Abstract
Alternative or regulated splicing can be applied to genes that are transcribed but whose products may be deleterious or unnecessary to the cell. In the yeast Saccharomyces cerevisiae, positive splicing regulation occurs during meiosis in which diploid cells divide to form haploid gametes. The Mer1 protein recruits the U1 snRNP to specific pre-mRNAs, permitting spliceosomal assembly and splicing. The mature transcripts are required for meiotic progression and, subsequently, sporulation. We have identified a novel allele (snu56-2) of the essential U1 snRNP protein Snu56p that exhibits a sporulation defect. Using a CUP1 reporter assay and reverse transcriptase PCR, we demonstrate that this allele specifically impairs Mer1p-activated splicing. This is not a reflection of a generally deficient spliceosome, as these cells splice vegetative transcripts efficiently. Furthermore, Snu56p depletion in vivo does not significantly impact mitotic splicing. Thus, its splicing function appears to be limited to Mer1p-activated meiosis-specific splicing. Two-hybrid studies indicate that Snu56p interacts with the other two U1 snRNP factors (Mer1p and Nam8p) required for this process. Interestingly, these two proteins do not interact, suggesting that Snu56p links pre-mRNA-bound Mer1p to Nam8p in the U1 snRNP. This work demonstrates that the Snu56 protein is required for splicing only during meiosis.
Precursor mRNAs (pre-mRNAs) in eukaryotes contain intervening sequences that must be removed to create a functional mRNA. The process of removing introns, or splicing, occurs in the nucleus and is catalyzed by a multisubunit complex termed the spliceosome (30). The spliceosome is built by the ordered association of the U1, U2, and U4/U5/U6 small nuclear ribonucleoprotein particles (snRNPs) and extrinsic protein factors with the pre-mRNA (reviewed in references 5, 6, and 25). Some metazoan pre-mRNAs can be alternatively spliced to produce mature mRNAs with different combinations of exons to encode distinct proteins (37). In metazoa and in yeast, splice site choice can be changed to switch from production of a functional mRNA to that of a nonfunctional one, in effect turning genes on or off at a posttranscriptional level (37). Thus, alternative splicing provides a genetic mechanism to increase both the protein coding capacity and regulatory flexibility of the eukaryotic genome.
In budding yeast, examples of regulated or alternative splicing are rare. Positive regulation of mRNA splicing occurs during meiosis. Mer1p, a U1 snRNP-associated protein, is expressed only during meiosis and activates the splicing of at least three pre-mRNAs (AMA1, HFM1/MER2, and REC107/MER3) (8, 9, 17, 38). Mer1p has a KH domain RNA-binding motif (46) and can specifically bind RNA that contains a Mer1 enhancer element (47). An emerging model is that transcript-bound Mer1p acts at the very first stage of spliceosome assembly to recruit the U1 snRNP to pre-mRNA (47, 48).
Mer1p fails to activate splicing in nam8 and snu17 mutant cells. Both genes encode nonessential spliceosomal factors that are expressed during both vegetative growth and meiotic development. NAM8 was originally identified as a multicopy suppressor of mitochondrial splicing deficiencies (14). An independent screen revealed that Nam8p is an RNA-binding protein that is essential for meiosis and required for efficient splicing of Mer1p-activated transcripts (38, 39). Finally, a U1 synthetic lethal screen and biochemical fractionations classified Nam8p as a yeast U1 snRNP-specific protein (24, 40). In contrast, Snu17p specifically binds to the U2 snRNP (23). Its role in Mer1p-activated splicing was identified by a systematic examination of strains in which nonessential spliceosomal genes are deleted (48).
Here we report that the splicing function of a constitutive U1 snRNP protein, Snu56p, is exclusive to Mer1p-activated meiosis-specific splicing. While screening for yeast mutants lethal in the absence of the HMT1 arginine methyltransferase gene, we identified a novel snu56 allele (snu56-2) that produces a sporulation-deficient phenotype in homozygous diploid cells. Utilizing a reporter assay system and reverse transcription (RT)-PCR, we show that Mer1p-activated splicing is impaired in this mutant whereas splicing during vegetative or mitotic growth appears unaffected. We also examined a previously reported SNU56 mutation, mud10-1, that is lethal in combination with a viable U1 snRNA mutation (24). Interestingly, this temperature-sensitive mutant exhibits neither constitutive nor Mer1p-activated splicing defects at the restrictive temperature. Furthermore, vegetative splicing efficiency is unaffected by Snu56p depletion, indicating that splicing is not the essential function of this protein. Finally, we demonstrate that Snu56p interacts with the other two U1 components (Mer1p and Nam8p) required for Mer1p-activated splicing. The fact that these proteins do not interact suggests that Snu56p may serve as the link between pre-mRNA-bound Mer1p and Nam8p in the U1 snRNP. Our characterization of Snu56p shows how a constitutive splicing factor combines with a cell-type-specific factor to regulate splicing during the developmental process of meiosis in yeast.
MATERIALS AND METHODS
Strains.
The yeast strains used in this study are listed in Table 1. Genetic manipulations were performed essentially as described previously (27). MHY361, the parental strain for the synthetic lethal screen, is a MATα sister spore of PSY1115. MHY360 is MHY361 cured of plasmid pMHY40. Strain MHY394 (snu56-2 hmt1Δ) was isolated following ethyl methanesulfonate (EMS) mutagenesis of MHY361 cells. Strain MHY400, used for genetic analysis, was created by mating strain MHY361 to strain MHY447, sporulating the resultant diploid, and exchanging plasmid pMHY40 for plasmid pRS316 in the appropriate spore clone. Four outcrosses of strain MHY394 to MHY400 yielded MHY689 (snu56-2 hmt1Δ). Three crosses of MHY394 into the W303a background generated two snu56-2 strains lacking the HMT1 deletion, MHY706 and MHY707. MHY679 was created by transforming strain 14299 with pMHY32.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| W303a | MATaura3-1 leu2-3,112 ade2-1 his3-11,15 trp1-1 | A. Tzagoloff |
| W303α | MATα ade2-1 trp1Δ63 ura3-1 leu2-3,112 his3-11,15 can1-100 | A. Tzagoloff |
| BMA64 | MATa/MATα ura3-1/ura3-1 ade2-1/ade2-1 leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 trp1Δ/trp1Δ can1-100/can1-100 | 2 |
| BMA64-1A | MATaura3-1 ade2-1 trp1Δ leu2-3,112 his3-11,15 can1-100 | 2 |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Saccharomyces Deletion Project |
| CH1305 | MATaura3 ade2 ade3 leu2 lys2 can | 3 |
| KH46 | MATα cup1::ura3-52 trp1 leu2-3,112 his3-1 lys2 ura3 ade2-101 | 47 |
| KH52 | MATacup1::ura3-52 trp1 leu2-3,112 his3-1 ura3 ade2-101 | 47 |
| Nam8Δ | MATα cup1::ura3-52 nam8::HIS3 trp1 leu2-3,112 his3-1 lys2 ura3 ade2-101 | 47 |
| PSY1115 | MATahmt1::HIS3 his3 leu2 lys−ura3 ade2 ade3 + pMHY40 (2μm HMT1 URA3 ADE3) | 43 |
| Y352 | MATaura3 trp1 leu2 ade2 U1::ADE2 cup1::hisG mud10-1 + pXL8 (PGAL1-U1-TRP1) | 24 |
| 14299 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 rmt2::KanMX | Saccharomyces Deletion Project |
| 23599 | MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 MET15/met15 LYS2/lys2 SNU56/snu56::KanMX | Saccharomyces Deletion Project |
| MHY360 | MATα hmt1::HIS3 his3 leu2 lys−ura3 ade2 ade3 | This study |
| MHY361 | MATα hmt1::HIS3 his3 leu2 lys−ura3 ade2 ade3 + pMHY40 (2μm HMT1 URA3 ADE3) | This study |
| MHY394 | MATα snu56-2 hmt1::HIS3 his3 leu2 lys−ura3 ade2 ade3 + pMHY40 (2μm HMT1 URA3 ADE3) | This study |
| MHY400 | MATahmt1::HIS3 his3 leu2 ura3 ade2 ade3 + pRS315 | This study |
| MHY408 | MATasnu56-2 hmt1::HIS3 his3 leu2 lys1 ura3 ade2 ade3 + pMHY41 (2μm LEU2 HMT1 ADE3) | This study |
| MHY447 | MATahmt1::HIS3 his3 leu2 lys1 ura3 ade2 ade8 | 51 |
| MHY472 | MATa/MATα hmt1::TRP1/HMT1 ade2-1/ade2-1 leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 trp1Δ/trp1Δ ura3-1/ura3-1 can1-100/can1-100 | This study |
| MHY477 | MATahmt1::TRP1 ade2-1 leu2-3,112 his3-11,15 trp1Δ ura3-1 can1-100 | This study |
| MHY573 | MATα snu56::KanMX his3Δ1 leu2Δ0 ura3D0 met15? lys2Δ0 + pMHY155 (2μm HIS3 LexA-SNU56) | This study |
| MHY679 | MATα his3D1 leu2Δ0 lys2Δ0 ura3Δ0 rmt2::KanMX + pMHY32 (PGAL1-HRP1-MYC LEU2) | This study |
| MHY687 | MATα SNU56-URA3 hmt1::HIS3 his3 leu2 lys−ura3 ade2 ade3 | This study |
| MHY689 | MATα snu56-2 Δhmt1::HIS3 his3 leu2 lys−ura3 ade2 ade3 + pMHY41 (2μm LEU2 HMT1 ADE3) | This study |
| MHY706 | MATα snu56-2 lys leu2 his3 ura3 ade2 | This study |
| MHY707 | MATasnu56-2 leu2 his3 ura3 ade2 | This study |
| MHY787 | MATα mud10-1 cup::hisG ade2 trp1 ura3 leu2 | This study |
| MHY853 | MATasnu56::KanMX his3Δ leu2Δ0 ura3Δ0 lys2 MET15 or met15 + pMHY275 (CEN URA3 SNU56) | This study |
| MHY1295 | MATaura3-1 ade2-1 leu2-3,112 his3-11,15 trp1Δ can1-100 + pSH18-34 (URA3 lacZ reporter) | This study |
| MHY1296 | MATahmt1::TRP1 ade2-1 leu2-3,112 his3-11,15 trp1Δ ura3-1 can1-100 + pSH18-34(URA3 lacZ reporter) | This study |
| MHY1350 | MATacup1::ura3-52 snu56-2 trp1 leu2 his3 ura3 ade2 | This study |
| MHY1357 | MATα cup1::ura3-52 snu56-2 leu2 his3 ura3 ade2 | This study |
| MHY1393 | MATacup1::ura3-52 nam8::HIS3 leu2 his3 ura3 trp1 ade2 | This study |
| MHY1397 | MATα cup1::ura3-52 nam8::HIS3 leu2 his3 ura3 ade2 lys1 | This study |
| MHY1407 | MATa/MATα cup1::ura3-52/cup1::ura3-52 leu2/leu2 nam8::HIS3/nam8::HIS3 his3/his3 ade2/ade2 ura3/ura3 TRP1/trp1 LYS1/lys1 | This study |
| MHY1410 | MATa/MATα cup1::ura3-52/cup1::ura3-52 leu2/leu2 snu56-2/snu56-2 his3/his3 ura3/ura3 ade2/ade2 TRP1/trp1 + pRS316 (CEN URA3) | This study |
| MHY1430 | MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15? ura3Δ0/ura3Δ0 snu56::KanMX/snu56::KanMX lys2Δ0/lysΔ0 + pMHY441 (CEN LEU2 snu56-2) | This study |
| MHY1413 | MATα cup1::ura3-52 mud10-1 leu2 his3 ura3 lys1 | This study |
| MHY1414 | MATacup1::ura3-52 mud10-1 leu2 his3 ura3 trp1 | This study |
| MHY1444 | MATa/MATα cup1::ura3-52/cup1::ura3-52 leu2/leu2 mud10-1/mud10-1 his3/his3 ura3/ura3 LYS1/lys1 TRP1/trp1 | This study |
| MHY1451 | MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15? ura3Δ0/ura3Δ0 snu56::KanMX/snu56::KanMX lys2Δ0/lysΔ0 + pMHY275 (CEN URA3 SNU56) | This study |
| MHY1455 | MATasnu56::KanMX his3Δ1 leu2Δ0 ura3D0 lys2 MET15 or met15 + pMHY155 (2μm HIS3 LexA-SNU56) | This study |
| MHY1456 | MATasnu56-2(T925C) his3Δ1 leu2Δ0 ura3Δ0 met15 | This study |
| MHY1458 | MATα snu56-2(T925) his3Δ1 leu2Δ0 ura3Δ0 met15 lys2 | This study |
| MHY1462 | MATamud10-1 his3 leu2 ura3 lys2 | This study |
| MHY1463 | MATα mud10-1 his3 leu2 ura3 trp1 | This study |
| MHY1467 | MATamud10-1 his3 leu2 ura3 trp1 | This study |
| MHY1468 | MATα his3 leu2 ura3 lys2 | This study |
| MHY1480 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 snu56-td URA3 | This study |
| MHY1481 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 snu56-td URA3 PGAL-Ub-Myc-UBR1 HIS3 | This study |
| MHY1482 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 PGAL-Ub-Myc-UBR1 HIS3 + pRS316 (CEN URA3) | This study |
An hmt1::TRP1 heterozygous diploid strain (MHY472) was constructed by PCR (2) of pUC19-TRP1 with the 5′ΔHMT1-3′ΔHMT1 hybrid primer set and subsequent transformation into strain BMA64 to disrupt one genomic copy of HMT1. The resulting diploid strain (MHY472) was sporulated to generate an hmt1Δ haploid (MHY477).
A mud10-1 (MHY787) mutant was constructed by mating strains MHY679 and Y352. Sporulation of strain 23599 transformed with pMHY275A or pMHY155 yielded strain MHY853 or MHY1455, respectively. Two-hybrid assay strains (MHY1295 and MHY1296) were created by transforming plasmid pSH18-34 into BMA64-1A and MHY477, respectively. Strains MHY1350 and MHY1357 are cup1::ura3-52 snu56-2 sister spores generated by crossing MHY706 to KH52. Strains MHY1393 and MHY1397 are cup1::ura3-52 nam8::HIS3 sister spores generated by crossing MHY707 to Nam8Δ. Strains MHY1413 and MHY1414 are cup1::ura3-52 mud10-1 sister spores generated by crossing MHY787 to MHY853. Homozygous diploid cup1Δ nam8Δ (MHY1407), cup1Δ snu56-2 (MHY1410), and cup1Δ mud10-1 (MHY1444) strains were created by mating MHY1350-MHY1357, MHY1393-MHY1397, and MHY1413-MHY1414 haploid pairs, respectively. A homozygous snu56Δ diploid strain (MHY1451) was created by transforming MHY573 with pMHY441, mating the resulting transformant with MHY853, and then screening for a diploid that only carried plasmid pMHY275. Strains MHY1462, MHY1463, MHY1467, and MHY1468 were created by crossing MHY1414 and MHY748.
A pair of isogenic snu56-2 strains (MHY1456 and MHY1458) was created by integration of BssHII-linearized pMHY562 into 23599 diploid cells. Transformants were sporulated, and URA+ Gentr haploid cells were streaked onto 5-fluoroorotic acid (5-FOA)-containing plates. FOAr cells were then screened for Gents. A snu56-td strain (MHY1480) was generated by integrating MscI-linearized pMHY497 into BY4741. To achieve complete Snu56p-td depletion, UBR1 was overexpressed in MHY1481 from a galactose promoter following integration of PmeI-linearized pKL54. A UBR1 overexpression control (MHY1482) was made by simultaneously transforming BY4741 with PmeI-linearized pKL54 and pRS316.
Plasmid constructs.
All of the plasmids and primers used in this study are listed in Table 2. Plasmid pMHY92, a yeast centromere URA3 plasmid carrying wild-type HMT1 and its flanking sequences, was created by inserting a 4.5-kb SacI chromosomal fragment encoding HMT1 into pRS316. Plasmid pMHY40 was generated by subcloning a 4.5-kb SacI fragment from pMHY92 into the SacI site of pPS719. Plasmid pPS794 was generated in Pam Silver's laboratory by inserting a 3,673-bp fragment containing the ADE3 gene into BamHI-SpeI-digested pRS425. Plasmid pMHY41 was generated by subcloning a 4.5-kb SacI fragment from pMHY92 into the SacI site of pPS794. Plasmid pMHY562 was created by subcloning a 2.1-kb XbaI-XhoI fragment containing snu56-2 from pMHY554 into pRS306.
TABLE 2.
Plasmids and primers used in this study
| Plasmid or primer | Description | Source or reference |
|---|---|---|
| pACTII | Yeast LEU2 two-hybrid vector carrying GAL4 AD | 12 |
| pACT2-BBP1 | Bbp1p fused to the Gal4 AD of pACTII | 48 |
| pACT2-LUC7 | Luc7p fused to the Gal4 AD of pACTII | 48 |
| pACT2-MER1 | Mer1p fused to the Gal4 AD of pACTII | 48 |
| pACT2-MUD2 | Mud2p fused to the Gal4 AD of pACTII | 48 |
| pACT2-NAM8 | Nam8p fused to the Gal4 AD of pACTII | 48 |
| pACT2-PRP11 | Prp11p fused to the Gal4 AD of pACTII | 48 |
| pACT2-SNU17 | Snu17p fused to the Gal4 AD of pACTII | 48 |
| pACT2-SNU71 | Snu71p fused to the Gal4 AD of pACTII | 48 |
| pEG202 BD | Yeast HIS3 two-hybrid vector carrying LexA DNA BD | 22 |
| pKL54 | HIS3 PGAL1-ubiquitin-Met-lacI-Myc-UBR1 integration vector | 34 |
| pPS719 | 2μm URA3 ADE3 vector | 45 |
| pPS794 | 2μm LEU2 ADE3 vector | Pam Silver laboratory |
| pPW66R | URA3 cdc28-td integration vector | 11 |
| pRS306 | Yeast integration vector URA3 | 44 |
| pRS313 | Yeast CEN cloning vector HIS3 | 44 |
| pRS315 | Yeast CEN cloning vector LEU2 | 44 |
| pRS316 | Yeast CEN cloning vector URA3 | 44 |
| pRS425 | Yeast 2μm cloning vector LEU2 | 7 |
| pMHY32 | CEN LEU2 vector carrying HRP1-myc under GAL1 promoter control | 27 |
| pMHY40 | 2μm URA3 ADE3 vector carrying a 4.5-kb chromosomal fragment containing HMT1 | This study |
| pMHY41 | 2μm LEU2 ADE3 vector carrying a 4.5-kb chromosomal fragment containing HMT1 | This study |
| pMHY92 | CEN URA3 vector carrying a 4.5-kb chromosomal fragment containing HMT1 | This study |
| pMHY154 | CEN URA3 vector carrying SNU56 with NcoI site inserted at start codon | This study |
| pMHY155 | SNU56 fused to the LexA DNA BD of pEG202 | This study |
| pMHY275A | CEN URA3 vector carrying SNU56 (SNU56 orientation 1) | This study |
| pMHY275B | CEN URA3 vector carrying SNU56 (SNU56 orientation 2) | This study |
| pMHY441 | CEN LEU2 vector carrying snu56-2 | This study |
| pMHY471 | snu56-2 fused to LexA DNA BD of pEG202 | This study |
| pMHY472 | CEN HIS3 vector carrying snu56-2 | This study |
| pMHY478 | CEN HIS3 vector carrying SNU56 | This study |
| pMHY480 | URA3 yeast integration vector carrying SNU56 | This study |
| pMHY496 | URA3 snu56-td (full-length) integration vector | This study |
| pMHY497 | URA3 snu56-td (N terminus) integration vector | This study |
| pMHY551 | CEN LEU2 vector carrying snu56-2 | This study |
| pMHY554 | CEN LEU2 vector carrying snu56-2 (S389F only) | This study |
| pMHY562 | URA3 integration vector carrying snu56-2 (S389F only) | This study |
| 5′ΔHMT1 | 5′-GCCGTTTCCAAAAAAGAGTTAGAACCGACAAATTCATCCAAAGAA-3′ | |
| 3′ΔHMT1 | 5′-TTTGTTTATTTGCTTTTCAAATTTTTTTCTTTCTCCAGCAAACAA-3′ | |
| ACT1exon1 | 5′-ACTGAATTAACAATGGATTCTG-3′ | |
| ACT1exon2 | 5′-AGCTTCATCACCAACGTAGGAG-3′ | |
| AMA1 RT-PCR 1 | 5′-GTACACTCTCGCCCG-3′ | |
| AMA1 RT-PCR 2 | 5′-GTCACCTGTCCCGAG-3′ | |
| ASC1 Lower | 5′-GTACATAGCCTTCTTAGCAGCC-3′ | |
| ASC1 Upper | 5′-GTCCGATGTTATGTCCGTTGAC-3′ | |
| HFM1 Lower | 5′-CATCAGGTGTCTGCTCTAAATCG-3′ | |
| HFM1 Upper | 5′-GTTTGATCGCCTCGGTACAGG-3′ | |
| HOP2 Lower RT-PCR | 5′-TCAAGAGCGTCTTTGGC-3′ | |
| HOP2 Upper RT-PCR | 5′-CGATAGAGCCATTCAGGC-3′ | |
| pRS316 (1719-1738) | 5′-TAGGGCGCTGGCAAGTGTAG-3′ | |
| REC107 Lower | 5′-CCGACTTTCTCTTCTTCTCGTCG-3′ | |
| REC107 Upper | 5′-GGAGAGTATGGAGCTGAGGG-3′ | |
| RPL28 Lower | 5′-CCAACTTGGAGACGAATCTAGC-3′ | |
| RPL28 Upper | 5′-TGCCTTCCAGATTCACTAAGAC-3′ | |
| SNU56 primer 1 | 5′-GAGCTGATACCGCTCGCCGCAG-3′ | |
| SNU56 primer 2 | 5′-CGCAGGCCATGGCATGATACTAATATTTTTTGC-3′ | |
| SNU56 primer 3 | 5′-GAACCCATGGCGCCCAAGAAGGAGAGGATTAGC-3′ | |
| SNU56 primer 4 | 5′-GGATGAGGGCTGCTCTCTTCTATAGC-3′ | |
| snu56-2 S389F StuI Upper | 5′-ATATAGGCCTTTATGGATGCAGTGAAAAAGAAGTCTTCG-3′ | |
| snu56-2 S389F StuI Lower | 5′-ATATAAGGCCTTTATGGTACCAATGCAATGTTG-3′ | |
| SPO70-300 LOWER | 5′-CCAGAGCAGCATGACTTCTTG-3′ | |
| SPO70-300 UPPER | 5′-CCTAGAATGGTTCAAACCAGGTG-3′ | |
| TUB1exon1 | 5′-AGAGAAGTTATTAGTATTAATG-3′ | |
| TUB1exon2 | 5′-GGAACGAACTTACCGTAGCCGG-3′ | |
| XBA-SNU56 | 5′-GTACTCTAGACACCGATGGCTTTCTAACGG-3′ | |
| Xho-SNU56 | 5′-CACACTCGAGCGCTCTTTGAATCCGCGATGA-3′ | |
| XHO-SNU56 no. 3 | 5′-ACCACTCGAGCATCTTCTTACACCGCATTTGATAATAT-3′ |
A snu56-td integration vector (pMHY497) was generated in several steps. First, pPW66R was digested with HindIII to release CDC28, blunted, and religated to generate pMHY483. Next, a 2.6-kb NcoI/XhoI SNU56 fragment from pMHY155 was inserted into NheI-linearized pMHY483 by blunt-end ligation to yield pMHY487. To shift the reading frame (pMHY496), an NheI (blunt)-XhoI fragment from pMHY487 was inserted into HindIII (blunt)-XhoI-digested pPW66R. To remove the C terminus of Snu56p, pMHY496 was digested with SphI-XhoI, blunted, and religated, yielding pMHY497.
A LexA-SNU56 fusion plasmid (pMHY155) was created in two steps. First, an NcoI site was introduced at the start codon of SNU56. This was accomplished by simultaneously ligating two PCR products encoding sequences flanking the start codon at a common NcoI site and inserting them into the XhoI and SphI sites of pMHY275A to yield pMHY154. The primer combinations used were SNU56 primer 1-SNU56 primer 2 and SNU56 primer 3-SNU56 primer 4. A ∼2,500-bp fragment encoding the SNU56 open reading frame and 3′ untranslated region sequences was excised from pMHY154 by NcoI-NotI digestion and inserted into pEG202 to yield pMHY155. To create a LexA-snu56-2 fusion plasmid (pMHY471), first, pMHY154 was amplified with primer pair SNU56 primer 3-snu56-2 S388F StuI Lower and pMHY478 was amplified with primer pair pRS316 (1719-1738)-snu56-S388F StuI upper. Next, these products were digested with NcoI/StuI and StuI/NotI, respectively, and simultaneously ligated into NcoI/NotI-digested pMHY155. All fusions were judged functional by the ability to rescue the snu56 null phenotype.
Isolation of mutants that require HMT1 for growth.
Strain MHY361 (hmt1::HIS3 ade2 ade3 ura3 plus pMHY40) served as the parental strain for the synthetic lethal screen. Cells were mutagenized to approximately 50% killing. About 1,778,000 cells survived the mutagenesis, most of them forming colonies with a distinct red-white sectoring phenotype. Two hundred fifty-eight nonsectoring red colonies were picked and restreaked on YEPD medium plates. Eleven candidates satisfied all of the criteria (28, 33) for synthetic lethality with hmt1Δ.
Cloning of SNU56.
MHY408, a snu56-2 hmt1::HIS3 strain carrying pMHY41 (2μ HMT1 LEU2 ADE3), was transformed with a YCp50-based genomic library (41). Following selection at 25°C on adenine-limited medium lacking uracil, 121 Sect+ colonies were identified among approximately 71,000 transformants. Colonies were then isolated from the white sectors of the Sect+ strains by restreaking onto Ura− dropout plates. Seventy-seven of these white colonies, cured of plasmid pHMY41 but retaining the library plasmid, were then tested for 5-FOA sensitivity (FOAs). Only 20 of the plasmid DNAs rescued from the 77 transformants restored sectoring when introduced back into the parent strain. Restriction analysis indicated that 5 of the plasmids contained HMT1 whereas the remaining 15 represented two distinct clones with common insert fragments. The plasmid with the smaller insert (A2 no. 8) was chosen for further analysis.
Deletion derivatives were used to define the region of plasmid A2 no. 8 required for SNU56-complementing activity. Digestion of A2 no. 8 with SnaBI produces fragments of approximately 16.0, 2.9, and 0.9 kb. The 2.9-kb fragment that complemented the snu56-2 allele was inserted in both orientations into the SmaI site of pRS316 to create pMHY275A and pMHY275B. A 2.1-kb fragment containing SNU56 was amplified from pMHY275A by using primers designed to introduce XhoI-XbaI ends (XHO-SNU56 no. 3 and XBA-SNU56) and inserted into pRS313 to produce pMHY478.
Chromosomal SNU56-URA3.
Genomic integration was used to mark the chromosomal SNU56 locus with URA3. Briefly, a URA3-based integrating plasmid carrying the 3′ end of SNU56 and downstream sequences (pMHY480) was created by subcloning a SalI fragment from pMHY275B into pRS306. This plasmid (pMHY480) was digested at a unique HindIII site within SNU56 and used to transform strain MHY360 (hmt1::HIS3). Potential integrants were selected by growth on Ura− dropout plates. Integration at the SNU56 locus of strain MHY687 was verified by Southern analysis (data not shown).
Snu56p depletion assays.
MHY1481 and MHY1482 cells were grown at 25°C in synthetic complete medium lacking uracil and histidine and supplemented with 2% raffinose and 0.1 mM CuSO4 (11) until the exponential growth phase was reached. The cells were then shifted to the same medium containing 2% galactose and lacking copper and grown at 25°C for 30 min before incubation at 37°C. Snu56p and intron-containing pre-mRNA levels were monitored by Western blotting and RT-PCR, respectively.
Immunoblot analysis.
Total cell extracts were prepared as previously described (4). Quick protein lysates were prepared by adapting a previously described protocol (29). Briefly, 3 × 108 mid-log-phase cells were harvested, washed with H2O, and resuspended in 500 μl sample buffer (10% glycerol, 2% sodium dodecyl sulfate [SDS], 5% 2-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM benzamidine, 0.0025% bromophenol blue). Glass beads were added, and cells were lysed by vortexing at 4°C for 3 min. The lysate was cleared by centrifugation and boiled, and 20-μl samples were examined. Proteins were resolved by SDS-polyacrylamide gel electrophoresis on 4 to 12% Bis-Tris polyacrylamide gels (Invitrogen), transferred to nitrocellulose, and immunoblotted as previously described (4). Mouse monoclonal antihemagglutinin (Roche), rabbit anti-Tub1p (a gift from Vincent Guacci), and rat anti-Tub1p (sc-53030; Santa Cruz) were used at 1:500, 1:2,000, and 1:1,000, respectively. Tandem affinity purification (TAP)-tagged proteins were detected with peroxidase antiperoxidase (Sigma) used at 1:2,000.
RNA and splicing assays.
Total RNA was prepared from mid-log-phase haploid cells or meiotic diploid cells collected 15 h after transfer to sporulation medium by using Tri-Reagent (Molecular Research Center). RT-PCR was performed on the samples by using the Titan One Tube System (Roche). One-fiftieth to 1/10 of the PCR product was analyzed on 3.5 or 5% polyacrylamide or 1.75% agarose gels. Splicing was also assessed by growth of yeast containing the AMA1-CUP1 fusion plasmid and pGMER1 vector at 25°C on SCD plates lacking uracil and tryptophan and containing 0.25 mM cupric sulfate (47).
Meiotic time courses.
Log-phase diploid cells grown in PSP2 medium (31) were shifted to SPM medium (31) for 36 h. At each time point, aliquots were removed and fixed in 70% ethanol and stained with 4′,6′-diamidino-2-phenylindole (DAPI). Fixed cells were wet mounted on slides, and cells and nuclei were visualized by Nomarski and fluorescence, respectively.
Two-hybrid assay.
LexA DNA-binding domain (BD) fusions were constructed in pEG202, whereas Gal4 activation domain (AD) fusions were constructed in pACTII. The two-hybrid (47) assay was performed as described previously (1). Briefly, both AD and BD plasmids were simultaneously introduced into either wild-type (BMA64-1A) or hmt1Δ mutant (MHY477) cells carrying the pSH18-34 reporter plasmid and selected on complete medium lacking uracil, histidine, and leucine. Individual transformants were patched onto the same medium, overlaid with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (13), and examined for blue color development after 4 h of incubation at 30°C. Interactions were quantitated by outgrowing a pool of colonies and assaying soluble β-galactosidase activity by the Microfuge protocol supplied with the Yeast β-Galactosidase Assay kit (Pierce).
Mapping of snu56-2.
Chromosomal snu56-2 was first PCR amplified (primers XHO-SNU56 no. 3 and XBA-SNU56) from MHY706 genomic DNA. The resulting product was XhoI/XbaI digested and inserted into pRS315 to yield pMHY441 (snu56-2). This plasmid was introduced into MHY853 (snu56::KanMX covered with URA3 SNU56 plasmid) and assayed for temperature sensitivity and growth rate phenotypes after loss of the wild-type plasmid by 5-FOA counterselection. Strain MHY1451 transformed with candidate snu56 plasmids and then cured of pMHY275 was assayed for sporulation. Sequencing was run in parallel with DNA amplified from a wild-type strain. Plasmid pMHY472 was produced by subcloning a 2.1-kb XhoI-XbaI restriction fragment containing snu56-2 from pMHY441 into pRS313.
The plasmid-borne snu56-2 point mutation responsible for the sporulation defect was mapped by swapping snu56 restriction fragments from plasmid pMHY472 (snu56-2 HIS3) or pmHY441 into pMHY478 (SNU56 HIS3). This plasmid was then exchanged with the wild-type plasmid in a homozygous diploid strain (MHY1451) otherwise disrupted for the chromosomal copies of SNU56. After loss of the plasmid encoding SNU56, sporulation ability was assayed. The plasmid identified by this strategy, pMHY551, is a pMHY441 derivative in which a 1.4-kb XhoI-HindIII snu56 fragment has been replaced with a corresponding wild-type fragment from pMHY478. Plasmid pMHY551 contains only the S389F mutation. S389F was then introduced singly into SNU56 by PCR with oligonucleotide pairs snu56-2 S389F StuI Upper-XBA-SNU56 and snu56-2 S389F StuI Lower-XHO-SNU56 no. 3. PCR products were simultaneously ligated into pRS315 to make pMHY554.
RESULTS
An hmt1Δ synthetic lethal screen identifies SNU56.
Hmt1p is a dispensable protein required for the arginine methylation of a number of RNA-binding proteins implicated in mRNA and rRNA processing (21, 26, 28, 43, 50, 51). In order to identify novel factors involved in these processes, a synthetic lethal screen was conducted with a strain that contains a deletion of the HMT1 gene and a wild-type copy of HMT1 on a URA3 plasmid. This strain was mutagenized with EMS to kill ∼50% of the cells. Cells containing mutations that are synthetically lethal with the HMT1 deletion were selected by a red-white colony sectoring assay (3). Eleven recessive mutants, encompassing four different complementation groups, were found to fulfill the criteria of being synthetic lethal with hmt1Δ (see Materials and Methods). In this paper, these groups are designated slh1-4, for synthetic lethal hmt1Δ. Of the 11 mutants, 6 were placed in the slh1 group, 3 were placed in the slh2 group, and the final 2 were slh3 and slh4. Genetic analysis indicated that the synthetic lethality and hmt1Δ allele cosegregated 2:2 in each case, demonstrating that the phenotype is caused by a single nuclear mutation in each of the candidates.
Mutations already known to be synthetic lethal with hmt1Δ include alleles of the npl3 (28) and cbp80 (43) RNA maturation genes. Consistent with these results, CBP80 and NPL3 were cloned by complementation of the slh1 and slh2 mutants, respectively (our unpublished results). For the present study, the single slh3 mutant strain (MHY394) was characterized further.
The corresponding wild-type gene was cloned from a yeast genomic library by screening for restoration of the sectoring phenotype (see Materials and Methods). Restriction mapping of a complementing plasmid, followed by retransformation with truncated inserts, localized the potential complementing region within a 2.9-kb SnaBI DNA fragment. DNA sequence analysis and BLASTn searches revealed two previously identified yeast genes: SNU56/MUD10 and BUD26. A SalI fragment carrying BUD26 was subcloned from the original clone and found to be insufficient for rescue of the Sect− phenotype, signifying that complementation was due to SNU56. Analogous to NPL3 and CBP80, SNU56 encodes a nuclear RNA-binding protein that associates with the yeast U1 snRNP (24, 52). The U1 snRNP is essential for recognition of the pre-mRNA 5′ splice site and subsequent assembly of the spliceosome.
Genomic integration was used to verify that SNU56 is the bona fide synthetic lethal gene rather than an unlinked suppressor. Wild-type SNU56 was marked on the chromosome with the URA3 gene in an hmt1Δ mutant strain (see Materials and Methods). The resulting haploid was crossed to the synthetic lethal mutant strain carrying a LEU2 HMT1 ADE3 plasmid. The diploid was sporulated, and tetrads were analyzed. Each spore carried the null allele of HMT1. Assuming linkage, the spores that carried the URA3-marked wild-type copy of SNU56 would be able to lose the LEU2 HMT1 ADE3 plasmid. Those that carried the mutant form of SNU56 from the synthetic lethal strain would require the plasmid to remain viable. To test for the ability to lose the plasmid, complete tetrads were monitored for the sectoring (Sect) phenotype. Sixteen tetrads were analyzed, and all displayed the behavior predicted for linkage, two Ura+ Sect+ and two Ura− Sect−. Furthermore, snu56-induced slow growth and hmt1Δ-induced synthetic lethality cosegregated, indicating that both phenotypes resulted from a mutation at a single locus. From this point on, we will refer to this allele as snu56-2.
The hmt1Δ-snu56 synthetic lethal interaction is allele specific.
SNU56 has been identified in two other synthetic lethality screens as MUD10 (mutant-U1 die) and LUC4 (lethal unless CBC produced) (20, 24). Since the luc4 allele does not confer a growth phenotype in a wild-type background (20), we have limited our analysis to the mud10-1 and snu56-2 alleles. Strains harboring either allele grow slowly at 30°C on rich medium on plates or in liquid culture (data not shown). The growth defects of snu56-2 mutant strains are similar at higher (36°C) or lower (25°C) temperatures (data not shown). In contrast, mud10-1 mutant strains are temperature sensitive for growth (24). To determine whether mud10-1 is synthetically lethal with the hmt1 null allele, an hmt1::KanMX strain (MHY671) was crossed to a mud10-1 strain to generate an hmt1::KanMX/HMT1 mud10-1/SNU56 diploid. The diploid cells were sporulated at 25°C, and tetrad progeny were scored for the presence of both the hmt1::KanMX null (Gentr) and mud10 (Ts−) alleles. In nine tetrads examined, 11 Gentr Ts− segregants of the presumed genotype hmt1::KanMX mud10-1 were obtained. When they were grown at 25°C, the growth of these double mutants was indistinguishable from that of the mud10-1 and hmt1Δ single-mutant spore clones (data not shown). From these results, it appears that the mud10-1 allele, unlike the snu56-2 allele, is not lethal in an hmt1Δ background.
SNU56 is not required for vegetative splicing.
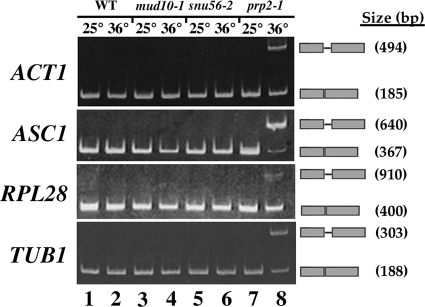
To determine whether Snu56p is required for splicing, RT-PCR assays were designed with intron-flanking primers to monitor specific pre-mRNA and mRNA transcripts in both snu56-2 and mud10-1 vegetative cells. S. cerevisiae intron-containing genes code for either nonribosomal or ribosomal proteins that differ by intron length. The latter group contains introns that are typically 300 to 500 nucleotides long, more than twice the size of nonribosomal protein gene introns (42). Thus, we chose representative ribosomal protein (ASC1 and RPL28) and nonribosomal protein (ACT1 and TUB1) transcripts for analysis (Fig. 1). All pre-mRNAs were efficiently spliced during vegetative growth in wild-type, mud10-1, and snu56-2 strains (Fig. 1, lanes 1 to 6). In contrast, the well-characterized splicing-defective prp2-1 mutant (49) accumulated unspliced transcripts following a shift to the restrictive temperature (Fig. 1, lanes 7 and 8). The presence of unspliced product in lane 8 confirms that both mRNA species are amplified in our assay system. These results demonstrate that the snu56-2 and mud10-1 mutations do not impair splicing in vegetative cells.
FIG. 1.
Vegetative mud10-1 and snu56-2 cells efficiently splice pre-mRNAs. Total RNA was prepared from exponential-phase wild-type (WT; lanes 1 and 2) and mud10-1 (lanes 3 and 4), snu56-2 (lanes 5 and 6), and prp2-1 (lanes 7 and 8) mutant cells grown at either 25°C or 36°C. ACT1, ASC1, RPL28, and TUB1 pre-mRNA splicing was analyzed by RT-PCR with the appropriate intron-flanking primer set. PCR products were resolved on 5% polyacrylamide gels and stained with ethidium bromide. The gene analyzed in each case is indicated to the left of the panel. Bands representing unspliced pre-mRNA and spliced mRNA are indicated to the right of each panel. PCR product sizes are also indicated on the right.
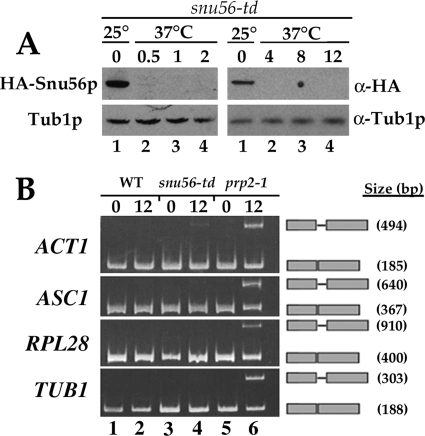
However, it remained possible that Snu56p plays a direct role in the splicing reaction but these two alleles affect other processes. To determine whether Snu56p is required for vegetative splicing, we depleted Snu56p in vivo by the degron strategy (11), which is based on integrating a heat-inducible degradation signal (ubiquitin-Arg-DHFRts) in frame with the N terminus of the gene of interest. This fusion also introduces a hemagglutinin epitope for detection of the fusion protein. The Snu56 protein is rapidly degraded after a shift to 37°C and is not detectable by 60 min (Fig. 2A, left panel). Interestingly, unspliced ACT1, ASC1, RPL28, and TUB1 transcripts do not accumulate significantly 12 h after Snu56p depletion (Fig. 2B). These results suggest that Snu56p is dispensable for splicing during vegetative growth.
FIG. 2.
In vivo depletion of Snu56p does not affect vegetative splicing. (A) Snu56p-td is rapidly depleted at 37°C. Western blot analysis was performed with extracts from snu56-td mutant cells (MHY1481) grown at 25°C or shifted to 37°C for the times indicated above the lanes (hours). The antibodies used are indicated on the right. Tub1p was used as a loading control. HA, hemagglutinin. (B) RT-PCR analysis of ACT1, ASC1, RPL28, and TUB1 pre-mRNA splicing. Total RNA was prepared from exponential-phase wild-type (WT; MHY1482; lanes 1 and 2) and snu56-td (MHY1481; lanes 3 and 4) and prp2-1 (P30-6D; lanes 5 and 6) mutant cells grown at either 25°C or shifted to 37°C for 12 h. RNA was analyzed by RT-PCR with the appropriate intron-flanking primer set. PCR products were resolved on 5% polyacrylamide gels and stained with ethidium bromide. The gene analyzed in each case is indicated to the left of the panel. Bands representing unspliced pre-mRNA and spliced mRNA are indicated to the right of each panel. PCR product sizes are also indicated on the right.
It was previously reported that the splicing of a reporter construct containing a mutant 5′ splice site was mildly impaired in mud10-1 cells (24). Given that natural nonconsensus 5′ splice sites are found in some yeast meiosis-specific transcripts (9), we reasoned that their splicing could be impaired in snu56 mutants.
Snu56p is required for Mer1p-activated splicing.
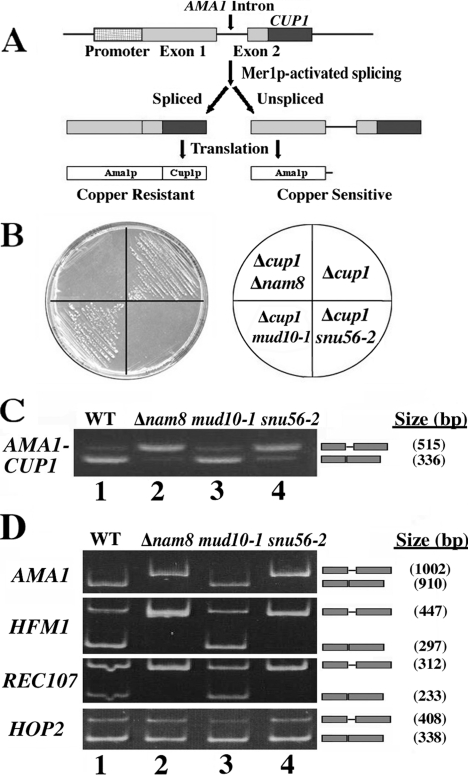
Mer1p activates the meiosis-specific splicing of at least three pre-mRNAs (REC107, HFM1, and AMA1) that contain a Mer1p splicing enhancer (8, 17, 47). Of the U1 and U2 snRNP genes not required for growth, only NAM8 and SNU17 are necessary for Mer1p-activated splicing (47, 48). To determine whether the essential SNU56 gene plays a role in this process, we utilized a copper reporter assay to measure Mer1p-activated splicing in snu56 mutant cells (47). Briefly, vegetative copper-sensitive cells (cup1Δ) defective for Snu56p activity were transformed with both a MER1 constitutive expression vector and an AMA1-CUP1 fusion plasmid that confers splicing-dependent growth of yeast on medium containing copper (Fig. 3A) (35, 47). Since Mer1p is normally absent during vegetative growth, cells containing the AMA1-CUP1 fusion plasmid only grow on plates containing copper if they also contain the MER1 expression plasmid (Fig. 3B). cup1Δ cells containing either the snu56-2 or the nam8Δ allele were not viable on these plates, whereas those harboring mud10-1 were viable (Fig. 3B).
FIG. 3.
Snu56p is required for Mer1p-activated splicing. (A) Schematic of the AMA1-CUP1 reporter assay used to monitor Mer1p-activated splicing in vegetative cells. The MER1 and AMA1-CUP1 fusion splicing reporter genes are constitutively expressed from separate plasmids. Mer1p-activated splicing results in the expression of an AMA1-CUP1 fusion protein conferring copper resistance. (B) Growth of cup1Δ mutant yeast on copper plates. Wild-type and snu56-2, mud10-1, and nam8Δ mutant cells that have had endogenous CUP1 knocked out and been transformed with MER1 and AMA1-CUP1 fusion splicing reporter plasmids are shown. Transformants were streaked onto synthetic complete medium containing 0.25 mM copper and lacking uracil and tryptophan (left). A plate schematic (right) indicates the mutant alleles of each strain. (C) RT-PCR analysis of AMA1-CUP1 pre-mRNA splicing. Total RNA was prepared from exponential-phase wild-type (WT; lane 1) and nam8Δ (lane 2), mud10-1 (lane 3), and snu56-2 (lane 4) mutant cells that have had endogenous CUP1 knocked out and been transformed with MER1 and AMA1-CUP1 fusion plasmids. RNA was analyzed by RT-PCR with primers complementary to the exons of the AMA1-CUP1 gene. PCR products were resolved on 1.75% agarose gels and stained with ethidium bromide. The gene analyzed in each case is indicated to the left of the panel. Bands representing unspliced pre-mRNA and spliced mRNA are indicated to the right of each panel. PCR product sizes are also indicated on the right. (D) AMA1, HFM1, and REC107 splicing during meiosis is Snu56p dependent. The RT-PCR products generated from total RNA samples were taken from wild-type (lane 1) and nam8Δ (lane 2), mud10-1 (lane 3), and snu56-2 (lane 4) mutant homozygous diploids 15 h after transfer to sporulation medium. PCR products were resolved on 5% acrylamide gels and stained with ethidium bromide. The gene analyzed in each case is indicated to the left of the panel. Bands representing unspliced pre-mRNA and spliced mRNA are indicated to the right of each panel. PCR product sizes are also indicated on the right.
RT-PCR analysis designed to distinguish spliced and unspliced AMA1-CUP1 transcripts shows that the copper-sensitive growth phenotypes of these cells reflect the absence of Mer1p splicing activation (Fig. 3C). Furthermore, endogenous AMA1 transcripts amplified from meiotic homozygous diploid cells exhibited similar splicing deficiencies (Fig. 3D). Likewise, the splicing of two other transcripts (HFM1 and REC107) that require Mer1p activity is inhibited (Fig. 3D). In contrast, a meiosis-specific transcript containing a nonconsensus 5′ splice site (HOP2) that is spliced independently of Mer1p and Nam8p is unaffected (Fig. 3D) (36). These results indicate that Snu56p is not simply required for weak 5′ splice site recognition but is specific for Mer1p-activated transcripts. Furthermore, the allele specificity suggests that SNU56 encodes a multifunctional protein and that only the snu56-2 mutation impairs Mer1p-activated splicing.
Homozygous diploid snu56-2 cells arrest during meiosis I and display sporulation defects.
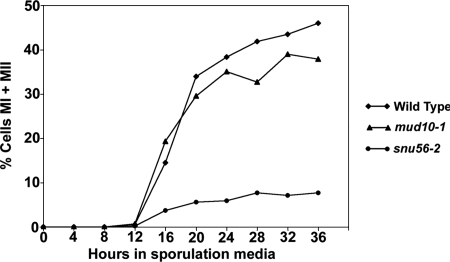
Mer1p-regulated splicing is required for chromosomal pairing, recombination, and separation during the first meiotic division (8, 15, 16). Since SNU56 is required for Mer1p-activated splicing, snu56-2 cells should exhibit a defect in meiotic progression. We monitored meiotic nuclear divisions of snu56-2 mutants by DAPI staining of fixed cells. This analysis indicated that by 36 h, 46% of the wild-type cells contained four individually staining nuclei diagnostic of the execution of meiosis I and meiosis II (Fig. 4). Comparable results were observed for mud10-1 mutants. However, by the same time point, only 8% of the snu56-2 cells progressed through both meiotic divisions (Fig. 4). These results indicate that Snu56p is necessary for the execution of meiosis and are further confirmation that Mer1p-activated splicing requires Snu56p.
FIG. 4.
Diploid snu56-2 mutant cells fail to execute meiosis. Meiotic nuclear divisions were examined by DAPI staining. Plotted is the percentage of cells that had undergone both nuclear divisions (MI and MII) at various times throughout sporulation. At least 200 cells from each strain were examined at each time point to determine the numbers of tri- and tetranucleate cells. Shown are averages of three independent experiments. All errors are within 5%.
We also tested the ability of these strains to form complete tetrads (Table 3). In the wild-type diploid strain, sporulation is completed 72 h following transfer to sporulation medium with an efficiency of about 42%. Interestingly, the mud10-1/mud10-1 diploid sporulated with a similar efficiency (41%) whereas the snu56-2/snu56-2 diploids did not sporulate (<0.5%). Both snu56-2 and mud10-1 heterozygous diploids and homozygous diploids carrying a plasmid-borne copy of SNU56 sporulated with efficiencies similar to that of the wild type. Thus, SNU56 is required for both meiotic nuclear division and sporulation.
TABLE 3.
Sporulation efficienciesa
| Diploidb | % Sporulation (72 h) |
|---|---|
| SNU56/SNU56 | 42 |
| SNU56/snu56-2 | 30 |
| snu56-2/snu56-2 | <0.5 |
| snu56-2/snu56-2 + (SNU56) | 30 |
| SNU56/mud10-1 | 40 |
| mud10-1/mud10-1 | 41 |
| mud10-1/mud10-1 + (SNU56) | 38 |
At least 200 cells of each strain were examined in triplicate. SNU56/SNU56 (MHY507 × MHY748), SNU56/snu56-2 (MHY507 × MHY1458), snu56-2/snu56-2 (MHY1456 × MHY1458), SNU56/mud10-1 (MHY1467 × MHY1468), and mud10-1/mud10-1 (MHY1462 × MHY1463) strains were tested.
(SNU56) signifies transformation with pMHY478.
Mapping of the snu56-2 allele.
The fact that the mud10-1 and snu56-2 mutants exhibit distinct growth, sporulation, and splicing phenotypes suggests that these alleles harbor different mutations. The mud10-1 allele has a C-to-T mutation at nucleotide 383 that results in the replacement of a serine with a phenylalanine at residue 124 (24). A comparison of plasmid-borne SNU56 and snu56-2 sequences identified four point mutations within the mutant allele that resulted in amino acid changes (see Materials and Methods). To identify the point mutation(s) responsible for the sporulation defect, we replaced restriction fragments of SNU56 with the corresponding snu56-2 segments. These hybrid plasmids containing one or more point mutations were then exchanged for the wild-type version in a homozygous diploid (MHY1451) with the chromosomal copies of SNU56 disrupted. The resulting strains were assayed for sporulation efficiency. This analysis defined the mutation responsible for the sporulation defect to a 700-bp HindIII-XbaI snu56-2 restriction fragment. This fragment contained a single T-to-C mutation at nucleotide 1165 changing serine 389 to phenylalanine.
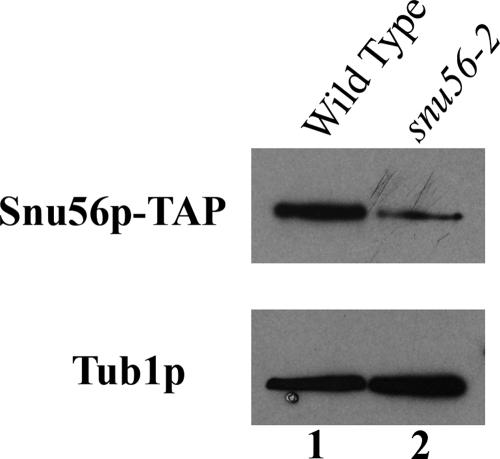
To test relative expression levels, we fused the TAP tag to the C termini of both SNU56 and snu56-2 and integrated these constructs at the SNU56 locus. Equal numbers of exponential-phase cells were harvested and lysed in SDS-polyacrylamide gel electrophoresis loading buffer to minimize the effects of protein instability. By Western blotting, the mutant protein is expressed at a level approximately half of that of the wild type (Fig. 5). However, the phenotype caused by snu56-2 is not simply due to reduced protein levels, as elevated copy number (2μm) and expression (ADH1 promoter) do not rescue the sporulation defect (our unpublished results).
FIG. 5.
Snu56 and snu56-2 protein expression levels. Western blot analysis was performed with exponential growth phase SNU56-TAP and snu56-2-TAP cell lysates. SNU56 alleles are indicated at the top of each lane, and the proteins visualized are indicated to the left. Tub1p was used as a loading control.
Snu56p protein-protein interactions.
Given that Snu56p is required for Mer1p splicing activation, we tested whether these two factors interact with a common set of proteins. In two-hybrid assays, Mer1p interacted with three U1 snRNP proteins (Snu56p, Snu71p, and Luc7p), two branch point binding proteins (Mud2p and Bbp1p), and a single U2 snRNP protein (Prp11) (48). We used this system to determine whether these proteins also interact with Snu56p. In our application, protein interactions were detected by activation of a β-galactosidase (lacZ) reporter gene through the interaction of a LexA DNA BD protein with a Gal4 transcriptional AD fusion (18). A yeast reporter strain was simultaneously transformed with plasmids encoding SNU56 fused to the BD and a test protein fused to the AD. The Snu56 fusion protein is functional in that it rescues the snu56 null lethal phenotype (our unpublished results). As a positive control, the AD-Snu56p fusion was tested against a BD-Mer1p fusion. Consistent with a previous report that tested a BD-Snu56p fusion against AD-Mer1p (48), an interaction between Snu56p and Mer1p was detected (Table 4). No interactions were observed with Snu71p, Luc7p, or Bbp1p (Table 4). However, Snu56p interacts strongly with Mud2p and weakly with Prp11p. In sum, these results indicate that Snu56p and Mer1p have at least two interactions in common and further support a role for Snu56p in Mer1p-dependent splicing.
TABLE 4.
Two-hybrid assay results obtained with SNU56 and snu56-2 mutant strainsa
| AD protein fusion | Colony color with BD-SNU56 | β-Galactosidase activity (U) | Colony color with BD-snu56-2 | β-Galactosidase activity (U) |
|---|---|---|---|---|
| Rna14p | White | 0 ± 0 | White | 0 ± 0 |
| Snu71p | White | 0 ± 0 | White | 0 ± 0 |
| Luc7p | White | 0 ± 0 | White | 0 ± 0 |
| Mud2p | Blue | 49 ± 3 (25 ± 6)b | White | 0 ± 0 |
| Bbp1p | White | 0 ± 0 | White | 0 ± 0 |
| Prp11p | Light blue | 13 ± 1 | White | 0 ± 0 |
| Mer1p | Light blue | 9 ± 4 | White | 0 ± 0 |
| Nam8p | Light blue | 4 ± 2 | White | 0 ± 0 |
| Snu17p | White | 0 ± 0 | White | 0 ± 0 |
Quantitative β-galactosidase assays were performed on strains expressing the indicated constructs. Values are from pooled colonies assayed in triplicate.
The value in parentheses represents AD-Mud2p activity in a Δhmt1 mutant strain.
We also assayed Snu56p for interactions with Nam8p and Snu17p, two additional factors necessary for Mer1p-activated splicing. Although two-hybrid interactions have not been detected between Mer1p and either Nam8p or Snu17p (48), we found Snu56p to interact weakly with Nam8p (Table 4). Interaction between Snu56p and Nam8p was measured at a strength approximately 1/10 of that seen between Snu56p and Mud2p. However, the signal was reproducibly above that observed for noninteractors.
We next tested whether the BD-snu56-2 protein interactions matched the wild type. Although BD-snu56-2 is expressed at a level comparable to that of BD-SNU56 and causes a sporulation defect equivalent to that caused by untagged mutant protein (our unpublished results), no interactions were observed (Table 4). This is consistent with the idea that the mutant protein is missing from the U1 snRNP and its absence may contribute to the Mer1p-activated splicing defect seen in snu56-2 cells.
HMT1 influences Snu56p protein-protein interactions.
To determine whether cellular arginine methylation influenced the Snu56p interactions, we compared two-hybrid interactions in wild-type and hmt1Δ haploid cells. The strongest Snu56p interactor, Mud2p, was examined. The interaction of this protein with Snu56p in hmt1Δ cells was measured at a strength approximately one-half of that seen in wild-type cells. This effect is specific, as a previously reported protein-protein interaction, Rna14p-Rna15p (32), is unaffected in hmt1Δ cells (our unpublished data).
The snu56-2 allele is lethal in nam8Δ mutant cells.
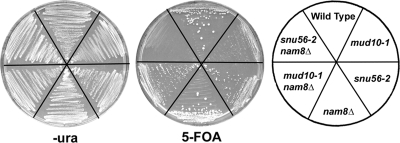
Although NAM8 is dispensable for vegetative growth, the overall U1 snRNP structure is destabilized in its absence. Specifically, the association of Snu56p and Snu71p with this complex is greatly reduced (24). We reasoned that NAM8 might become essential in vegetative cells if the activity of Snu56p is also compromised. To test this, we constructed snu56-2 nam8Δ and mud10-1 nam8Δ double mutants expressing wild-type SNU56 from a URA3 plasmid. Cells containing a lethal mutant allele combination would require the URA3 SNU56 plasmid to remain viable, whereas nonlethal combinations would be able to lose the plasmid. To test for the ability to lose the plasmid, cells were streaked onto plates containing 5-FOA. The presence of 5-FOA on the plates selects against cells that carry the URA3 plasmid. Indicative of a synthetic lethal relationship, snu56-2 nam8Δ cells were 5-FOA sensitive (Fig. 6). Interestingly, synthetic lethal effects of the snu56-nam8Δ combination exhibited allele specificity. The mud10-1 nam8Δ mutant cells were 5-FOA resistant (Fig. 6). The wild type and snu56-2 and mud10-1 single mutants were also 5-FOA resistant (Fig. 6).
FIG. 6.
Synthetic lethality of snu56 alleles. Wild-type and mud10-1, snu56-2, nam8Δ, mud10-1 nam8Δ, and snu56-2 nam8Δ mutant cells bearing a CEN URA3 SNU56 plasmid are shown. Cells were streaked onto synthetic complete medium lacking uracil (left) or containing 5-FOA (center). The mutant allele(s) present in each strain is indicated by the plate schematic (right). Plates were incubated at 25°C for 2 days.
DISCUSSION
In this work, we demonstrate that a constitutive U1 snRNP component, Snu56p, combines with a cell type-specific factor (Mer1p) to activate splicing during the developmental process of meiosis in yeast. The evidence that Snu56p splicing activity is required for Mer1p-activated splicing is based on the analysis of constitutive and meiosis-specific transcripts. As judged by both a CUP1 reporter assay of vegetative haploids and RT-PCR analysis of homozygous meiotic diploids, snu56-2 mutants accumulate unspliced meiosis-specific Mer1p-activated transcripts. As expected, these cells are also deficient in meiotic progression and sporulation. The association of Snu56p with U1 snRNP components required for this process was confirmed by two-hybrid analysis, and these protein-protein interactions are summarized in Fig. 7. Interestingly, the snu56-2 protein does not interact with any tested spliceosomal proteins, suggesting its absence from the U1 snRNP. The Snu56p requirement for Mer1p-activated splicing is specific rather than a general reflection of an inefficient splicing apparatus as Mer1p-independent splicing during meiosis appears unaffected by this mutation. Finally, spliced transcript examination in temperature-sensitive mud10-1 and Snu56p-depleted cells suggests that this U1 snRNP component may be dispensable for mitotic splicing.
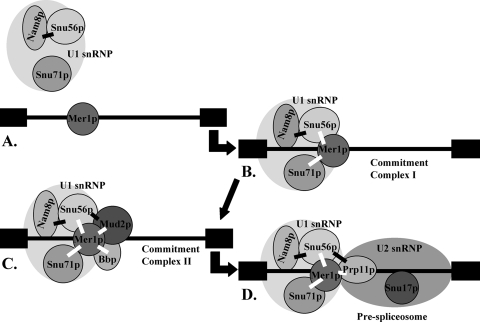
FIG. 7.
Snu56p protein-protein interactions during early spliceosome formation. Snu56p stabilizes Nam8p within the U1 snRNP (A) and associates with Mer1p to aid U1 snRNP recruitment to Mer1p-activated introns during commitment complex I formation (B). Later, interactions with Mud2p during commitment complex II assembly (C) and Prp11p during prespliceosome formation (D) may help stabilize these spliceosomal intermediates. Black boxes represent exons, and the black line represents a Mer1p-activated intron. Black bars between proteins represent two-hybrid interactions identified in this work, and white bars denote those previously identified (48).
In this study, we have taken advantage of a unique Snu56p mutation (snu56-2) to test the role of this constitutive U1 snRNP protein in Mer1p-activated splicing. This point mutation specifically impairs the nonessential meiotic activity of this protein without significantly impacting its essential function, which remains unknown. It is possible that other essential U1 snRNP proteins also play a role in this process. However, distinguishing between their roles in general and Mer1p-dependent splicing may be difficult. Two likely candidates include Snu71p and Snu65p. One of these proteins (Snu71p) is lost from the purified U1 snRNP in the absence of Nam8p, while the interaction of the other (Snu65p) is stabilized with the U1 snRNP (24).
Alternative or regulated splicing increases genomic coding capacity while providing a layer of posttranscriptional regulation. In yeast, the cooperation of constitutive snRNP members Snu56p, Nam8p, and Snu17p with meiosis-specific Mer1p activates the splicing of at least three transcripts (AMA1, HFM1, and REC107) required for meiotic progression (47, 48). This process is analogous to the positive splicing regulation of human fibroblast growth factor receptor 2, the apoptotic regulator Fas, and the Drosophila sex determinant male-specific lethal 2 (10, 19). In these cases, the TIA-1 protein binds to intronic U-rich sequences and facilitates U1 snRNP recruitment to nonconsensus 5′ splice sites. Although not a constitutive U1 snRNP member, TIA-1 shows structural (26% identity) and functional (intron recognition) similarities to Nam8p. This raises the possibility that other constitutive yeast U1 snRNP factors, including Snu56p, may have functional homologues in mammals that are not constant U1 snRNP members.
The fact that SNU56 is an essential gene whereas Mer1p-dependent meiosis-specific splicing is a nonessential process in vegetative cells suggests that Snu56p has another cellular activity. This conclusion is supported by our observation that Snu56p depletion does not significantly impair vegetative splicing. One idea consistent with our results is that SNU56 does not directly affect the splicing reaction itself but is either directly or indirectly necessary for communication between the splicing and RNA export machinery.
What is the relationship between Snu56p and arginine methylation? Our working model is that Npl3p arginine methylation enhances its interaction with Snu56p. In this and previous work, we have identified both SNU56 and NPL3 mutant alleles that synergize with a HMT1 null allele to produce a lethal phenotype (28). Npl3p is a substrate for arginine methylation (28) and, like Snu56p, associates with the U1 snRNP (24). We have observed that arginine methylation enhances the interaction between Npl3p and Snu56p in a two-hybrid analysis (our unpublished results). The fact that the Mer1p-dependent splicing function of Snu56p appears unaffected by cellular arginine methylation status implies that it may be required for its essential function. This is consistent with our finding that snu56-2 is lethal in an hmt1Δ background.
Mutational mapping provides an opportunity to identify Snu56p functional domains. Although Snu56p exhibits RNA-binding affinity both in vitro (24) and in vivo (52), the sequence does not contain any known consensus RNA-binding motifs. Furthermore, no other known motifs that would suggest a function for this protein have been detected in the sequence. In this work, we have determined that the mud10-1 (S124F) and snu56-2 (S389F) point mutations map to diverse locations within Snu56p and impair distinct cellular functions, indicating that this protein consists of at least two functional regions. Based on our results, one is involved in an essential vegetative function whereas the other is necessary for Mer1p-activated splicing. Our results do not preclude the possibility that these functional domains could overlap.
Our work fits with previous data linking Snu56p to Nam8p and Mer1p-activated splicing. Nam8p contains three RNA recognition motifs and has been cross-linked to a nonconserved intron region just downstream of the 5′ splice site (40, 52). Interestingly, this is near the position where the Mer1p splicing enhancer is found in Mer1p-activated transcripts. Snu56p appears to map adjacent to Nam8p on pre-mRNA (52), consistent with genetic experiments suggesting that this pair of proteins might function in a concerted manner (24). Although Nam8p is not essential for splicing, its absence destabilizes the overall structure of the U1 snRNP and prevents Mer1p-activated splicing. The association of Mer1p with the U1 snRNP is not disrupted in nam8Δ cells, despite the fact that activated splicing is blocked (47). This suggests that splicing activation by Mer1p may require stable association of Snu56p with the U1 snRNP.
Our results favor a model in which Snu56p serves to bridge intron-bound Mer1p to Nam8p in the U1 snRNP (Fig. 7). Consistent with this, we have shown that Snu56p interacts with both Nam8p and Mer1p in two-hybrid experiments. Furthermore, the snu56-2 protein does not interact with any of the spliceosomal partners tested, suggesting that it is no longer a U1 snRNP member. To elucidate the role of Snu56p in Mer1p-activated splicing, we conducted TAP of the U1 snRNP by using TAP-tagged Snu56p. We found that although Snu56p-TAP could pull down the U1 snRNP, the mutant was too unstable in these extracts to be purified (our unpublished results). It is possible that other members of the U1 snRNP remain stably in a complex that is still capable of constitutive splicing but not competent for Mer1p-activated splicing in snu56-2 mutant cells.
Acknowledgments
We are grateful to Michael Rosbash, Ren-Jang Lin, Vincent Guacci, and Edward Silverman for plasmids and strains. We are especially grateful to Marc Spingola for his quick response to several reagent requests. We thank Kimberly Brown for her work initiating the hmt1Δ synthetic lethal screen and Katrina Cooper and Raymond O'Keefe for providing primer sequences. We also thank Randy Strich for his inspired experimental insight and critical reading of the manuscript. Finally, we thank Eric Moss for constructive discussions.
This work was supported by grants from the National Institutes of Health (GM58493) and the Foundation of the University of Medicine and Dentistry of New Jersey to M.F.H.
Footnotes
Published ahead of print on 11 February 2008.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 2.Baudin-Baillieu, A., E. Guillemet, C. Cullin, and F. Lacroute. 1997. Construction of a yeast strain deleted for the TRP1 promoter and coding region that enhances the efficiency of the polymerase chain reaction-disruption method. Yeast 13353-356. [DOI] [PubMed] [Google Scholar]
- 3.Bender, A., and J. R. Pringle. 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 111295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bossie, M. A., C. DeHoratius, G. Barcelo, and P. Silver. 1992. A mutant nuclear protein with similarity to RNA binding proteins interferes with nuclear import in yeast. Mol. Biol. Cell 3875-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36333-360. [DOI] [PubMed] [Google Scholar]
- 6.Burge, C. B., T. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNA by the spliceosome, p. 525-560. In R. F. Gesteland (ed.), The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110119-122. [DOI] [PubMed] [Google Scholar]
- 8.Cooper, K. F., M. J. Mallory, D. B. Egeland, M. Jarnik, and R. Strich. 2000. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc. Natl. Acad. Sci. USA 9714548-14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, C. A., L. Grate, M. Spingola, and M. Ares, Jr. 2000. Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 281700-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Gatto-Konczak, F., C. F. Bourgeois, C. Le Guiner, L. Kister, M. C. Gesnel, J. Stevenin, and R. Breathnach. 2000. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 206287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dohmen, R. J., P. Wu, and A. Varshavsky. 1994. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 2631273-1276. [DOI] [PubMed] [Google Scholar]
- 12.Durfee, T., K. Becherer, P. L. Chen, S. H. Yeh, Y. Yang, A. E. Kilburn, W. H. Lee, and S. J. Elledge. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7555-569. [DOI] [PubMed] [Google Scholar]
- 13.Duttweiler, H. M. 1996. A highly sensitive and non-lethal beta-galactosidase plate assay for yeast. Trends Genet. 12340-341. [DOI] [PubMed] [Google Scholar]
- 14.Ekwall, K., M. Kermorgant, G. Dujardin, O. Groudinsky, and P. P. Slonimski. 1992. The NAM8 gene in Saccharomyces cerevisiae encodes a protein with putative RNA binding motifs and acts as a suppressor of mitochondrial splicing deficiencies when overexpressed. Mol. Gen. Genet. 233136-144. [DOI] [PubMed] [Google Scholar]
- 15.Engebrecht, J., and G. S. Roeder. 1990. MER1, a yeast gene required for chromosome pairing and genetic recombination, is induced in meiosis. Mol. Cell. Biol. 102379-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engebrecht, J., and G. S. Roeder. 1989. Yeast mer1 mutants display reduced levels of meiotic recombination. Genetics 121237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engebrecht, J. A., K. Voelkel-Meiman, and G. S. Roeder. 1991. Meiosis-specific RNA splicing in yeast. Cell 661257-1268. [DOI] [PubMed] [Google Scholar]
- 18.Fields, S., and R. Sternglanz. 1994. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 10286-292. [DOI] [PubMed] [Google Scholar]
- 19.Förch, P., O. Puig, N. Kedersha, C. Martinez, S. Granneman, B. Seraphin, P. Anderson, and J. Valcarcel. 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 61089-1098. [DOI] [PubMed] [Google Scholar]
- 20.Fortes, P., J. Kufel, M. Fornerod, M. Polycarpou-Schwarz, D. Lafontaine, D. Tollervey, and I. W. Mattaj. 1999. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol. Cell. Biol. 196543-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gary, J. D., W. J. Lin, M. C. Yang, H. R. Herschman, and S. Clarke. 1996. The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J. Biol. Chem. 27112585-12594. [DOI] [PubMed] [Google Scholar]
- 22.Golemis, E. A., I. Serebriiskii, R. L. Finley, M. G. Kolonin, J. Gyrusis, and R. Brent. 1999. Interaction trap/two-hybrid system to identify interacting proteins, p. 20.1.1-20.1.40. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 4. John Wiley/Greene, New York, NY. [Google Scholar]
- 23.Gottschalk, A., C. Bartels, G. Neubauer, R. Luhrmann, and P. Fabrizio. 2001. A novel yeast U2 snRNP protein, Snu17p, is required for the first catalytic step of splicing and for progression of spliceosome assembly. Mol. Cell. Biol. 213037-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottschalk, A., J. Tang, O. Puig, J. Salgado, G. Neubauer, H. V. Colot, M. Mann, B. Seraphin, M. Rosbash, R. Luhrmann, and P. Fabrizio. 1998. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4374-393. [PMC free article] [PubMed] [Google Scholar]
- 25.Grainger, R. J., and J. D. Beggs. 2005. Prp8 protein: at the heart of the spliceosome. RNA 11533-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green, D. M., K. A. Marfatia, E. B. Crafton, X. Zhang, X. Cheng, and A. H. Corbett. 2002. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 2777752-7760. [DOI] [PubMed] [Google Scholar]
- 27.Henry, M., C. Z. Borland, M. Bossie, and P. A. Silver. 1996. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics 142103-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henry, M. F., and P. A. Silver. 1996. A novel methyltransferase (Hmt1p) modifies poly(A)+-RNA-binding proteins. Mol. Cell. Biol. 163668-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvath, A., and H. Riezman. 1994. Rapid protein extraction from Saccharomyces cerevisiae. Yeast 101305-1310. [DOI] [PubMed] [Google Scholar]
- 30.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 125-14. [DOI] [PubMed] [Google Scholar]
- 31.Kassir, Y., and G. Simchen. 1991. Monitoring meiosis and sporulation in Saccharomyces cerevisiae. Methods Enzymol. 19494-110. [DOI] [PubMed] [Google Scholar]
- 32.Kessler, M. M., M. F. Henry, E. Shen, J. Zhao, S. Gross, P. A. Silver, and C. L. Moore. 1997. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 112545-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koepp, D. M., D. H. Wong, A. H. Corbett, and P. A. Silver. 1996. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J. Cell Biol. 1331163-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labib, K., J. A. Tercero, and J. F. Diffley. 2000. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science 2881643-1647. [DOI] [PubMed] [Google Scholar]
- 35.Lesser, C. F., and C. Guthrie. 1993. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics 133851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leu, J.-Y., and G. S. Roeder. 1999. Splicing of the meiosis-specific HOP2 transcript utilizes a unique 5′ splice site. Mol. Cell. Biol. 197933-7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matlin, A. J., F. Clark, and C. W. Smith. 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6386-398. [DOI] [PubMed] [Google Scholar]
- 38.Nakagawa, T., and H. Ogawa. 1997. Involvement of the MRE2 gene of yeast in formation of meiosis-specific double-strand breaks and crossover recombination through RNA splicing. Genes Cells 265-79. [DOI] [PubMed] [Google Scholar]
- 39.Ogawa, H., K. Johzuka, T. Nakagawa, S. H. Leem, and A. H. Hagihara. 1995. Functions of the yeast meiotic recombination genes, MRE11 and MRE2. Adv. Biophys. 3167-76. [DOI] [PubMed] [Google Scholar]
- 40.Puig, O., A. Gottschalk, P. Fabrizio, and B. Seraphin. 1999. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev. 13569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rose, M. D., P. Novick, J. H. Thomas, D. Botstein, and G. R. Fink. 1987. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60237-243. [DOI] [PubMed] [Google Scholar]
- 42.Rymond, B. C., and M. Rosbash. 1992. Yeast pre-mRNA splicing, p. 143-192. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 43.Shen, E. C., M. F. Henry, V. H. Weiss, S. R. Valentini, P. A. Silver, and M. S. Lee. 1998. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 12679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silberstein, S., G. Schlenstedt, P. A. Silver, and R. Gilmore. 1998. A role for the DnaJ homologue Scj1p in protein folding in the yeast endoplasmic reticulum. J. Cell Biol. 143921-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siomi, H., M. J. Matunis, W. M. Michael, and G. Dreyfuss. 1993. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 211193-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spingola, M., and M. Ares, Jr. 2000. A yeast intronic splicing enhancer and Nam8p are required for Mer1p-activated splicing. Mol. Cell 6329-338. [DOI] [PubMed] [Google Scholar]
- 48.Spingola, M., J. Armisen, and M. Ares, Jr. 2004. Mer1p is a modular splicing factor whose function depends on the conserved U2 snRNP protein Snu17p. Nucleic Acids Res. 321242-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vijayraghavan, U., M. Company, and J. Abelson. 1989. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 31206-1216. [DOI] [PubMed] [Google Scholar]
- 50.Xu, C., and M. F. Henry. 2004. Nuclear export of hnRNP Hrp1p and nuclear export of hnRNP Npl3p are linked and influenced by the methylation state of Npl3p. Mol. Cell. Biol. 2410742-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu, C., P. A. Henry, A. Setya, and M. F. Henry. 2003. In vivo analysis of nucleolar proteins modified by the yeast arginine methyltransferase Hmt1/Rmt1p. RNA 9746-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, D., and M. Rosbash. 1999. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev. 13581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]