 independent (data not shown), indicating that Snf1 independence is not a unique property of the Med3-Adr1 fusion.
TABLE 5.
Snf1-independent activation by artificial recruitment of Mediator
| Gene | Growth conditiona | Relative expression level with activatorb:
|
|||
|---|---|---|---|---|---|
| Med3-Adr1 DBDc
|
Adr1 WT
|
||||
| SNF1 | snf1 | SNF1 | snf1 | ||
| ADH2 | R | 120 | 100 | 0.5 | 0.5 |
| DR | 1,700 | 300 | 126 | 0.4 | |
| ATO3 | R | 41 | 67 | 6.3 | 3.4 |
| DR | 266 | 150 | 50 | 4.5 | |
| ALD4 | R | 4.0 | 3.5 | 0.1 | 0.0 |
| DR | 110 | 33 | 9.7 | 0.2 | |
| ACS1 | R | 20 | 6.0 | 1.1 | 0.9 |
| DR | 950 | 120 | 190 | 3.9 | |
| FDH | R | 0.2 | 0.2 | 0.2 | 0.2 |
| DR | 56 | 4.6 | 2.3 | 0.4 | |
| ADY2 | R | 1.5 | 1.6 | 0.5 | 0.4 |
| DR | 2,900 | 100 | 240 | 0.7 | |
| ICL2 | R | 0.9 | 0.8 | 0.9 | 0.7 |
| DR | 18 | 0.9 | 7.3 | 1.2 | |
| FBP1 | R | 0.2 | 0.2 | 0.2 | 0.1 |
| DR | 17 | 0.3 | 130 | 0.6 | |
| ICL1 | R | 1.5 | 1.7 | 1.1 | 0.8 |
| DR | 4.9 | 1.5 | 87 | 1.4 | |
| MLS1 | R | 0.3 | 0.2 | 0.2 | 0.2 |
| DR | 12 | 0.4 | 87 | 0.4 | |
| MDH2 | R | 4.3 | 3.0 | 1.9 | 1.5 |
| DR | 58 | 12 | 43 | 3.9 | |
R, repression (5% glucose); DR, derepression for 6 h (0.05% glucose).
mRNA levels were calculated by qRT-PCR as described in Materials and Methods. The quantities determined for each primer pair were divided by the quantity determined for the control ACT1. WT, wild type.
Strains were ECY10, CTYTY75, CTYTY61, and CTYTY66.
The Snf1-independent activation of Adr1-dependent genes suggested that the Mediator-Adr1 fusion might be causing a transcription enhancement of all glucose-repressed genes. To test this possibility, the transcript levels of several Snf1- and Cat8-dependent genes (FBP1, MLS1, ICL1, and MDH2) were measured. These genes were expected to be relatively unaffected by Med3-Adr1, since Adr1 makes a minor contribution to their derepression (57). The data in Table 5 show that under repressing conditions, Med3-Adr1 had no effect on the expression of FBP1, ICL1, and MLS1 and activated MDH2 about twofold. As expected, the derepression of these genes was strongly Snf1 dependent. Only MDH2 derepression had a significant SNF1-independent component when Med3-Adr1 was present, suggesting that Med3-Adr1 can activate MDH2 expression in a Snf1-independent manner. Med3-Adr1 reduced derepression of FBP1, ICL1, and MLS1 about eightfold (Table 5), and an array analysis of gene expression in cells with Adr1-Med15 revealed both activating and repressing effects on some Cat8-dependent genes (unpublished data). These results are consistent with previous reports that Med15 can function in both repression and activation (35). With regard to the overall effects of the Adr1-Mediator fusions on glucose-repressed genes, the data indicated that the fusions were not acting as a nonspecific activator. They could, however, bypass the SNF1 requirement for the activation of some Adr1-dependent genes.
If Adr1-Mediator fusions are able to overcome glucose repression in a Snf1-independent manner, promoter occupancy should be independent of Snf1. In agreement with this interpretation, quantitative ChIP showed that while the Adr1-TBP fusion was SNF1 dependent like wild-type Adr1, Med3-Adr1 fusions occupied the ADH2 promoter in SNF1 wild-type and snf1 deletion strains under repressing and derepressing conditions (Fig. 1B and D).
DISCUSSION
The binding site for the Zn finger activator Adr1 is in a nucleosome-free region at several Adr1-regulated promoters, yet binding is not detected by ChIP under glucose-repressing conditions. Nonetheless, Adr1 appears to be competent to bind DNA, even when the glucose level is high. We tested binding by the DBD by ChIP analysis and were unable to detect promoter occupancy, suggesting that an activation domain is needed for stable promoter binding. We hypothesized that in the presence of glucose, Adr1 can bind its cognate promoters weakly and transiently but that its binding is not sufficiently stable to be detected by ChIP. Since Adr1 activation domains have been found to require SAGA components to be able to function and to interact with them in vitro (12), one possible stabilizing factor could be interaction with coactivator complexes that are recruited under derepressing conditions. We tested this possibility using an artificial recruitment assay. We fused the Adr1 DBD to coactivator subunits and found that Mediator tail fusions bound and activated several Adr1-dependent genes in the presence of the repressing carbon source glucose. Fusion to the Mediator head and middle components yielded weaker activators, possibly because of incorrect orientation or steric hindrance when the Adr1 DBD was fused to this region of Mediator. Nonetheless, since the Mediator head and middle component fusions and the Gcn5 and Snf1 fusions were able to activate, albeit weakly, the stabilizing effect may extend to the rest of Mediator and possibly to other coactivators, any one of which can facilitate Adr1 binding when recruited to the promoter.
Fusion of the Adr1 DBD to Mediator creates a different kind of activator than the fusion of the Adr1 DBD to Tbp. Adr1-Mediator fusions are able to overcome the repressive mechanism at the ADH2 promoter and bind to the promoter, subsequently activating transcription. Adr1-Tbp, on the other hand, is still subject to glucose repression. The ability of Adr1-Tbp to activate the ADH2 promoter is surprising in light of a report that artificial recruitment of Tbp could not activate transcription at several promoters at which the TATA element is in a nucleosomal location (37). The ADH2 promoter requires extensive chromatin remodeling that is Adr1 dependent, and one of the remodeled nucleosomes contains the TATA element (51). Nonetheless, Adr1-Tbp acts like a classical activator at the ADH2 promoter. One possibility is that Adr1-Mediator binding is accompanied by or immediately recruits chromatin-modifying activities, whereas Adr1-Tbp may be unable to recruit the necessary activities to allow stable binding under repressing growth conditions. Alternatively, Adr1-Mediator could be part of a holoenzyme complex that brings RNA polymerase II to the promoter, regardless of chromatin structure.
Adr1 DBD-Mediator tail fusions were strong constitutive binders and activators. The fusions appeared to associate with the rest of the Mediator complex, supporting the hypothesis that activator binding can be stabilized at the promoter by the recruitment of a functional coactivator. Surprisingly, perturbing Mediator by MED15 deletion did not affect the binding of wild-type Adr1. Since ADH2 activation was noticeably delayed by coactivator deletions, a possible explanation is that Mediator, SAGA, and Swi/Snf all play important roles in ADH2 expression but that individual coactivator subunits do not have important roles in Adr1 binding. Instead, the coactivators might be redundant with regard to Adr1 binding. In fact, we have found that individual subunits of SAGA and Swi/Snf can also be deleted without strongly affecting Adr1 binding (R. Biddick et al., unpublished data). Also consistent with the explanation of coactivator redundancy, we found that the Adr1-Med15 fusion could activate ADH2 in the absence of SAGA or Swi/Snf subunits. Together, these results suggest that strong binding to Mediator can replace contacts with multiple coactivators.
Binding and activation by Adr1 DBD-Mediator tail fusions at several promoters were Snf1 independent in the presence of glucose and showed a reduced requirement for Snf1 in derepression. The mechanism by which Snf1 regulates Adr1 binding is unknown, but the ability of the fusions to bypass the Snf1 requirement suggests that Snf1 may aid coactivator recruitment, either directly or indirectly. In particular, Snf1 might be involved in Mediator recruitment, since there is both genetic and physical evidence for an interaction of Snf1 with Mediator (33, 43, 58). In summary, our model is that the binding of Adr1 to the ADH2 promoter under normal repressing conditions is not detectable, because without the signals for derepression, Adr1 lacks stabilizing interactions with recruited coactivators. Our data with Adr1-Mediator fusions suggest that one factor in the stabilization of an activator and the subsequent initiation complex formation might be the interaction between the activator and Mediator at activated promoters. The fact that at least one Adr1-Mediator fusion can activate ADH2 in mutants of Swi/Snf or SAGA subunits suggests that coactivators may be redundant with regard to the stabilization of factor binding.
Acknowledgments
This work was supported by research grant GM-26079 from the National Institutes of Health to E.T.Y. and NIGMS grant PHS NRSA T32 GM07270 to R.B.
We thank other members of the lab for their support, J. Hopper for the plasmid containing MED15, and S. Hahn for the plasmid containing SPT15 (TBP) and for thoughtful comments on the manuscript.
Footnotes
Published ahead of print on 4 February 2008.
REFERENCES
- 1.Agricola, E., L. Verdone, B. Xella, E. Di Mauro, and M. Caserta. 2004. Common chromatin architecture, common chromatin remodeling, and common transcription kinetics of Adr1-dependent genes in Saccharomyces cerevisiae. Biochemistry 438878-8884. [DOI] [PubMed] [Google Scholar]
- 2.Beier, D. R., A. Sledziewski, and E. T. Young. 1985. Deletion analysis identifies a region, upstream of the ADH2 gene of Saccharomyces cerevisiae, which is required for ADR1-mediated derepression. Mol. Cell. Biol. 51743-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biddick, R., and E. T. Young. 2005. Yeast mediator and its role in transcriptional regulation. C. R. Biol. 328773-782. [DOI] [PubMed] [Google Scholar]
- 4.Biddick, R. K., G. L. Law, and E. T. Young. 2008. Adr1 and Cat8 mediate coactivator recruitment and chromatin remodeling at glucose-regulated genes. PLoS One 3e1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumberg, H., A. Eisen, A. Sledziewski, D. Bader, and E. T. Young. 1987. Two zinc fingers of a yeast regulatory protein shown by genetic evidence to be essential for its function. Nature 328443-445. [DOI] [PubMed] [Google Scholar]
- 6.Blumberg, H., T. A. Hartshorne, and E. T. Young. 1988. Regulation of expression and activity of the yeast transcription factor ADR1. Mol. Cell. Biol. 81868-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryant, G. O., and M. Ptashne. 2003. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell 111301-1309. [DOI] [PubMed] [Google Scholar]
- 8.Carlson, M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2202-207. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee, S., and K. Struhl. 1995. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature 374820-822. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, J. X., M. Gandolfi, and M. Ptashne. 2004. Activation of the Gal1 gene of yeast by pairs of ‘non-classical’ activators. Curr. Biol. 141675-1679. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, J. X., J. Nevado, Z. Lu, and M. Ptashne. 2002. The TBP-inhibitory domain of TAF145 limits the effects of nonclassical transcriptional activators. Curr. Biol. 12934-937. [DOI] [PubMed] [Google Scholar]
- 12.Chiang, Y. C., P. Komarnitsky, D. Chase, and C. L. Denis. 1996. ADR1 activation domains contact the histone acetyltransferase GCN5 and the core transcriptional factor TFIIB. J. Biol. Chem. 27132359-32365. [DOI] [PubMed] [Google Scholar]
- 13.Collart, M. A., and S. Oliviero. 1993. Preparation of yeast RNA, p. 13.12.1-13.12.5. In V. B. Chanda (ed.), Current protocols in molecular biology, vol. 2. John Wiley and Sons, New York, NY. [DOI] [PubMed] [Google Scholar]
- 14.Cook, W. J., D. Chase, D. C. Audino, and C. L. Denis. 1994. Dissection of the ADR1 protein reveals multiple, functionally redundant activation domains interspersed with inhibitory regions: evidence for a repressor binding to the ADR1c region. Mol. Cell. Biol. 14629-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denis, C. 1987. The effects of ADR1 and CCR1 gene dosage on the regulation of the glucose-repressible alcohol dehydrogenase from Saccharomyces cerevisiae. Mol. Gen. Genet. 208101-106. [DOI] [PubMed] [Google Scholar]
- 16.Denis, C. L., S. C. Fontaine, D. Chase, B. E. Kemp, and L. T. Bemis. 1992. ADR1c mutations enhance the ability of ADR1 to activate transcription by a mechanism that is independent of effects on cyclic AMP-dependent protein kinase phosphorylation of Ser-230. Mol. Cell. Biol. 121507-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Mauro, E., S. G. Kendrew, and M. Caserta. 2000. Two distinct nucleosome alterations characterize chromatin remodeling at the Saccharomyces cerevisiae ADH2 promoter. J. Biol. Chem. 2757612-7618. [DOI] [PubMed] [Google Scholar]
- 18.Dombek, K. M., S. Camier, and E. T. Young. 1993. ADH2 expression is repressed by REG1 independently of mutations that alter the phosphorylation of the yeast transcription factor ADR1. Mol. Cell. Biol. 134391-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dombek, K. M., and E. T. Young. 1997. Cyclic AMP-dependent protein kinase inhibits ADH2 expression in part by decreasing expression of the transcription factor gene ADR1. Mol. Cell. Biol. 171450-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donoviel, M. S., N. Kacherovsky, and E. T. Young. 1995. Synergistic activation of ADH2 expression is sensitive to upstream activation sequence 2 (UAS2) orientation, copy number, and UAS1-UAS2 helical phasing. Mol. Cell. Biol. 153442-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorris, D. R., and K. Struhl. 2000. Artificial recruitment of TFIID, but not RNA polymerase II holoenzyme, activates transcription in mammalian cells. Mol. Cell. Biol. 204350-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisen, A., W. E. Taylor, H. Blumberg, and E. T. Young. 1988. The yeast regulatory protein ADR1 binds in a zinc-dependent manner to the upstream activating sequence of ADH2. Mol. Cell. Biol. 84552-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guarente, L. 1983. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101181-191. [DOI] [PubMed] [Google Scholar]
- 24.Haurie, V., M. Perrot, T. Mini, P. Jeno, F. Sagliocco, and H. Boucherie. 2001. The transcriptional activator Cat8p provides a major contribution to the reprogramming of carbon metabolism during the diauxic shift in Saccharomyces cerevisiae. J. Biol. Chem. 27676-85. [DOI] [PubMed] [Google Scholar]
- 25.Hedges, D., M. Proft, and K.-D. Entian. 1995. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 151915-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huh, J. R., J. M. Park, M. Kim, B. A. Carlson, D. L. Hatfield, and B. J. Lee. 1999. Recruitment of TBP or TFIIB to a promoter proximal position leads to stimulation of RNA polymerase II transcription without activator proteins both in vivo and in vitro. Biochem. Biophys. Res. Commun. 25645-51. [DOI] [PubMed] [Google Scholar]
- 27.Irani, M., W. E. Taylor, and E. T. Young. 1987. Transcription of the ADH2 gene in Saccharomyces cerevisiae is limited by positive factors that bind competitively to its intact promoter region on multicopy plasmids. Mol. Cell. Biol. 71233-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1441425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang, Y. W., P. R. Dohrmann, and D. J. Stillman. 1995. Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcriptional regulation. Genetics 14047-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang, J. S., S. H. Kim, M. S. Hwang, S. J. Han, Y. C. Lee, and Y. J. Kim. 2001. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 27642003-42010. [DOI] [PubMed] [Google Scholar]
- 31.Karnitz, L., M. Morrison, and E. T. Young. 1992. Identification and characterization of three genes that affect expression of ADH2 in Saccharomyces cerevisiae. Genetics 132351-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15963-972. [DOI] [PubMed] [Google Scholar]
- 33.Kuchin, S., I. Treich, and M. Carlson. 2000. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 977916-7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemieux, K., and L. Gaudreau. 2004. Targeting of Swi/Snf to the yeast GAL1 UAS G requires the Mediator, TAF IIs, and RNA polymerase II. EMBO J. 234040-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishizawa, M. 2001. Negative regulation of transcription by the yeast global transcription factors, Gal11 and Sin4. Yeast 181099-1110. [DOI] [PubMed] [Google Scholar]
- 36.Ptashne, M., and A. Gann. 1997. Transcriptional activation by recruitment. Nature 386569-577. [DOI] [PubMed] [Google Scholar]
- 37.Ryan, M. P., G. A. Stafford, L. Yu, and R. H. Morse. 2000. Artificially recruited TATA-binding protein fails to remodel chromatin and does not activate three promoters that require chromatin remodeling. Mol. Cell. Biol. 205847-5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakurai, H., and T. Fukasawa. 2003. Artificial recruitment of certain Mediator components affects requirement of basal transcription factor IIE. Genes Cells 841-50. [DOI] [PubMed] [Google Scholar]
- 39.Schuller, H. J. 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43139-160. [DOI] [PubMed] [Google Scholar]
- 40.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 1943-21. [DOI] [PubMed] [Google Scholar]
- 41.Shuster, J., J. Yu, D. Cox, R. V. L. Chan, M. Smith, and E. Young. 1986. ADR1-mediated regulation of ADH2 requires an inverted repeat sequence. Mol. Cell. Biol. 61894-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sloan, J. S., K. M. Dombek, and E. T. Young. 1999. Post-translational regulation of Adr1 activity is mediated by its DNA binding domain. J. Biol. Chem. 27437575-37582. [DOI] [PubMed] [Google Scholar]
- 43.Song, W., I. Treich, N. Qian, S. Kuchin, and M. Carlson. 1996. SSN genes that affect transcriptional repression in Saccharomyces cerevisiae encode SIN4, ROX3, and SRB proteins associated with RNA polymerase II. Mol. Cell. Biol. 16115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steger, D. J., E. S. Haswell, A. L. Miller, S. R. Wente, and E. K. O'Shea. 2003. Regulation of chromatin remodeling by inositol polyphosphates. Science 299114-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1183-93. [DOI] [PubMed] [Google Scholar]
- 46.Tachibana, C., R. Biddick, G. L. Law, and E. T. Young. 2007. A poised initiation complex is activated by SNF1. J. Biol. Chem. 28237308-37315. [DOI] [PubMed] [Google Scholar]
- 47.Tachibana, C., J. Y. Yoo, J.-B. Tagne, N. Kacherovsky, T. I. Lee, and E. T. Young. 2005. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol. Cell. Biol. 252138-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka, M. 1996. Modulation of promoter occupancy by cooperative DNA binding and activation-domain function is a major determinant of transcriptional regulation by activators in vivo. Proc. Natl. Acad. Sci. USA 934311-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor, W. E., and E. T. Young. 1990. cAMP-dependent phosphorylation and inactivation of yeast transcription factor ADR1 does not affect DNA binding. Proc. Natl. Acad. Sci. USA 874098-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thukral, S. K., M. A. Tavianini, H. Blumberg, and E. T. Young. 1989. Localization of a minimal binding domain and activation regions in yeast regulatory protein ADR1. Mol. Cell. Biol. 92360-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verdone, L., G. Camilloni, E. Di Mauro, and M. Caserta. 1996. Chromatin remodeling during Saccharomyces cerevisiae ADH2 gene activation. Mol. Cell. Biol. 161978-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verdone, L., J. Wu, K. van Riper, N. Kacherovsky, M. Vogelauer, E. T. Young, M. Grunstein, E. Di Mauro, and M. Caserta. 2002. Hyperacetylation of chromatin at the ADH2 promoter allows Adr1 to bind in repressed conditions. EMBO J. 211101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voth, W. P., Y. W. Jiang, and D. J. Stillman. 2003. New ‘marker swap’ plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast 20985-993. [DOI] [PubMed] [Google Scholar]
- 54.Wang, X., and C. A. Michels. 2004. Mutations in SIN4 and RGR1 cause constitutive expression of MAL structural genes in Saccharomyces cerevisiae. Genetics 168747-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson, W., D. Hardie, and S. Hawley. 1996. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 61426-1434. [DOI] [PubMed] [Google Scholar]
- 56.Xiao, H., J. D. Friesen, and J. T. Lis. 1995. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol. Cell. Biol. 155757-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young, E. T., K. M. Dombek, C. Tachibana, and T. Ideker. 2003. Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem. 27826146-26158. [DOI] [PubMed] [Google Scholar]
- 58.Young, E. T., N. Kacherovsky, and K. Van Riper. 2002. Snf1 protein kinase regulates Adr1 binding to chromatin but not transcription activation. J. Biol. Chem. 27738095-38103. [DOI] [PubMed] [Google Scholar]
- 59.Yu, J., M. S. Donoviel, and E. T. Young. 1989. Adjacent upstream activation sequence elements synergistically regulate transcription of ADH2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 934-42. [DOI] [PMC free article] [PubMed] [Google Scholar]