Abstract
The diverse biological actions of retinoic acid (RA) are mediated by RA receptors (RARs) and retinoid X receptors (RXRs). Modulation of transcription by RARs/RXRs is achieved through two activation functions, ligand-independent AF-1 and ligand-dependent AF-2, located in the A/B and E domains, respectively. While the coregulatory proteins that interact with the E domain are well studied, the A/B domain-interacting partners and their influence(s) on the function of RARs are poorly understood. Acinus-S′ is an ubiquitous nuclear protein that has been implicated in inducing apoptotic chromatin condensation and regulating mRNA processing. Our data demonstrate that Acinus-S′ can specifically repress ligand-independent and ligand-dependent expression of a DR5 RA response element(RARE)-dependent reporter gene and several endogenous RAR-regulated genes in a dose-dependent and gene-specific manner. Chromatin immunoprecipitation assays show that Acinus-S′ associates with RAREs within the promoters of endogenous genes independent of RA treatment. Furthermore, the C-terminal end of Acinus-S′ and the B domain of RARβ interact independently of ligand, and the C-terminal end of Acinus-S′ is sufficient for the repression of RAR-regulated gene expression. Finally, histone deacetylase activity only partially accounts for the repressive effect of Acinus-S′ on RAR-dependent gene expression. These findings identify Acinus-S′ as a novel RAR-interacting protein that regulates the expression of a subset of RAR-regulated genes through direct binding to the N-terminal B domains of RARs.
The biological activities of retinoic acid (RA) and its synthetic analogues are mediated through binding to specific nuclear receptors termed RA receptors (RARs) and retinoid X receptors (RXRs). Like all members of the steroid/thyroid hormone superfamily, RARs and RXRs share a common domain architecture containing five or six structurally and functionally distinct regions, termed domains A to F. The two domains that share the most homology among nuclear receptor superfamily members and are the best studied are the central C domain, which is involved in DNA binding, and the E domain, which is responsible for both ligand binding and ligand-dependent transactivation activity (AF-2). The most variable in amino acid sequence is the amino-terminal A/B domain, which contains a ligand-independent transactivation function (AF-1). The D domain represents a linker region between domains C and E. Carboxyl-terminal region F is not present in RXRs and is not well studied in RARs (for a review, see reference 9).
The current understanding of the mechanism of ligand-dependent (AF-2) transcriptional activation by RARs comes from numerous structural and functional studies and postulates that unliganded RARs interact with transcriptional corepressors such as nuclear receptor corepressors and SMRT (silencing mediator for RAR and TR) through their E domains, resulting in silencing of RA-responsive genes. Upon RA binding, structural changes in the E domain induce dissociation of corepressors and subsequent recruitment of transcriptional coactivator proteins such as transcriptional mediators/intermediary factor 1 and 2, steroid receptor coactivator 1, and CREB binding protein/p300, causing transcriptional activation (for a review, see reference 5). In addition, AF-1 activity located in the A/B domains can be either constitutive or ligand independent, can synergize with AF-2, and can be modulated by protein-protein interactions (8, 21) or through posttranslational modifications such as phosphorylation (6, 7, 15, 29). Since AF-1 activity is also promoter and cell specific, this domain may be responsible for specific functions of different RAR or RXR isoforms.
Acinus (apoptotic chromatin condensation inducer in the nucleus) is a nuclear protein that has been reported to induce apoptotic chromatin condensation in the nucleus and possibly to mediate nuclear structural changes in normal cells. Both mouse and human Acinus proteins are found expressed as three isoforms (termed Acinus-L, Acinus-S′, and Acinus-S) that most likely arise due to alternative splicing (30). Acinus-L is the longest isoform, containing 1,341 amino acids, while Acinus-S′ and Acinus-S consist of 614 and 583 amino acids, respectively. Structurally, all three Acinus isoforms share a common central domain that shows homology to the RNA recognition motif (RRM) of Drosophila Sxl protein and a C-terminal end rich in Arg/Glu, Arg/Asp, and Arg/Ser repeats (see Fig. S1 in the supplemental material). Acinus-L contains an additional N-terminal SAP domain that has been implicated in non-sequence-specific DNA binding and also in mediating structural chromatin changes (1).
Functionally, Acinus-S, Acinus-S′, and Acinus-L can induce chromatin condensation during apoptosis (16, 30). Additionally, all forms of Acinus were identified as a component of a splicing complex termed ASAP (apoptosis- and splicing-associated protein) (33), as well as a member of a multiprotein exon-junction complex (36). Proteomic analysis revealed that Acinus localizes to particular nuclear compartments called the interchromatin granule clusters (27, 31, 44) that are known to be responsible for the accumulation and assembly of splicing factors. Since Acinus contains an RRM found in several splicing proteins and localizes to the splicing compartment, it has been hypothesized that it can function as an mRNA-processing factor. In agreement with this was a study that suggested that Acinus inhibits RNA processing in vitro (33).
In the present work, we identified Acinus-S′ as a protein that binds via its C-terminal domain to the N-terminal A/B domain of RARs. The functional interaction between RARs and Acinus-S′ occurs on the promoters of RAR-regulated genes and results in ligand-dependent and ligand-independent repression of RAR-regulated gene expression. Histone deacetylation appears to contribute in part to the repressive effect of Acinus-S′ on RAR-regulated gene expression.
MATERIALS AND METHODS
Cell culture.
CV-1, Cos-7, and P19 embryonal carcinoma cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 μg/ml penicillin, and 100 units/ml streptomycin. The cells were maintained in an incubator at high humidity, 5% CO2, and 37°C. All transfections were performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's recommendations.
Yeast two-hybrid screening.
Yeast two-hybrid screening was performed using Yeastmaker yeast transformation system 2 (Clontech Laboratories, Inc.) according to manufacturer's instructions. The bait plasmid contained the A domain and the first seven amino acids of the B domain of mouse RARβ3 (amino acids 1 to 94) cloned into the EcoRI site of pGBKT7 in frame with the GAL4 DNA binding domain. An 11-day mouse embryo cDNA library prepared in pACT2 (Clontech) was used as the prey. Plasmid (pACT2) DNA from positive clones was isolated using the QIAprep spin miniprep kit (Qiagen) and rescued by transformation in Escherichia coli DH5α. Isolated plasmid DNAs were sequenced and the cDNA sequences submitted for nucleotide and translated protein BLAST searches. Full-length human Acinus-S′ cDNA was purchased from Origene.
GST pull-down assays.
Glutathione S-transferase (GST)-pull-down assays were performed essentially as previously described (24). GST-Acinus-S′ (amino acids 1 to 614), GST-Acinus-S′ N terminus (amino acids 1 to 286), GST-Acinus-S′ RRM (amino acids 287 to 420), GST-Acinus-S′ C terminus (amino acids 421 to 614), and GST-SMRT were expressed in BL21-DE3 E. coli cells and purified using glutathione-agarose beads (Sigma). Full-length mouse RARα1, RARβ1, RARβ2, RARβ3, RARβ4, RARγ2,RXRα, RARβ deletion mutants [RARβ3(CDEF), amino acids 115 to 482; RARβ3(DEF), amino acids 181 to 482; RARβ1(AB), amino acids 1 to 81; RARβ2(AB), amino acids 1 to 74; RARβ3(AB), amino acids 1 to 98; RARβ4(BCD), amino acids 1 to 136, and RARβ4(BC), amino acids 1 to 81], estrogen receptor (ER), thyroid receptor (TR), and peroxisome proliferative activated receptor gamma (PPARγ) were in vitro transcribed and translated using TnT kit (Promega) and [35S]methionine (1,175 Ci/mmol; Perkin-Elmer Life and Analytical Sciences) following the manufacturer's protocol.
Transactivation assays.
Transactivation assays were performed essentially as previously described (32, 35). CV-1 or Cos-7 cells were transfected with 0.5 μg DR5 RA response element (RARE)-chloramphenicol acetyltransferase (CAT) or RARE-Luc reporter DNA (Panomics), 0.25 μg pCMV-β-galactosidase DNA, or 0.1 μg pRL DNA; 0.5 μg RAR expression construct DNA; and 5 μg pcDNA3.1/His-Acinus S′ DNA or empty vector DNA (pcDNA3.1). For the SP1 and PPARγ transactivation assays, the transfections were performed with 1 μg SP1-Luc reporter DNA (Panomics) or PPARγ-Luc reporter DNA (Panomics), 0.1 μg pRL DNA, 0.5 μg RARβ3 expression construct DNA (for SP1 assay) or pCMX-PPARγ DNA (for PPARγ assay), and 5 μg pcDNA3.1/His-Acinus-S′ DNA or empty vector DNA (pcDNA3.1). After 4 h of incubation, the transfected cells were treated with 1 μM RA, 1 μM ciglitazone, 100 nM trichostatin A (TSA), or ethanol carrier for 24 h and then harvested. For each experiment, the CAT or firefly luciferase activity was normalized to β-galactosidase or Renilla luciferase activity, respectively. The change in normalized CAT or firefly luciferase activity was calculated relative to that for cells that were transfected with empty vector DNA and treated with ethanol, which was set as 1. Values are the means ± standard deviations for three experiments performed in triplicate.
RNA isolation and real-time PCR.
For quantitative reverse transcription-PCR (RT-PCR), P19 cells were plated on 100-mm tissue culture dishes and transfected with the indicated amounts of pEGFP-Acinus DNA or empty vector DNA (pEGFP). At 24 h posttransfection, the cells were treated with ethanol or 1 μM RA for 4 h and collected for RNA isolation. Total RNA was isolated using RNAzol B RNA isolation reagent (Tel-Test, Inc.) according to the manufacturer's instructions. One microgram of total RNA was used in the reverse transcription reaction with oligo(dT) primers supplied in the Advantage RT-for-PCR kit (Clontech). Subsequently, 10 μl of RT reaction mixture was used for quantitative real-time PCR using SYBR green PCR chemistry (Applied Biosystems) according to manufacturer's instructions. Specific PCR primers (see Table S1 in the supplemental material) were synthesized and optimized for amplification of each gene. Changes in gene expression were calculated using relative quantitation of a target against an endogenous, unchanged GAPDH (glyceraldehyde-3-phosphate dehydrogenase) standard. The cycling parameters were 95°C for 10 min and then 40 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 90 s. Values are the means ± standard deviations for triplicate samples.
Coimmunoprecipitation and Western blot analysis.
P19 cells were plated on 100-mm tissue culture dishes and transfected with 10 μg of pEGFP-Acinus-S′ or empty vector DNA (pEGFP) and 10 μg of pOPRSVICAT-RARβ3 DNA. At 24 h posttransfection, the cells were treated with ethanol or 1 μM RA for 4 h and total cell protein lysates prepared. Coimmunoprecipitation and Western blot analysis were performed essentially as previously described previously (26, 37, 41) using pan-RAR antibody (Santa Cruz), anti-mouse immunoglobulin G (IgG) (Santa Cruz), or polyclonal green fluorescent protein (GFP) whole antiserum (Abcam). Immunoblotting for coimmunoprecipitation experiments was performed using the One-Step Complete IP-Western kit (Genscript) according to the manufacturer's protocol. Antibodies used were anti-GFP (Abcam) and anti-pan-RAR antibody (Santa Cruz).
Western blot analysis using nuclear protein extracts was performed as previously described (26). Anti-GFP (Abcam), anti-lamin B (Santa Cruz), or anti-His (Santa Cruz) antibody was used as the primary antibody with the corresponding horseradish peroxidase-conjugated IgG secondary antibody (Amersham), followed by visualization with ECL-plus (Amersham).
Chromatin immunoprecipitation (ChIP) assay.
P19 cells were plated on 100-mm tissue culture dishes and transfected with 10 μg of pEGFP-Acinus-S′ DNA. At 24 h posttransfection, the cells were incubated in the presence or absence of 1 μM RA for 4 h. Chromatin was prepared using the modified two-step cross-linking method as described by Nowak et al. (23). DNA/protein complexes were immunoprecipitated using 4 μg of antibody against pan-RAR (Santa Cruz), acetylated histone H3 (Upstate), RNA polymerase (RNAP) (Santa Cruz), or IgG (Santa Cruz) or 5 μl of anti-GFP antibody (to immunoprecipitate GFP-tagged Acinus S′) (Abcam). PCR amplification of immunoprecipitated DNA was performed using the primers indicated in Table S2 in the supplemental material. PCR cycling conditions were as follows: 94°C for 5 min; 28 to 35 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 1 min; and finally 72°C for 8 min.
RESULTS
Identification of Acinus as an RARβ3 N terminus-interacting protein.
To isolate proteins that bind to and potentially regulate the functional activity of the N termini of RARs, we performed a yeast two-hybrid screen of a 11-day mouse embryo cDNA library with the N-terminal 94 amino acids of RARβ3 containing the full A domain and the first seven amino acids of the B domain. Among several potential candidates, we identified a clone that displayed 98% sequence homology to transcript variant 2 (NM_023190.1) and transcript variant 1 (NM_019567.1) of mouse apoptotic chromatin condensation inducer (Acinus). The clone contained a ∼800-bp-long cDNA insert carrying the 3′-terminal portion of the Acinus open reading frame (nucleotides 4020 to 4169 of transcript variant 2 and nucleotides 1716 to 1867 of transcript variant 1) along with the 3′ untranslated end. Since all three Acinus isoforms share identical C-terminal ends (see Fig. S1 in the supplemental material) and there are no reported functional differences between the three isoforms, we obtained the full-length human Acinus-S′ cDNA (Origene) and performed our studies with the Acinus-S′ isoform exclusively.
Effect of Acinus-S′ on the expression of RAR-regulated genes.
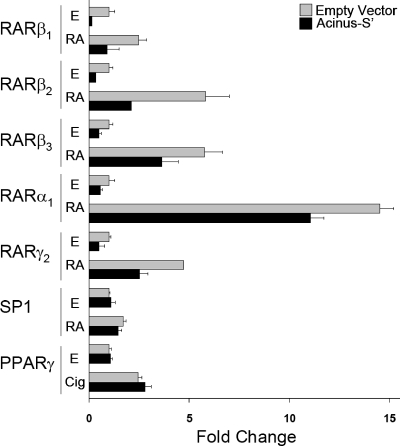
First, we assessed the effect of Acinus-S′ on the activity of a CAT reporter gene whose expression was under the control of a minimal thymidine kinase promoter and DR5 RARE. Acinus-S′ overexpression decreased both ligand-independent (ethanol treated) and ligand-dependent (RA treated) CAT reporter gene activity when CV-1 cells were cotransfected with RARβ1, RARβ2, RARβ3, RARα1, and RARγ2, by 25 to 75% (Fig. 1). In contrast, Acinus-S′ overexpression did not have an effect on the expression of luciferase reporter gene activity regulated by either SP1 or PPARγ in both the presence and absence of exogenous ligand. These results suggest that Acinus-S′ represses transcription with specificity for genes whose expression is under the regulatory control of a RARE and RARs. Furthermore, the repression of RAR-regulated genes by Acinus-S′ is not dependent on the presence of exogenous RA.
FIG. 1.
Effect of Acinus-S′ on reporter gene activity. The bar graph indicates relative reporter gene activity levels from transfected CV-1 cells treated with ethanol (E), 1 μM RA, or 1 μM ciglitazone (Cig) for 24 h. The change in normalized CAT or Luc activity was calculated relative to cells that were transfected with empty vector DNA (no Acinus-S′) and treated with ethanol (set as 1). Error bars indicate standard deviations.
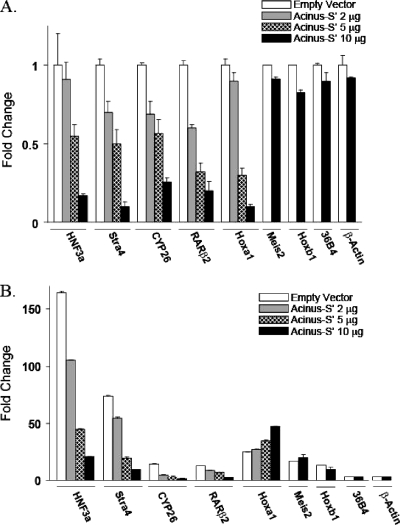
To test the effect of Acinus-S′ on the expression of endogenous RAR-responsive genes, P19 cells were transfected with increasing concentrations of Acinus-S′ DNA and treated with either ethanol or 1 μM RA for 4 h. We observed a stepwise, dose-dependent decrease in the mRNA levels of HNF3a, CYP26A1, RARβ2, and Stra4 in both ethanol-treated (Fig. 2A) and RA-treated (Fig. 2B) cells compared to mock-transfected cells. The maximal repression was observed when the cells were transfected with the highest concentration of Acinus-S′ DNA. On the other hand, the mRNA levels of two genes, those for Meis2 and Hoxb1, were not changed even in the presence of the highest concentration of Acinus-S′ DNA. Interestingly, Hoxa1 showed an unexpected behavior, where overexpression of Acinus-S′ in ethanol-treated cells resulted in a decrease in the basal Hoxa1 mRNA level, while RA-induced expression of Hoxa1 mRNA was moderately increased by Acinus-S′. Finally, overexpression of Acinus-S′ did not change the mRNA levels of two RAR-independent genes, those for 36B4 and β-actin. Taken together, these data indicate that Acinus-S′ can affect the ligand-independent and ligand-dependent expression of RAR-regulated genes in vivo in a gene-specific manner.
FIG. 2.
Effect of Acinus-S′ on the expression of early RAR-regulated genes in P19 cells in the absence (A) and in the presence (B) of RA. P19 cells were plated on 100-mm tissue culture dishes and transfected with 2, 5, or 10 μg of pEGFP-Acinus-S′ DNA or mock transfected with 10 μg of empty pEGFP vector. At 24 h after transfection, the cells were treated with ethanol (A) or 1 μM RA (B) for 4 h. The expression levels of a panel of early RAR-responsive genes (HNF3a, Stra4, CYP26A1, RARβ2, Hoxa1, Meis2, and Hoxb1 genes) and control genes (36B4 and β-actin genes) were measured by real-time RT-PCR. The expression level of each gene examined was normalized to the endogenous GAPDH levels and expressed relative to the cells transfected with empty vector and treated with ethanol (set to 1). Error bars indicate standard deviations.
Characterization of the physical interaction between Acinus-S′ and nuclear receptors.
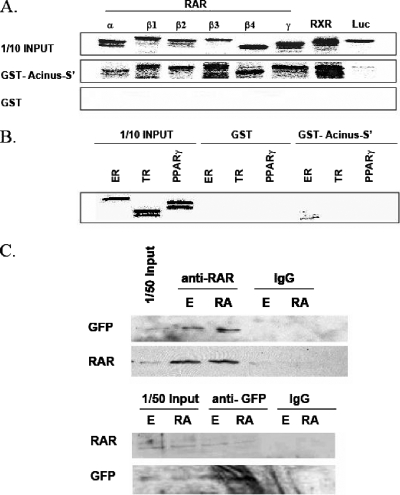
To confirm the physical interaction between Acinus-S′ and RARs, we performed in vitro GST-pull down assays with GST-tagged full-length Acinus-S′ and in vitro-transcribed and -translated [35S]methionine-labeled full-length RARs (RARα1, RARβ1, RARβ2, RARβ3, RARβ4, and RARγ2), RXRα, and luciferase. As seen in Fig. 3A, all RAR isotypes and isoforms tested, as well as RXRα, were able to bind full-length GST-Acinus-S′ and did not bind GST alone. As expected for a negative control, luciferase did not interact with either GST-Acinus-S′ or GST alone. Additionally, we performed in vitro binding assays with GST-tagged full-length Acinus-S′ and in vitro-transcribed and -translated [35S]methionine labeled ER, TR, and PPARγ as representative members of the steroid/thyroid hormone receptor superfamily (Fig. 3B). GST pull-down assays revealed that ER, TR, and PPARγ were not able to interact with GST-Acinus-S′ or GST alone. These results demonstrate that Acinus-S′ is a protein that specifically interacts with RARs and RXRα and not with the entire nuclear receptor superfamily.
FIG. 3.
(A) Interaction of Acinus-S′ with RARs and RXR. GST pull-down assays were performed with purified GST-Acinus-S′ and in vitro-transcribed and -translated [35S]methionine-labeled RARs (RARα1, RARβ1, RARβ2, RARβ3, RARβ4, and RARγ2), RXRα, and luciferase. (B) Interaction of Acinus-S′ with members of steroid/thyroid hormone receptor superfamily. The interaction was tested in GST pull-down assays with GST-Acinus-S′ and in vitro-transcribed and -translated labeled ER, TR, and PPARγ. (C) In vivo interaction of Acinus-S′ with RARβ. P19 cells were plated on 100-mm tissue culture dishes and transfected with 10 μg of pEGFP-Acinus-S′ DNA and 10 μg of pOPRSVICAT-RARβ3 DNA. Twenty-four hours later, the cells were treated with ethanol (E) or 1 μM RA for 4 h. Protein complexes were immunoprecipitated from whole-cell lysates using anti-pan-RAR, anti-GFP or mouse IgG. Western blot analysis was performed with either GFP antibody or pan-RAR antibody using the One-Step Complete IP-Western kit (Genscript).
We next assessed whether RA (RAR/RXR ligand) had an effect on the interaction between Acinus-S′ and RARs. Either 1 μM RA or ethanol was added to the GST-pull-down reaction mixtures containing purified GST-Acinus-S′ and in vitro-transcribed and -translated full-length [35S]methionine-labeled RARs (RARα1, RARβ1, RARβ2, and RARβ3,) or RXRα. We did not observe any differences in binding between RARs/RXRα and Acinus-S′ in the presence or absence of RA (see Fig. S2 in the supplemental material).
To investigate the in vivo interaction of Acinus-S′ and RARβ, we carried out coimmunoprecipitation studies with overexpressed GFP-Acinus-S′ and RARβ3 in P19 cells treated with ethanol or 1 μM RA (Fig. 3C). The protein complexes were immunoprecipitated from whole-cell lysates using anti-pan-RAR, anti-GFP, or mouse IgG. Subsequent Western blots using antibodies directed against RAR or GFP revealed that the RAR antibody precipitated both RAR and GFP-Acinus-S′ and that the GFP antibody precipitated both GFP-Acinus-S′ and RAR. IgG did not precipitate either RAR or GFP-Acinus-S′. No significant differences were observed in the amount of precipitated interacting proteins (RAR or Acinus-S′) between the ethanol- and RA-treated cells. Consistent with the in vitro binding assays, coimmunoprecipitation experiments confirm that Acinus-S′ physically interacts with RARβ in vivo and that the interaction was not affected by the RAR ligand RA.
Interaction of RARβ with Acinus-S′ occurs in vivo on the promoters of RAR-regulated genes.
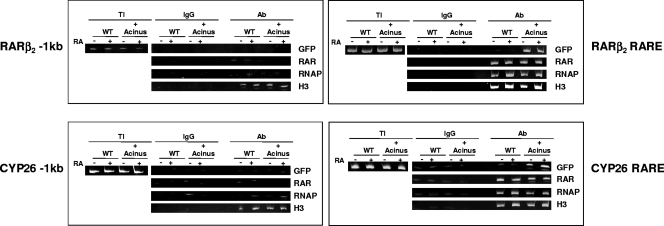
Next, we performed ChIP assays to test the in vivo association of overexpressed GFP-Acinus-S′ on the promoters of two RAR-regulated genes (RARβ2 and CYP26A1 genes) in P19 cells treated with either ethanol or RA. ChIPs were carried out with mouse IgG as a negative control and antibodies to GFP (to precipitate Acinus-S′), pan-RAR, acetylated histone H3, and RNA polymerase. We detected the presence of GFP-Acinus-S′ on the RAREs of both the RARβ2 and CYP26A1 genes (Fig. 4) but not in the region that is located ∼1 kb upstream of both the RARβ2 RARE and CYP26A1 RARE (Fig. 4). Association of RNA polymerase and acetylated histone H3 in the vicinity of the RAREs of both the RARβ2 and CYP26A1 genes was slightly increased upon RA treatment. On the other hand, GFP-Acinus-S′ and RARs were recruited to the RARβ2 and CYP26A1 promoters irrespective of the presence of RA. Altogether, the data indicate that Acinus-S′ is recruited to the RAREs of endogenous RAR-regulated promoters and that the interaction occurs regardless of the presence of RA.
FIG. 4.
Interaction of Acinus-S′ on the promoters of the RAR-regulated RARβ2 and CYP26 genes. P19 cells were transfected with pEGFP-Acinus-S′ DNA (+Acinus), and untransfected P19 cells were used as a control (WT). At 24 h after transfection, the cells were treated with ethanol or 1 μM RA for 4 h. Chromatin was immunoprecipitated from cells in each experimental condition using antibodies to GFP, pan-RAR, RNA polymerase (RNAP), and acetylated histone H3. Mouse IgG was used as a negative control. The precipitated DNA was amplified by PCR using specific primers flanking the RAREs of RARβ2 and CYP26A1 and a region located ∼1 kb upstream of the RARβ2 and CYP26A1 RAREs. PCR products were separated by 10% polyacrylamide gel electrophoresis and visualized using ethidium bromide staining.
Mapping domains that are important for the interaction of Acinus-S′ with RARβ.
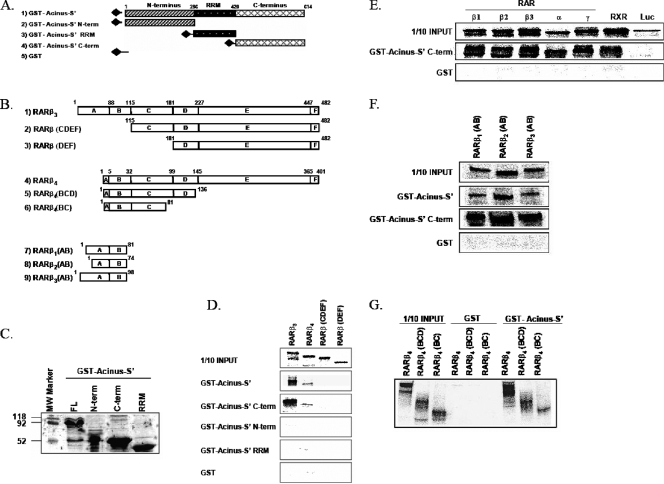
To map the interacting regions of Acinus-S′ and RARβ, we constructed various Acinus-S′ (Fig. 5A) and RARβ (Fig. 5B) deletion mutants and assessed their binding properties in a series of GST pull-down experiments. First, we performed a GST pull-down assay with equal amounts of full-length GST-Acinus-S′, GST-Acinus-S′ N terminus, GST-Acinus-S′ RRM, or GST-Acinus-S′ C terminus (Fig. 5C) and in vitro-transcribed and -translated [35S]methionine labeled full-length RARβ3 or RARβ4 (contains an A domain of only 4 amino acids), RARβ deleted of the A/B domain [RARβ(CDEF)], and RARβ lacking domains A through C [RARβ(DEF)]. As seen from Fig. 5D, full-length GST-Acinus-S′ and GST-Acinus-S′ C terminus bind full-length RARβ3 and RARβ4 but cannot bind the other RAR deletion mutants, RARβ(CDEF) and RARβ(DEF). On the other hand, GST-Acinus-S′ N terminus and GST-Acinus-S′ RRM did not bind any of the RAR proteins. This result indicates that C terminus of Acinus-S′ is important for binding to the N-terminal end of RARβ. To further test the importance of the C terminus of Acinus-S′ for the interaction with RARs, we assessed binding of the isolated GST-Acinus-S′ C terminus with full-length RARs (RARα1, RARβ1, RARβ2, RARβ3, and RARγ2) and RXRα. As expected, we observed strong binding between C terminus of Acinus-S′ and all RARs and RXR tested (Fig. 5E).
FIG. 5.
(A and B) Schematic representation of Acinus-S′ (A) and RARβ (B) deletion mutants used in GST pull-down assays to map the interaction domains. (C) Purification of GST-Acinus-S′ deletion mutants. GST fusion Acinus-S′ plasmid DNAs representing the full-length GST-Acinus-S′, GST-Acinus-S′ N terminus, GST-Acinus-S′ C terminus, and GST-Acinus-S′ RRM were expressed in E. coli BL21, purified using a glutathione-agarose affinity purification protocol, resolved by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and visualized using Coomassie blue staining. Equal amounts of purified proteins were used for GST-pull down assays. (D) The C terminus of Acinus-S′ binds to the N terminus of RARβ. GST-Acinus-S′, GST-Acinus-S′ N terminus, GST-Acinus-S′ RRM, and GST-Acinus-S′ C terminus were incubated with in vitro-transcribed and -translated full-length [35S]methionine-labeled RARβ3, RARβ4, RARβ(CDEF), and RARβ(DEF). (E) Interaction of the C-terminal region of Acinus-S′ with RARs and RXRα. GST pull-down assays were performed using GST-Acinus-S′ C terminus and in vitro-transcribed and -translated [35S]methionine-labeled RARs (RARα1, RARβ1, RARβ2, RARβ3, RARγ2), RXRα, and luciferase. (F) The C terminus of Acinus-S′ binds to the A/B domain of RARβ. Full-length GST-Acinus-S′ and GST-Acinus S′ C terminus were incubated with in vitro-transcribed and -translated [35S]methionine-labeled A/B domains of RARβ isoforms [(RARβ1(AB), RARβ2(AB), and RARβ3(AB)]. (G) The C terminus of Acinus-S′ interacts with the B domain of RARβ. Interaction assays were performed with full-length GST-Acinus-S′ and in vitro-transcribed and -translated [35S]methionine-labeled RARβ4 and RARβ4 deletion mutants lacking the EF domain [RARβ4(BCD)] or lacking the DEF and partial C domains [RARβ4(BC)].
Furthermore, to confirm that the interaction occurs between the C terminus of Acinus-S′ and the N-terminal A/B domain of RARβ, we performed GST pull-down experiments with in vitro-transcribed and -translated A/B domains of RARβ1, RARβ2, and RARβ3 and GST fused to the C terminus of Acinus-S′. Figure 5F shows that not only the full-length GST-Acinus-S′ but also the C-terminal end of Acinus-S′ (GST-Acinus-S′ C terminus) interacts strongly with isolated A/B domains of all three RARβ isoforms.
Since we demonstrated that Acinus-S′ was able to bind to isolated A/B domains of various RARβ isoforms and was also able to interact with RARβ4, which lacks the majority of the A domain, we hypothesized that the B domain of RARβ contains the binding site for Acinus-S′. Therefore, we performed GST pull-down assays with C-terminal deletion mutants of RARβ4 (for a schematic representation, see Fig. 5B) and GST-Acinus-S′. As seen in Fig. 5G, full-length GST-Acinus-S′ was able to bind RARβ4 as well as all C-terminal deletion mutants of RARβ4 [RARβ4(BCD) and RARβ4(BC)]. Therefore, the minimal portion of RARβ that still binds Acinus-S′ contains amino acids 1 to 81 of RARβ4, representing the B domain and partial C domain of RARβ. However, Fig. 5D demonstrates that CDEF domains alone cannot bind GST-Acinus-S′. Taken together, these data strongly suggest that Acinus-S′ binds to the B domains of RARs.
Importance of the C-terminal end of Acinus-S′ for the expression of RAR-regulated genes.
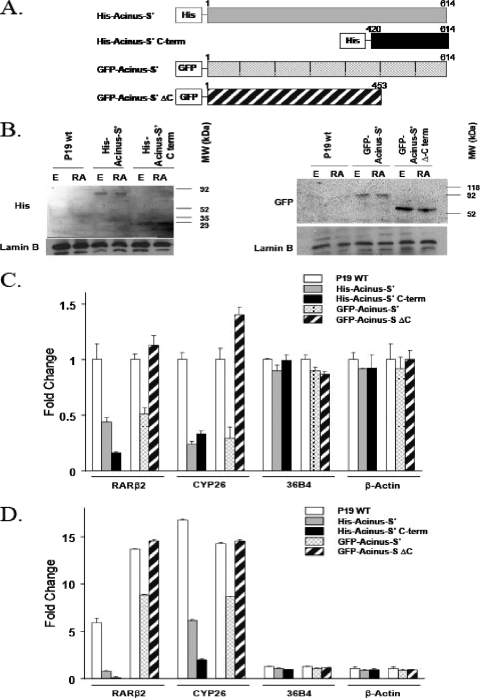
Since we demonstrated using in vitro binding assays that the C-terminal end of Acinus-S′ is responsible for the interaction with RARs and RXR, we asked whether the C-terminal end of Acinus-S′ is sufficient to inhibit the expression of RAR-regulated genes. We constructed a His-tagged C-terminal end of Acinus-S′ containing amino acids 420 to 614 and GFP-fused Acinus-S′ ΔC, comprised of amino acids 1 to 453 (Fig. 6A). Overexpression of the 194 C-terminal amino acids of Acinus-S′ (His-Acinus-S′ C terminus) in P19 cells (Fig. 6B) was sufficient to repress the basal (Fig. 6C) and RA-induced (Fig. 6D) expression of the RAR-dependent RARβ2 and CYP26A1 genes but did not have an effect on the expression of the 36B4 and β-actin control genes. On the other hand, the mRNA levels of RARβ2 and CYP26A1 did not differ between P19 cells overexpressing Acinus-S′ lacking the C-terminal end (GFP-Acinus-S′ ΔC) (Fig. 6B) and mock-transfected P19 cells (Fig. 6C and D). Therefore, Acinus-S′ with the C-terminal RAR-interacting domain deleted (GFP-Acinus-S′ ΔC) lost the ability to repress the expression of RARβ2 and CYP26A1. Taken together, these results demonstrate that the C-terminal end of Acinus-S′ (amino acids 420 to 614) is sufficient for the repression of genes regulated by RARs.
FIG. 6.
(A) Schematic representation of Acinus-S′ constructs used to determine the functional importance of the Acinus-S′ C-terminal domain. (B) Expression of Acinus-S′ C-terminal deletion mutants in P19 cells. P19 cells were plated on 100-mm tissue culture dishes and transfected with 10 μg of one of the following DNAs: His-Acinus-S′, His-Acinus-S′ C terminus, GFP-Acinus-S′, or GFP-Acinus-S′ ΔC. At 24 h after transfection, the cells were treated with ethanol (E) or 1 μM RA for 4 h and nuclear extracts were prepared. Western blot analysis was performed using primary anti-His or anti-GFP antibody followed by a secondary horseradish peroxidase-anti-rabbit antibody. Anti-lamin B primary antibody was used as a loading control. (C and D) Effect of the C-terminal end of Acinus-S′ on the expression of RAR-regulated genes in the absence (C) and presence (D) of RA. P19 cells were transfected and treated as described above. The expression levels of RAR-dependent genes (RARβ2 and CYP26A1) and control genes (36B4 and β-actin) were measured by real-time RT-PCR. The expression level of each gene examined was normalized to the endogenous GAPDH levels and expressed relative to the ethanol-treated P19 cells. Error bars indicate standard deviations.
Histone deacetylation contributes in part to the repression of RAR-regulated gene expression by Acinus-S′.
Since histone deacetylation is often associated with transcriptional repression by corepressors (40), we examined the effect of TSA, a potent histone deacetylase (HDAC) inhibitor, on the repression of RAR-regulated gene activity by Acinus-S′. Treatment of the Acinus-S′ transfected cells with TSA resulted in an increase in the level of luciferase reporter gene activity; however, this increase was only approximately 70% of that observed in cells that were not transfected with Acinus-S′ (Fig. 7). This suggests that histone deacetylation accounts for only part of the Acinus-S′-mediated repression of RAR-dependent transcription.
FIG. 7.
Role of histone deacetylation on repression of RARE reporter activity by Acinus-S′. The bar graph indicates relative RARE reporter gene activity levels from transfected Cos-7 cells treated with ethanol (EtOH), 1 μM RA, or 100 nM TSA for 24 h. The change in normalized Luc activity was calculated relative to cells that were transfected with empty vector DNA (no Acinus-S′) and treated with ethanol and vehicle (set as 1). Error bars indicate standard deviations.
DISCUSSION
The studies described in this paper have identified Acinus-S′ as a novel, gene-specific mediator of both ligand-independent and ligand-dependent repression of RAR-regulated gene expression. Binding assays demonstrate that the C-terminal end of Acinus-S′ physically interacts with the N-terminal B domain of RARβ, while ChIP assays demonstrate that Acinus-S′ associates with the RAREs of two genes, those for RARβ2 and CYP26A1. In addition, the C-terminal region of Acinus-S′ that contains Arg/Asp, Arg/Glu, and Arg/Ser repeats is sufficient to reduce the mRNA levels of two RAR-regulated genes. Finally, histone deacetylation partially accounts for the repressive effect of Acinus-S′ on RAR-regulated gene expression.
In recent years, an ever-growing amount of evidence has emerged that clearly demonstrates the ability of the N-terminal A/B domains of nuclear receptors to engage in direct, physical interaction with various proteins. N-terminal A/B domain-interacting proteins have the ability to modulate the function of nuclear receptors by either enhancing or repressing the transcriptional activation of the interacting receptor. In addition to components of basal transcriptional machinery and general coactivators and corepressors, N-terminal A/B domains can interact with proteins involved in processes such as mRNA processing and splicing (p68, SRA, ANT-1, and p54nrb), with chaperones (BAG-1) or with a number of proteins containing specific enzymatic activities such as acetyl- and methyltransferases (12, 17, 25, 34, 39, 43).
In the specific case of RARs, even though numerous studies have suggested the functional interaction of AF-1 of RARs with various pathways including kinase and ubiquitination, only one protein (vinexin-β) has been reported to interact exclusively with the N-terminal end of RARγ and repress RARγ-mediated transcription (8). This interaction requires the presence of nonphosphorylated Ser77/Ser79 or Ser66/Ser68 residues in the B domain of RARg1 or RARγ2, respectively (8). In contrast to vinexin-β, Acinus-S′ interacts with all three RAR isotypes while not interacting with at least three other members of the steroid/thyroid hormone superfamily. Unlike vinexin-β, which specifically binds RARγ by interacting with an amino acid which is not conserved among all three RAR isotypes (Ser77/Ser66 of RARγ1/RARγ2), a conserved sequence in the B domain of RARs and RXRα is most likely necessary for the binding of Acinus-S′. An alignment of the 27 amino acids in the B domains of RARα, RARβ, RARγ, and RXRα shows that five amino acid residues are absolutely conserved (Q69, S72, V77, S79, and P85 of RARα1) and 19 residues show more than 75% conservation. Furthermore, 20 of 27 amino acid residues are absolutely conserved among the three RAR subtypes (Fig. 8). Since the bait used in the yeast two-hybrid studies contained the A domain and the first seven amino acids of the B domain of RARβ3, it is most likely that the conserved residues Q69 and/or S72 (of RARα1) are important for the interaction between Acinus-S′ and RARs/RXRα.
FIG. 8.
Alignment of the B domains of RARs and RXRα. Amino acids conserved in all three RARs and RXRα are indicated in bold.
Although our studies demonstrate that the B domain is critical for the binding of Acinus-S′ and RARs, we also observed that the in vitro binding of Acinus-S′ to RARβ3 appeared to be stronger than that of RARβ4. Since the A domain of RARβ4 contains only 4 amino acid residues while that of RARβ3 is 87 amino acids in length, it is possible that the A domain may also contribute either directly or indirectly to the binding of RARs with Acinus-S′. Since the amino-terminal AF-1 containing A/B domains of RARs have been demonstrated to be targets for phosphorylation that may induce modifications in the conformation of the amino-terminal portion of the receptor (28), it is possible that changes in the phosphorylation status of the A/B domains of RARs may contribute to the binding of Acinus-S′ and RARs. On the other hand, it is also possible that transient, low-specificity interactions may occur between the A domains of RARs and Acinus-S′, since it has been suggested that the A domains of nuclear receptors are flexible and contain naturally disordered regions (20).
Transcriptional activation of gene expression by nuclear receptors is known to involve both ligand-dependent (AF-2) and ligand-independent (AF-1) activities. The current model of transcriptional regulation by RARs suggests that ligand-dependent transcriptional activation involves the binding of transcriptional corepressors through their E domains, resulting in silencing of RA-responsive genes in the absence of RA. Upon RA binding, structural changes in the E domain induce dissociation of corepressors and subsequent recruitment of transcriptional coactivator proteins. On the other hand, a distinct group of coregulatory proteins, including RIP-140, LCoR, and PRAME have been demonstrated to bind to the AF-2 region and to confer transcriptional repression of gene expression by RA-bound RARs (13, 14, 38). This model is consistent with the ligand binding domain crystal structure, in which major conformational changes occur upon binding of RA. In contrast, no crystal structure data are available for the N-terminal A/B domain of any nuclear receptor, including RARs, and the mechanism underlying ligand-independent transcription is poorly understood. Our data show that Acinus-S′ binds to the B domains of RARs and represses transcriptional activity of RAR-regulated genes in both the presence and absence of exogenous RA. This is consistent with the fact that AF-1 functions in a ligand-independent fashion, suggesting that there are different pools of RAR protein complexes that interact with AF-2 versus AF-1.
Interestingly, the effect of Acinus-S′ on the expression levels of endogenous RAR-regulated genes was gene specific. Within a group of seven well-studied RAR-regulated genes examined, four (the HNF3a, Stra4, CYP26A1, and RARβ2 genes) were significantly repressed by the overexpression of Acinus-S′, the levels of expression of two genes (the Meis2 and Hoxb1 genes) did not change, and one gene (the Hoxa1 gene) showed ligand-independent down-regulation and ligand-dependent up-regulation. Using ChIP assays, we confirmed that Acinus-S′ associates on the RAREs of CYP26A1 and RARβ2 in vivo. However, we did not explore Acinus-S′ binding to the RAREs of Meis2 and Hoxb1. It is possible that the architecture of the promoter regions of genes whose expression is unaffected by Acinus-S′ is not permissive for the association of Acinus-S′. Alternatively, Acinus-S′ may not be an effective regulator of Meis2 and Hoxb1 expression in the context of the other transcriptional regulatory proteins that regulate the expression of these two genes. Additionally, Hoxa1, which showed differential regulation by Acinus-S′ depending on the presence of exogenous RA, has a complex RA-inducible RAIDR5 enhancer sequence located in the 3′ end of its gene (18, 19). This enhancer region contained a DR5 RARE and two conserved DNA elements termed CE1 and CE2. It is possible that the protein complexes associated with CE1 and/or CE2 in the presence and absence of RA contribute to the differential effect of Acinus-S′ on Hoxa1 expression. Taken together, these results indicate that Acinus-S′ can affect the response of RAR-regulated genes in a promoter-specific manner.
The precise mechanism by which Acinus-S′ represses RAR-regulated gene expression is not yet known. Based on our data, Acinus-S′-mediated repression of gene expression is partially due to HDAC activity. Acinus has been reported to be in a complex, termed ASAP, which can potentially recruit HDAC (30, 33). ASAP consists of a stable Acinus-RNPS1 heterodimer which can associate with SAP18 (30, 33). While RNPS1 functions as a general activator of RNA splicing, SAP18 is a component of the SIN3-HDAC complex that represses transcription through deacetylation of histones in promoter regions (10, 42). Therefore, on the promoters of RAR-regulated genes, RARs could recruit the Acinus-RNPS1-SAP18 complex and ultimately associate with HDAC, resulting in the deacetylation of histones in the promoter region and leading to the repression of transcription. However, since TSA treatment does not fully restore RAR-dependent transcription in the presence of Acinus-S′, Acinus-S′ is likely to repress transcription by an additional mechanism(s). One likely mechanism may involve the control of mRNA processing by Acinus-S′ because of its reported association with components of the mRNA-processing machinery (27, 31, 33, 36, 44). Several coregulatory proteins, including PGC-1, CoAA, and CAPER, have been demonstrated to have a dual function in the regulation of gene expression by regulating transcription and mRNA processing (2, 3, 4, 11, 22).
Supplementary Material
Acknowledgments
We thank Andrea Tyukody for her expert technical assistance. We also thank Ronald Evans, The Salk Institute, La Jolla, CA, and Pierre Chambon, ICS-IGBMC, Illkirch, France, for the generous gift of the nuclear receptor clones used in these studies.
This work was supported by National Institutes of Health grants DK44517 (to D.R.S.) and DK67558 (to D.R.S.) and a grant from the Pennsylvania Department of Health (to D.R.S.).
The Pennsylvania Department of Health specifically disclaims responsibilities for any analyses, interpretations, and conclusions.
Footnotes
Published ahead of print on 4 February 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 2000. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25112-114. [DOI] [PubMed] [Google Scholar]
- 2.Auboeuf, D., E. Batsche, M. Dutertre, C. Muchardt, and B. W. O'Malley. 2007. Coregulators: transducing signal from transcription to alternative splicing. Trends Endocrinol. Metab. 18122-129. [DOI] [PubMed] [Google Scholar]
- 3.Auboeuf, D., D. H. Dowhan, M. Dutertre, N. Martin, S. M. Berget, and B. W. O'Malley. 2005. A subset of nuclear receptor coregulators act as coupling proteins during synthesis and maturation of RNA transcripts. Mol. Cell. Biol. 255307-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auboeuf, D., D. H. Dowhan, X. Li, K. Larkin, L. Ko, S. M. Berget, and B. W. O'Malley. 2004. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol. Cell. Biol. 24442-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastien, J., and C. Rochette-Egly. 2004. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 3281-16. [DOI] [PubMed] [Google Scholar]
- 6.Bastien, J., S. Adam-Stitah, T. Riedl, J. M. Egly, P. Chambon, and C. Rochette-Egly. 2000. TFIIH interacts with the retinoic acid receptor gamma and phosphorylates its AF-1-activating domain through cdk7. J. Biol. Chem. 27521896-21904. [DOI] [PubMed] [Google Scholar]
- 7.Bour, G., E. Gaillard, N. Bruck, S. Lalevee, J. L. Plassat, D. Busso, J. P. Samama, and C. Rochette-Egly. 2005. Cyclin H binding to the RARalpha activation function (AF)-2 domain directs phosphorylation of the AF-1 domain by cyclin-dependent kinase 7. Proc. Natl. Acad. Sci. USA 10216608-16613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bour, G., J. L. Plassat, A. Bauer, S. Lalevee, and C. Rochette-Egly. 2005. Vinexin beta interacts with the non-phosphorylated AF-1 domain of retinoid receptor gamma (RARgamma) and represses RARgamma-mediated transcription. J. Biol. Chem. 28017027-17037. [DOI] [PubMed] [Google Scholar]
- 9.Chambon, P. 1996. A decade of molecular biology of retinoic acid receptors. FASEB J. 10940-954. [PubMed] [Google Scholar]
- 10.Cheng, S. Y., and J. M. Bishop. 2002. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc. Natl. Acad. Sci. USA 995442-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowhan, D. H., E. P. Hong, D. Auboeuf, A. P. Dennis, M. M. Wilson, S. M. Berget, and B. W. O'Malley. 2005. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol. Cell 17429-439. [DOI] [PubMed] [Google Scholar]
- 12.Endoh, H., K. Maruyama, Y. Masuhiro, Y. Kobayashi, M. Goto, H. Tai, J. Yanagisawa, D. Metzger, S. Hashimoto, and S. Kato. 1999. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol. Cell. Biol. 195363-5372. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Epping, M. T., L. Wang, M. J. Edel, L. Carlee, M. Hernandez, and R. Bernards. 2005. The human tumor antigene PRAME is a dominant repressor of retinoic acid receptor signaling. Cell 122835-847. [DOI] [PubMed] [Google Scholar]
- 14.Fernandes, I., Y. Bastien, T. Wai, K. Nygard, R. Lin, O. Cormier, H. S. Lee, F. Eng, N. R. Bertos, N. Pelletier, S. Maden, V. K. Han, X. J. Yang, and J. H. White. 2003. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol. Cell 11139-150. [DOI] [PubMed] [Google Scholar]
- 15.Gianni, M., A. Tarrade, E. A. Nigro, E. Garattini, and C. Rochette-Egly. 2003. The AF-1 and AF-2 domains of RAR gamma 2 and RXR alpha cooperate for triggering the transactivation and the degradation of RAR gamma 2/RXR alpha heterodimers. J. Biol. Chem. 27834458-34466. [DOI] [PubMed] [Google Scholar]
- 16.Hu, Y., J. Yao, Z. Liu, X. Liu, H. Fu, and K. Ye. 2005. Akt phosphorylates acinus and inhibits its proteolytic cleavage, preventing chromatin condensation. EMBO J. 243543-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishitani, K., T. Yoshida, H. Kitagawa, H. Ohta, S. Nozawa, and S. Kato. 2003. p54nrb acts as a transcriptional coactivator for activation function 1 of the human androgen receptor. Biochem. Biophys. Res. Commun. 306660-665. [DOI] [PubMed] [Google Scholar]
- 18.Langston, A. W., and L. J. Gudas. 1992. Identification of a retinoic acid responsive enhancer 3′ of the murine homeobox gene Hox-1.6. Mech. Dev. 38217-227. [DOI] [PubMed] [Google Scholar]
- 19.Langston, A. W., J. R. Thompson, and L. J. Gudas. 1997. Retinoic acid-responsive enhancers located 3′ of the Hox A and Hox B homeobox gene clusters. Functional analysis. J. Biol. Chem. 2722167-2175. [DOI] [PubMed] [Google Scholar]
- 20.Lavery, D. N., and I. J. McEwan. 2005. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem. J. 391449-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, P. J., V. Lardeux, and P. Lefebvre. 2005. The proliferating cell nuclear antigen regulates retinoic acid receptor transcriptional activity through direct protein-protein interaction. Nucleic Acids Res. 334311-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monsalve, M., Z. Wu, G. Adelmant, P. Puigserver, M. Fan, and B. M. Spiegelman. 2000. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6307-316. [DOI] [PubMed] [Google Scholar]
- 23.Nowak, D. E., B. Tian, and A. R. Brasier. 2005. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. BioTechniques 39715-725. [DOI] [PubMed] [Google Scholar]
- 24.Purev, E., A. Giordano, D. R. Soprano, and K. J. Soprano. 2006. Interaction of PP2A catalytic subunit with Rb2/p130 is required for all-trans retinoic acid suppression of ovarian carcinoma cell growth. J. Cell. Physiol. 206495-502. [DOI] [PubMed] [Google Scholar]
- 25.Qi, C., J. Chang, Y. Zhu, A. V. Yeldandi, S. M. Rao, and Y. J. Zhu. 2002. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor alpha. J. Biol. Chem. 27728624-28630. [DOI] [PubMed] [Google Scholar]
- 26.Qin, P., J. M. Haberbusch, K. J. Soprano, and D. R. Soprano. 2004. Retinoic acid regulates the expression of PBX1, PBX2, and PBX3 in P19 cells both transcriptionally and post-translationally. J. Cell. Biochem. 92147-163. [DOI] [PubMed] [Google Scholar]
- 27.Rappsilber, J., U. Ryder, A. I. Lamond, and M. Mann. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 121231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rochette-Egly, C. 2003. Nuclear receptors: integration of multiple signaling pathways through phosphorylation. Cell. Signal. 15355-366. [DOI] [PubMed] [Google Scholar]
- 29.Rochette-Egly, C., S. Adam, M. Rossignol, J. M. Egly, and P. Chambon. 1997. Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell 9097-107. [DOI] [PubMed] [Google Scholar]
- 30.Sahara, S., M. Aoto, Y. Eguchi, N. Imamoto, Y. Yoneda, and Y. Tsujimoto. 1999. Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature 401168-173. [DOI] [PubMed] [Google Scholar]
- 31.Saitoh, N., C. S. Spahr, S. D. Patterson, P. Bubulya, A. F. Neuwald, and D. L. Spector. 2004. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 153876-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scafonas, A., C. L. Wolfgang, J. L. Gabriel, K. J. Soprano, and D. R. Soprano. 1997. Differential role of homologous positively charged amino acid residues for ligand binding in retinoic acid receptor alpha compared with retinoic acid receptor beta. J. Biol. Chem. 27211244-11249. [DOI] [PubMed] [Google Scholar]
- 33.Schwerk, C., J. Prasad, K. Degenhardt, H. Erdjument-Bromage, E. White, P. Tempst, V. J. Kidd, J. L. Manley, J. M. Lahti, and D. Reinberg. 2003. ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol. Cell. Biol. 232981-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shatkina, L., S. Mink, H. Rogatsch, H. Klocker, G. Langer, A. Nestl, and A. C. Cato. 2003. The cochaperone Bag-1L enhances androgen receptor action via interaction with the NH2-terminal region of the receptor. Mol. Cell. Biol. 237189-7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tairis, N., J. L. Gabriel, M. Gyda 3rd, K. J. Soprano, and D. R. Soprano. 1994. Arg269 and Lys220 of retinoic acid receptor-beta are important for the binding of retinoic acid. J. Biol. Chem. 26919516-19522. [PubMed] [Google Scholar]
- 36.Tange, T. O., T. Shibuya, M. S. Jurica, and M. J. Moore. 2005. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA 111869-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vuocolo, S., E. Purev, D. Zhang, J. Bartek, K. Hansen, D. R. Soprano, and K. J. Soprano. 2003. Protein phosphatase 2A associates with Rb2/p130 and mediates retinoic acid-induced growth suppression of ovarian carcinoma cells. J. Biol. Chem. 27841881-41889. [DOI] [PubMed] [Google Scholar]
- 38.Wei, L. N. 2004. Retinoids and receptor interacting protein 140 (RIP140) in gene regulation. Curr. Med. Chem. 111527-1532. [DOI] [PubMed] [Google Scholar]
- 39.Wu, X., H. Li, and J. D. Chen. 2001. The human homologue of the yeast DNA repair and TFIIH regulator MMS19 is an AF-1-specific coactivator of estrogen receptor. J. Biol. Chem. 27623962-23968. [DOI] [PubMed] [Google Scholar]
- 40.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9140-147. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, D., S. Vuocolo, V. Masciullo, T. Sava, A. Giordano, D. R. Soprano, and K. J. Soprano. 2001. Cell cycle genes as targets of retinoid induced ovarian tumor cell growth suppression. Oncogene 207935-7944. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, Y., Z. W. Sun, R. Iratni, H. Erdjument-Bromage, P. Tempst, M. Hampsey, and D. Reinberg. 1998. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell 11021-1031. [DOI] [PubMed] [Google Scholar]
- 43.Zhao, Y., K. Goto, M. Saitoh, T. Yanase, M. Nomura, T. Okabe, R. Takayanagi, and H. Nawata. 2002. Activation function-1 domain of androgen receptor contributes to the interaction between subnuclear splicing factor compartment and nuclear receptor compartment. Identification of the p102 U5 small nuclear ribonucleoprotein particle-binding protein as a coactivator for the receptor. J. Biol. Chem. 27730031-30039. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419182-185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.