Abstract
Human renal clear cell carcinoma (RCC) is frequently associated with loss of the von Hippel-Lindau (VHL) tumor suppressor (pVHL), which inhibits ubiquitylation and degradation of the alpha subunits of hypoxia-inducible transcription factor. pVHL also ubiquitylates the large subunit of RNA polymerase II, Rpb1, phosphorylated on serine 5 (Ser5) within the C-terminal domain (CTD). A hydroxylated proline 1465 within an LXXLAP motif located N-terminal to the CTD allows the interaction of Rpb1 with pVHL. Here we report that in RCC cells, pVHL regulates expression of Rpb1 and is necessary for low-grade oxidative-stress-induced recruitment of Rpb1 to the DNA-engaged fraction and for its P1465 hydroxylation, phosphorylation, and nondegradative ubiquitylation. Egln-9-type prolyl hydroxylases, PHD1 and PHD2, coimmunoprecipitated with Rpb1 in the chromatin fraction of VHL+ RCC cells in response to oxidative stress, and PHD1 was necessary for P1465 hydroxylation while PHD2 had an inhibitory effect. P1465 hydroxylation was required for oxidative-stress-induced Ser5 phosphorylation of Rpb1. Importantly, overexpression of wild-type Rpb1 stimulated formation of kidney tumors by VHL+ cells, and this effect was abolished by P1465A mutation of Rpb1. These data indicate that through this novel pathway involving P1465 hydroxylation and Ser5 phosphorylation of Rbp1, pVHL may regulate tumor growth.
pVHL is the main tumor suppressor for which loss of activity is causatively linked to renal clear cell carcinoma (RCC), the most malignant and common form of kidney cancer. The VHL gene is mutated or hypermethylated in about 40 to 70% of sporadic RCC. Hereditary loss of pVHL function in von Hippel-Lindau (VHL) disease also results in highly vascularized RCC and capillary tumors of other organs, such as hemangioblastoma of the central nervous system and retinal angioma (19, 20). A body of experimental evidence, based on a subcutaneous xenograft model system, supports the idea that accumulation of the alpha subunit of the hypoxia-inducible transcription factor (HIF) HIF-2α and induction of HIF target gene products, resulting from the loss of pVHL-mediated ubiquitylation, are necessary and sufficient to promote growth of RCC tumors (22, 23). HIF activation has also been demonstrated as an early tumorigenesis event in kidneys from VHL patients (32). Biochemically, pVHL is the substrate-recognizing component of a multiprotein E3 ubiquitin ligase complex containing elongins C and B, Cullin 2, and the RING-H2 finger protein Rbx-1 (for a review, see reference 19). pVHL-dependent ubiquitylation of HIF-αs is preceded by hydroxylation of conserved proline residues located within LXXLAP motifs (16, 17) by the O2-, Fe(II)-, and oxyglutarate-regulated Egl-9-type proline hydroxylases (PHDs) (7). Thus, an important aspect of pVHL's tumor suppressing activity is the prevention of HIF-α accumulation, which in turn suppresses induction of the HIF target genes. Clearly, however, pVHL activity is not limited to regulation of HIFs. Other targets of pVHL-associated E3 ligase activity are atypical protein kinase C λ (46), deubiquitylating enzyme 1 (28), and two subunits of the RNA polymerase II complex (RNAPII), Rpb1 (25) and Rpb7 (40). Because of the crucial role of RNAPII in gene expression, this article is focused on the function of pVHL in regulating Rpb1.
Rpb1 is the largest subunit of RNAPII and carries the fundamental enzymatic activity of the complex, synthesizing all cellular mRNAs. An important aspect of RNAPII regulation occurs through phosphorylation and dephosphorylation of serine residues within 52 YSPTSPS heptad repeats of the C-terminal domain (CTD) of Rpb1. In the preinitiation complex, the CTD is hypophosphorylated. Upon transition from transcription initiation to elongation, the Ser5 residues within multiple heptad repeats of the CTD become phosphorylated (21, 33, 47, 54). Subsequently, during processive elongation, Rpb1 becomes hyperphosphorylated on Ser2 residues within the heptad repeats (21, 33, 47, 54). Eventually, the CTD needs to be dephosphorylated to reenter transcription. Phosphorylation of the CTD is an important, although not well understood, regulatory step, resulting in binding of specific protein factors coupling transcription with processing of the primary transcripts and export and translation of mRNA (4, 33, 47).
RNAPII activity is also regulated by ubiquitylation, which, depending on the context, can lead to degradation or be nondegradative. Rpb1 is ubiquitylated and degraded in response to DNA lesions induced by UV light (2, 25, 51) and high, millimolar concentrations of H2O2 (15), possibly due to stalling of the RNAPII complexes. Ubiquitylation of Rpb1 also occurs during ongoing transcription without apparent degradation (26, 27, 39). Polyubiquitylation of Rpb1 during active transcription involves lysine K48- and K63-linked ubiquitin chains (27). Ubiquitylation of both of these lysines has been reported to regulate protein activity without targeting proteins for proteasomal degradation (8, 56, 57). Phosphorylation of Ser5 of Rpb1 is often a prerequisite for Rpb1 ubiquitylation.
We have previously reported that pVHL binds Rpb1 hyperphosphorylated on Ser5 residues of the CTD, leading to its ubiquitylation (25). pVHL binding occurs through an LGQLAP motif on Rpb1 that bears sequence and structural similarity to a pVHL-binding domain within HIF-1α. This motif is located in the pocket between Rpb1 and another subunit of the RNAPII complex, Rpb6, on the surface of the complex and N-terminal to the beginning of the CTD (25). Similar to its interaction with HIF-α, binding of pVHL to Rpb1 requires hydroxylation of the proline P1465 within the LGQLAP motif (25).
In this article we report that pVHL regulated constitutive steady-state levels of Rpb1 in RCC cells. We also show that pVHL stimulated translocation of Rpb1 from the soluble to the chromatin-engaged fraction and induced Rpb1 P1465 hydroxylation, Ser5 phosphorylation, and nondegradative ubiquitylation in response to low-grade oxidative stress. Consistent with these findings, we observed that all three Egl-9-type proline hydroxylases (PHD1 to PHD3) and pVHL were present and induced in the chromatin fraction in response to the oxidative stress. PHD1 and PHD2 were coimmunoprecipitated with Rpb1 in response to oxidative stress in a pVHL-dependent manner. Functional experiments revealed that PHD1 was necessary for oxidative-stress-induced P1465 hydroxylation while surprisingly, PHD2 had an inhibitory effect on this modification. We further discovered that P1465 hydroxylation was necessary for subsequent Ser5 phosphorylation of Rpb1 in response to oxidative stress. Finally, we found that expression of wild-type Rpb1 stimulated formation of tumors by pVHL+ cells and that this effect was prevented by P1465A mutation. These data implicate that hydroxylation of P1465 may have oncogenic effects. In view of the role of pVHL in promoting P1465 hydroxylation, the results shed new light on the role of pVHL in renal cancer tumorigenesis.
(Part of the work presented in this article has been submitted as patent application no. 107 38-83.)
MATERIALS AND METHODS
Antibodies and constructs.
The following antibodies were used: H14, which recognizes Rpb1 phosphorylated on Ser5, and 8GW16, which recognizes total Rpb1 (Covance); N20 against the N terminus of Rpb1 (Santa Cruz); anti-pVHL (Ig32), antihemagglutinin (anti-HA) tag (12CA5), and antiubiquitin (StressGen); antihistidine (Invitrogen; also H1029 from Sigma); anti-PHD1/2/3 and anti-HIF-2α (Novus); and anti-H3 (Abcam). The antibody against hydroxylated proline within the Rpb1 peptide (HP) was custom made by Alpha Diagnostic, Inc. (San Antonio, TX). This antibody was used at a concentration of 1:500 for Western blots (see Fig. 1Aii). Secondary antibodies were obtained from Sigma, Cell Signaling, or Amersham.
FIG. 1.
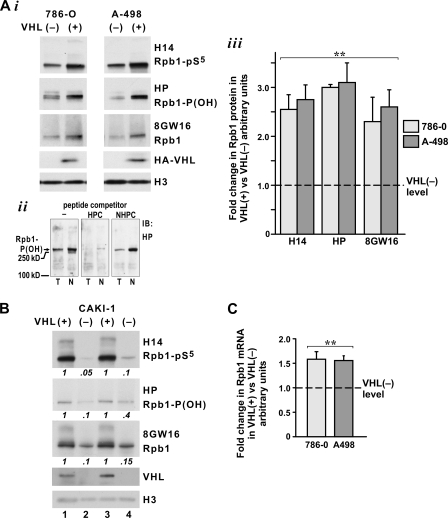
pVHL regulates constitutive levels of Rpb1 in RCC cells. (A) Western blot analysis (i) and quantification of four independent experiments (iii) demonstrating a two- to threefold induction in the steady-state levels of Rpb1 in total chromatin-enriched nuclear extracts from 786-O and A-498 RCC cells with reconstituted pVHL compared to those in VHL− cells. (ii) Specificity of HP antibody in detecting Rpb1 hydroxylated on P1465 [Rpb1-P(OH)]. HP antibody detects a band of 250 kDa in total cellular (T) and nuclear (N) fractions in the absence of the competitor peptide and in the presence of nonhydroxylated peptide competitor (NHPC) but not in the presence of the same amount of hydroxylated peptide competitor (HPC). H14 antibody detects Ser5-phosphorylated Rpb1 (Rpb1-pS5); 8GW16 detects total Rpb1. The H3 antibody detects histone 3 to demonstrate equal loading. The HA-tagged-pVHL level (HA-VHL) was detected with an anti-HA antibody (12CA5). (B) Western blot showing a decrease in Rpb1 in two independent clones of Caki-1 cells with pVHL knockdown obtained using two different shRNA constructs and measured with the indicated Rpb1 antibodies. pVHL levels were detected using an anti-VHL antibody (Ig32). (C) RT-PCR analysis of Rpb1 mRNA in 786-O and A-498 cells with reconstituted pVHL compared to levels in VHL− cells; n = 4. (A and C) Steady-state levels of the Rpb1 protein or mRNA in VHL− cells have been assigned a value of 1 (dashed line across the graph). **, P < 0.01; IB, immunoblot.
The Rpb1 construct was based on the construct pAT7h1 (45). In this construct, Rpb1 is labeled with a six-histidine tag in the 3′ end. Mutation of P1465A was performed in this expression vector by mutating codon CCG to GCG. A 5.8-kb HindIII-XbaI fragment of Rpb1 was cut out of the pAT7h1 vector and inserted into the HindIII/AlfII sites of pcDNA3.1/hygro(−). The 5′ end was amplified with the 5′ primer containing the Flag tag and inserted into NheI/HindIII sites. Both NheI and AlfII sites were blunted. Subsequently, the NheI/XhoI fragment of Rpb1 containing the mutation from the pAT7h vector was used to replace this fragment in pcDNA3.1 containing wild-type Rpb1. This construct contains the same histidine tag as the original one. The quality of all constructs was confirmed by sequencing. Transfections were performed using Lipofectamine (Invitrogen).
Cell cultures and treatments.
786-O and A-498 RCC cells were grown as described previously (25). A-498 cells were purchased from ATCC. Stable transfections of human HA-VHL or Rpb1 cDNAs were obtained using selection with G418 and hygromycin. Mouse embryonic fibroblasts (MEFs) were isolated from E12.5 PHD knockout (PHD1−/− or PHD2−/−) or respective wild-type fetuses as follows: briefly, after removal of all internal organs, embryonic carcasses were minced and digested with trypsin solution (0.25%) containing 0.1 μg/ml DNase and the digested tissue homogenized and washed in EF medium (Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum [FBS], 2 mM glutamine, 1× minimal essential medium nonessential amino acids, 1 mM pyruvate, 1× β-mercaptoethanol, and penicillin-streptomycin). Single-cell suspensions were cultured and passaged in the medium described. For immortalization, MEFs were cotransfected with a simian virus 40 large-T-antigen construct and pNT-hygro (carrying the hygromycin resistance gene) using SuperFect lipofection (Qiagen). Starting 48 h posttransfection, stably transfected cells underwent hygromycin selection. For experiments, immortalized MEFs were used at a density of 2 × 104 to 3 × 104/cm2.
For H2O2 treatment, growth medium was removed and stored and cells were washed with phosphate-buffered saline. Treatment was performed with 25 μM H2O2 in serum-free RPMI 1640 medium for 15 min or, for controls, with RPMI 1640 without H2O2. Immediately after, H2O2-containing medium was removed and cells were returned to their original growth medium. With the exception of time courses, in most experiments cells were collected 4 h after reconstitution of the growth medium.
Intracellular H2O2 was measured using the 2,7′-dichlorofluorescein diacetate method (Molecular Probes) as described previously (24). Briefly, cells were grown and exposed to exogenous H2O2 as described above or treated with cell-permeating polyethylene glycol catalase (5,000 U/ml) for 15 min. Cells were then washed twice with loading buffer (24), incubated with the same buffer containing 10 μM 2,7′-dichlorofluorescein diacetate for 10 min at 37°C, and then washed again with the same buffer and analyzed by using a spectrofluorometer at an excitation wavelength of 485 nm and an emission wavelength of 530 nm. After the measurements, the cells were lysed and the protein concentration was determined. The data presented on dichlorofluorescein fluorescence are normalized to the protein concentration.
Fourteen samples of fresh snap-frozen human clear cell carcinoma and paired normal kidney control samples were obtained from the tissue bank of the Department of Pathology and Laboratory Medicine at the University of Cincinnati or purchased from NDRI under exemption from the IRB protocol.
Preparation of extracts.
Total cellular extracts were obtained by lysing cell pellets in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], and 1% NP-40).
To obtain total nuclear chromatin-enriched extracts, cell pellets were resuspended in 3 volumes of cell lysis buffer (10 mM Tris [pH 7.5], 10 mM NaCl, 3 mM MgCl2, 0.5% Triton), lysed on ice for 2 min, and centrifuged at 5,000 × g for 2 min. Pelleted nuclei were resuspended in 3 volumes PH buffer (100 mM KCl, 20 mM Tris [pH 7.8], 0.05% Tween 20 containing protease inhibitors and dithiothreitol). The NaCl concentration was adjusted to 0.3 M, and the nuclei were extracted for 30 min at 4°C. The nuclei were then digested with DNase and micrococcal nuclease (10 U and 37.5 U, respectively, per 100 μl of pellet volume) for 1 h to release DNA-bound RNAPII complexes. Following the digestion, NP-40 was added to a final concentration of 0.5% and NaCl was adjusted to a final concentration of 0.5 M. Extracts were incubated at 4°C for 30 min and centrifuged at 14,000 rpm for 20 min, and then glycerol was added to a final concentration of 5%. Examples of Western blots using these extracts are shown among others in Fig. 1A and B and 2A.
FIG. 2.
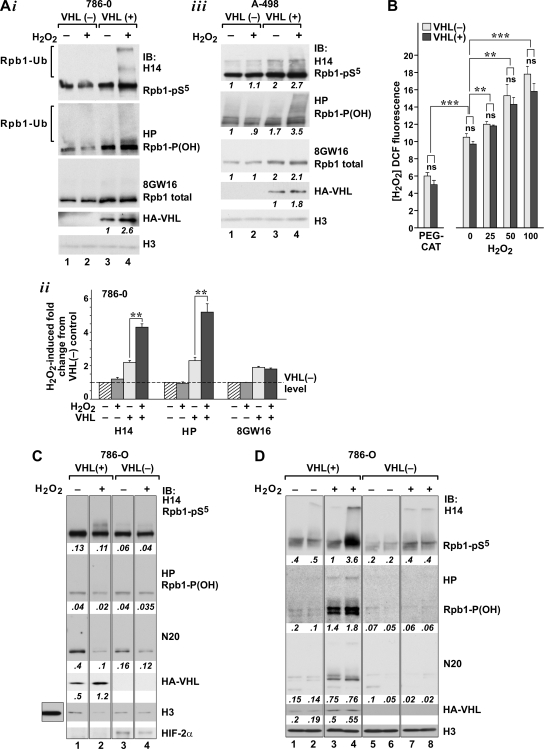
Oxidative stress induces pVHL-dependent P1465 hydroxylation, Ser5 phosphorylation, and ubiquitylation of Rpb1. (A) Western blot analysis (i) and quantification (ii) (n = 4; **, P < 0.01) of changes in Rpb1 in the total chromatin-enriched nuclear extracts from 786-O cells and A-498 cells (iii). For quantification, the optical density measurements were performed with each individual lane using a rectangular box of the same size. The measurements were normalized to the level of Rpb1 detected with each antibody in control VHL− cells, which was assigned a value of 1 (dashed line). pVHL and H3 were detected as for Fig. 1. In panels i and iii, pVHL quantification of the pVHL expression in VHL+ cells under oxidative stress is shown as normalized to constitutive expression. (B) Measurements of H2O2 levels using dichlorofluorescein (DCF) fluorescence in 786-O VHL− and VHL+ cells, untreated or treated with the indicated concentrations of H2O2 or polyethylene glycol-catalase (PEG-CAT) (n = 6). **, P < 0.01; ***, P < 0.001. (C) Western blot analysis of Rpb1 and pVHL in the soluble nuclear fraction. Immunoblotting for HIF-2α demonstrates a lack of HIF induction by oxidative stress in VHL+ cells. The additional square to the left of the main panel in the lane probed with H3 antibody is shown to emphasize the difference in H3 concentrations between the soluble and engaged fractions. (D) Western blot analysis of Rpb1 and pVHL in the chromatin fractions corresponding to the soluble fraction shown in panel C (left). For quantification in panels C and D, optical density measurements were performed; the data are expressed in arbitrary units not normalized to any particular conditions.
To obtain the fraction of chromatin enriched specifically for Rpb1 engaged on the DNA, we followed the protocol described in reference 38 with modifications. The nuclear pellets, obtained as described above, were extracted for 30 min with 3 volumes of PH buffer with 0.3 M NaCl. The soluble nuclear fraction was collected by centrifugation. This fraction is shown in Fig. 2C. Pellets were then digested with DNase and micrococcal nuclease for 1 h in 4°C. After digestion and centrifugation, the remaining pellets were extracted with 3 volumes of PH buffer containing 0.5 mM NaCl, twice for 15 min or once for 30 min at 4°C. These fractions are shown in Fig. 2D.
To obtain the combined chromatin fraction, the soluble fraction was separated as described above, the remaining pellet was digested with nucleases (see above), and then the entire digest was extracted with a final concentration of 0.5 M NaCl for 30 min at 4°C. Combined chromatin extracts of human tumors were obtained as follows: small pieces of frozen tumor were first allowed to swell in cell lysis buffer for 10 min and then were minced in the Dounce homogenizer in the same buffer to complete homogeneity. The homogenates were centrifuged at 14,000 rpm for 20 min, and the remaining pellets were extracted with 0.3 M NaCl for 30 min at 4°C and centrifuged. The pellets were digested with the nucleases as described above. NP-40 was added to a final concentration of 0.5% and NaCl to a final concentration of 0.5 M, and the pellets were extracted for 30 min at 4°C.
Denatured lysates were obtained by boiling indicated fractions in 3 volumes of SDS lysis buffer (1% SDS, 10 mM Na2HPO4-NaH2PO4, pH 7.2, 150 mM NaCl, 5 mM EDTA, 2 mM EGTA) for 10 min. The lysates were diluted with immunoprecipitation buffer (see next section), centrifuged at 21,000 × g for 30 min, and used for immunoprecipitations with H14 antibody.
Western blotting was performed according to standard protocols, with equal amounts of protein loaded in each lane.
Immunoprecipitations.
For H14 immunoprecipitations, agarose beads with preconjugated goat anti-immunoglobulin M antibodies (Sigma) were incubated with the extract (see above section) in buffer containing 50 mM HEPES, pH 7.8, 150 mM NaCl, 5 mM MgCl2, 5% (vol/vol) glycerol, and 0.1% Triton (IP buffer). For those experiments where we analyzed only covalent modifications of Rpb1, the beads were washed for 15 min each in IP buffer and then in IP buffer containing 0.5% Triton X-100, 0.5% Igepal, and 0.5% sodium deoxycholate, and finally twice in the IP buffer. For those experiments where we analyzed proteins coimmunoprecipitated with Rpb1, the beads were washed with the IP buffer containing 0.5% NP-40. Immunoprecipitated proteins were eluted by boiling in SDS sample buffer, resolved by SDS-polyacrylamide gel electrophoresis on 4 to 22% or 10% gradient gels, and detected by immunoblotting. The coimmunoprecipitations of Rpb1 using anti-HA antibody detecting the VHL tag or the coimmunoprecipitations using H14 antibody were performed as described previously (25). Antihistidine immunoprecipitations of Rpb1 were performed under the same conditions as anti-H14 immunoprecipitations.
RNA interference assays.
All short hairpin RNA (shRNA) lentivirus vectors based on the pLKO.1 vector with the U6 promoter and puromycin selection marker were purchased from Open Biosystems. The lentivirus constructs against PHD1 were TRC-22325 and TRC-22324, that against PHD2 was TRC-1045, and those against pVHL were TRC-39623 and TRC-39624. DNA constructs were vesicular stomatitis virus G (VSV-G) envelope packaged (Cincinnati Children's Hospital Medical Center Viral Vector Core) and used to infect cells. Stably transfected pools and clones were selected using 3.5 μg/ml of puromycin starting 72 h after transfections. Empty vector and nontargeting shRNA constructs were used as controls in these experiments. On Target plus Smartpool small interfering RNA (siRNA) against human PHD3 or siCONTROL were purchased from Dharmacon. Cells were seeded at low confluence in antibiotic-free medium 24 h before transfection. Transfections were performed twice using Lipofectamine RNAiMAX (Invitrogen) and 100 nM of siRNA with a 48-h interval between transfections, according to the manufacturer's protocol. Inhibition of the PHD3 protein, as measured by Western blotting, was measured 72 h after the second transfection.
RT-PCR.
Equal fractions of RNA were employed for first-strand cDNA synthesis using a SuperScript III First-strand cDNA synthesis system for reverse transcription-PCR (RT-PCR) (Invitrogen) according to the manufacturer's protocol. The following forward and reverse primers were used for cDNA amplification: for human endogenous Rpb1, forward, 5′-GAA CCC GGT TAC TTA TTT ATT CGT TAC CCT-3′, and backward, 5′-ACA TGG AAC TGG AGG AGC TTC ACA-3′; for the human exogenous Rpb1, forward, 5′-TGG ATT ACA AGG ATG ACG ATG ACA AGC A-3′; the backward primer was the same as in the case of the endogenous Rpb1. The annealing temperature was 56°C in both cases.
Orthotopic xenografts in nude mice.
Fifty thousand cells resuspended in Matrigel to a final volume of 50 μl were slowly injected into the parenchyma of kidneys, approximately 1 to 0.5 mm from the surface, of 4- to 5-week-old athymic nude mice, similar to protocols described by others (41). Mice were sacrificed after 12 weeks, and the tumors, including the entire kidney, were collected and weighed. The noninjected kidney was used as a negative control. Tumors were dissected, fixed in formalin, and paraffin embedded. Sections were prepared for immunocytochemistry and stained with antihistidine antibody (H1029), followed by secondary biotinylated antibody (M.O.M kit; Vector Laboratories) and the ABC reagent (Vector Laboratories). Positive staining was detected with the DAB substrate (Sigma), and sections were counterstained briefly with hematoxylin. All procedures involving animals were approved by the Institutional Animal Care and Use Committee of the University of Cincinnati and are consistent with guidelines provided by the National Institutes of Health.
2-Dimensional gel electrophoresis, image analysis, and protein identification.
Extracts from VHL− or VHL+ 786-O cells, untreated or treated with H2O2, were separated by two-dimensional (2-D) gel electrophoresis and visualized by silver staining, all as described previously (18). Briefly, 150 μg of protein from two to three replicates of each extract was loaded onto an 18-cm, pH 4 to 7 immobilized-pH-gradient strip (GE Health Care). After electrophoretic separation based on isoelectric point, the immobilized-pH-gradient strips were reduced and alkylated, equilibrated in SDS-polyacrylamide gel electrophoresis gel buffer, and then overlaid onto the second-dimension precast 10% Tricine gel (Genomic Solutions). After electrophoretic separation in the second dimension, the proteins were visualized by silver staining and the images captured on a FujiFilm FLA5100 digital image scanner. Quantification of protein changes across replicates of the four conditions analyzed were captured via image analysis using Progenesis/SameSpot image analysis software (NonLinear Dynamic, Ltd.). Proteins that showed a significant change in abundance between one or more of the conditions were subsequently excised from the gel, digested with trypsin, and identified by mass spectrometry on an Applied Biosystems 4800 matrix-assisted laser desorption ionization-time-of-flight /time-of-flight instrument using methods originally described in reference 53 but with modifications detailed in reference 18.
Quantification.
Optical densities of Western blots were measured using the ImageQuant 5.2 (Molecular Dynamics) software program. Averaged data are expressed as means ± standard errors of the means where n is >2 or as means and standard deviations of the mean where n is 2.
RESULTS
pVHL regulates steady-state levels of the Rpb1 protein in RCC cells.
In our previous work analyzing the effects of pVHL on Rpb1 in the rat pheochromocytoma cell line PC12, we found that overexpression of human pVHL resulted in an overall decreased level of Rpb1, while the knockdown of endogenous pVHL augmented steady-state levels of Rpb1 (25). These effects seemed to be consistent with the role of pVHL in ubiquitylation of Rpb1 and the expected resulting degradation (25). To our surprise, reconstitution of pVHL in two different RCC cell lines increased steady-state levels of Rpb1 (Fig. 1A). We found a two- to threefold increase in Rpb1 protein levels in total chromatin-enriched nuclear extracts prepared following a protocol that enriches for Rpb1 engaged on the DNA by digestion of nuclei with DNase and micrococcal nuclease followed by high-salt extraction (Fig. 1Ai). Quantitatively similar increases were measured using antibodies detecting all forms of Rpb1, such as 8GW16; antibodies specific for Rpb1 phosphorylated mainly on Ser5, such as H14; and an HP antibody specifically detecting Rpb1 hydroxylated on P1465 (Fig. 1Aiii). This indicated that an increase in the total amount of the Rpb1 protein was paralleled by increased levels of Rpb1 phosphorylation and hydroxylation. The HP antibody was generated against an Rpb1 peptide containing hydroxylated P1465 (Fig. 1Aii). Because reconstitution of pVHL in RCC cells may potentially have some nonphysiological effects, such as those resulting from a very high level of expression, we also examined whether the knockdown of pVHL levels in RCC cells that express endogenous pVHL, such as Caki-1 cells, could decrease levels of Rpb1 expression (Fig. 1B). Indeed, the stable knockdown of pVHL using two different shRNA constructs resulted in severalfold decreases in steady-state levels of Rpb1, as measured with the indicated antibodies. The corresponding changes in the steady-state levels of Rpb1 mRNA between VHL+ and VHL− 786-0 or A-498 RCC cells were measured only in the range of 0.5-fold (Fig. 1C). Thus, it is likely that full induction of Rpb1 expression mediated by pVHL in RCC cells includes other levels of regulation, such as an increased translation rate or stabilization of the Rpb1 protein. Overall, these data indicate that pVHL is a regulator of constitutive levels of Rpb1 expression in RCC cells and that this constitutive regulation is specific for RCC cells, since pVHL had an opposite effect on Rpb1 steady-state levels in PC12 cells. The molecular mechanisms and functional implications of this constitutive regulation will be the subject of a separate study.
Oxidative stress stimulates pVHL-dependent phosphorylation on Ser5, P1465 hydroxylation, and nondegradative ubiquitylation of Rpb1.
Treatment of 786-O cells with a low dose (25 μM) of H2O2 for 15 min resulted in a twofold, significant induction of Rpb1 phosphorylation and P1465 hydroxylation and the appearance of a higher-molecular-weight ladder, measured in total chromatin-enriched nuclear extracts obtained from cells with reconstituted pVHL but not from the cells without pVHL (Fig. 2Ai and Aii). This response started immediately after treatment, reached a plateau approximately 4 h later, and lasted for up to 16 h (data not shown). However, oxidative stress did not influence total steady-state levels of the Rpb1 protein, as measured with 8GW16 (Fig. 2A) or N20 (not shown) antibody. The levels of pVHL were increased in the total nuclear fraction in response to oxidative stress. A very similar result was obtained when the A-498 cell line was used (Fig. 2Aiii). The concentration of H2O2 used in these experiments increased the intracellular concentration of H2O2 by approximately 10% in both VHL+ and VHL− cells (Fig. 2B). Expression of pVHL did not have any effects on the constitutive or induced levels of H2O2 under these experimental conditions (Fig. 2B).
The total nuclear chromatin-enriched extracts, such as those shown in Fig. 2A, contained Rpb1 that included soluble Rpb1, as well as Rpb1 engaged on the DNA that is released from the DNA by DNase/micrococcal digestion. Because the engaged Rpb1 is likely to represent the transcriptionally involved RNAPII complexes (either actively transcribing or paused), we were particularly interested in the effects of pVHL in oxidative-stress-induced phosphorylation, hydroxylation, and ubiquitylation of engaged Rpb1. Thus, the total chromatin-enriched nuclear extract was separated into the soluble fraction, obtained by standard extraction of nuclei with buffer containing 0.3 M NaCl (Fig. 2C), and the engaged fractions, obtained by DNase and micrococcal nuclease digestion of the pellet remaining after extraction with 0.3 M NaCl, followed by two high-salt (0.5 M NaCl) extractions of the nuclease digest (Fig. 2D) (38). The soluble nuclear fraction of VHL− cells, as expected, contained the HIF-2α protein (Fig. 2C, lane 3), and expression of HIF-2α was decreased in response to oxidative stress (lane 4). In contrast, the chromatin fraction of VHL− cells did not contain the HIF-2α protein (data not shown). Oxidative stress did not induce HIF-2α in VHL+ cells. The soluble fraction also contained a very low concentration of histones compared to that of the engaged fractions (Fig. 2C). The digested and high-salt-extracted fractions did not contain DNA detectable by ethidium bromide staining on agarose gels and were highly enriched in histones (Fig. 2D and C) and histone methyltransferases (not shown here) and thus represented a crude chromatin fraction.
Exposure of VHL+ cells to H2O2 led to an induction of the total amount of Rpb1 (N20 antibody) within the chromatin fraction (Fig. 2D, lanes 3 and 4), which was accompanied by a comparable decrease in total Rpb1 in the soluble fraction, as shown in Fig. 2C (compare lane 2 to lane 1). Rpb1 recruited to the chromatin fraction was also significantly more hydroxylated (HP antibody) and phosphorylated (H14 antibody). Both of these modifications were induced at a level higher than could be accounted for by total Rpb1 induction alone (Fig. 2D, right). This indicates an additional direct effect of oxidative stress on hydroxylation and phosphorylation of Rpb1. Rpb1 recruited to the engaged fraction also demonstrated a higher-migrating smear and bands, an indication of ubiquitylation (see also Fig. 3A). In contrast, in the VHL− cells, there was no recruitment of Rpb1 to the engaged fractions (Fig. 2D, lanes 7 and 8). Oxidative stress stimulated phosphorylation of Rpb1 in the engaged fractions, but this was substantially less than that in VHL+ cells (Fig. 2D, compare lanes 7 and 8 with lanes 3 and 4). Importantly, oxidative stress failed to induce P1465 hydroxylation in these chromatin fractions of VHL− cells. Although some ubiquitylation of Rpb1 was detected using H14 antibody in the soluble nuclear fraction of VHL+ cells but not VHL− cells, it was not accompanied by any changes in P1465 hydroxylation (Fig. 2B). Importantly, pVHL was present in both soluble and chromatin fractions, and its levels were induced in both fractions in response to oxidative stress (Fig. 2C and D; also compare with panel A).
FIG. 3.
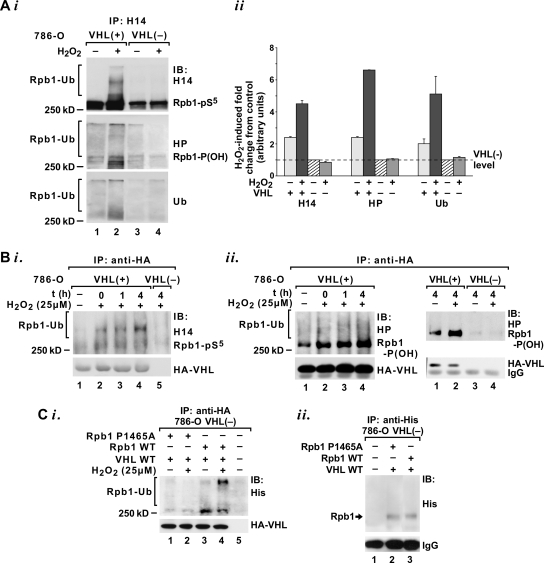
pVHL directly ubiquitylates phosphorylated and hydroxylated Rpb1 in response to oxidative stress. (A) Immunoblot (i) and quantification (ii) (n = 2) of Rpb1-pS5 immunoprecipitated from a denatured chromatin fraction using H14 antibody and then probed with H14, HP, and antiubiquitin (Ub) antibodies. The measurements were normalized to the levels of Rpb1 detected with each antibody in control VHL− cells, which were assigned a value of 1 (dashed line). (B) Immunoblot of Rpb1-pS5 (i) (H14) or Rpb1-P(OH) (ii) (HP) coimmunoprecipitated with the 12CA5 antibody against the HA tag on pVHL. Time point “0” indicates collection immediately following a 15-min exposure; 1 and 4 indicate hours after time zero. The 12CA5 antibody does not coimmunoprecipitate hydroxylated Rpb1 in VHL− cells (i, lane 5; ii, right blot). (Ci) Immunoblot of wild-type (lanes 3 and 4) or P1465A mutant (lanes 1 and 2) histidine-tagged Rpb1 coimmunoprecipitated using the 12CA5 antibody against the HA tag on pVHL and detected with antihistidine antibody. Lane 5 is a negative control using extract without histidine-tagged Rpb1. (Cii) Immunoblot of wild-type (lane 3) or P1465A mutant (lane 2) histidine-tagged Rpb1 coimmunoprecipitated using antihistidine antibody and detected with the same antibody to demonstrate equal expression of both forms of Rpb1. Lane 1 is a negative control using extract without histidine-tagged Rpb1. IP, immunoprecipitation; IB, immunoblot.
To confirm that a pool of engaged Rpb1 was simultaneously phosphorylated on Ser5, P1465 hydroxylated, and polyubiquitylated, we immunoprecipitated Rpb1, using the H14 antibody, from the denatured second chromatin fraction corresponding to the extracts shown in Fig. 2D in lanes 2, 4, 6, and 8 (Fig. 3A). Immunoblotting with HP and antiubiquitin antibodies demonstrated that Rpb1 hyperphosphorylated on Ser5 was, indeed, hydroxylated and ubiquitylated in response to oxidative stress and that this response depended on the presence of functional pVHL (Fig. 3A). These data show that the higher-migrating forms of Rpb1 generated in response to H2O2 result from polyubiquitylation.
A direct role for pVHL in the oxidative-stress-induced ubiquitylation of Rpb1 was further supported by the finding that treatment with H2O2 stimulated rapid and sustained binding of hyperphosphorylated (Fig. 3Bi) and hydroxylated (Fig. 3Bii) Rpb1 to pVHL in total chromatin-enriched nuclear extracts. There was no coimmunoprecipitation of hydroxylated Rpb1 with VHL in VHL− cells (Fig. 3Bii, right). This induction was detectable immediately following exposure (time point zero) and additionally augmented over a subsequent 4-h exposure (Fig. 3B). Clearly, after exposure to oxidative stress, the Rpb1 bound to pVHL shifted, over time, toward higher-migrating molecular forms, which indicates active in vivo ubiquitylation, but not degradation, of Rpb1.
To directly confirm that P1465 hydroxylation is required for the induction of Rpb1 binding to pVHL and Rpb1 ubiquitylation in response to oxidative stress, we expressed wild-type and P1465A mutant forms of Rpb1 cDNA in combination with VHL cDNA in 786-O cells (Fig. 3C). The two forms of Rpb1 were expressed at comparable levels (Fig. Cii). The wild-type histidine-tagged exogenous Rpb1 was coimmunoprecipitated with an antibody against the hemagglutinin tag on pVHL from total chromatin-enriched nuclear extracts (Fig. 3Ci, lanes 3 and 4). In contrast, the P1465A mutant was coimmunoprecipitated only minimally (Fig. 3Ci, lanes 1 and 2). Importantly, wild-type Rpb1 coimmunoprecipitated with pVHL exhibited more of the higher-molecular-weight forms of Rpb1 in response to H2O2 than was the case with the control conditions (Fig. 3Ci, lane 4), indicating ubiquitylation.
Altogether, these results show that reconstitution of pVHL in RCC cells has profound effects on Rpb1 in the chromatin fraction. Oxidative stress triggers recruitment of Rpb1 and pVHL into specific chromatin fractions where Rpb1 undergoes extensive P1465 hydroxylation, phosphorylation, and pVHL-mediated nondegrading ubiquitylation.
Egl-9-type proline hydroxylases regulate P1465 hydroxylation.
The region of Rpb1 containing the LGQLAP motif, which includes hydroxylated P1465, is analogous to motifs on HIF-αs (25). Thus, it is possible that similar proline hydroxylases are involved in the hydroxylation of both prolines. To determine if PHD1 to PHD3 (7) are involved in the hydroxylation of Rpb1, we first analyzed the chromatin fractions containing hydroxylated Rpb1, such as those shown in Fig. 2D, for the presence of PHDs. All three PHDs were present and induced by oxidative stress within the chromatin fractions that contained the highest levels of hydroxylated Rpb1, as demonstrated in Fig. 2D (Fig. 4A). However, this oxidative-stress-induced increase in PHDs was entirely absent in the corresponding fractions from VHL− cells. A similar result was observed in the combined chromatin fraction from A-498 cells, where the levels of all three were increased in response to oxidative stress (Fig. 4B). In contrast to the effects of oxidative stress on the PHD levels within the chromatin fraction, the effects on the total steady-state levels of PHDs in total cellular lysates were very minimal, with a small increase in PHD2 and PHD3 levels in control VHL− cells (Fig. 4C). These data demonstrate that all three PHDs are present within the chromatin fraction, in physical proximity to Rpb1, and are regulated by oxidative stress.
FIG. 4.
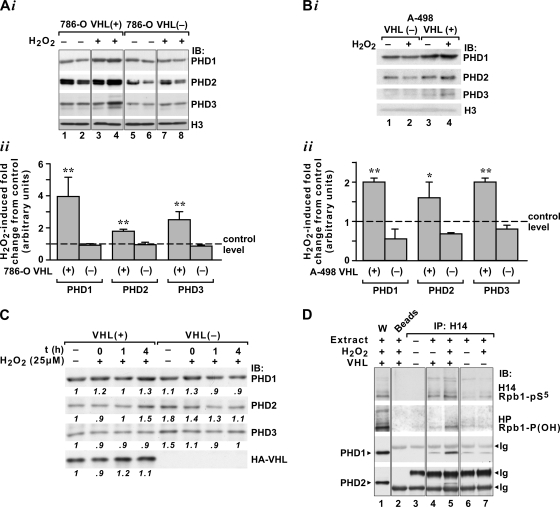
Oxidative stress stimulates induction of PHDs in the chromatin fraction and binding of PHD1 and PHD2 to Rpb1. (A) Western blot analysis (i) and quantification (ii) of levels of PHDs in the indicated chromatin fractions from VHL+ or VHL− 786-O cells. Because the blot shown in this figure is from the same experiment as the blot shown in Fig. 2D, the same H3 Western blot is shown in both. (B) Western blot analysis (i) and quantification (ii) of PHD levels in the combined chromatin fraction from A-498 cells. In the quantifications (n = 4; **, = P < 0.01; *, P < 0.05), the data are presented as changes induced by oxidative stress compared to the control levels, which are given a value of 1 and marked by a dashed line. IB, immunoblot. (C) Western blot analysis of total cellular lysates for PHD1 to PHD3 and VHL in VHL+ or VHL− 786-O cells. Extracts were obtained at the indicated time points after the end of H2O2 treatment. (D) Immunoblot of a coimmunoprecipitation of PHD1 and PHD2 with Rpb1 using H14 antibody in chromatin fractions from 786-O VHL+ (lanes 4 and 5) or VHL− (lanes 6 and 7) cells. Lane 1 shows a Western blot of indicated extracts; lane 2 shows a negative control using beads coated with secondary antibody but without H14 antibody; lane 3 shows H14 antibody alone. IP, immunoprecipitation; Ig, immunoglobulin.
Most importantly, oxidative stress induced binding of two PHDs, PHD1 and PHD2, to Rpb1 in the chromatin fraction enriched for engaged Rpb1 (such as shown in Fig. 2D, lanes 3, 4, 7, and 8) in VHL+ cells but not in VHL− cells (Fig. 4D). Immunoprecipitation of Rpb1 using H14 antibody demonstrated that PHD1 coimmunoprecipitated with Rpb1 under constitutive conditions in both VHL+ and VHL− cells at approximately similar levels. This interaction was substantially induced in response to H2O2 treatment but only in the VHL+ cells. PHD2 did not bind to the phosphorylated Rpb1 constitutively, but its binding was induced by oxidative stress in VHL+ cells. We did not detect PHD3 in the complex with Rpb1. These data indicate that oxidative stress induces pVHL-dependent formation of an Rpb1-PHD1/2 complex within the chromatin fraction and point toward the possibility that either PHD1 or PHD2 regulates Rpb1 hydroxylation.
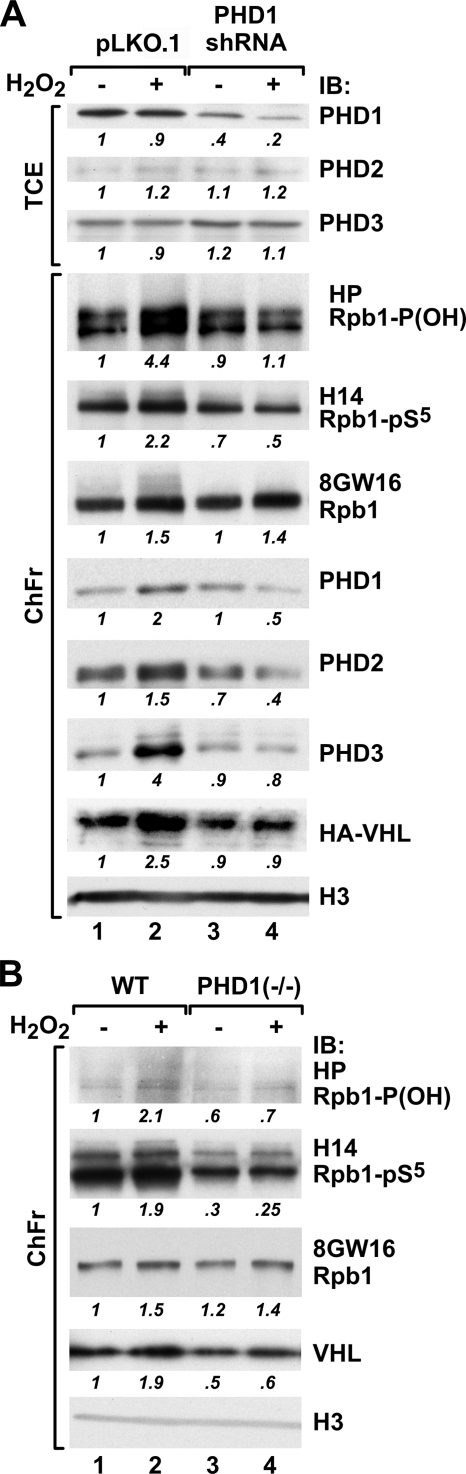
To functionally determine the role of individual PHDs in the hydroxylation of Rpb1, we performed knockdowns of individual PHDs using either stable expression of shRNA constructs or transient expression of siRNAs and analyzed the combined chromatin fractions (Fig. 5). An shRNA construct against PHD1 mRNA reduced the total levels of the PHD1 protein by 60% in the nontreated 786-O VHL+ cells, but it was 80% efficient in cells treated with H2O2, suggesting changes in the PHD1 protein or mRNA half-life under these conditions (Fig. 5A). Knockdown of PHD1 had a minimal effect on the total cellular levels of PHD2 and PHD3. Clearly, the knockdown of PHD1 suppressed oxidative-stress-induced accumulation of all three PHDs and pVHL in the combined chromatin fraction, as well as oxidative-stress-induced hydroxylation and ubiquitylation of Rpb1. Most interestingly, however, the knockout of PHD1 completely inhibited induction of Ser5 phosphorylation of Rpb1 in response to oxidative stress, without any major changes in the total amount of engaged Rpb1, as measured using the total Rpb1 8GW16 antibody (Fig. 5A). Because we were unable, despite multiple trials and additional use of siRNA approaches, to reduce constitutive levels of the PHD1 protein in the chromatin fraction below 40% of the constitutive level, we used MEFs from PHD1 knockout mice (Fig. 5B, lanes 3 and 4). Wild-type MEFs responded to oxidative stress with a substantial induction of Rpb1 hydroxylation and induction of Ser5 phosphorylation, as well as accumulation of pVHL in the chromatin fraction (Fig. 5B, lanes 1 and 2), all of which were absent in the PHD1 knockout MEFs. These data indicate that PHD1 is a necessary proline hydroxylase for P1465 hydroxylation in response to oxidative stress.
FIG. 5.
Effects of PHD1 knockdown on P1465 hydroxylation and Ser5 phosphorylation of Rpb1 in response to oxidative stress. (A) Western blot of total cellular extracts (TCE) or combined chromatin fractions (ChFr) from 786-O VHL+ cells stably transfected with control pLKO.1 vector (lanes 1 and 2) or with the same vector containing shRNA against human PHD1 (lanes 3 and 4). (B) Western blot analysis of combined chromatin fractions from wild-type or PHD1−/− MEFs. Blots in both panels were probed with the indicated antibodies. WT, wild type; IB, immunoblot.
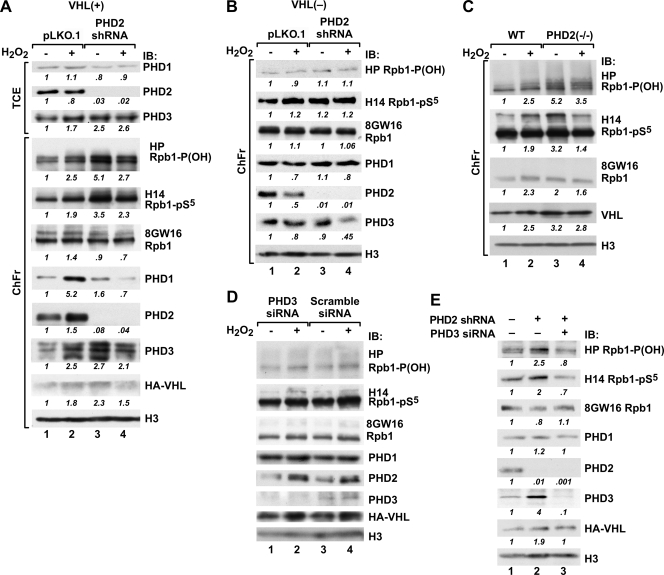
An shRNA construct against PHD2 mRNA very efficiently knocked down the levels of PHD2 protein in 786-O VHL+ cells (Fig. 6A, lanes 3 and 4) and VHL− cells (Fig. 6B, lanes 3 and 4), with a more than 99% reduction under both control and oxidative stress conditions. It was also accompanied by a small increase in the PHD3 level in the total cellular fraction, most likely due to the induction of HIF (49). Surprisingly, the knockdown of PHD2 substantially induced P1465 hydroxylation and Ser5 phosphorylation of Rpb1 under control conditions, and oxidative stress did not cause further stimulation of hydroxylation or Ser5 phosphorylation but actually decreased both of them (Fig. 6A, top, lanes 3 and 4). This was accompanied by minimal changes in the total amount of engaged Rpb1 as measured using the 8GW16 antibody. PHD2 knockdown had a small inducing effect on constitutive PHD1 accumulation and caused a substantial shift in the accumulation of PHD3 and pVHL into the chromatin extract under control conditions. However, decreased levels of both PHDs and pVHL were observed in response to oxidative stress compared with the control levels. These results were confirmed using PHD2 knockout MEFs, where constitutive hydroxylation and Ser5 phosphorylation were also induced and there was no further induction by oxidative stress, compared to results for wild-type MEFs (Fig. 6C, lanes 3 and 4). Similar to the case with 786-O cells, there was an increased accumulation of pVHL in the chromatin fraction of PHD2 knockout MEFs under control conditions.
FIG. 6.
The effects of PHD2 and PHD3 knockdown on Rpb1 hydroxylation and phosphorylation. (A) Western blot of total cellular extracts (TCE) or combined chromatin fractions (ChFr) from 786-O VHL+ cells stably transfected with control pLKO.1 vector (lanes 1 and 2) or with the same vector containing shRNA against human PHD2 (lanes 3 and 4). IB, immunoblot. (B) Western blot as in panel A but showing PHD2 knockdown performed in VHL− cells. (C) Western blot analysis of combined chromatin fractions from wild-type (WT) or PHD2−/− MEFs. (D) Western blot of total chromatin-enriched nuclear extracts from VHL+ 786-O cells transfected with siRNAs against human PHD3 (lanes 1 and 2) or scrambled siRNAs (lanes 3 and 4). (E) Western blot of control (no oxidative stress) total chromatin-enriched nuclear extracts from VHL+ 786-O cells with stable PHD2 knockdown (lanes 2 and 3) transfected with scrambled siRNAs (lane 2) or siRNAs against human PHD3 (lane 3). Lane 1 shows the same type of extract but from VHL+ 786-O cells stably transfected with empty pLKO.1 vector and serves as a control for lane 2 to demonstrate the effects of PHD2 knockdown on Rpb1 hydroxylation and phosphorylation.
To determine whether pVHL is required for PHD2-knockdown-induced constitutive Rpb1 hydroxylation and hydroxylation-associated Ser5 phosphorylation, we analyzed the effects of PHD2 knockdown in 786-O VHL− cells (Fig. 6B). Clearly, loss of PHD2 had no effect on Rpb1 P1465 hydroxylation, Ser5 phosphorylation, or accumulation of PHD3 in the chromatin fraction. This indicates that induction of P1465 hydroxylation and Ser5 phosphorylation that is constitutively induced by loss of PHD2 activity is also strictly dependent on the presence of pVHL.
In contrast to the knockdown of PHD1 and PHD2, knockdown of PHD3 using siRNA did not result in any changes in Rpb1 hydroxylation or phosphorylation (Fig. 6D). However, the simultaneous knockdown of PHD3 and knockout of PHD2 substantially reduced Rpb1 hydroxylation in 786-O cells (Fig. 6E).
Taken together, these data show that both PHD1 and PHD3 but not PHD2 can hydroxylate Rpb1. PHD1 is mainly necessary for P1465 hydroxylation in response to oxidative stress, and this activity remains under constitutive inhibition from PHD2, which is also present in the chromatin fraction. PHD3 activates Rpb1 hydroxylation only in certain circumstances, such as, for example, when present at an increased concentration in the chromatin.
Hydroxylation of P1465 regulates phosphorylation of Rpb1 on Ser5.
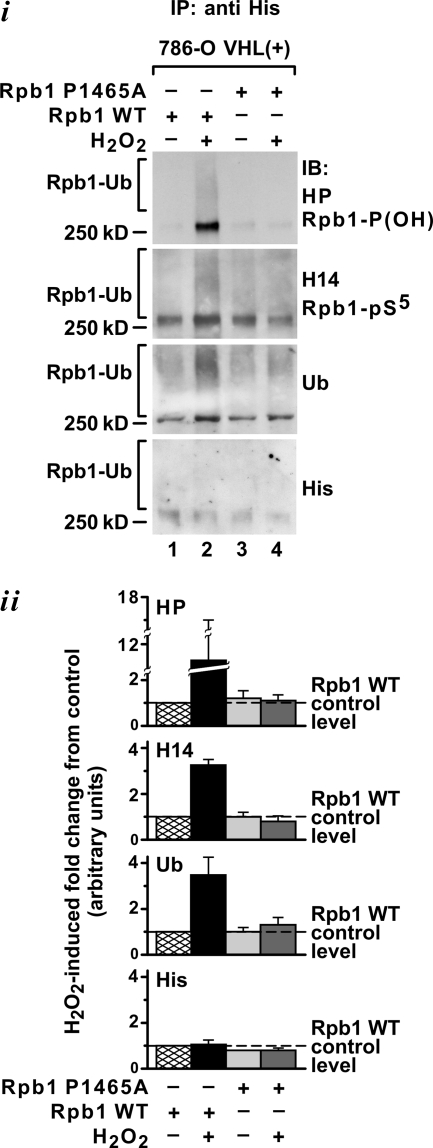
The most intriguing observation in the above experiments is that Ser5 phosphorylation is regulated by P1465 hydroxylation. Thus, pVHL is a regulator not only of Rpb1 ubiquitylation and hydroxylation but ultimately of its phosphorylation on Ser5 in response to oxidative stress. To further confirm that hydroxylation of Rpb1 is required for its phosphorylation in response to H2O2, we expressed histidine-tagged Rpb1, either the wild type or the P1465A mutant, which does not undergo hydroxylation and does not bind pVHL (see Fig. 3Ci), and analyzed Ser5 phosphorylation of both forms of Rpb1 in total chromatin-enriched nuclear extracts. Wild-type Rpb1 was hydroxylated, Ser5 phosphorylated, and ubiquitylated in response to oxidative stress (Fig. 7). However, phosphorylation of Ser5 was not induced in the P1465A Rpb1 mutant in response to oxidative stress, although, like wild-type Rpb1, this mutant was constitutively phosphorylated on Ser5 (Fig. 7). As expected, the P1465A mutant form of Rpb1 was not ubiquitylated in response to oxidative stress (Fig. 7; see also Fig. 3Ci). These modifications were not accompanied by changes in the protein expression level of the Rpb1 wild-type or mutant form (see also Fig. 3Cii). These findings further support the conclusion that P1465 hydroxylation functions in regulating Ser5 phosphorylation induced by exposure of cells to low-grade oxidative stress. Thus, by regulation of Rpb1 phosphorylation, P1465 hydroxylation may regulate gene expression.
FIG. 7.
The P1465A mutant of Rpb1, which does not undergo hydroxylation, is not Ser5 phosphorylated in response to oxidative stress. Immunoblotting (i) and quantification (ii) (n = 2) of an immunoprecipitation of wild-type (lanes 1 and 2) or P1465A mutant (lanes 3 and 4) histidine-tagged Rpb1 using antihistidine antibody are shown. Immunoprecipitations (IP) were performed using total chromatin-enriched nuclear extracts. Blots were probed with the indicated antibodies. Data were normalized to values in lane 1, which were measured in the presence of wild-type Rpb1 under control conditions (dashed line).
Analysis of pVHL-dependent protein expression patterns correlating with Rpb1 modifications in response to oxidative stress.
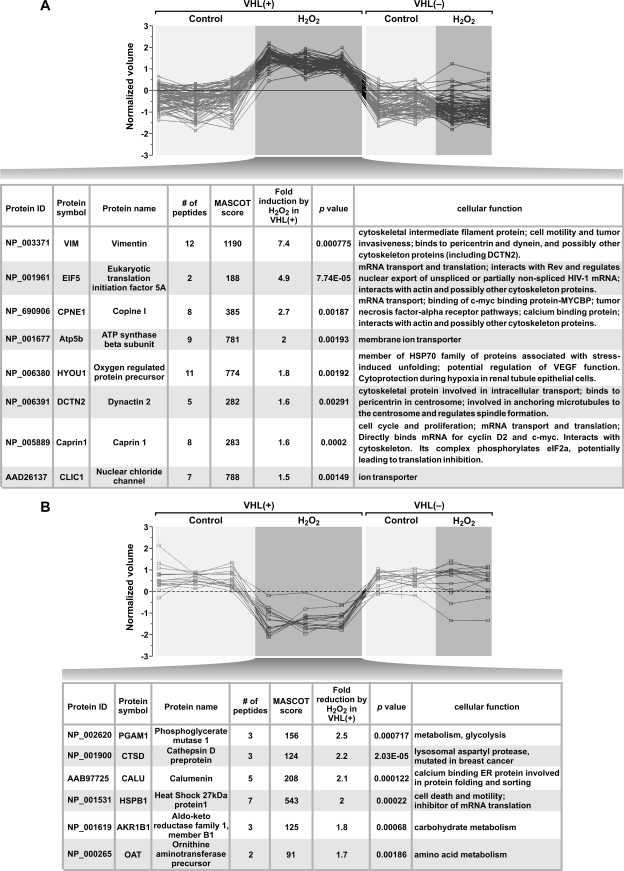
Phosphorylation of Rpb1 engaged on the DNA, particularly on Ser5 of the CTD, could function as a fine tuner of RNAPII activity. This regulation could occur at different levels, such as transcription elongation and processivity, splicing, mRNA stability and export, or protein translation (33, 52). Thus, to interrogate the functional role of the observed biochemical effects on Rpb1 in the regulation of gene expression, we used proteomic analysis of extracts from untreated and H2O2-treated VHL+ and VHL− cells. We reasoned that changes detected in the levels of proteins will represent final readouts of the effects of Rpb1 modifications, regardless of the specific process affected by Ser5 phosphorylation. We evaluated those proteins whose expression changed with oxidative stress in a manner dependent on functional pVHL. In particular, we identified 318 spots whose intensities did not differ between VHL+ and VHL− cells under baseline conditions. From that group, we selected those spots whose intensities were significantly changed by at least 50% in VHL+ cells but were not simultaneously affected in VHL− cells in response to H2O2 treatment. We found that intensities of 22% (72 spots) were significantly increased by H2O2 treatment in VHL+ cells (Fig. 8A). Interestingly, of the eight spots in this category for which we were able to identify protein by mass spectrometry-based peptide sequencing, six proteins are involved in cell proliferation by regulating mRNA transport and translation in the context of RNA “stress granules” that contain multiple cytoskeletal and RNA-binding proteins (Fig. 8A). The intensities of only 4% (14) of spots were decreased by H2O2 treatment in VHL+ cells (Fig. 8B). Of these, six proteins which we were able to identify by mass spectrometry did not suggest any specific common functional pattern; however, two of the inhibited proteins, HSPB1 and CALU, are also involved in regulation of mRNA translation and endoplasmic reticulum-mediated protein folding and thus were within a functional platform similar to that of proteins induced by H2O2 in VHL+ cells. These data were gathered under oxidative-stress conditions that did not induce HIF accumulation (see Fig. 2C) but which correlated with oxidative-stress-induced hydroxylation, phosphorylation, and ubiquitylation of Rpb1 (Fig. 2D). Thus, these results imply that the oxidative-stress-induced modifications of Rpb1 induce an increase in the steady-state levels of a subset of proteins which share some functional similarity in the context of regulation of cell proliferation, mRNA transport, and translation.
FIG. 8.
Analysis of protein expression patterns induced by oxidative stress in VHL+ but not VHL− cells. (A) Quantification of protein spots that are significantly induced by oxidative stress in VHL+ cells but are not affected in VHL− cells. The y axis represents the normalized volume of individual spots on the logarithmic scale. The relative quantification of each spot was captured by comparing all of the silver-stained 2-D gel images using Progenesis/SameSpots image analysis software. The table lists individual identified proteins and their ontological connection for proteins induced by oxidative stress in VHL+ cells. (B) Analysis of the patterns of protein expression and quantification of protein spots that are significantly decreased by oxidative stress in VHL+ cells but are not affected in VHL− cells. All labeling is as in panel A.
Role of P1465 in oncogenesis.
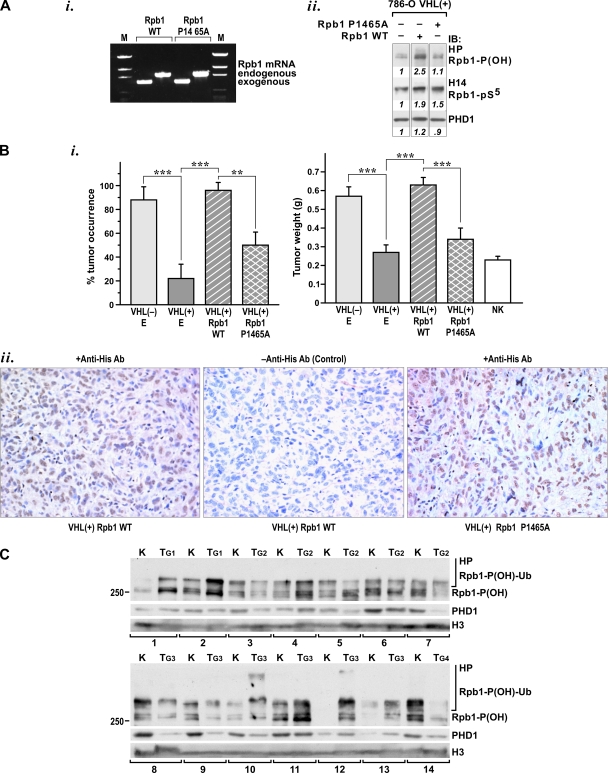
To begin to understand the functional role of P1465 in cell biology, particularly in terms of tumor-suppressing activities of pVHL, we examined the effects of expression of wild-type or P1465A Rpb1 on the oncogenic potential of 786-O cells. To do this, pools of 768-O VHL+ cells stably expressing comparable levels of wild-type Rpb1 or Rpb1 P1465A (Fig. 9Ai and Bii) were injected into the kidneys of nude mice and tumor formation was assessed (Fig. 9Bi). Despite trying multiple approaches, we were not able to obtain VHL− cells exogenously expressing either form of Rpb1. Expression of both forms increased the amount of Rpb1 phosphorylated on Ser5, as measured in the total chromatin nuclear extracts (Fig. 9Aii). However, expression of wild-type Rpb1 resulted in higher levels of phosphorylated Rpb1 and substantially increased levels of P1465 hydroxylation (Fig. 9Aii). Interestingly, expression of wild-type Rpb1 significantly stimulated formation of tumors by VHL+ cells, which, when transfected with an empty vector, formed infrequent and very small tumors (Fig. 9Bi). In contrast, expression of Rpb1 P1465A decreased the frequency of tumor formation and tumor size compared to results with wild-type Rpb1 (Fig. 9Bi). Expression of the two exogenous forms of Rpb1 in the tumor cells was very comparable and, in both cases, was localized to the nuclei. This was demonstrated by immunocytochemistry using antihistidine antibody on sections from isolated tumors (Fig. 9Bii). A thorough histopathological analysis of these tumors will be the subject of another study, but compared to the VHL− tumors, the VHL+ wild-type Rpb1 tumors showed a high level of spindle-shaped sarcomatoid histology and were less vascular. In addition, they did not show any changes in HIF-α mRNA levels, which could be expected due to increased RNAPII activity (data not shown). These data indicate that increased levels of Rpb1 in VHL+ 786-O cells have an oncogenic effect that is primarily mediated by P1465 hydroxylation, as evidenced by the finding that expression of the P1465A mutant inhibited this oncogenic activity.
FIG. 9.
Role of P1465 hydroxylation in formation of renal cancer tumors. (A) (i) RT-PCR of mRNA levels in tumorigenic cells showing exogenous wild-type (WT) Rpb1 and P1465A Rpb1 as well as endogenous human Rpb1. (ii) Western blot analysis, using the indicated antibody, of total chromatin-enriched nuclear extracts from pools of 786-O VHL+ cells transfected with an empty vector (lane 1), Rpb1 WT (lane 2), or Rpb1 P1465 (lane 3). (B) (i) Frequency (left) and size (right) of tumors resulting from orthotopic xenografts involving injections of 786-O VHL+ cells stably expressing Rpb1 WT, P1465A, or empty vector (E). Formation of tumors by VHL− cells transfected with an empty vector is shown as a positive control for tumor formation. In each group a total of 15 mice were injected in three independent series of five; **, P < 0.01; ***, P < 0.001. In each case, tumor size includes the injected kidney. NK, average size of normal mouse kidney based on weight of 20 noninjected kidneys. (ii) Photomicrograph (magnification, ×200) of immunostaining for exogenous histidine-tagged Rpb1 (WT, left; P1465A, right; negative control without primary antibody [Ab], middle) using H1029 antibody. (C) Western blot analysis, using indicated antibodies, of combined chromatin fractions from 14 RCC tumors (T) or adjacent normal kidney (K). G number describes the grade of each tumor.
Next, we wanted to determine the status of P1465 hydroxylation in human RCC. We analyzed the status of P1465 hydroxylation in crude chromatin fractions obtained from 14 human RCC samples and compared these results to those for normal kidney specimens from the same patients (Fig. 9C). The tissue used for preparation of the extract contained more than 90% clear cells as determined by hematoxylin and eosin staining of representative sections (not shown). We found that in 7 out of 14 tumors there was an increase in P1465 hydroxylation, while in the remaining 7 there was actually a decrease (Fig. 9C). Most interestingly, in 11 out of 14 cases the status of P1465 hydroxylation correlated with the status of PHD1 accumulation in the chromatin fraction, i.e., a decrease in Rpb1 hydroxylation was accompanied by decreased PHD1 levels compared to those in the normal kidney and vice versa (Fig. 9C). In two cases, the status of PHD1 did not differ between the tumor and matched kidney, and in one case the correlation was reversed. While the analysis of correlations between pVHL status (mutations) and Rpb1 levels and P1465 hydroxylation is under way, it is clear that at least some cases of human RCCs demonstrate increased P1465 hydroxylation. These correlative data also support our cell culture results that PHD1 is the relevant hydroxylase responsible for P1465 hydroxylation.
DISCUSSION
The research presented here opens several avenues regarding novel activities of pVHL and Egl-9-type proline hydroxylases that may be fundamental to their function. On the basis of these results, we propose a mechanism whereby pVHL and PHDs modulate gene expression by regulating RNAPII activity. We further propose that this mechanism is independent of proline hydroxylation and pVHL-dependent ubiquitylation of HIF and that the effects of pVHL and PHDs on RNAPII are critical factors in the regulation of tumorigenesis.
We have shown that reconstitution of pVHL in RCC cells stimulates total steady-state levels of Rpb1 which is Ser5 phosphorylated and P1465 hydroxylated. Furthermore, we have demonstrated that pVHL is necessary for oxidative-stress-stimulated recruitment of Rpb1 to the DNA and P1465 hydroxylation, CTD Ser5 phosphorylation, and ubiquitylation of the fraction of Rpb1 that is engaged on the DNA. Interestingly, this ubiquitylation does not cause Rpb1 degradation but rather correlates with accumulation of engaged Rpb1. While the necessity of P1465 hydroxylation for the subsequent pVHL-dependent ubiquitylation of Rpb1 has been recognized before (25), in the present work we have significantly expanded our understanding of the role of pVHL by showing that pVHL also regulates P1465 hydroxylation of the DNA-engaged Rpb1 and that this hydroxylation involves the PHDs. One of the most important and novel findings of our study is that this pVHL-dependent P1465 hydroxylation of Rpb1 is required for oxidative-stress-induced Ser5 phosphorylation of Rpb1 engaged on the DNA. Thus, it can be concluded that pVHL regulates the amount of Rpb1 phosphorylated on Ser5 engaged on the DNA under oxidative-stress conditions. The finding that a CTD-associated E3 ubiquitin ligase regulates CTD phosphorylation is independently supported by the recent demonstration that the yeast Rpb1 E3 ligase, Rsp5, stimulates CTD phosphorylation in an in vitro assay (35). We also found that overexpression of wild-type Rpb1 stimulates the oncogenic potential of VHL+ cells and that this effect can be inhibited by P1465A mutation of Rpb1. This finding implicates the pVHL-mediated hydroxylation of Rpb1 as an oncogenic mechanism and is supported by the data from human RCCs. Although the number of samples studied is small and potential correlations with tumor grade or stage and the status of pVHL are not yet established, 50% of human RCCs demonstrated an increase in P1465. These data are consistent with results published by other laboratories in which under some circumstances, such as in teratomas or fibrosarcomas derived from VHL+/− versus VHL−/− cells, the presence of pVHL has tumor-promoting activities (30, 31). The potential growth-promoting activity of pVHL is also supported by the role of pVHL in embryonic development (11) and by clinical evidence that the prognosis is worse for RCC associated with a wild-type VHL gene than for tumors with mutated pVHL, although the mechanism of this phenomenon is not understood (44, 48, 58).
A direct evaluation of the role of pVHL-dependent modifications of Rpb1 in the transcriptional or posttranscriptional regulation of specific genes is likely to be very complex and will require further studies. However, our analysis of protein changes occurring in response to H2O2 treatment, as the final readout correlating with the pVHL-dependent modification of Rpb1, indicates an overall stimulatory effect of oxidative stress on subsets of proteins in VHL+ cells, consistent with increased levels of a potentially active fraction of Rpb1 on the DNA. It is striking that the majority of proteins explicitly identified in our proteomic screen, including EIF5, CPNE1, and Caprin 1, play a role in the transport and regulation of translation of specific mRNAs. It is particularly interesting that Caprin 1 binds G3BP1 (RasGAP SH3 domain binding protein), which associates with mRNAs for two Rpb1 kinases, cdk7 and cdk9, increasing levels of the cdk7 protein while decreasing those of cdk9 (29, 55). Potential effects of pVHL in the regulation of protein translation have a precedent in the reported role of pVHL in the stimulation of p53 translation (9). The effects regarding induction of cytoskeletal proteins are also consistent with the previously reported role of pVHL in binding and stabilizing microtubules (14). Our study potentially expands this role into regulation of expression levels of vimentin and dynactin, at least in response to some stressors.
Perhaps one of our most interesting observations is that proline hydroxylases have a strong presence and activity within the chromatin fraction and that PHD1 and PHD2 bind to Rpb1 in response to oxidative stress in a pVHL-dependent manner. The biochemical nature of these interactions and the role of pVHL in the formation of such a complex will be determined in future studies. Because in the in vitro hydroxylation experiments using Rpb1 peptides and purified enzymes none of the PHDs hydroxylates Rpb1 (data not shown), it is very likely that PHDs and Rpb1 are part of a much bigger protein complex. The formation of protein complexes in which individual PHDs homodimerize and heterodimerize has been reported in the context of PHD3 (43). We identified PHD1 as the enzyme necessary for hydroxylation of Rpb1 and found that its knockdown inhibited Rpb1 hydroxylation in response to oxidative stress. This is consistent with previous reports showing expression of PHD1 predominantly in the nuclei of cells in different organs (36) and that PHD2 and PHD3 but not PHD1 are the main HIF-α hydroxylases (1, 42). The mechanism by which oxidative stress induces PHD1 activity is currently not understood but clearly must differ from the inhibition of PHD2 activity by oxidative stress reported in the case of HIF-α hydroxylation (3, 5, 10, 12, 13, 34). This difference is potentially due to oxidative-stress-induced PHD1 activity taking place in the context of a multiprotein complex associated with RNAPII.
Surprisingly, we found that a knockdown of PHD2 stimulated constitutive hydroxylation and Ser5 phosphorylation of Rpb1, which occurred in a pVHL-dependent manner. This was accompanied by a strong induction of PHD3 in the chromatin fraction, and indeed, Rpb1 hydroxylation under these conditions required PHD3 activity. It remains to be determined whether the effect of PHD2 knockout is mediated through the loss of physical interaction of PHD2 with PHD1 and Rpb1. The PHD2 knockout may allow more PHD1 and PHD3 binding to Rpb1 and stronger hydroxylating activity. In addition, knockout of PHD2 induces HIF-2α (1), which in turn may strongly stimulate PHD3 expression and its chromatin association. This raises the interesting possibility that HIF might also regulate P1465 hydroxylation and Ser5 phosphorylation of Rpb1, thus controlling gene expression at a different level in addition to stimulating transcription initiation.
Several different kinases phosphorylate Ser5, of which cdk7, cdk8, cdk9, and ERKs are the most thoroughly investigated (15, 47). Ser5 is also subject to dephosphorylation by different phosphatases that when inhibited may affect steady-state levels of Ser5 phosphorylation (52) and gene expression. The roles of these individual kinases in the pVHL-dependent phosphorylation of Rpb1 remain to be determined. We do not expect ERKs to be involved in this regulation, because the phosphorylation of Ser5 resulting from their activity occurs on soluble Rpb1 and functions as an adaptation to severe oxidative stress to prevent reentry of Rpb1 molecules into transcription (47). However, it is important to note that with our experimental model system, the effect of oxidative stress on the pVHL-dependent induction of P1465 hydroxylation, Ser5 phosphorylation, and ubiquitylation of Rpb1 occurs on engaged Rpb1 and is long lasting, starting immediately after exposure to H2O2 but reaching a plateau at approximately 4 h after stimulation and then persisting for several additional hours. This regulation is clearly different from previously reported responses to higher doses (0.25 to 10 mM) of hydrogen peroxide (15; M. L. Ignacak and M. F. Czyzyk-Krzeska, unpublished results), where fast (within minutes of exposure) and pVHL-independent phosphorylation of Rpb1 Ser5 phosphorylation occurs and where such doses of H2O2 result in significant cell death (15). We chose low doses of H2O2, in the range between 10 and 50 μM, to mimic the subtle changes in intracellular H2O2 concentrations that may occur during physiological or pathophysiological variations in the endogenous metabolism but which do not lead to cell death.
Unlike the effects of pVHL on HIF-αs, we did not detect any apparent degradation of Rpb1 in response to hydroxylation/ubiquitylation. These conclusions differ from the previously suggested role of pVHL-mediated polyubiquitylation in the degradation of Rpb1 as an adaptation to UV-induced DNA damage in PC12 cells (25). Thus, pVHL-mediated ubiquitylation of Rpb1 may have different regulatory activities toward Rpb1 that function in different contexts and can either lead to or prevent its degradation. pVHL not only directly targets proteins for ubiquitylation but also ubiquitylates and targets for degradation the deubiquitylating enzymes (28), a process which, in a secondary manner, may affect ubiquitylation of some currently unidentified substrates. Recent evidence has demonstrated that pVHL can induce assembly of polyubiquitin chains on the HIF-1α substrate, not only through ubiquitin K48 but also through other lysines (37). This strongly supports the possibility that the role of pVHL in protein ubiquitylation may extend beyond targeting for proteasomal degradation. The differences in pVHL-mediated effects on constitutive levels of Ser5-phosphorylated Rpb1 in PC12 and RCC cells may also result from tissue-specific characteristics. There is support for this idea in the literature. For example, a lack of correlation between pVHL-dependent tumor formation and HIF accumulation has been described in the case of type 2C VHL disease, where certain mutations encoded within the VHL gene, such as V84L and L188V, cause pheochromocytoma tumors without promoting RCC (6, 50). However, the effects of oxidative stress on P1465 hydroxylation, Ser5 phosphorylation, and ubiquitylation of Rpb1 were very similar in PC12 (data not shown) and RCC cells, an indication that this is a more general mechanism of regulation.
In conclusion, our data demonstrate that pVHL and PHDs regulate Rpb1 in an HIF-independent manner. By stimulating hydroxylation of P1465, phosphorylation of CTD Ser5, and nondegradative ubiquitylation, pVHL and PHDs could fine-tune RNAPII activities and gene expression. These effects on RNAPII could then participate in the tumor-suppressing or growth-promoting activities of pVHL in renal cancer.
Acknowledgments
This work was supported in part by the following grants: NIH HL58687 and HL66312, NCI CA122346, and DoD W81XWH-07-02-0026 (to M.F.C.-K.) and a British Heart Foundation program grant (to P.H.M.).
We thank T. Knyushko and Y. Stratton for insightful discussion during preparation of the manuscript, V. Zimmerman for help with the intrakidney injections, A. Gibson for general technical assistance, K. Rask for protein identification by mass spectrometry, G. Doerman for preparing the figures, and M. Daston for editorial assistance. The clone of Rpb1 was originally provided by Marc Vigneron.
Footnotes
Published ahead of print on 19 February 2008.
REFERENCES
- 1.Berra, E., E. Benizri, A. Ginouves, V. Volmat, D. Roux, and J. Pouyssegur. 2003. HIF prolyl hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1 alpha in normoxia. EMBO J. 224082-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bregman, D. B., R. Halaban, A. J. van Gool, K. A. Henning, E. C. Friedberg, and S. L. Warren. 1996. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc. Natl. Acad. Sci. USA 9311586-11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burnelle, J. K., E. L. Bell, N. M. Quaseda, K. Vercautern, V. Tiranti, M. Zeviani, R. C. Scarpulla, and N. S. Chandel. 2005. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metabol. 1409-414. [DOI] [PubMed] [Google Scholar]
- 4.Carty, S. M., and A. L. Greenleaf. 2002. Hyperphosphorylated C-terminal repeat domain-associating proteins in the nuclear proteome link transcription to DNA/chromatin modification and RNA processing. Mol. Cell Proteomics 1598-610. [DOI] [PubMed] [Google Scholar]
- 5.Chandel, N. S., E. Maltepe, E. Goldwasser, C. E. Mathieu, M. C. Simon, and P. T. Schumacker. 1998. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. USA 9511715-11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clifford, S. C., M. E. Cockman, A. C. Smallwood, D. R. Mole, E. R. Woodward, P. H. Maxwell, P. J. Ratcliffe, and E. R. Maher. 2001. Contrasting effects on HIF-1alpha regulation by disease-causing pVHL mutations correlate with patterns of tumorigenesis in von Hippel-Lindau disease. Hum. Mol. Genet. 101029-1038. [DOI] [PubMed] [Google Scholar]
- 7.Epstein, A. C., J. M. Gleadle, L. A. McNeill, K. S. Hewitson, J. O'Rourke, D. R. Mole, M. Mukherji, E. Metzen, M. I. Wilson, A. Dhanda, Y. M. Tian, N. Masson, D. L. Hamilton, P. Jaakkola, R. Barstead, J. Hodgkin, P. H. Maxwell, C. W. Pugh, C. J. Schofield, and P. J. Ratcliffe. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 10743-54. [DOI] [PubMed] [Google Scholar]
- 8.Flick, K., I. Ouni, J. A. Wohlschlegel, C. Capati, W. H. McDonald, J. B. Yates, and P. Kaiser. 2004. Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat. Cell Biol. 6634-641. [DOI] [PubMed] [Google Scholar]
- 9.Galbán, S., J. L. Martindale, K. Mazan-Mamczarz, I. López de Silanes, J. Fan, W. Wang, J. Decker, and M. Gorospe. 2003. Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol. Cell. Biol. 237083-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerald, D., E. Berra, Y. M. Frapart, D. A. Chan, A. J. Giaccia, D. Mansuy, J. Pouyssegur, M. Yaniv, and F. Mechta-Grigoriou. 2004. JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell 118781-794. [DOI] [PubMed] [Google Scholar]
- 11.Gnarra, J. R., J. M. Ward, F. D. Porter, J. R. Wagner, D. E. Devor, A. Grinberg, M. R. Emmert-Buck, H. Westphal, R. D. Klausner, and W. M. Linehan. 1997. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc. Natl. Acad. Sci. USA 949102-9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzy, R. D., B. Hoyos, E. Robin, H. Chen, L. Liu, K. D. Mansfield, M. C. Simon, U. Hammerling, and P. T. Schumacker. 2005. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 1401-408. [DOI] [PubMed] [Google Scholar]
- 13.Guzy, R. D., B. Sharma, E. Bell, N. S. Chandel, and P. T. Schumacker. 2008. Loss of SdhB, but not SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol. Cell. Biol. 28718-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hergovich, A., J. Lisztwan, R. Barry, P. Ballschmieter, and W. Krek. 2003. Regulation of microtubule stability by the von Hippel-Lindau tumor suppressor protein pVHL. Nat. Cell Biol. 564-70. [DOI] [PubMed] [Google Scholar]
- 15.Inukai, N., Y. Yamaguchi, I. Kuraoka, T. Yamada, S. Kamijo, J. Kato, K. Tanaka, and H. Handa. 2004. A novel hydrogen peroxide-induced phosphorylation and ubiquitination pathway leading to RNA polymerase II proteolysis. J. Biol. Chem. 2798190-8195. [DOI] [PubMed] [Google Scholar]
- 16.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIF-alpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292464-468. [DOI] [PubMed] [Google Scholar]
- 17.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292468-472. [DOI] [PubMed] [Google Scholar]
- 18.Jarrold, B., J. DeMuth, K. Greis, T. Burt, and F. Wang. 2005. An effective skeletal muscle pre-fractionation method to remove abundant structural proteins for optimized two dimensional gel electrophoresis. Electrophoresis 262269-2278. [DOI] [PubMed] [Google Scholar]
- 19.Kaelin, W. G., Jr. 2005. Proline hydroxylation and gene expression. Annu. Rev. Biochem. 74115-128. [DOI] [PubMed] [Google Scholar]
- 20.Kaelin, W. G., Jr. 2002. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer 2673-682. [DOI] [PubMed] [Google Scholar]
- 21.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Gene. Dev. 142452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo, K., J. Klco, E. Nakamura, M. Lechpammer, and W. G. Kaelin, Jr. 2002. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell 1237-246. [DOI] [PubMed] [Google Scholar]
- 23.Kondo, K., W. Y. Kim, M. Lechpammer, and W. G. Kaelin, Jr. 2003. Inhibition of HIF2-alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 1439-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kroll, S. L., and M. F. Czyzyk-Krzeska. 1998. The role of H2O2 and heme-containing oxygen sensors in O2-dependent regulation of tyrosine hydroxylase gene expression. Am. J. Physiol. 274C167-C174. [DOI] [PubMed] [Google Scholar]
- 25.Kuznetsova, A. V., J. Meller, P. O. Schnell, J. A. Nash, M. L. Ignacak, Y. Sanchez, J. W. Conaway, R. C. Conaway, and M. F. Czyzyk-Krzeska. 2003. von Hippel-Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination. Proc. Natl. Acad. Sci. USA 1002706-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, K. B., D. Wang, S. J. Lippard, and P. A. Sharp. 2002. Transcription-coupled and DNA damage-dependent ubiquitination of RNA polymerase II in vitro. Proc. Natl. Acad. Sci. USA 994239-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, K. B., and P. A. Sharp. 2004. Transcription-dependent polyubiquitination of RNA polymerase II requires lysine 63 of ubiquitin. Biochemistry 4315223-15229. [DOI] [PubMed] [Google Scholar]
- 28.Li, Z., X. Na, D. Wang, S. R. Schoen, E. M. Messing, and G. Wu. 2002. Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 2774656-4662. [DOI] [PubMed] [Google Scholar]
- 29.Lypowy, J., I.-Y. Chen, and M. Abdellatif. 2005. An alliance between RAS GTPase activating protein, filamin C and RAS GTPAse-activating protein SH3 domain binding protein regulates myocyte growth. J. Biol. Chem. 28025717-25728. [DOI] [PubMed] [Google Scholar]
- 30.Mack, F. A., W. K. Rathmell, A. M. Arsham, J. Gnarra, B. Keith, and M. C. Simon. 2003. Loss of pVHL is sufficient to cause HIF dysregulation in primary cells but does not promote tumor growth. Cancer Cell 375-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mack, F. A., J. H. Patel, M. P. Biju, V. H. Haase, and M. C. Simon. 2005. Decreased growth of Vhl−/− fibrosarcomas is associated with elevated levels of cyclin kinase inhibitors p21 and p27. Mol. Cell. Biol. 254565-4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandriota, S. J., K. J. Turner, D. R. Davies, P. G. Murray, N. Y. Morgan, H. M. Sowter, C. C. Wykoff, E. R. Maher, A. L. Harris, P. J. Ratcliffe, and P. H. Maxwell. 2002. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in nephron. Cancer Cell 1459-468. [DOI] [PubMed] [Google Scholar]
- 33.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416499-506. [DOI] [PubMed] [Google Scholar]
- 34.Mansfield, K. D., R. D. Guzy, Y. Pan, R. M. Young, T. P. Cash, P. T. Schumacker, and M. C. Simon. 2005. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-α activation. Cell. Metab. 1393-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Max, T., Sogaard, M., and J. Q. Svejstrup. 2007. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with Mediator. J. Biol. Chem. 28214113-14120. [DOI] [PubMed] [Google Scholar]
- 36.Metzen, E., U. Berchner-Pfannschmidt, P. Stengel, J. H. Marxsen, I. Stolze, M. Klinger, W. Q. Huang, C. Wotzlaw, T. Hellwig-Burgel, W. Jelkmann, H. Acker, and J. Fandrey. 2003. Intracellular localization of human HIF-1 alpha hydroxylases: implications for oxygen sensing. J. Cell Sci. 1161319-1326. [DOI] [PubMed] [Google Scholar]
- 37.Miller, F., A. Kentsis, R. Osman, and Z.-Q. Pan. 2005. Inactivation of VHL by tumorigenic mutations that disrupt dynamic coupling of the pVHL-hypoxia-inducible transcription factor-1alpha complex. J. Biol. Chem. 2807985-7996. [DOI] [PubMed] [Google Scholar]
- 38.Mintz, P. J., S. D. Patterson, A. F. Neuwald, C. S. Spahr, and D. L. Spector. 1999. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 184308-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitsui, A., and P. A. Sharp. 1999. Ubiquitination of RNA polymerase II large subunit signaled by phosphorylation of carboxyl-terminal domain. Proc. Natl. Acad. Sci. USA 966054-6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Na, X., H. O. Duan, E. M. Messing, S. R. Schoen, C. K. Ryan, P. A. di Sant'Agnese, E. A. Golemis, and G. Wu. 2003. Identification of the RNA polymerase II subunit hsRPB7 as a novel target of the von Hippel-Lindau protein. EMBO J. 224249-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naito, S., A. C. von Eischenbach, R. Giavazzi, and I. J. Fidler. 1986. Growth and metastasis of tumor cells isolated from human renal cell carcinoma implanted into different organs of nude mice. Cancer Res. 464109-4115. [PubMed] [Google Scholar]
- 42.Nakayama, K., I. J. Frew, M. Hagensen, M. Skals, H. Habelhah, A. Bohumik, T. Kadoya, H. Erdjument-Bromage, P. Tempst, P. B. Frapell, D. D. Bowtell, and Z. Ronai. 2004. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell 117941-952. [DOI] [PubMed] [Google Scholar]
- 43.Nakayama, K., S. Gazdoiu, R. Abraham, Z. Q. Pan, and Z. Ronai. 2007. Hypoxia-induced assembly of prolyl hydroxylase PHD3 into complexes: implications for its activity. Biochem. J. 401217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumann, H. P. H., B. U. Bender, D. P. Berger, J. Laubenberger, W. Schultze-Seemann, U. Wetterauer, F. J. Ferstl, E. W. Herbest, G. Schwarzkopf, F. J. Hes, C. J. M. Lips, J. M. Lamiell, O. Masek, P. Riegler, B. Mueller, D. Glavac, and H. Brauch. 1998. Prevelance, morphology and biology of renal cell carcinoma in von Hippel-Lindau disease compared to sporadic renal cell carcinoma. J. Urol. 1601248-1254. [PubMed] [Google Scholar]
- 45.Nguyen, V. T., F. Giannoni, M. F. Dubois, S. J. Seo, M. Vigneron, C. Kedinger, and O. Bensaude. 1996. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 242924-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okuda, H., K. Saitoh, S. Hirai, K. Iwai, Y. Takaki, M. Baba, N. Minato, S. Ohno, and T. Shuin. 2001. The von Hippel-Lindau tumor suppressor protein mediates ubiquitination of activated atypical protein kinase C. J. Biol. Chem. 27643611-43617. [DOI] [PubMed] [Google Scholar]
- 47.Palancade, B., and O. Bensaude. 2003. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 2703859-3870. [DOI] [PubMed] [Google Scholar]
- 48.Parker, A. S., J. C. Cheville, C. M. Lohse, T. Igel, B. C. Lebovich, and M. L. Blute. 2005. Loss of expression of von Hippel-Lindau tumor suppressor protein associated with early-stage clear cell renal carcinoma. Urology 651090-1095. [DOI] [PubMed] [Google Scholar]
- 49.Pescador, N., Y. Cuevas, S. Naranjo, M. Alcaide, D. Villar, M. O. Landazuri, and L. Del Peso. 2005. Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd3) gene. Biochem. J. 390189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rathmell, W. K., M. M. Hickey, N. A. Bezman, C. A. Chmielecki, N. C. Carraway, and M. C. Simon. 2004. In vitro and in vivo models analyzing von Hippel-Lindau disease-specific mutations. Cancer Res. 648595-8603. [DOI] [PubMed] [Google Scholar]
- 51.Ratner, J. N., B. Balasubramanian, J. Corden, S. L. Warren, and D. B. Bregman. 1998. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J. Biol. Chem. 2735184-5189. [DOI] [PubMed] [Google Scholar]
- 52.Reyes-Reyes, M., and M. Hampsey. 2007. Role of the SSu72 C-terminal domain phosphatase in RNA polymerase II transcription elongation. Mol. Cell. Biol. 27926-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68850-858. [DOI] [PubMed] [Google Scholar]
- 54.Shilatifard, A. 2004. Transcriptional elongation control by RNA polymerase II: a new frontier. Biochim. Biophys. Acta 167779-86. [DOI] [PubMed] [Google Scholar]
- 55.Solomon, S., Y. Xu, B. Wang, M. D. David, P. Schubert, D. Kennedy, and J. W. Schrader. 2007. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2α, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 272324-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spence, J., R. R. Gali, G. Dittmar, F. Sherman, M. Karin, and D. Finley. 2000. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 10267-76. [DOI] [PubMed] [Google Scholar]
- 57.Sun, L., and Z. J. Chen. 2004. The novel functions of ubiquitination in signaling. Curr. Opin. Cell Biol. 16119-126. [DOI] [PubMed] [Google Scholar]
- 58.Yao, M., M. Yoshida, T. Kishida, N. Nakaigawa, M. Baba, K. Kobayashi, T. Miura, M. Moriyama, Y. Nagashima, Y. Nakatani, Y. Kubota, and K. Kondo. 2002. VHL tumor suppressor gene alterations associated with good prognosis in sporadic clear-cell renal carcinoma. J. Natl. Cancer Inst. 941569-1575. [DOI] [PubMed] [Google Scholar]