Abstract
The genes transcribed by RNA polymerase III (Pol III) generally have intragenic promoter elements. One of them, the yeast U6 snRNA (SNR6) gene is activated in vitro by a positioned nucleosome between its intragenic box A and extragenic, downstream box B separated by ∼200 bp. We demonstrate here that the in vivo chromatin structure of the gene region is characterized by the presence of an array of positioned nucleosomes, with only one of them in the 5′ end of the gene having a regulatory role. A positioned nucleosome present between boxes A and B in vivo does not move when the gene is repressed due to nutritional deprivation. In contrast, the upstream nucleosome which covers the TATA box under repressed conditions is shifted ∼50 bp further upstream by the ATP-dependent chromatin remodeler RSC upon activation. It is marked with the histone variant H2A.Z and H4K16 acetylation in active state. In the absence of H2A.Z, the chromatin structure of the gene does not change, suggesting that H2A.Z is not required for establishing the active chromatin structure. These results show that the chromatin structure directly participates in regulation of a Pol III-transcribed gene under different states of its activity in vivo.
In the budding yeast, ∼300 genes transcribed by RNA polymerase III (Pol III) yield the short noncoding RNAs like tRNA, U6 snRNA, and 5S rRNA, for example (25). The basal transcription factor of Pol III, TFIIIC, binds to the two intragenic promoter elements, boxes A and B, and recruits the initiation factor TFIIIB 30 bp upstream of the transcription start site, which recruits Pol III in turn (18, 52). U6 snRNA is conserved from yeast to mammals and is transcribed by RNA Pol III (9). Unlike other Pol III-transcribed genes, box B in the U6 snRNA gene (SNR6) of Saccharomyces cerevisiae is found 120 bases downstream of the terminator (10), positioning it ∼200 bp away from box A, in contrast to the optimal 50- to 60-bp separation of the two found in most tDNAs. The TATA box at the −30-bp position can drive the transcription in vitro on naked DNA templates, but box B and TFIIIC are required to relieve the chromatin-mediated repression of SNR6 (10, 11, 17), indicating that TFIIIC helps alter the chromatin structure in favor of TFIIIB binding.
Active participation of chromatin in the transcription by Pol II is well documented (30). In contrast, little is known about the role of chromatin in regulation of Pol III-transcribed genes. This may be partly because of the prevailing notion that Pol III-transcribed genes are devoid of nucleosomes (37, 66). Recently, several genome-wide studies have reported association of chromatin modifying complexes with Pol III-transcribed genes. The chromatin remodeling complex of yeast, Isw2, localizes to a large number of tDNAs (20) while RSC is targeted to virtually all Pol III-transcribed genes (38). Histone chaperone Asf1 also localizes to many Pol III genes including SNR6 (53). Global nucleosome depletion activates transcription of some unusual Pol III genes that are not transcribed under normal conditions (23), showing a repressive role of chromatin on Pol III transcription. All these studies suggest that various chromatin remodeling and modification machineries may be required even for the Pol III transcription in vivo.
Transcription by Pol III is tightly regulated in response to growth stage, growth conditions, and stress by a central regulator, Maf1 (19, 65). Nutrient deprivation is known to down-regulate Pol III transcription as well as Pol I transcription. Nitrogen and carbon starvation especially leads to immediate downregulation of transcription by Pol III (13, 42). Nutrient availability is sensed by the TOR (target of rapamycin) pathway, and the effects of starvation can be mimicked by the drug rapamycin, which inhibits the TOR pathway (5). However, there are no studies available on the response of chromatin structure of Pol III-transcribed genes to repression.
The reported in vivo chromatin structure of SNR6, which is lost in the absence of TFIIIC binding, has one nucleosome downstream of the B box, an array of nucleosomes upstream of the TATA box, and a subnucleosomal size (∼100 bp) protection between boxes A and B (21, 35), leading to the idea that the DNA between boxes A and B is occupied not by a nucleosome but by a non-histone protein like Nhp6 (21, 29, 34). Our previous in vitro studies have shown that binding of TFIIIC and TFIIIB to SNR6 leads to the positioning of a nucleosome between boxes A and B as well as upstream of the TATA box (56, 57). As these results strongly advocate for the presence of a nucleosome between boxes A and B in vivo, we used histone depletion and repression conditions to find the chromatin structure of the SNR6 gene region in vivo. The results presented in this study unambiguously show the presence of a nucleosome between the A and B boxes in vivo and explain the previously reported subnucleosomal size protection (21) seen between the A and B boxes in vivo. We identify RSC as the remodeler, which keeps the TATA box nucleosome free, and show the presence of the histone variant H2A.Z in the nucleosome upstream of the TATA box. Our results suggest that this variant does not affect gene expression but may be required for demarcating the chromatin structure of a gene.
MATERIALS AND METHODS
Yeast strains and media.
Yeast strains used in this study were gifts from different sources, as listed in Table 1. Cells were grown in YEP (yeast extract and peptone) medium containing either 2% glucose or 2% galactose. Cells were nutritionally deprived by shifting to 0.15× YEP without any carbon source (47, 48) and allowed to grow for 1 h longer, unless otherwise stated.
TABLE 1.
Yeast strains used in this study
| Yeast strain | Genotype | Reference or source |
|---|---|---|
| UKY403 | MATaade2-101 his3-Δ200 leu2-3,112 lys2-80 trp1-Δ901 ura3-52 GAL+ thr tyr arg4-1 Δh4-1 [HIS3+] Δh4-2 [LEU2+]/pUK421(TRP+, GAL1-H4-2+) | 24 |
| MHY308 | MATaade2-101 his3-Δ200 leu2-3,112, lys2-801 trp1-Δ901 ura3-52 GAL+ thr tyr arg4-1 Δh4-1 [HIS3+] Δh4-2 [LEU2+]/pUK499(URA3+ H4-2+) | 24 |
| FLAG-H2B | MATahta1-htb1Δ::LEU2 hta2-htb2Δ::TRP1 leu2-Δ1 ura3-52 trp1-Δ63 his3-Δ200/pFB1251 (HIS3 CEN ARS HTA1 FLAG-HTB1) | 39 |
| RSC2-Myc | MATaura3-52 trp1-Δ63 his3-Δ200 leu2::PET56 RSC2-9Myc:TRP1 | 38 |
| MW671-Myc | MATα ade2-101 his3Δ-200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 rpc160::HIS3 MAF1-13Myc:KanMX6 pC160-240(TRP1, 3HA-RPC160) | 41 |
| YM1730 | MATα his3-Δ0 leu2-Δ0 ura3-Δ0 lys2-Δ0 htz1::Kan | 36 |
| SWR1-TAP | MATahis3-1 leu2-0 met15-0 ura3-0 SWR1-TAP:HIS | 16 |
| MW3993 | MATα ura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 rsc4-Δ4::HIS3 | 58 |
| MW4019 | MATα ura3-52 his3-Δ200 ade2-101 trp1-Δ63 lys2-801 leu2-Δ1 rsc4-Δ4::HIS3 STH1-13Myc:KanMX6 | 58 |
| YBL467 | MATaura3-1 lys2Δ::hisG trp1-1 his3-11,15 leu2-3,112 can1-100 Hta1-Flag:LoxP/Hta2-3FLAG:Kan | 31 |
| YBL325 | MATaura3-1 lys2Δ::hisG trp1-1 his3-11,15 leu2-3,112 can1-100 Htz1-3xFlagP:LoxP | 31 |
| YBL556 | MATahis3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 Htz1-TAP:HIS | 31 |
| YBL557 | MATahis3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 SWR1Δ::KANMX6 Htz1-TAP:HIS | 31 |
| YJW253 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 sas2Δ::TRP1 | 54 |
| YJW458 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 SAS4-13Myc:kanMX6, SAS2-TAP:TRP1 | 54 |
| YWJS069 | MATahis3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 HTZ1-TAP:His3MX6 sas2Δ::kanMX | 54 |
| YTT196 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 isw2::LEU2 | 63 |
| W3031a | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | Laboratory stock |
In vivo chromatin structure analysis.
Chromatin structure in vivo was analyzed by the indirect end-labeling (IEL) method (27), wherein instead of pure nuclei, the spheroplasts were directly digested with micrococcal nuclease (MNase). This is a rapid protocol which concludes in ∼8 min, thus reducing the chance for any change in chromatin structure due to prolonged stress. This is more important here because Pol III transcription responds to cell wall stress, and the transcription is repressed within a short time (64, 65). Cells were harvested and spheroplasted for 1 min (cells grown in galactose) or 40 s (cells grown in glucose). Spheroplasts were incubated with 15, 30, or 45 U of MNase (GE Healthcare) at 37°C for 4 min. DNA was extracted, digested with Pst1 and Nde1, and resolved on a 1.5% agarose gel. Southern blots were probed with a 230-bp PCR-amplified, 32P-labeled DNA, located 822 bp downstream of the transcription initiation site of SNR6, abutting the Pst1 end. A 100-bp ladder run on the same gel was transferred and probed separately for size-marking purposes.
Antibodies.
Antibodies antihemagglutinin (HA), anti-H4K16Ac (Abcam), anti-Myc (9E10) (Santa Cruz), FLAG-M2 agarose (Sigma), and immunoglobulin G Sepharose (GE Healthcare) were purchased.
ChIP assays.
For a better resolution in chromatin immunoprecipitation (ChIP) assays, the formaldehyde cross-linked cells were digested with MNase to get fragments predominantly of mono- or dinucleosomal size (33) with the following modifications. Cells were fixed in 1% formaldehyde for 15 min in the case of histones and Pol III and for 30 min for other factors. The chromatin was digested with 50 U of MNase (GE Healthcare) by incubation for 40 min at 37°C. Digestion was stopped by the addition of 10 mM EDTA, and the extract after centrifugation was incubated with antibodies overnight at 4°C. An extract equivalent to 20 optical density units of cells was used for each immunoprecipitation. For ChIP assay using anti-Myc antibody, chromatin was fragmented by sonication while TAP-tagged cells were disrupted using glass beads instead of zymolyase (3), and chromatin was digested with MNase. As ChIP assays with different tags show different relative enrichment values for the same target, probably due to the difference in the tag and the protocol followed, results of most of the experiments were confirmed by using different tags for the same protein. As shown in Fig. 1B (upper panel), three sets of primer pairs were used. The primer pair Upstream amplifies the region from bp −174 to −96 upstream of the TATA box, TATA-A box amplifies the region from bp −120 to +10 encompassing the TATA box and transcription start site (+1), and the primer pair A-B boxes amplifies the region from bp +61 to +187 between boxes A and B.
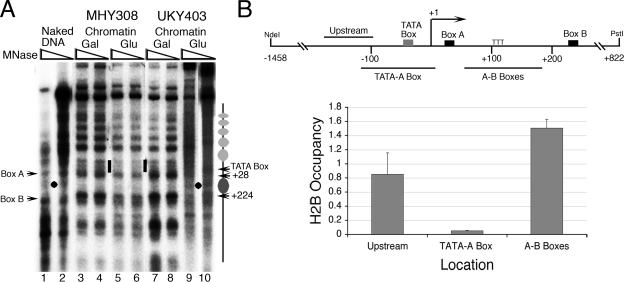
FIG. 1.
Yeast SNR6 is nucleosomal in vivo. (A) IEL analysis of the chromatin structure in strain UKY403 and its isogenic strain MHY308. The cells grown in YEP-galactose medium to an A600 of 0.7 were shifted to YEP medium containing glucose and harvested after 3 h. Gray ovals denote the nucleosomal size protections. The dot denotes the single MNase cut between boxes A and B in the naked DNA. The short bar marks the exposed region around the TATA box in the chromatin. (B) Nucleosome mapping on SNR6 by the ChIP assay. Chromatin with Flag-tagged histone H2B was immunoprecipitated after MNase digestion using FLAG-M2 agarose and analyzed by real-time PCR for different regions of SNR6. The upper panel shows the positions of different amplicons on the gene region. Numbers refer to the transcription initiation site (+1). Occupancies were calculated as relative enrichment over the TEL VIR region, and the averages from three independent experiments, with error bars, are shown.
Real-time PCR and quantification.
ChIP DNA was quantified in 10-μl reaction mixtures by a real-time PCR method based on Sybr green chemistry (3) using an ABI Prism 7900HT RT-PCR machine. Transcription by Pol III is absent from the subtelomeric region of chromosome VIR (TEL VIR), which is nucleosomal in structure. Either TEL VIR or the nonchromosomal, mitochondrial gene Cox3 was used as control regions for ChIP. Occupancies were calculated as the relative enrichment above the control values and normalized against the value for mock immunoprecipitation.
Expression analysis.
Total RNA was isolated according to Schmitt et al. (51). Primer extension to give cDNAs for U6 (98 bp) and a Pol II-transcribed U4 (116 bp) RNA was done using radioactively labeled primers, and products were resolved on a 10% polyacrylamide-urea gel (57). Bands were visualized by phosphorimaging and quantified using Image Gauge (Fuji) software.
RESULTS
The yeast U6 snRNA gene is nucleosomal in vivo.
To resolve the nature of the reported subnucleosomal size protection between boxes A and B of SNR6 in vivo, we used the yeast strain UKY403 in which histone H4 can be depleted when the cells are shifted from galactose to glucose for growth (24). The IEL technique (62) was used to study the chromatin structure of SNR6 under normal and histone depletion conditions (Fig. 1A). Since IEL is a low-resolution technique, the protections due to TFIIIB or TFIIIC binding to the boxes cannot be seen, and mappings can give an error of ∼20 bp. Nevertheless, compared to the naked DNA (lanes 1 and 2), we could see a protection of 190- to 200-bp size between boxes A and B (Fig. 1A, dark gray oval, lanes 3 to 4 and 7 to 8), flanked by two hypersensitive sites, one close to the A box (mapped to approximately bp +28) and one just upstream of the B box (mapped to approximately bp +224). This protection persists in the control strain MHY308 (lanes 5 to 6) when shifted to glucose. But in UKY403, the MNase cut between boxes A and B on the naked DNA (black dot) reappears in the chromatin lanes (lanes 9 to 10) under the histone depletion conditions, showing that the observed protection is nucleosomal. The region upstream of the A box, constituting the start site and TATA box, remains exposed to MNase digestion under all conditions, indicating that the whole stretch is devoid of nucleosomes (short bars). Compared to the naked DNA digestion pattern in lanes 1 and 2, the region upstream of the TATA box in both of the strains grown in galactose show an array of positioned nucleosomes from bp −70 (Fig. 1A, array of gray ovals) upward. This mapping agrees with the previously reported chromatin structure of SNR6, where high-resolution mapping showed the spread of the nucleosome array starting from bp −56 upward (35).
The IEL results shown in Fig. 1A indicate a nucleosome-free region of ∼100 bp covering the start site and TATA box flanked by two positioned nucleosomes, one between boxes A and B on the 3′ side and one upstream of the TATA box. We confirmed these results by ChIP assay on the cells expressing FLAG-tagged histone H2B. As shown in Fig. 1B, the region between boxes A and B shows a high level of occupancy by H2B, showing unambiguously that the region between the A and B boxes is nucleosomal. The H2B occupancy over the region covering the TATA box and start site was very low while that over the region upstream of the TATA box was high, in accordance with the IEL result. Thus, our ChIP assays with MNase treatment could reproduce the IEL data quite well. Under normal growth conditions, SNR6 is expressed at high levels. The chromatin structure of SNR6 participates in activation of the gene (57) and may change under repressive conditions. Therefore, we analyzed the in vivo chromatin structure of the gene under repressed conditions.
Repression of Pol III transcription changes the chromatin structure of the SNR6 gene in vivo.
Transcription by Pol III is severely curtailed under all stress conditions. The common repressor under all stress conditions, Maf1, localizes to yeast SNR6 in vivo (41, 48) and represses SNR6 transcription in vitro (14, 44). We subjected yeast cells to nutritional deprivation by shifting them to 0.15× YEP medium lacking a carbon source for various periods of time. As U6 snRNA is highly stable (12, 58), no significant reduction is seen in RNA level even after 4 h (see Fig. 4E) although repression can be established within 30 min. Therefore, we monitored SNR6 repression by analyzing the status of the Pol III transcription machinery, expecting a loss after repression. We checked for the occupancy of RPC160, the largest subunit of Pol III, which is reported to decrease under repression on tRNA genes (41). The ChIP assays on a strain harboring the RPC160 gene with three copies of an N-terminal HA tag showed that RPC160 occupancy drops drastically under repression for 40 min over both the TATA-A box region and the A-B boxes (Fig. 2A).
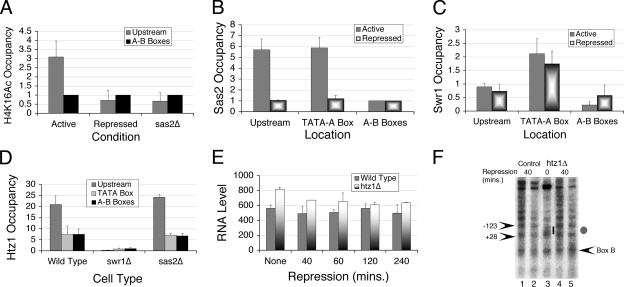
FIG. 4.
Deposition of Htz1 in the upstream nucleosome does not affect SNR6 expression. Occupancies were calculated as relative enrichment over the control region, and the averages from three independent experiments with error bars are shown in each of the panels. (A) Effect of repression and Sas2 deletion on H4K16 acetylation levels in the upstream nucleosome with respect to the TEL VIR region. (B) Occupancy of Sas2-TAP over SNR6 relative to the TEL VIR region decreases under repression. For both panels A and B, values are compared with the occupancy over the A-B box region. (C) Swr1-TAP occupancy over SNR6 relative to the TEL VIR region. (D) Htz1-TAP occupancy in wild-type, swr1Δ, and sas2Δ cells compared to the Cox3 region. (E) Time course analysis of the effect of repression on the U6 RNA level in the absence of Htz1. Total RNA was isolated from wild-type and htz1Δ cells and reverse transcribed with radioactively labeled primers specific for U6 snRNA and Pol II-transcribed U4 snRNA. (F) In vivo IEL analysis of chromatin before (lane 3) and after shifting the htz1Δ cells to 0.15× YEP medium for 1 h (lanes 4 and 5). Control (lanes 1 and 2) shows chromatin of wild-type cells under repression. Gray oval denotes the position of a nucleosomal protection region, while the bar shows the exposed region around the TATA box under the active state in lane 3. Arrowheads mark the positions of the MNase cut sites.
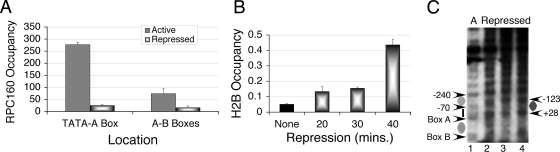
FIG. 2.
Chromatin structure of SNR6 under repression. Nutrient deprivation in 0.15× YEP medium lacking glucose was used to repress Pol III transcription. Occupancies were calculated as relative enrichment over the TEL VIR region, and the averages from three independent experiments, with error bars, are shown. (A) ChIP assays showing occupancy of the HA-tagged RPC160 over SNR6 in wild-type cells without (active) or with repression for 40 min (repressed). (B) Progressive increase in occupancy of the FLAG-H2B over TATA-A box region under repressed condition. (C) In vivo IEL analysis of chromatin before (lane 1) and after shifting the cells to 0.15× YEP medium for 1 h (lanes 2 to 4). Lane A, control sample under active state without repression. Gray ovals denote the position of a nucleosomal protection region. Arrowheads with numbers indicate the positions of MNase cut sites.
In contrast to Pol III occupancy, a time course study of H2B (FLAG tagged) levels over the TATA-A box region under repression shows a steady increase with time, demonstrating an approximately 10-fold increase after 40 min of repression (Fig. 2B). Therefore, we analyzed the SNR6 chromatin structure under repression by the IEL method (Fig. 2C). The upstream nucleosome (Fig. 2C, gray oval) (bp −70 to −240) is lost, and the TATA-A box region, which is exposed (short bar) in the active growth state (lane 1), shows a nucleosomal size protection (+28 to −123) (Fig. 2C, dark gray oval, lanes 2 to 4) with a 5′ hypersensitive boundary under repressed conditions. The nucleosome between boxes A and B is not lost as TFIIIC continues to occupy the gene under repression (47), in agreement with our previous observation that positioning of this nucleosome is TFIIIC dependent (57). No other change in nucleosomal array in the region further upstream is visible. This observation suggests that the response of the chromatin structure to repressive signals consists of altering the position of only one nucleosome in the upstream region of the gene out of a long array of positioned nucleosomes.
The nucleosome upstream of the TATA box consists of the histone variant H2A.Z.
In contrast to an increase in H2B levels over the TATA-A box region (Fig. 2B) after 40 min of repression, no difference is seen in the H2B levels over the upstream region while the level goes up between boxes A and B by 1.8-fold (Fig. 3A). Similarly, ChIP assays using a strain carrying a FLAG tag on both of the H2A genes (Fig. 3B) shows an increase of approximately twofold over the A-B box region and ∼10-fold over the TATA-A box region, comparable to the increase in H2B levels under repression. Surprisingly, H2A occupancy for the upstream nucleosome increased threefold upon repression, unlike H2B (compare Fig. 3A and B). This observation suggested a different dynamics for the upstream region and a possibility that the upstream nucleosome contains a variant H2A under active conditions, which is replaced by the canonical H2A under repression. Moreover, there is a significant overlap between the upstream and TATA-A box primer positions (Fig. 1B) as well as two alternative positions of the upstream nucleosome (Fig. 2C), which may be the reason for continued histone signals in this region under repression.
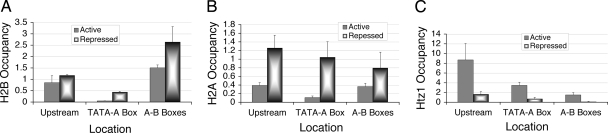
FIG. 3.
Enrichment of the upstream nucleosome with the histone variant H2A.Z. Occupancies were calculated as relative enrichment over the TEL VIR region. Results from ChIP assays showing averages from three independent experiments with error bars are shown. (A) Occupancy of the FLAG-H2B over upstream, TATA-A box, and A-B box regions under active or repressed state. (B) Occupancy of the FLAG-H2A over upstream, TATA-A box, and A-B box region in YEP medium with glucose or after 1 h of repression. (C) Occupancy of Htz1-FLAG on the upstream and A-B box regions of SNR6 after 1 h of repression.
A recent global study (2) has shown the presence of H2A.Z, the only H2A variant in budding yeast, coded by the HTZ1 gene, in the flanking regions of tRNA genes and in a single nucleosome upstream of SNR6 (http://nucleosomes.sysbio.bx.psu.edu/yeast-h2az/?chrom=12&feature=366236). To check for the presence of H2A.Z in the upstream nucleosome by ChIP assay, yeast strains harboring FLAG-tagged (Fig. 3C), HA-tagged, or TAP-tagged Htz1 (data not shown) were used. Though the absolute values were different, all three ChIP assays for Htz1 show low signal over the A-B box region and high enrichment in the upstream nucleosome (Fig. 3C). Our ChIP assay thus shows the presence of Htz1 containing the nucleosome on SNR6 at the same position as found in the whole-genome map (2). Further, ChIP assays for Htz1-FLAG (Fig. 3C) and Htz1-HA (data not shown) in the upstream nucleosome revealed a drastic drop of Htz1 occupancy upon repression. These results show that Htz1 in the upstream nucleosome is replaced by H2A under repression, as evident from the decrease in Htz1 (Fig. 3C) and increase in H2A (Fig. 3B) under repression. Deposition of H2A.Z in this nucleosome is probably not related to the transcription of the solo δ element present in the upstream region of U6, as discussed later.
The upstream nucleosome is acetylated at H4K16 by the SAS complex.
The relationship between Htz1 and histone acetylation is well documented. Acetylation of lysine at the 16th position (K16) of histone H4 is required for blocking the spread of heterochromatin near telomeres (28, 60), and H4K16 acetylation is required for subtelomeric incorporation of Htz1 (54). Therefore, we measured the H4K16 acetylation level in the upstream region of SNR6. A ChIP assay with antibodies specific for acetylated H4K16 shows that, compared to the A-B boxes, the upstream nucleosome is enriched with acetylation at H4K16, and, similar to Htz1, this modification is lost under repressive conditions (Fig. 4A), suggesting that it may be closely related to association of Htz1 with the upstream nucleosome in the active state of the gene. Acetylation of H4K16 in yeast is carried out by the SAS complex (55, 61). ChIP assays for H4K16 acetylation in a sas2Δ strain did not show any enrichment in the upstream nucleosome, indicating that the SAS complex is responsible for this acetylation (Fig. 4A).
To confirm that SAS is recruited to SNR6, we performed a ChIP assay for two subunits of SAS complex using the strains harboring Sas2-TAP or Sas4-myc. Compared to the region between the A and B boxes, the results show a clear enrichment of Sas2 (Fig. 4B) as well as Sas4 (data not shown) over both the TATA-A box and the upstream regions. Similar to acetylated H4K16, SAS occupancy over both of these regions is also reduced under repressed conditions (Fig. 4B). Results suggest that in the active state, the upstream nucleosome is marked with H4K16 acetylation by the SAS complex.
H2A.Z in the upstream nucleosome is deposited by the SWR1 complex.
The results shown in Fig. 3B and C suggest that the histone H2A in the upstream nucleosome is replaced by the variant H2A.Z under the active state. The SWR1 complex in yeast is reported to exchange Htz1 with H2A in an ATP-dependent manner (46). A ChIP assay for the TAP-tagged Swr1 subunit of SWR1 complex shows an enrichment in the TATA-A box as well as the upstream region of the gene, probably due to the overlapping region of the two amplicons (Fig. 1B). Nevertheless, that Swr1 occupancy is approximately twofold higher over the TATA-A box region than over the upstream nucleosome region (Fig. 4C) suggests that Swr1 is localized close to the TATA box. Thus, it is possible that the Pol III machinery sitting at the TATA box recruits SWR1. As expected, the occupancy was low over the A-B box region. No significant difference in Swr1 occupancy upon repression is seen anywhere (Fig. 4C). However, compared to the wild type, no Htz1-TAP enrichment in the swr1Δ background is seen in the upstream nucleosome (Fig. 4D), confirming that the SWR1 complex is responsible for the Htz1 deposition near SNR6. These results show that active transcription of SNR6 marks the region upstream of the TATA box with a positioned nucleosome containing Htz1 in an Swr1-dependent manner.
H2A.Z deposition in the upstream nucleosome does not require H4K16 acetylation.
Acetylation of H4K16 is required for subtelomeric incorporation of Htz1 (54). We asked whether H4K16 acetylation is a prerequisite for Htz1 deposition in the upstream nucleosome of SNR6. Since we see the loss of H4K16 acetylation in the upstream nucleosome in a sas2Δ strain (Fig. 4A), we checked for Htz1-TAP occupancy in the sas2Δ background. However, since the loss of SWR1 or H4K16 acetylation impairs the Htz1 deposition on the subtelomeric region in sixth chromosome (TEL VIR), we used the mitochondrial Cox3 gene to normalize the Htz1-TAP occupancy in swr1Δ and sas2Δ strains. Surprisingly, sas2Δ cells do not show a decrease in the enrichment of Htz1 in the upstream nucleosome (Fig. 4D). These results with sas2Δ thus suggest that H4K16 acetylation is not a prerequisite for Htz1 deposition in the upstream nucleosome.
H2A.Z deposition is not required for SNR6 expression.
Htz1 has a wide variety of roles from transcription regulation to chromosomal stability (26, 45). To know the significance of the presence and dynamics of Htz1 in SNR6 transcription, we quantified the U6 transcript levels relative to U4 RNA levels in both wild-type and htz1Δ cells under active as well as repressed states (Fig. 4E). The results show that though RNA levels do not change due to repression, the presence of Htz1 appears to be slightly repressive for active transcription by Pol III in the wild-type cells. A small increase in the U6 transcript levels in htz1Δ is maintained even after 2 h of repression, suggesting that the chromatin structure in the absence of Htz1 may be different.
We compared the in vivo chromatin structure of the htz1Δ cells with that of wild-type cells under active or repressed conditions by the IEL method (Fig. 4F). Chromatin structure in both types of cells was indistinguishable under repressed conditions, showing the presence of a nucleosome covering the TATA box (Fig. 4F, gray oval, compare lanes 4 and 5 to lanes 1 and 2). The TATA box region remains exposed (short bar, lane 3) in the absence of H2A.Z under active conditions. Results shown in Fig. 4E and F suggest that H2A.Z must be regulating SNR6 expression through an unknown, indirect mechanism without having a direct effect on the chromatin organization.
Chromatin remodeling by RSC in the upstream region of SNR6.
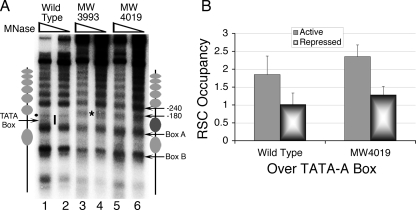
The demonstration that an ATP-dependent remodeling activity is required in vitro to generate a chromatin structure (56, 57) similar to the in vivo structure as reported here and elsewhere (35) prompted us to explore the effect of different ATP-dependent chromatin remodelers on SNR6 chromatin structure in vivo. Two chromatin remodelers, RSC and Isw2, are recruited to Pol III-transcribed genes (38, 20). Unlike results with tRNA genes (4), we did not find any effect of either Isw1 or Isw2 on SNR6 chromatin (data not shown). A deletion of 4 amino acids from the C terminus of the RSC4 subunit (RSC4-Δ4) of the RSC complex abolishes RSC interaction with Pol III, and this mutant is defective in SNR6 transcription also (58). Therefore, we explored the chromatin structure of SNR6 in two RSC4-Δ4 mutant strains, MW3993 and MW4019, by the in vivo IEL method. Strikingly, we could find that the entire region encompassing the TATA box and start site in both the mutant strains is protected from MNase cleavage (Fig. 5A). This region, exposed in the wild-type strain, becomes nucleosomal in the mutants (Fig. 5A, dark gray oval, lanes 3 to 6 versus lanes 1 to 2). Similar to results under repressed conditions (Fig. 2C), mapping of the cut sites measured the protection as ∼150 bp, from position +28 to −123, indicating the presence of a positioned nucleosome covering the TATA box and start site. The region upstream of the bp −125 position encompasses a hypersensitive region of ∼57 bp size (Fig. 5A, asterisk) (from bp −123 to −180). Therefore, the ∼60-bp region further upstream (from bp −180 to −240) is probably nonnucleosomal. The nucleosome between boxes A and B is retained, and no other change is seen in the nucleosomal array further upstream of bp −240. This observation reveals the possibility that during activation in the wild-type cells, RSC slides a positioned nucleosome from the TATA box to the upstream region. We had reported earlier a similar chromatin remodeling after TFIIIB binding to the TATA box in vitro that results in positioning of a nucleosome upstream of the TATA box (56). Similarly, recruitment of Isw2p by TFIIIB is also known to affect the chromatin upstream of some tRNA genes (4). To prove that this remodeling may indeed be due to RSC action, we confirmed its presence on the gene by the ChIP method. As previously reported (58), ChIP of myc-tagged RSC2 in the wild-type or myc-tagged Sth1 in the RSC4-Δ4 strain (MW4019) shows that both wild-type and mutant versions of RSC are recruited to the gene (Fig. 5B). Absence of RSC interaction with Pol III in the RSC4-Δ4 mutant suggests that Pol III is not required for recruitment of RSC but that the remodeling activity needs the interaction with Pol III. Earlier studies from our laboratory have shown a two-step chromatin remodeling mechanism wherein a similar remodeling in the upstream region required the recruitment of a remodeler by TFIIIB. Thus, it is possible that RSC is recruited by TFIIIB, but its contact with Pol III is required for the remodeling of the chromatin in the upstream gene region.
FIG. 5.
Chromatin structure of SNR6 in RSC4-Δ4 mutants. (A) The strain MW4019 differs from MW3993 in having the RSC subunit Sth1 as Myc tagged. IEL analysis shows the shift of an upstream nucleosome with an RSC4 mutation. Yeast cells were grown in YEP medium containing glucose to an A600 of 0.7. Gray ovals denote the nucleosomal size protections, and the bar marks the exposed region in lanes 1 and 2. The dot denotes a cut by MNase at the bp −70 position, and the asterisk denotes the new cut site spanning the bp −123 to −178 position. (B) Effect of repression on the occupancy of the Myc-tagged RSC subunits, RSC2 (RSC4 wild type), and Sth1 (RSC4 mutant MW4019).
The ChIP data have shown that H2A and H2B occupancies over the TATA-A box region increase upon repression (Fig. 3A and B), while the nucleosome positions over the TATA box in the RSC4-Δ4 mutant (Fig. 5A) and repressed wild-type cells (Fig. 2C) are similar. These observations suggest that under repressive conditions, the RSC-mediated remodeling over SNR6 may be abolished. A ChIP assay shows that RSC occupancy over SNR6 under repression goes down by twofold in both wild-type and RSC4-Δ4 mutant strains (Fig. 5B). This result confirms that the loss in remodeling activity of RSC under the repression conditions is responsible for the increased occupancy of histones over the TATA region under repression. Results also show that loss of RSC from the gene after TFIIIC binding probably helps repression set in, as TFIIIB alone is not enough to transcribe SNR6 chromatin (56).
Taken together, results presented in this study have shown that the chromatin structure of yeast SNR6 in vivo has a positioned nucleosome between boxes A and B and that another nucleosome is positioned upstream of its proximal sequence element (PSE)-like element due to chromatin remodeling in the TATA box-A box region by RSC. The gene is regulated by controlling the availability of the TATA box for the active preinitiation complex formation through mobility of this upstream nucleosome under various conditions, which is marked by the presence of the histone variant H2A.Z and acetylation at H4K16. These markings and chromatin modifiers are lost when the gene is repressed whereas the chromatin remodeling by RSC is independent of the H2A.Z deposition in the upstream nucleosome. The results suggest that while the downstream nucleosome in the transcribed region may provide the necessary template structure for the transcription, the upstream nucleosome may be regulatory in nature.
DISCUSSION
DNA between the A and B boxes is compacted by a nucleosome in vivo.
A non-histone protein like Nhp6 and not a nucleosome was suggested to occupy the DNA between boxes A and B of SNR6 (21, 29). However, Nhp6 deletion does not show any change in the MNase digestion pattern between boxes A and B (34), and recent reports suggest that the major role for Nhp6 in Pol III transcription may be in stabilization of the TFIIIB-DNA complex (7). Evidence provided by our studies for the presence of a nucleosome between the boxes A and B of yeast SNR6 in vivo has revealed the possibility that Pol III transcribes the DNA wound over a nucleosome in SNR6. According to our results, approximately one turn of the nucleosomal DNA (5′ half) is constituted by the transcribed region of the gene. The read-though and transcription of this nucleosome by Pol III during active transcription may result in the disruption of the histone-DNA contacts only in the transcribed region. This probably generates an altered structure in its 5′ half and makes the nucleosome less static, which may be the reason for the subnucleosomal size protection toward the 3′ half in high-resolution footprinting studies reported by others (21). Stabilization of this nucleosome under repressive conditions may be the reason for the observed twofold increase in H2A and H2B occupancy in the A-B box region upon repression (Fig. 3). The loss of the H2A-H2B dimer or an altered nucleosome structure during active transcription can lead to a more exposed nucleosomal DNA. The possibility of a role for FACT (facilitates chromatin transcription) or other factors involved in the exchange of the H2A-H2B dimer during transcription, as in the case of Pol II genes, cannot be excluded at this stage.
A positioned nucleosome in the upstream region of the U6 snRNA genes.
In vertebrates, U6 promoter is completely upstream without any B box element. The role of a positioned nucleosome in the upstream region of human U6 gene transcription is well documented (59, 67). The positioned nucleosome on the human U6 brings together two regulatory elements, PSE and distal sequence element, located upstream at positions −70 and −220 bp, respectively (67), while in case of yeast the positioned nucleosome brings boxes A and B together in the region downstream of the start site (57). Yeast has a PSE-like element at bp −48 to −59, although it is dispensable for transcription (17). Interestingly, we now find that, similar to the human U6, a nucleosome is positioned upstream of the region from bp −60 to −70 in the yeast U6 gene. These observations indicate that though the promoter structures of yeast and human U6 are entirely different, they have probably evolved through similar chromatin regulatory mechanisms.
Nutrient deprivation reduces total transcription activity, including that due to Pol II. In yeast, a Ty1 long terminal repeat (YLRCδ5) is located at bp −91 to −425 upstream of the transcription initiation site of the U6 snRNA gene. Solo δ elements lack the internal epsilon (ɛ) region (49), which constitutes the downstream activating sequences necessary for active Ty1 transcription. The solo δ transcription is reduced to low-level basal transcription if no enhancer elements are present nearby (6, 32). There are no Pol II enhancer elements near YLRCδ5, ruling out the possibility of its active transcription. In the absence of any regulatory regions, no changes in the basal transcription of YLRCδ5 are expected under starvation conditions, eliminating the possibility that Htz1 deposition is related to transcription of the long terminal repeat. Thus, Htz1 deposition in the upstream nucleosome may be directly related to U6 transcription.
Expression-related chromatin remodeling of yeast SNR6 in vivo.
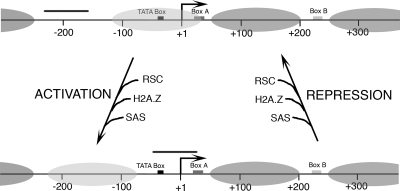
Binding of TFIIIC and TFIIIB to SNR6 chromatin in vitro leads to ATP-dependent chromatin remodeling whereby one nucleosome is positioned between boxes A and B and one nucleosome over the TATA box is shifted further upstream (56). This study shows that a similar mechanism operates in vivo and identifies RSC as the remodeler, which shifts the nucleosome from the TATA box region to a position further upstream, resulting in stabilization of the TFIIIB-DNA complex and transcriptional activation. As depicted in Fig. 6, the chromatin structure of SNR6 under repressed conditions has an array of positioned nucleosomes with only a short gap (∼80 bp long) in a nucleosome-free region upstream of its nonfunctional PSE-like element (Fig. 6, upper panel, bar). Under the active state, several chromatin modifying activities are recruited in the upstream region, most probably by the transcription preinitiation complex, which result in the upward shift of the nucleosome covering the TATA box. As the position of only one nucleosome in the upstream region, out of a long array of positioned nucleosomes, is changed, it appears that this nucleosome may be regulatory in nature and may require unique markings. In concurrence with this requirement, acetylation of histone H4K16 and replacement of histone H2A with its variant H2A.Z are found only in this nucleosome (Fig. 6, light gray oval). With repression of the gene, these markings, which are not found on any other nucleosome on the gene, are lost, and the nucleosome returns to the ground state, making the TATA box and start site unavailable. However, the nucleosome between boxes A and B is not lost under repression as it is TFIIIC dependent (57), and TFIIIC does not leave the gene under repression (47). This observation is also in agreement with an earlier report that the continuous presence of a DNA-binding protein is required to maintain chromatin structure (43). Under stress conditions, reduced/altered activity of TFIIIC or TFIIIB is proposed to be the reason behind the decline in Pol III transcript levels (13). Our studies show that the chromatin structure directly participates in the activation process of this Pol III-transcribed gene.
FIG. 6.
Changes in the chromatin structure of SNR6 under active or repressed conditions. For clarity, Pol III transcription machinery, which remains on the gene even under repression, is omitted. The bar denotes the DNA region exposed under these conditions. The position of the nucleosome between the A and B boxes (dark gray oval) does not change while the upstream nucleosome (shown in light gray) moves. When the gene is activated, RSC, SWR1, and SAS complexes are recruited to the gene. RSC slides one nucleosome to a position upstream of the TATA box, and H2A in this nucleosome is exchanged with H2A.Z by the SWR1 complex. The SAS complex acetylates H4K16. When the gene is repressed, RSC, SAS, and H2A.Z are lost, and this nucleosome goes back to cover the TATA box and transcription start site.
Role of H2A.Z in expression of Pol III-transcribed genes.
The transcription machinery of Pol III can influence the local chromatin structure (40). A recent global study has shown the presence of H2A.Z containing nucleosomes in the flanking regions of tRNA genes (2). Our ChIP assay confirms the data from the whole-genome mapping study that a nucleosome on SNR6 contains H2A.Z (2). All of these observations suggest that the presence of H2A.Z may also have relevance to the transcription of the genes transcribed by RNA Pol III. H2A.Z is reported to regulate Pol II transcription (1, 46, 50) as well as mark the repressed Pol II-transcribed genes for rapid induction (8, 22). As H2A.Z is also known to prevent ectopic spreading of silent heterochromatin (36), the reported block of heterochromatin spread by tRNA genes (15) may also be related to the presence of an H2A.Z-containing nucleosome in the flanking regions of these genes (2). The presence of H2A.Z in the upstream nucleosome of SNR6, in contrast, does not appear to have a similar function. Our results show that the presence of H2A.Z is not required for SNR6 expression, but its presence gives lower RNA levels under the active state. It appears that the deposition of H2A.Z in the upstream nucleosome, which would eventually be the target of remodeling under repression, is opposite from the effect seen on the Pol II-transcribed genes and may be to keep the transcript levels under check in the active state. Unlike Pol II-transcribed genes, the A-B box nucleosome in the transcribed region does not have H2A.Z, again suggesting that Pol III-transcribed genes may be following different pathways to regulate chromatin-related issues. Thus, the results in this study have shown for the first time that there is active participation of chromatin structure in the regulation of a small gene like SNR6 transcribed by Pol III and that the histone variant H2A.Z annotates the gene in its active state.
Acknowledgments
We thank Michael Grunstein, Olivier Lefebvre, Hiten Madhani, Frank Pugh, Kevin Struhl, Toshio Tsukiyama, Michel Werner, and Jerry Workman for kind gifts of the yeast strains used in this study.
Financial support from the Council of Scientific and industrial Research (CSIR), Government of India, is acknowledged. A.G.A. is a recipient of a CSIR senior research fellowship.
Footnotes
Published ahead of print on 11 February 2008.
REFERENCES
- 1.Adam, M., F. Robert, M. Larochelle, and L. Gaudreau. 2001. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 216270-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert, I., T. N. Mavrich, L. P. Tomsho, J. Qi, S. J. Zanton, S. C. Schuster, and B. F. Pugh. 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446572-576. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio, O., J. G. Geisberg, E. Sekinger, A. Yang, Z. Moqtaderi, and K. Struhl. 2005. Unit 21.3. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Cell Biol. doi: 10.1002/0471142727.mb2103s69. [DOI] [PubMed]
- 4.Bachman, N., M. E. Gelbart, T. Tsukiyama, and J. D. Boeke. 2005. TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev. 19955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 725-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeke, J. D., and S. B. Sandmeyer. 1991. Yeast transposable elements, p. 193-261. In J. R. Broach, J. R. Pringle, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 7.Braglia, P., S. L. Dugas, D. Donze, and G. Dieci. 2007. Requirement of Nhp6 proteins for transcription of a subset of tRNA genes and heterochromatin barrier function in Saccharomyces cerevisiae. Mol. Cell. Biol. 271545-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brickner, D. G., I. Cajigas, Y. Fondufe-Mittendorf, S. Ahmed, P. C. Lee, J. Widome, and J. H. Brickner. 2007. H2A.Z mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5704-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brow, D. A., and C. Guthrie. 1988. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature 334213-218. [DOI] [PubMed] [Google Scholar]
- 10.Brow, D. A., and C. Guthrie. 1990. Transcription of a yeast U6 snRNA gene requires a polymerase III element in a novel position. Genes Dev. 41345-1356. [DOI] [PubMed] [Google Scholar]
- 11.Burnol, A. F., F. Margottin, J. Huet, G. Almouzni, M. N. Prioleau, M. Mechali, and A. Sentenac. 1993a. TFIIIC relieves repression of U6 snRNA transcription by chromatin. Nature 362475-477. [DOI] [PubMed] [Google Scholar]
- 12.Burnol, A. F., F. Margottin, P. Schultz, M. C. Marsolier, P. Oudet, and A. Sentenac. 1993b. Basal promoter and enhancer element of yeast U6 snRNA gene. J. Mol. Biol. 233644-658. [DOI] [PubMed] [Google Scholar]
- 13.Clarke, E. M., C. L. Peterson, A. V. Brainard, and D. L. Riggs. 1996. Regulation of RNA polymerase I and III transcription systems in response to growth conditions. J. Biol. Chem. 27122189-22195. [DOI] [PubMed] [Google Scholar]
- 14.Desai, N., J. Lee, R. Upadhya, Y. Chu, R. D. Moir, and I. M. Willis. 2005. Two steps in Maf1-dependent repression of transcription by RNA polymerase III. J. Biol. Chem. 2806455-6462. [DOI] [PubMed] [Google Scholar]
- 15.Donze, D., and R. T. Kamakaka. 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20520-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durant, M., and B. F. Pugh. 2007. NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 275327-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eschenlauer, J. B., M. W. Kaiser, V. L. Gerlach, and D. A. Brow. 1993. Architecture of a yeast U6 RNA gene promoter. Mol. Cell. Biol. 133015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geiduschek, E. P., and G. A. Kassavetis. 2001. The RNA polymerase III transcription apparatus. J. Mol. Biol. 3101-26. [DOI] [PubMed] [Google Scholar]
- 19.Geiduschek, E. P., and G. A. Kassavetis. 2006. Transcription: adjusting to adversity by regulating RNA polymerase. Curr. Biol. 16R849-R851. [DOI] [PubMed] [Google Scholar]
- 20.Gelbart, M. E., N. Bachman, J. Delrow, J. D. Boeke, and T. Tsukiyama. 2005. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 19942-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerlach, V. L., S. K. Whitehall, E. P. Geiduschek, and D. A. Brow. 1995. TFIIIB placement on a yeast U6 RNA gene in vivo is directed primarily by TFIIIC rather than by sequence-specific DNA contacts. Mol. Cell. Biol. 151455-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gligoris, T., G. Thireos, and D. Tzamarias. 2007. The Tup1 corepressor directs Htz1 deposition at a specific promoter nucleosome marking the Gal1 gene for rapid activation. Mol. Cell. Biol. 274198-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guffanti, E., R. Percudani, O. Harismendy, J. Soutourina, M. Werner, M. G. Iacovella, R. Negri, and G. Dieci. 2006. Nucleosome depletion activates poised RNA polymerase III at unconventional transcription sites in Saccharomyces cerevisiae. J. Biol. Chem. 28129155-29164. [DOI] [PubMed] [Google Scholar]
- 24.Han, M., and M. Grunstein. 1988. Nucleosome loss activates yeast downstream promoters in vivo. Cell 551137-1145. [DOI] [PubMed] [Google Scholar]
- 25.Hani, J., and H. Feldmann. 1998. tRNA genes and retroelements in yeast genome. Nucleic Acids Res. 26689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamakaka, R. T., and S. Biggins. 2005. Histone variants: deviants? Genes Dev. 19295-316. [DOI] [PubMed] [Google Scholar]
- 27.Kent, N. A., and J. Mellor. 1995. Chromatin structure snapshots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 233786-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura, A., T. Umehara, and M. Horikoshi. 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32370-377. [DOI] [PubMed] [Google Scholar]
- 29.Kruppa, M., R. D. Moir, D. Kolodrubetz, and I. M. Willis. 2001. Nhp6, an HMG1 protein, functions in SNR6 transcription by RNA polymerase III in S. cerevisiae. Mol. Cell 7309-318. [DOI] [PubMed] [Google Scholar]
- 30.Li, B., M. Carey, and J. W. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 31.Li, B., S. G. Pattenden, D. Lee, J. Gutierrez, J. Chen, C. Seidel, J. Gerton, and J. L. Workman. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 10218385-18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao, X. B., J. J. Clare, and P. J. Farabaugh. 1987. The upstream activation site of a Ty2 element of yeast is necessary but not sufficient to promote maximal transcription of the element. Proc. Natl. Acad. Sci. USA 848520-8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, C. L., T. Kaplan, M. Kim, S. Buratowski, S. L. Schreiber, N. Friedman, and O. Rando. 2005. Single nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 31753-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez, S., M. Livingstone-Zatchej, S. Jourdain, F. Thoma, A. Sentenac, and M. C. Marsolier. 2001. High-mobility-group proteins NHP6A and NHP6B participate in activation of the RNA polymerase III SNR6 gene. Mol. Cell. Biol. 213096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsolier, M. C., S. Tanaka, M. Livingstone-Zatchej, M. Grunstein, F. Thoma, and A. Sentenac. 1995. Reciprocal interferences between nucleosomal organization and transcriptional activity of the yeast SNR6 gene. Genes Dev. 9410-422. [DOI] [PubMed] [Google Scholar]
- 36.Meneghini, M. D., M. Wu, and H. D. Madhani. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112725-736. [DOI] [PubMed] [Google Scholar]
- 37.Morse, R. H., S. Y. Roth, and R. T. Simpson. 1992. A transcriptionally active tRNA gene interferes with nucleosome positioning in vivo. Mol. Cell. Biol. 124015-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2002b. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 16806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng, H. H., R. Xu, Y. Zhang, and K. Struhl. 2002a. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 27734655-34657. [DOI] [PubMed] [Google Scholar]
- 40.Noma, K., H. P. Cam, R. J. Maraia, and S. I. Grewal. 2006. A role for TFIIIC transcription factor complex in genome organization. Cell 125859-872. [DOI] [PubMed] [Google Scholar]
- 41.Oficjalska-Pham, D., O. Harismendy, W. J. Smagowicz, A. Gonzalez de Peredo, M. Boguta, A. Sentenac, and O. Lefebvre. 2006. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated phosphorylation of Maf1. Mol. Cell 22623-632. [DOI] [PubMed] [Google Scholar]
- 42.Oliver, S. G., and C. S. McLaughlin. 1977. The regulation of RNA synthesis in yeast. I: Starvation experiments. Mol. Gen. Genet. 154145-153. [DOI] [PubMed] [Google Scholar]
- 43.Pazin, M. J., P. Bhargava, E. P. Geiduschek, and J. T. Kadonaga. 1997. Nucleosome mobility and the maintenance of nucleosome positioning. Science 276809-812. [DOI] [PubMed] [Google Scholar]
- 44.Pluta, K., O. Lefebvre, N. C. Martin, W. J. Smagowicz, D. R. Stanford, S. R. Ellis, A. K. Hopper, A. Sentenac, and M. Boguta. 2001. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol. Cell. Biol. 215031-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pusarla, R., and P. Bhargava. 2005. histones in functional diversification core histone variants. FEBS J. 2725149-5168. [DOI] [PubMed] [Google Scholar]
- 46.Raisner, R. M., and H. D. Madhani. 2006. Patterning chromatin: form and function for H2A.Z variant nucleosomes. Curr. Opin. Genet. Dev. 16119-124. [DOI] [PubMed] [Google Scholar]
- 47.Roberts, D. N., A. J. Stewart, J. T. Huff, and B. R. Cairns. 2003. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc. Natl. Acad. Sci. USA 10014695-14700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts, D. N., B. Wilson, J. T. Huff, A. J. Stewart, and B. R. Cairns. 2006. Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol. Cell 22633-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roeder, G. S., and G. R. Fink. 1980. DNA rearrangements associated with a transposable element in yeast. Cell 21239-249. [DOI] [PubMed] [Google Scholar]
- 50.Santisteban, M. S., T. Kalashnikova, and M. M. Smith. 2000. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103411-422. [DOI] [PubMed] [Google Scholar]
- 51.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acid Res. 183091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schramm, L., and N. Hernandez. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 162593-2620. [DOI] [PubMed] [Google Scholar]
- 53.Schwabish, M. A., and K. Struhl. 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22415-422. [DOI] [PubMed] [Google Scholar]
- 54.Shia, W. J., B. Li, and J. L. Workman. 2006. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 202507-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shia, W. J., S. Osada, L. Florens, S. K. Swanson, M. P. Washburn, and J. L. workman. 2005. Characterization of the yeast trimeric-SAS acetyl transferase complex. J. Biol. Chem. 28011987-11994. [DOI] [PubMed] [Google Scholar]
- 56.Shivaswamy, S., and P. Bhargava. 2006. Positioned nucleosomes due to sequential remodeling of the yeast U6 small nuclear RNA chromatin are essential for its transcriptional activation. J. Biol. Chem. 28110461-10472. [DOI] [PubMed] [Google Scholar]
- 57.Shivaswamy, S., G. A. Kassavetis, and P. Bhargava. 2004. High-level activation of transcription of the yeast U6 snRNA gene in chromatin by the basal RNA polymerase III transcription factor TFIIIC. Mol. Cell. Biol. 243596-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soutourina, J., V. Bordas-Le Floch, G. Gendrel, A. Flores, C. Ducrot, H. Dumay-Odelot, P. Soularue, F. Navarro, B. R. Cairns, O. Lefebvre, and M. Werner. 2006. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol. Cell. Biol. 264920-4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stunkel, W., I. Kober, and K. H. Seifart. 1997. A nucleosome positioned in the distal promoter region activates transcription of the human U6 gene. Mol. Cell. Biol. 174397-4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suka, N., K. Luo, and M. Grunstein. 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine 16 and spreading of heterochromatin. Nat. Genet. 32378-383. [DOI] [PubMed] [Google Scholar]
- 61.Sutton, A., W. J. Shia, D. Band, P. D. Kaufman, S. Osada, J. L. Workman, and R. Sternglanz. 2003. Sas4 and Sas5 are required for the histone acetyl transferase activity of Sas2 in the SAS complex. J. Biol. Chem. 27816887-16892. [DOI] [PubMed] [Google Scholar]
- 62.Thoma, F., L. W. Bergman, and R. T. Simpson. 1984. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J. Mol. Biol. 177715-733. [DOI] [PubMed] [Google Scholar]
- 63.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Upadhya, R., J. Lee, and I. M. Willis. 2002. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol. Cell 101489-1494. [DOI] [PubMed] [Google Scholar]
- 65.Willis, I. M., and R. D. Moir. 2007. Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem. Sci. 3251-53. [DOI] [PubMed] [Google Scholar]
- 66.Wittig, S., and B. Wittig. 1982. Function of a functional tRNA gene promoter depends on nucleosome position. Nature 29731-38. [DOI] [PubMed] [Google Scholar]
- 67.Zhao, X., P. S. Pendergrast, and N. Hernandez. 2001. A positioned nucleosome on the human U6 promoter allows recruitment of SNAPc by the Oct-1 POU domain. Mol. Cell 7539-549. [DOI] [PubMed] [Google Scholar]