Abstract
Wnt signaling activates gene expression through the induced formation of complexes between DNA-binding T-cell factors (TCFs) and the transcriptional coactivator β-catenin. In colorectal cancer, activating Wnt pathway mutations transform epithelial cells through the inappropriate activation of a TCF7L2/TCF4 target gene program. Through a DNA array-based genome-wide analysis of TCF4 chromatin occupancy, we have identified 6,868 high-confidence TCF4-binding sites in the LS174T colorectal cancer cell line. Most TCF4-binding sites are located at large distances from transcription start sites, while target genes are frequently “decorated” by multiple binding sites. Motif discovery algorithms define the in vivo-occupied TCF4-binding site as evolutionarily conserved A-C/G-A/T-T-C-A-A-A-G motifs. The TCF4-binding regions significantly correlate with Wnt-responsive gene expression profiles derived from primary human adenomas and often behave as β-catenin/TCF4-dependent enhancers in transient reporter assays.
Physiological Wnt signaling is required for the maintenance of the crypt progenitor phenotype and controls the proliferation/differentiation switch in the adult, self-renewing intestinal epithelium (33). A constitutively active Tcf/β-catenin transcription complex, resulting from mutations in adenomatous polyposis coli (APC), Axin, or β-catenin, is the primary transforming factor in colorectal cancer (CRC) (25, 26, 32); aberrant Tcf/β-catenin activity results in a transcriptional profile in CRC cells similar to that which is physiologically driven by Tcf/β-catenin in the crypt stem/progenitor cells of the intestine (49). Through candidate gene approaches and microarray technology, a large number of genes have been uncovered whose expression levels are altered upon abrogation or activation of the Wnt pathway (for references, see http://www.stanford.edu/∼rnusse/pathways/targets.html). It remains unclear whether the affected genes are direct or indirect targets of the Tcf/β-catenin transcription factor complex. cis-regulatory elements directly bound by Tcf have been identified for only a few candidate genes. Such studies have been mostly limited to regulatory regions close to the transcription start site (TSS) of candidate genes (e.g., see reference 17). A comprehensive identification of regulatory elements is essential for a more complete understanding of the transcriptional repertoire driven by the Wnt pathway and the elucidation of the molecular mechanisms by which Tcf and β-catenin control the transcription of their target genes.
A recent approach taken to achieve such goals is chromatin immunoprecipitation (ChIP)-coupled DNA microarray analysis (ChIP-on-chip), which couples the immunoprecipitation of chromatin-bound transcription factors with the identification of the bound DNA sequences through hybridization on DNA microarrays (35). This approach has been used to generate, among others, a comprehensive map of active, preinitiation complex-bound promoters in human fibroblast cells (24). Microarrays covering the nonrepetitive sequence of chromosomes 21 and 22 have allowed the study of histone H3 methylation and acetylation patterns in human hepatoma cells (5) and estrogen receptor binding sites in breast cancer cells (8). The latter study revealed selective binding of estrogen receptor (ER) to a limited number of sites, most of which were distant from the TSSs of ER-regulated genes (8). Similar conclusions were put forth by work examining the in vivo binding of transcription factors Sp1, c-Myc, and p53 along chromosomes 21 and 22: most binding sites identified do not correspond to the proximal promoters of protein-coding genes but rather lie within or immediately 3′ to well-characterized genes or are significantly correlated with noncoding RNAs (10). Collectively these studies point to the necessity of interrogating entire genomes for the comprehensive determination of in vivo-occupied binding sites (9, 23, 52, 54).
In the present work, we used a combination of ChIP and location analysis with genome-wide tiling arrays to generate a genome-wide binding profile of TCF4, the T-cell factor (TCF) family member most prominently expressed in the mammalian intestine (1, 26).
MATERIALS AND METHODS
ChIP.
LS174T cells were cross-linked with 1% formaldehyde for 20 min at room temperature. The reaction was quenched with glycine at a final concentration of 0.125 M. The cells were successively washed with phosphate-buffered saline, buffer B (0.25% Triton-X 100, 10 mM EDTA, 0.5 mM EGTA, 20 mM HEPES [pH 7.6]) and buffer C (0.15 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES [pH 7.6]) at 4°C for 10 min each. The cells were then resuspended in ChIP incubation buffer (0.3% sodium dodecyl sulfate [SDS], 1% Triton-X 100, 0.15 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES [pH 7.6]) and sheared using a Bioruptor sonicator (Cosmo Bio Co., Ltd.) with six pulses of 30 s each at the maximum setting. The sonicated chromatin was centrifuged for 15 min and incubated for 12 h at 4°C with either a polyclonal anti-TCF4 antibody (sc-8631; Santa Cruz Biotechnology, Inc.) or a monoclonal anti-TCF4 antibody (1) (05-511; Upstate) at 1 μg of antibody per 106 cells with protein G beads (Upstate). The beads were successively washed 2 times with buffer 1 (0.1% SDS, 0.1% deoxycholate, 1% Triton-X 100, 0.15 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES [pH 7.6]), one time with buffer 2 (0.1% SDS, 0.1% sodium deoxycholate, 1% Triton-X 100, 0.5 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES [pH 7.6]), one time with buffer 3 (0.25 M LiCl, 0.5% sodium deoxycholate, 0.5% NP-40, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES [pH 7.6]), and two times with buffer 4 (1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES [pH 7.6]) for 5 min each at 4°C. The precipitated chromatin was eluted by incubation of the beads with elution buffer (1% SDS, 0.1 M NaHCO3) at room temperature for 20 min, de-cross-linked by incubation at 65°C for 5 h in the presence of 200 mM NaCl, extracted with phenol-chloroform, and precipitated.
For sequential ChIP, the eluted chromatin was diluted with ChIP incubation buffer without SDS to the incubation conditions of the first ChIP. Half the amount of antibody was added to the second ChIP and processed as for the first.
Ligation-mediated PCR amplification, labeling, and hybridization.
The ChIP material was amplified for labeling as described previously (35). Labeling of the material, hybridization, and scanning of the arrays were performed by Nimblegen, Inc.
Quantitative PCR (qPCR).
ChIP experiments were analyzed with quantitative PCR in an iCycler iQ real-time PCR detection system (Bio-Rad), using iQ Sybr green supermix (Bio-Rad). Specific primers were designed using Beacon Designer software (Premier Biosoft International) and verified for specificity by in silico PCR (http://genome.cse.ucsc.edu/cgi-bin/hgPcr). ChIP values were normalized as a percentage of input. The specificity of ChIP values was expressed as the change from respective values for control regions (i.e., exon 2 of the nonexpressed myoglobin gene). Based on TCF4 occupancy values over a number of such negative control regions, we defined as positive those regions whose change in occupancy over the control region was greater than threefold.
Reporter assays.
Genomic fragments encompassing typically about 1 kb of genomic sequence encompassing a TCF4 peak were amplified by PCR from human genomic DNA and cloned in front of the firefly luciferase gene in pGL3b or pGL4.10, in the case of TSS-proximal regions, or in front of a minimal fragment encompassing the TATA box of the adenovirus major late promoter cloned in front of the firefly luciferase gene in pGL3b or a minimal TATA box cloned in front of the firefly luciferase gene in pGL4.10, in the case of non-TSS-proximal regions. For the control experiment, human genomic DNA was digested with KpnI and 15 fragments of approximately 1 kb cloned in front of the firefly luciferase gene in pGL3b and 15 fragments cloned in front of a minimal fragment encompassing the TATA box of the adenovirus major late promoter cloned in front of the firefly luciferase gene in pGL3b were used in the reporter assays. The reporters were transfected with Fugene 6 (Roche Diagnostics) in LS174T or LS174T/ΔNTCF4 cells (the latter inducibly overexpress ΔNTCF4 upon doxycycline treatment) with Renilla luciferase as a transfection control and appropriate expression vectors, and their activity was measured using a dual-luciferase reporter assay system (Promega).
Array design.
The genome-wide hybridization was performed on a NimbleGen Systems, Inc., set of 36 arrays containing a total of 13,787,634 oligonucleotides of 50 bp covering the repeat-masked portion of the human genome for chromosomes 1 to 22p at 100-bp resolution (NCBI35/HG17 genome build).
To verify the peaks obtained by the genome-wide array, two sets of triplicate experiments were performed, a dedicated array for chromosomes 1 to 22p and the tiling array for chromosomes 22q/X/Y. The dedicated array contained 1,251,695 oligonucleotides covering the putative TCF-4-bound sequences extracted from the genome-wide array (chromosomes 1 to 22p), plus a tiled region from chromosome 21 (chromosome 21: 33206900 to 46800000) at 100-bp resolution for normalization purposes. The dedicated array was divided over two slides, both containing the full tiled region. The replicates for the tiling array for chromosomes 22q/X/Y contained 769,784 oligonucleotides on two slides.
Identification of TCF-4-binding regions.
Three different peak identification software packages were used to extract putative peaks from the genome-wide scans, Mpeak (MP) (http://www.stat.ucla.edu/∼zmdl/mpeak), TileMap (TM) (21), and NimbleGen Peakdetection (NP) (NimbleGen, Inc.), to maximize the inclusion of putative TCF-4-binding regions on the dedicated array. MP (version 2.0) was used with default settings and a threshold of 2.5S.D.; TM was used with HMM (posterior probability of >0.5; maximal gap allowed, 100; UMS on; G0 p%, 0.01; G1 q%, 0.05; selection offset on; grid size, 1,000; expected hyblength, 50; no repeat filter; no test statistics) to combine neighboring probes. The Nimblegen program (version 2) was used with a 1% FPR cutoff. Identified peaks were extended 1,000 bp on either site from the center of the peaks, resulting in 67,838 peak areas. Probes for inclusion on the dedicated array were filtered using BLAT software (22), excluding probes aligning more than 10 times in the genome. Following three replicate hybridizations on the dedicated arrays and on the array covering chromosomes 22q, X, and Y, application of Tukey's biweight analysis on the chromosome 21 tile path was used to normalize and scale each slide (http://mathworld.wolfram.com/tukeysbiweight.html). The mean ratio signal and variance were calculated for each probe, and peak recognition with the same peak recognition algorithms as described above was performed with the mean ratio signal track. The gap parameter for both MP and TM was set to 250 bp, i.e., allowing a maximum of 250 bp between probes that constitute a peak. The rest of the parameter settings for the programs were adjusted to call approximately the same number of peaks with each method. Using a 2.5-standard-deviation cutoff for MP, a total of 15,282 or 1,176 peaks were called in the dedicated design or the chromosome 22q/X/Y set, respectively. Using these numbers as a reference, both NP and TM were tried iteratively with increasing or decreasing thresholds for peak detection to achieve a peak set of approximately 15,000 peaks in the dedicated design set and 1,100 peaks in the chromosome 22q/X/Y set. The final peak thresholds for NP were 0.14 (dedicated design) and 0.02 (chromosome 22q/X/Y). The final peak thresholds for TM were 0.10 (dedicated design) and 0.95 (chromosome 22q/X/Y). The overlap of peaks found with the different programs was determined by defining overlaps as peaks positioned within 1,000 bp of each other. The set of peaks found by TM which overlapped with both MP and NP peaks was chosen as the final peak set. The final peak set contains 11,912 peaks in the regions of the dedicated design set and 555 peaks in chromosome 22q/X/Y set (see Table S8 in the supplemental material). The final peak set was divided in four confidence groups by the mean signal and variance of the probes within a peak. A total of nine probes around the peak position were used to calculate the mean signal and variance for each peak. The peak confidence sets were divided around the median mean signal and the median variance of the dedicated array (set A, mean peak signal of >1.5 and mean peak variance of <0.5; set B, mean peak signal of >1.5 and mean peak variance of >0.5; set C, mean peak signal of <1.5 and mean peak variance of >0.5; set D, mean peak signal of <1.5 and mean peak variance of <0.5).
Comparison between TCF-bound region and random genomic regions.
A randomization test was preformed in order to compare properties of TCF-bound regions with those of other genomic regions. One hundred or 250 (where indicated) random sets were sampled from the human genome assembly to retain the same region size and distribution between chromosomes as with the original 6,868 TCF-bound sites. All random peaks were chosen from the unmasked sequence that was interrogated by the ChIP-on-chip experiment. The analyses of TCF-bound region properties with respect to gene structure, CpG islands, capped analysis of gene expression (CAGE) tags, clustering of sites around TSS, presence of the TCF motif, and evolutionary conservation were performed for real and random sets.
Evolutionary conservation of TCF-bound regions and motifs.
Pairwise nucleotide BlastZ-net human-mouse, human-rat, human-chicken, and human-dog alignments were taken from the Ensembl database (19). Total conservation at consensus TCF motifs, TCF-bound regions, and random regions (200 bp around the center of the peak in both cases) were calculated. Insertions/deletions and unaligned segments were excluded from this calculation.
Identification of transcription factor-binding sites in TCF4-binding regions.
Matrices from the Transfac database (version 11.1) were searched for using the matrix scanning program Storm (37) with a per-match P value cutoff of 0.0001 and an Hg17 intergenic 8mer word table. The matches for each matrix were tabulated across the foreground (500 bp around peak centers) and background (1,000-bp flanking sequence around peak centers) sets. A proportion test was then performed using the statistical computing language R, specifically, the prop.test function of R version 2.6.1. To derive sequence logos from Transfac matrices, a custom program was used. To generate logos from the Storm output, the WebLogo software program, version 2.8.2 (http://weblogo.berkeley.edu/), was used.
Biological function of TCF-bound genes.
Genes upregulated in human primary adenomas and bound by TCF4 within 100 kb of their TSSs were interrogated for gene ontology category and KEGG (Kyoto encyclopedia of genes and genomes) pathway enrichment using the web-based tool g:Profiler (http://biit.cs.ee/gprofiler/) (34).
Microarray data accession numbers.
The microarray data can be accessed at http://www.ebi.ac.uk/arrayexpress/, experiment code E-TABM-402.
RESULTS
Genome-wide profile of TCF4 binding in CRC cells.
To identify in vivo TCF4-binding sites in a comprehensive manner, we optimized sequential chromatin immunoprecipitations using a goat polyclonal antibody raised against the N terminus of the TCF4 protein. The increase in specific enrichment attained by the sequential immunoprecipitations (12) should allow the comprehensive identification of even “weak” TCF4-binding sites in the genome of CRC cells. All experiments were performed with the diploid, β-catenin mutant human colon cancer cell line LS174T, which expresses a TCF4-dependent transcriptional program similar to that which is physiologically driven by Tcf/β-catenin in the proliferative compartment of intestinal crypts (49). As shown in Fig. 1a (see also Fig. S1 in the supplemental material), the proximal promoter of the SP5 gene, a previously described Wnt target (41, 43), was enriched >100-fold after one round of immunoprecipitation and was enriched >1,000-fold after two sequential immunoprecipitations using the anti-TCF4 antibody. Consistent with c-Myc being Wnt responsive in LS174T cells (49), the previously described TCF response element in the c-Myc promoter (17) was also enriched (Fig. 1a) (see Fig. S1 in the supplemental material), albeit to a lower extent. These observations were independently confirmed using an anti-TCF4 monoclonal antibody raised in our laboratory (1) (not shown).
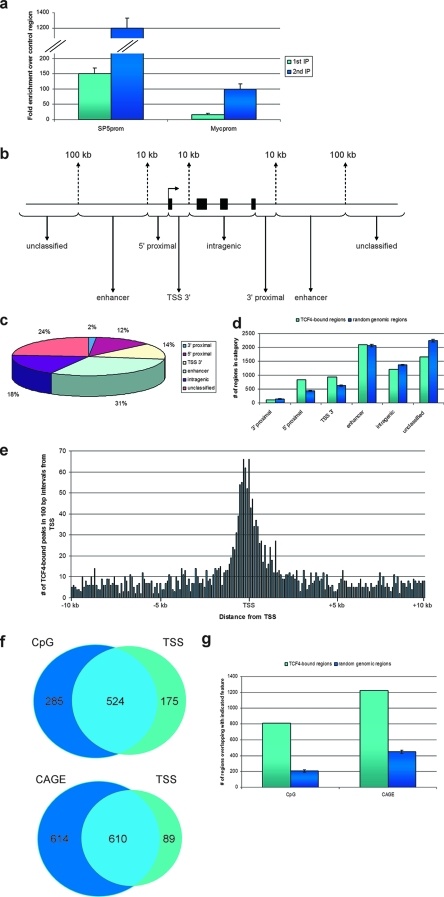
FIG. 1.
ChIPs over regions bound by TCF4 and genomic distribution of TCF4-bound regions. (a) Association of TCF4 with the proximal promoters of SP5 and c-Myc was determined by single (light blue) or sequential (dark blue) ChIP followed by qPCR and expressed as relative enrichment over the nonbound exon 2 of the myoglobin gene. Error bars represent standard deviations for three independent experiments. (b) Schematic illustration delineating the criteria for binding-site classification with respect to a gene locus. (c) Localization of TCF4-binding sites in relation to annotation to nearest transcription units. Shown are percentages of binding sites in the different location categories as defined in panel b. (d) Distribution in categories, defined as in panel b, of TCF4-bound regions (light blue) or random genomic regions (dark blue). Error bars represent standard deviations of 100 random groups. (e) Distribution in 100-bp intervals of TCF4-bound regions located within 10 kb of annotated TSSs. (f) Venn diagrams depicting the number of TCF4-bound regions within 1 kb of CpG islands, annotated transcription start sites of protein-coding genes, or both (top) and the number of TCF4-bound regions within 1 kb of CAGE tags, annotated transcription start sites of protein-coding genes, or both (bottom). (g) Distribution, in categories defined as in panel f, of TCF4-bound regions (light blue) or random genomic regions (dark blue). Error bars represent standard deviations of 250 random groups.
DNA (either from input chromatin or from sequential ChIP material) was amplified by ligation-mediated PCR and labeled with Cy3 and Cy5, respectively. The probe samples were then hybridized to a set of 36 microarrays covering the repeat-masked regions of the human genome at 100-bp resolution (NimbleGen Systems, Inc.) (apart from the q arm of chromosome 22 and chromosomes X and Y; see below). Three different algorithms, MP (http://www.stat.ucla.edu/∼zmdl/mpeak), TM (21), and NP (NimbleGen Systems, Inc.), were used to predict a total of 67,838 putative TCF4-binding sites. The application of all three programs redundantly aimed at the inclusion in the peak count of the greatest possible number of putative TCF4-binding sites and the minimization of false negatives. To verify the binding sites predicted from the genome-wide hybridization, we designed dedicated arrays covering regions of 2 kb around each detected peak (chromosomes 1 to 21 and 22p). ChIP-on-chip experiments were performed on the dedicated arrays with three biological replicates (independent TCF4 chromatin immunoprecipitates, independently amplified and labeled). The same replicates were used to probe in triplicate the 100-bp-resolution tiling path array covering the remaining chromosomes, 22q, X, and Y.
The peak detection procedure performed for both the replicates of the dedicated arrays and the replicates of the chromosome 22q/X/Y tiling path array was the following: The three biological replicates were merged into one data set by calculating the mean ratio signal for each probe. The three peak recognition algorithms were applied to the mean ratio signal track, and only peaks found by all three algorithms were retained to extract 11,912 binding regions from the dedicated arrays and 555 binding regions from the chromosome 22q/X/Y array. By requiring three out of three programs to detect each peak, we increased the stringency of peak prediction to minimize the inclusion of false positives in the final set of TCF4-binding sites. Prior to validation by quantitative PCR analysis, the detected peaks were further subdivided into four groups according to mean peak signal values and mean peak variance over a region of nine probes surrounding the peak center (set A, mean peak signal of >1.5 and mean peak variance of <0.5; set B, mean peak signal of >1.5 and mean peak variance of >0.5; set C, mean peak signal of <1.5 and mean peak variance of >0.5; set D, mean peak signal of <1.5 and mean peak variance of <0.5). For both the dedicated and chromosome 22q/X/Y binding sites, 15 randomly selected peaks from each of the 4 groups were validated by quantitative PCR. All 60 peaks from both sets A and B from the dedicated design, as well as chromosome 22q/X/Y, were positive. Only 8/15 and 6/15 peaks from set C and 7/15 and 9/15 peaks from set D for the dedicated design and chromosome 22q/X/Y, respectively, were positive in the qPCR assays (see Fig. S2 and S3 and Table S1 in the supplemental material). The accuracy rate for both the dedicated design and the chromosome 22q/X/Y sets of binding sites is 75%; this indicates that the three biological replicates on our dedicated design maintain the same specificity as the three biological replicates on the chromosome 22q/X/Y tiling array, validating the dedicated array approach, in agreement with other studies (23, 24).
Sets A and B gave an accuracy rate of 100%. Since sets C and D yielded accuracy rates between 40% and 60% and contained peaks of mostly lower levels of specific enrichment than A and B, we continued our analyses with the binding regions of sets A and B only. Merging of peaks within 1,000 bp of each other in these two groups resulted in 6,868 high confidence TCF4-binding sites (see Table S2 in the supplemental material). We estimated that this approach may miss up to 2,150—mostly low-enrichment—binding sites but should increase the specificity of subsequent analyses.
As expected, the high-confidence peak set included prominent binding sites over the proximal promoters of the SP5 and c-Myc genes (not shown). An additional 44 TCF4-binding sites from peak sets A and B near known target genes of the pathway (45, 48, 49) were all confirmed by qPCR (see Fig. S4 and Table S3 in the supplemental material), further underscoring the specificity of the generated TCF4-binding profile.
We also proceeded to investigate the presence of validated TCF4-binding sites in other CRC cell lines. To this effect, chromatin immunoprecipitations with the goat polyclonal antibody against TCF4 were performed with HCT116 and DLD1 cells, and 25 randomly selected binding sites were tested by qPCR (see Fig. S5 in the supplemental material). Of the 25 tested binding regions, 20 (80%) were positive in HCT116 cells and 24 (96%) were positive in DLD1 cells. The high percentage of TCF4-binding sites bound in all three cell lines further stresses the relevance of the generated TCF4-binding profile for the investigation of TCF4-mediated transcriptional regulation in CRC.
Distribution of TCF4-binding sites with respect to gene structure.
To evaluate the distribution of the TCF4-binding sites along the genome, we annotated these with respect to the TSS of the nearest gene (based on Ensembl v34 (6). Peaks were defined as either 5′-proximal (10 kb upstream of the TSS), TSS 3′ (10 kb downstream of the TSS), intragenic (within gene bodies, from 10 kb 3′ from the TSS to the gene end), 3′ proximal (within 10 kb downstream of the gene), or distal “enhancer” (10 to 100 kb either up- or downstream of gene boundaries). Peaks located more than 100 kb away from the nearest gene were annotated as unclassified (Fig. 1b).
Eight hundred thirty-nine (12%) of peaks were found within 5′-proximal locations, 941 (14%) were located in TSS 3′ positions, and 117 (2%) within 3′-proximal locations. One thousand two hundred nine (18%) peaks were found within genes, further than 10 kb from the TSS. Two thousand ninety-eight (31%) peaks were located in putative long-range “enhancer” positions (up to 100 kb up- or downstream of a gene). One thousand six hundred sixty-four (24%) peaks were not located within 100 kb of the boundaries of the nearest gene (unclassified) (Fig. 1c). When this distribution of peaks was compared to that of random genomic fragments, it became apparent that there was a striking bias for TCF4-binding sites within 10 kb both up- and downstream of TSSs (Fig. 1d). The pronounced clustering of TCF4-bound regions around TSSs can be prominently observed in Fig. 1e, a plot of the distribution of binding sites relative to the distance from the TSS. Despite this conspicuous pattern observed for peaks near TSS, more than 70% of TCF4-bound regions are located at distances greater than 10 kb from the nearest annotated transcription starts, a distribution which is similar to that determined using similar global approaches for other sequence-specific DNA-binding transcription factors, such as Oct4 and Nanog (29), p53 (51), and ER (8).
We also analyzed the overlap of TCF4-bound regions with respect to CpG islands and found 809 of them to be within 1,000 bp of annotated CpG islands (Fig. 1f), a number much greater than that observed for random genomic regions (Fig. 1g). Significantly, 285 (35%) of the TCF4-bound regions overlapping CpG islands were not in similar proximity (within 1 kb) to TSSs of protein coding genes (Fig. 1f).
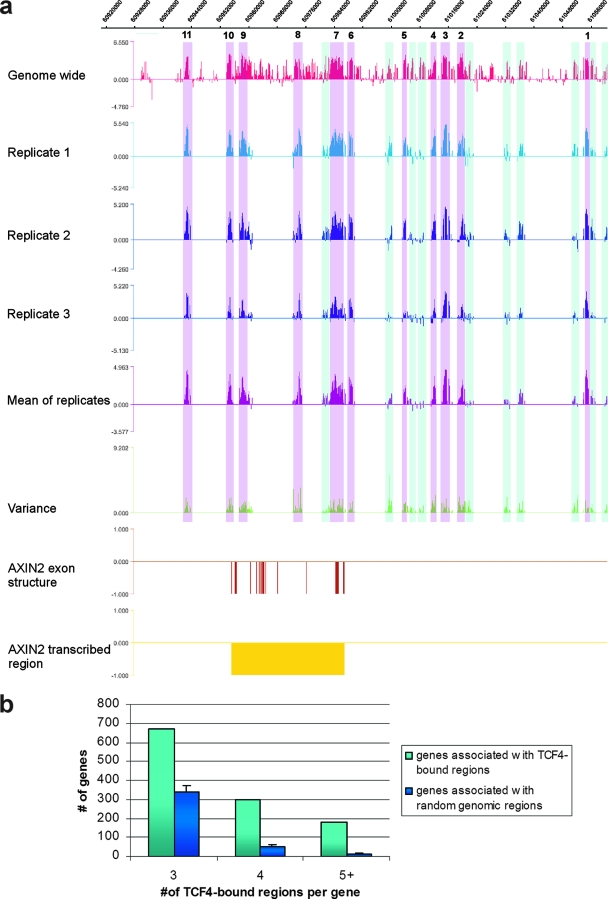
Visual inspection of the distribution of the TCF4-binding regions revealed another interesting observation: peaks frequently cluster around putative target genes. An extreme example was provided by AXIN2, a well-known target gene of the Wnt pathway (31), which associates with no fewer than 11 peaks within 100 kb of its TSS (Fig. 2a). We explored whether this clustered distribution of peaks around genes was nonrandom by comparing it to the distribution expected for randomly selected genomic regions. The analysis shown in Fig. 2b demonstrates that the distribution was indeed not random, since there were significantly more genes that associate with three or more TCF4-binding sites than expected, providing statistical validation to this striking phenomenon.
FIG. 2.
TCF4-binding-site clustering around target genes. (a) TCF4-binding-site distribution around the AXIN2 gene. Depicted is the binding pattern of TCF4 around AXIN2 as revealed by the genome-wide experiment and the three independent biological replicates on the dedicated array, including the mean and variance tracks from the three replicates. High-confidence peaks are highlighted in magenta and numbered 1 to 11, low-confidence peaks in light blue. (b) Numbers of genes bound within 100 kb of their TSSs by three, four, or five or more TCF4-binding sites (light blue) or random genomic regions (dark blue). Error bars represent standard deviations of 100 random groups.
Determination and conservation of the TCF4-binding DNA motif.
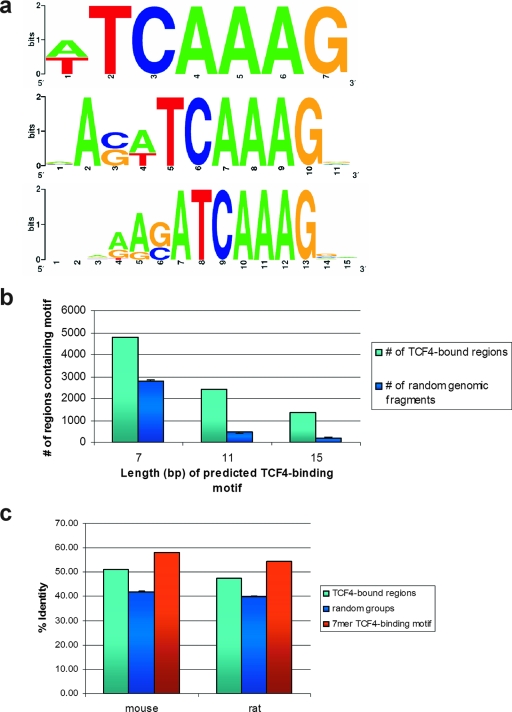
In in vitro selection-based assays, we have previously defined the optimal TCF-binding motif as AAGATCAAAGG (44, 46). Using a different in vitro approach, Hallikas and colleagues defined a slightly shorter optimal TCF4 binding motif: CATCAAAGG (14). We proceeded to mine the underlying sequence of the TCF4-bound peaks to determine the cis element(s) which mediates TCF4 binding in vivo. We applied MDscan, a de novo motif discovery algorithm (28), using random samples of the peaks validated by qPCR for program training. The most common motifs discovered within windows of different lengths bore a strong resemblance to the consensus motif identified in the in vitro studies. Three examples of these—the most common motif within a 7-bp, 11-bp, or 15-bp window—are depicted in Fig. 3a. Seventy percent (4,793/6,868) of the sites bound by TCF4 contained at least the shortest (7 nucleotides) motif uncovered by our method, and this ratio was much greater than that expected for random genomic fragments. This statistical significance held true also for the occurrence of the longer motifs (Fig. 3b). We further examined the evolutionary conservation of the TCF4-binding regions and DNA-binding motifs with respect to the genomes of rat and mouse (Fig. 3c), as well as dog and chicken (not shown): Both the sequences surrounding the centers of the peaks and the TCF4-binding motifs contained within the sequences were significantly more conserved compared to random genomic segments, as expected for functional transcriptional regulatory regions (chi-square, P < 0.01). These observations further underscored the validity of our TCF4-binding sites.
FIG. 3.
(a) Sequence logos illustrating the nucleotide distribution for the in vivo TCF4 consensus sites of 7, 11, and 15 bp, as defined by MDscan. (b) Number of TCF4-bound (light blue) or random genomic (dark blue) regions containing the indicated TCF4-binding motif, as depicted in panel a. Error bars represent standard deviations of 100 random groups. (c) Percent identities of TCF4-bound regions (light blue), random genomic regions (dark blue), and the 7-mer TCF4-binding motif (red, as depicted in panel a) for mouse-human and rat-human pairwise genomic comparisons. Error bars represent standard deviations of 100 random groups.
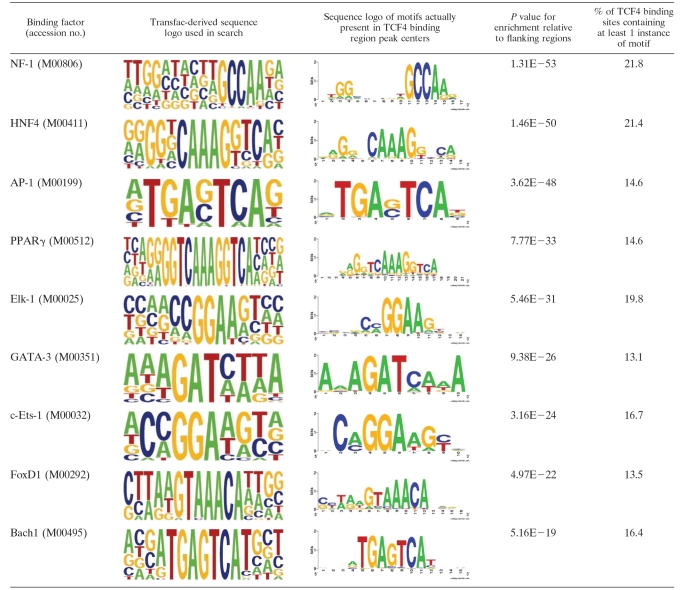
We further mined the sequences encompassing the TCF4-binding regions to identify binding sites for other transcription factors using the Transfac database. Matrices were searched for using the program Storm (37) in the 500-bp genomic regions surrounding the TCF4-binding-region centers, and their incidence was compared to incidence in the 1,000-bp genomic regions flanking the peak centers on either side. This analysis revealed the presence of a number of motifs for transcription factors, such as NF1, HNF4, PPARγ, and others, specifically enriched in TCF4-binding regions (Table 1). These factors potentially coregulate transcription of TCF4 target genes.
TABLE 1.
Transfac matrices enriched around TCF4 binding-site centersa
Depicted are sequence logos of transcription factor binding sites identified as significantly enriched when sequences surrounding TCF4 binding-site centers (500 bp around each center) are compared to the 1,000-bp regions flanking the binding-site centers on either side.
Correlation between TCF4 occupancy and Wnt-dependent transcriptional regulation.
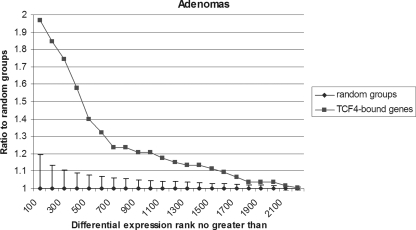
We have previously described the global transcriptional program driven by the Wnt pathway in CRC (36, 45, 48, 49). In references 36 and 45, we and collaborators performed an exhaustive array-based comparison of the Wnt target gene program with colorectal cell lines and primary human adenomas. These expression data sets were used to investigate the potential correlation between Wnt-mediated transcriptional effects and the genome-wide TCF4 binding profile. A stepwise differential expression rank analysis showed a significant correlation between TCF4 occupancy of target-gene regulatory regions and genes upregulated in human primary adenomas compared to normal colonic mucosa; genes with TCF4-binding regions within 100 kb of their TSS were more likely to show significantly upregulated expression than genes without (Fig. 4). Significant correlation between TCF4 occupancy and expression profile changes was also observed in LS174T cells inducibly overexpressing an N-terminally truncated dominant-negative mutant form of TCF4 (ΔNTCF4) (see Fig. S6 in the supplemental material). These data demonstrate that Wnt-dependent transcriptional changes correlate strongly with direct TCF4 occupancy of regulatory regions, even when the sources of the binding and expression profiles are different (CRC cell lines versus primary adenomas).
FIG. 4.
Correlation of TCF4 binding and TCF4/β-catenin-controlled gene expression. Differential expression rank analysis for genes bound within 100 kb of TSS by TCF4 or random groups from genes upregulated in human primary adenomas, using a step size of 100. Error bars represent standard deviations of 100 random groups.
To examine the possibility that TCF4-binding sites have the potential to regulate RNA species not profiled by the above expression microarray experiments, we overlapped our TCF4-bound regions with CAGE tags, generated by the FANTOM3 (functional annotation of mouse 3) consortium (http://fantom.gsc.riken.go.jp/) (7), to define often previously unknown TSSs. We found that 1,224 TCF4-bound regions were within 1 kb and 3,324 were within 10 kb of human CAGE tags, a colocalization much greater than expected for random genomic regions (Fig. 1f and g; also data not shown). Significantly, 614 of the 1,224 (50%) and 1,729 of the 3,224 (54%) of TCF4-bound regions overlapping CAGE tags within 1 and 10 kb, respectively, did not overlap TSSs of known protein coding genes within the same distances (Fig. 1f; also data not shown). This provides an indication that many TCF4-bound regions may regulate transcription of novel RNA species not profiled by conventional expression microarrays.
Biological functions of TCF4 target genes.
Functional categorization of TCF4 target genes (genes upregulated in human primary adenomas and bound by TCF4 within 100 kb of TSSs) revealed enrichment of genes involved in a broad spectrum of functions, such as cell proliferation (P = 4.34 × 10−9), transcription (P = 5.3 × 10−7), cell adhesion (P = 6.19 × 10−6), and the proteasome complex (P = 5.09 × 10−8) (see Table S4 in the supplemental material). Further examination of genes bound by TCF4 within 10 kb of TSS (irrespective of whether they were upregulated in human adenomas) revealed additional enriched categories, including negative regulation of programmed cell death (P = 9.6 × 10−6) and establishment and maintenance of chromatin (P = 7.7 × 10−7) (see Table S4 in the supplemental material). Promotion of cell proliferation and the negative regulation of apoptosis are functions consistent with the activity of a transcription factor at the end point of the Wnt pathway, which is involved in maintaining the proliferative compartment of the mammalian intestinal crypt and in carcinogenesis. The list of bound genes also contains a large number of sequence-specific transcription factors, many of which were not previously known to be targets of the Wnt signaling pathway. The abundance of sequence-specific transcription factors among the TCF4-bound genes should clarify regulatory relationships that will help distinguish direct from indirect targets of the pathway. It is noteworthy that these targets include three members of the TCF family, LEF1, TCF7 (TCF1), and TCF7L2 (TCF4) itself. It should further be noted that KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways with components enriched in the TCF4-bound gene set included the Wnt pathway itself (P = 7.7 × 10−6) and axon guidance (P = 8.9 × 10−6) (see Table S4 in the supplemental material). The latter contains the previously identified targets EPHB2 and EPHB3 (3, 4), which serve to position cells in the intestinal epithelium along the crypt/villus axis. Other genes in this category may be also involved in similar processes.
Transcriptional regulatory activity of TCF4-bound regions.
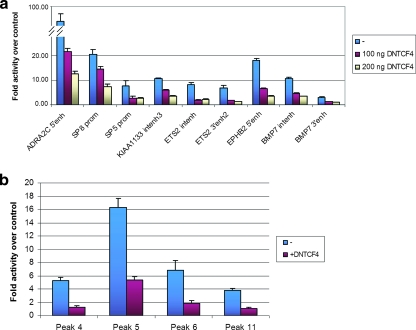
We next investigated whether the identified TCF4-bound genomic regions exert transcriptional regulatory activity. Fragments of approximately 1,000 bp surrounding 22 peaks (see Table S5 in the supplemental material) were cloned either as promoters (in the case of peaks that were located in the vicinity of the TSSs of target genes) or as enhancers upstream of a minimal fragment encompassing the TATA box of the adenovirus major late promoter. The resulting plasmids were transiently transfected into LS174T cells. Ten of the 22 regions enhanced transcription of the luciferase reporter in this assay. These included the proximal promoter of SP5, a region far downstream of the ADRA2C gene, which was the strongest enhancer tested at more than 90-fold the activity of the control, as well as the 3′ and intronic peaks associated with the BMP7 gene. Cotransfection of ΔNTCF4 led to downregulation of the activity of nine elements (Fig. 5a). As a control experiment, we cloned 15 random genomic regions as promoters and 15 random genomic regions as enhancers in front of the same luciferase reporter. Of these, only three were transcriptionally active and none was regulated by cotransfection of ΔNTCF4 (data not shown).
FIG. 5.
Transcriptional activity of TCF4-bound regions in CRC cells. (a) TCF4-binding regions were cloned into the pGL3b or pGL3/AdMLTATA vector, in the case of TSS-proximal or non-TSS-proximal regions, respectively, and transfected into Ls174T with cotransfection of the CMV-Renilla vector as the normalizing control and with or without cotransfection of ΔNTCF4. Values are expressed as activity relative to that of the respective empty pGL3 vectors. Error bars represent standard deviations for three independent experiments. (b) Eleven TCF4-binding regions surrounding the AXIN2 gene within 100 kb of the TSS were cloned into the pGL4.10 or pGL4.10/TATA vector, in the case of TSS-proximal or non-TSS-proximal regions, respectively, and transfected into LS174T/ΔNTCF4 cells with cotransfection of the CMV-Renilla vector as the normalizing control and with or without doxycycline treatment to induce ΔNTCF4 expression. Values are expressed as activity relative to that of the respective empty pGL4.10 vectors. Error bars represent standard deviations for three independent experiments.
We further investigated the transcriptional activity of all peaks surrounding the AXIN2 gene. The latter is a well-known target of the pathway (31) and is surrounded by at least 11 TCF4-binding regions within 100 kb of its TSS (Fig. 2a) (see Table S5 in the supplemental material). Using a similar strategy to that described above, we found that of those 11 peaks, 4 enhanced the transcriptional activity of a luciferase reporter and were downregulated by overexpression of ΔNTCF4 (Fig. 5b). The regions active in these experiments include a peak near the TSS of the gene (peak 6) in a region previously shown to display Wnt-regulated transcriptional activity (20), two peaks 5′ of the TSS, and 1 peak 3′ to the end of the transcription unit.
These experiments demonstrated that a significant subset of TCF4-bound regions uncovered by the ChIP-on-chip approach score as Wnt-responsive transcriptional regulatory regions in transient reporter gene assays. The subset of Wnt-regulated regions included both peaks near TSSs (i.e., the SP5, SP8, and AXIN2 proximal promoters and an EPHB2 5′ peak) and binding regions further away from TSS (i.e., an ADRA2C 3′ peak, the AXIN2 far-upstream peaks, and the BMP7 and ETS2 intronic peaks), consistent with the Wnt pathway having the ability to regulate transcription of target genes from large distances.
DISCUSSION
Genome-wide approaches for the identification of transcription factor-binding sites are increasingly becoming the tools of choice for the elucidation of the transcriptional circuitries governing development, homeostasis, stem cell biology, or the genesis of cancer (9-11, 29, 51). The ChIP-on-chip approach we have employed here has allowed us to comprehensively map genome-wide chromatin occupancy by TCF4, the transcription factor at the end point of Wnt signaling in the mammalian intestine. Our experiments reveal that the binding profile of TCF4 resembles that of other studied transcription factors, such as ER and p53 (29, 51), in that TCF4 binding is observed both in the vicinity and also at sites located at great distances from the TSSs of annotated genes. An interesting and novel observation to emerge from our results is that TCF4-binding sites frequently cluster in the vicinity of putative target genes. AXIN2 is a characteristic example, with TCF4-binding sites located in intronic, far-upstream, and downstream locations. This multitude of binding sites around genes like AXIN2 may serve multiple regulatory purposes; while as demonstrated in this study, some of the binding sites identified act as classical transcriptional regulatory elements, including regions both upstream and downstream of the gene, the multiple TCF4-binding regions around AXIN2 may serve other purposes, such as maintaining an open chromatin domain or providing a more accurate sensor for the intranuclear β-catenin concentration. It should in the least provide a source of mechanistic insight in future studies of Wnt-dependent transcriptional regulation.
Our study correlates the global profile of TCF4 binding with differential expression array-based data to provide a view of the direct targets of the Wnt pathway in the mammalian intestine. The correlation between the primary adenoma-derived expression data and the cell-line-derived TCF4-binding pattern is particularly striking in that the two data sets are derived from different, albeit both Wnt-driven, sources. It should be noted here that only 12.5% (282/2,248) of the genes upregulated in adenomas were bound by TCF4 within 10 kb and only 20.5% (462/2,248) were bound within 100 kb of the transcription start, the limit of annotation applied to these analyses. Many indirect targets are likely to exist in the upregulated genes, since a number of genes bound by TCF4 encode transcription factors themselves, as well as more-direct targets, with TCF4-binding sites further away from the TSS. Conversely, only 12.5% (462/3,676) of the genes bound by TCF4 within 100 kb of the transcription start site were significantly upregulated in adenomas. This is in line with what has been reported in previous studies (52, 54) and most likely has both technical and biological reasons: slight expression level changes below the limit of detection of these analyses may contribute to the underdetection of valid TCF4 targets. Furthermore, functional redundancy in enhancer and transcription factor action may contribute to the lack of detectable transcriptional changes at some TCF4-occupied genes. Additionally, TCF4-binding sites located at greater distances from transcription start sites and annotated to the closest gene may in fact be exerting their regulatory function elsewhere, including on other genes further away or even on other chromosomes (40) or on noncoding regulatory RNAs not profiled in these studies; the last is also suggested by the significant overlap between TCF4-binding sites and CAGE tags.
Our approach has also allowed us to use the sequence underlying the TCF4 peaks to determine the in vivo TCF4-binding motif. The motif thus generated is very similar to motifs determined through in vitro experiments. Moreover, the motif is statistically overrepresented in the TCF4 peaks compared to occurrence in random genomic fragments, as expected for functional TCF4-binding sites, and both the TCF4-binding motifs and the underlying sequence of the TCF4-bound regions are evolutionarily conserved. It should be noted that some TCF4-binding regions do not contain a recognizable TCF motif (2,075/6,868; 30%). TCF4 may be recruited to these sites by an atypical binding motif not identified by our analyses or through protein-protein interactions with other factors directly recruited to these regions. More likely, TCF4 association with these sites may be indirect, mediated by enhancer “looping” effects: recruitment may be mediated by physical association of distinct genomic regions in cis looping out the intervening DNA (15, 16, 39) or between regions located on other chromosomes (30, 40). Additional experiments are under way to distinguish between these possibilities.
In a previously published study, the Enhancer Element Locator (EEL) computational tool developed by Hallikas and colleagues integrated conservation of in vitro-determined binding sites along with affinity and clustering information to predict TCF4-controlled enhancers (14). EEL predicted 130 putative Wnt-responsive enhancers containing 2 or more TCF4-binding sites, only 10 of which overlap (are within 1,000 bp of each other) with our experimentally validated set of 6,868 peaks. This overlap is slightly greater than random coincidence would allow (see Fig. S7 and Table S6 in the supplemental material). In order to exclude the possibility that the limited overlap between our data sets was caused by a failure of our ChIP-on-chip approach to uncover these binding sites, 10 randomly selected EEL-predicted enhancers (see Table S6 in the supplemental material) were tested by quantitative PCR on TCF4-ChIP material from LS174T cells. All sites tested were negative (enriched <2-fold over a control region in qPCR assays; data not shown), excluding the possibility that EEL-predicted enhancers are missed as false negatives. This means that the EEL bioinformatics tool predicts <0.15% of sites occupied by TCF4 in CRC cells, despite the significantly higher-than-random sequence conservation of our peaks. Of course, it is not unlikely that some of the remaining predicted enhancers not occupied in our CRC cells may represent authentic Wnt-responsive regulatory elements in other contexts. Comparison of the two studies does, however, underscore the fact that current computational tools are limited in their ability to predict the full complement of sites occupied by a transcription factor in a tissue of interest.
While this article was in preparation, a study was published identifying β-catenin-binding sites in the human CRC cell line HCT116, using serial analysis of chromatin occupancy (53). Of the 412 binding sites identified by Yochum et al., 293 binding sites are represented on the NimbleGen genome-wide arrays used in this study and are possible candidates for overlap with the TCF4-binding sites identified here. Of those 293 β-catenin-binding sites, 52 (18%) overlapped with our 6,868 TCF4-binding regions, a proportion which, albeit relatively small, was much greater than that determined for random genomic sequences (see Fig. S8 and Table S7 in the supplemental material). The overlap calculated for the 252 β-catenin-binding sites that contained a consensus TCF4-binding motif within 5 kb and the 4,793 TCF4-binding regions containing ≥1 TCF4 motif within 1 kb was similar (38 binding regions; 16%) and still significant (see Fig. S8 in the supplemental material). The incomplete overlap between the two sets of locational information may be due to the different experimental approaches (ChIP-on-chip versus serial analysis of chromatin occupancy, immunoprecipitations against TCF4 versus β-catenin, respectively).
A number of TCF4-binding regions act as Wnt-responsive promoters or enhancers in transient-transfection experiments, including regions both in the vicinity of and at great distances from transcription start sites. However, more than half (20/33) of TCF4-bound regions were inactive or nonregulated in this assay. Some regions may exert their regulatory activity through effects on the surrounding chromatin template, effects that may be difficult to recapitulate on transiently transfected templates. In the case of the 5′ hypersensitive sites of the β-globin locus control region, the enhancer activity of only 5′ HS2 is detectable in transient-transfection experiments whereas that of HS3 and -4 only becomes apparent when these are integrated into chromatin (27). In this respect, the binding of TCF4 may serve to regulate histone modifications and/or chromatin structure over these regions, since it has been demonstrated to interact through β-catenin both with chromatin remodelers, such as Brg1 (2), and with the histone modifiers MLL and p300/CBP (18, 38, 42). Interestingly, TCFs have also been shown to exert potent intrinsic DNA-bending activity (13, 47, 50). These actions, rather than impinging directly on preinitiation complex formation on promoters of regulated genes, may serve a chromatin opening function, maintaining chromatin domains in a “poised” conformation and facilitating subsequent events involved in transcriptional activation. This model would be compatible with the multiplicity of sites, only some of which act as classical transcriptional regulatory elements, surrounding some target genes, such as AXIN2. Intriguingly, these potential activities of the TCF4/β-catenin complex might be modulated—facilitated or repressed—by other transcription factors which may bind with them on the same genomic regions, as predicted by the enrichment of the TCF4-binding regions in relevant transcription factor-binding matrices.
In conclusion, the current study provides a genome-wide binding profile of TCF4, the major transcription factor at the end point of Wnt signaling in the intestine. Combination of this locational information and differential expression data allows the delineation of the direct transcriptional targets of TCF4 in the human intestine and unveils Wnt-responsive cis elements by which their expression is controlled.
Supplementary Material
Acknowledgments
We thank members of the Clevers and Stunnenberg laboratories for help and discussions, Tokameh Mahmoudi for critical reading of the manuscript, Thanasis Margaritis for assistance with bioinformatics. and Andrea Haegebarth for help with figure preparation.
P.H. is supported by successive European Molecular Biology Organization and Human Frontier Science Program Organization long-term fellowships. M.A.V.D. is supported by EU-FP6 IP EPITRON and STREP X-TRA-NET. S.D. is supported by EU-FP6 IP HEROIC.
Footnotes
Published ahead of print on 11 February 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Barker, N., G. Huls, V. Korinek, and H. Clevers. 1999. Restricted high level expression of Tcf-4 protein in intestinal and mammary gland epithelium. Am. J. Pathol. 15429-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker, N., A. Hurlstone, H. Musisi, A. Miles, M. Bienz, and H. Clevers. 2001. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 204935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batlle, E., J. Bacani, H. Begthel, S. Jonkheer, A. Gregorieff, M. van de Born, N. Malats, E. Sancho, E. Boon, T. Pawson, S. Gallinger, S. Pals, and H. Clevers. 2005. EphB receptor activity suppresses colorectal cancer progression. Nature 4351126-1130. [DOI] [PubMed] [Google Scholar]
- 4.Batlle, E., J. T. Henderson, H. Beghtel, M. M. van den Born, E. Sancho, G. Huls, J. Meeldijk, J. Robertson, M. van de Wetering, T. Pawson, and H. Clevers. 2002. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 111251-263. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120169-181. [DOI] [PubMed] [Google Scholar]
- 6.Birney, E., D. Andrews, M. Caccamo, Y. Chen, L. Clarke, G. Coates, T. Cox, F. Cunningham, V. Curwen, T. Cutts, T. Down, R. Durbin, X. M. Fernandez-Suarez, P. Flicek, S. Graf, M. Hammond, J. Herrero, K. Howe, V. Iyer, K. Jekosch, A. Kahari, A. Kasprzyk, D. Keefe, F. Kokocinski, E. Kulesha, D. London, I. Longden, C. Melsopp, P. Meidl, B. Overduin, A. Parker, G. Proctor, A. Prlic, M. Rae, D. Rios, S. Redmond, M. Schuster, I. Sealy, S. Searle, J. Severin, G. Slater, D. Smedley, J. Smith, A. Stabenau, J. Stalker, S. Trevanion, A. Ureta-Vidal, J. Vogel, S. White, C. Woodwark, and T. J. Hubbard. 2006. Ensembl 2006. Nucleic Acids Res. 34D556-D561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carninci, P., A. Sandelin, B. Lenhard, S. Katayama, K. Shimokawa, J. Ponjavic, C. A. Semple, M. S. Taylor, P. G. Engstrom, M. C. Frith, A. R. Forrest, W. B. Alkema, S. L. Tan, C. Plessy, R. Kodzius, T. Ravasi, T. Kasukawa, S. Fukuda, M. Kanamori-Katayama, Y. Kitazume, H. Kawaji, C. Kai, M. Nakamura, H. Konno, K. Nakano, S. Mottagui-Tabar, P. Arner, A. Chesi, S. Gustincich, F. Persichetti, H. Suzuki, S. M. Grimmond, C. A. Wells, V. Orlando, C. Wahlestedt, E. T. Liu, M. Harbers, J. Kawai, V. B. Bajic, D. A. Hume, and Y. Hayashizaki. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38626-635. [DOI] [PubMed] [Google Scholar]
- 8.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 12233-43. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 381289-1297. [DOI] [PubMed] [Google Scholar]
- 10.Cawley, S., S. Bekiranov, H. H. Ng, P. Kapranov, E. A. Sekinger, D. Kampa, A. Piccolboni, V. Sementchenko, J. Cheng, A. J. Williams, R. Wheeler, B. Wong, J. Drenkow, M. Yamanaka, S. Patel, S. Brubaker, H. Tammana, G. Helt, K. Struhl, and T. R. Gingeras. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116499-509. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, A. S., V. X. Jin, M. Fan, L. T. Smith, S. Liyanarachchi, P. S. Yan, Y. W. Leu, M. W. Chan, C. Plass, K. P. Nephew, R. V. Davuluri, and T. H. Huang. 2006. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol. Cell 21393-404. [DOI] [PubMed] [Google Scholar]
- 12.Denissov, S., M. van Driel, R. Voit, M. Hekkelman, T. Hulsen, N. Hernandez, I. Grummt, R. Wehrens, and H. Stunnenberg. 2007. Identification of novel functional TBP-binding sites and general factor repertoires. EMBO J. 26944-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giese, K., J. Cox, and R. Grosschedl. 1992. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell 69185-195. [DOI] [PubMed] [Google Scholar]
- 14.Hallikas, O., K. Palin, N. Sinjushina, R. Rautiainen, J. Partanen, E. Ukkonen, and J. Taipale. 2006. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 12447-59. [DOI] [PubMed] [Google Scholar]
- 15.Hatzis, P., I. Kyrmizi, and I. Talianidis. 2006. Mitogen-activated protein kinase-mediated disruption of enhancer-promoter communication inhibits hepatocyte nuclear factor 4α expression. Mol. Cell. Biol. 267017-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatzis, P., and I. Talianidis. 2002. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell 101467-1477. [DOI] [PubMed] [Google Scholar]
- 17.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science 2811509-1512. [DOI] [PubMed] [Google Scholar]
- 18.Hecht, A., K. Vleminckx, M. P. Stemmler, F. van Roy, and R. Kemler. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 191839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbard, T. J., B. L. Aken, K. Beal, B. Ballester, M. Caccamo, Y. Chen, L. Clarke, G. Coates, F. Cunningham, T. Cutts, T. Down, S. C. Dyer, S. Fitzgerald, J. Fernandez-Banet, S. Graf, S. Haider, M. Hammond, J. Herrero, R. Holland, K. Howe, K. Howe, N. Johnson, A. Kahari, D. Keefe, F. Kokocinski, E. Kulesha, D. Lawson, I. Longden, C. Melsopp, K. Megy, P. Meidl, B. Ouverdin, A. Parker, A. Prlic, S. Rice, D. Rios, M. Schuster, I. Sealy, J. Severin, G. Slater, D. Smedley, G. Spudich, S. Trevanion, A. Vilella, J. Vogel, S. White, M. Wood, T. Cox, V. Curwen, R. Durbin, X. M. Fernandez-Suarez, P. Flicek, A. Kasprzyk, G. Proctor, S. Searle, J. Smith, A. Ureta-Vidal, and E. Birney. 2007. Ensembl 2007. Nucleic Acids Res. 35D610-D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jho, E. H., T. Zhang, C. Domon, C. K. Joo, J. N. Freund, and F. Costantini. 2002. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 221172-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji, H., and W. H. Wong. 2005. TileMap: create chromosomal map of tiling array hybridizations. Bioinformatics 213629-3636. [DOI] [PubMed] [Google Scholar]
- 22.Kent, W. J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, T. H., Z. K. Abdullaev, A. D. Smith, K. A. Ching, D. I. Loukinov, R. D. Green, M. Q. Zhang, V. V. Lobanenkov, and B. Ren. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 1281231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, T. H., L. O. Barrera, M. Zheng, C. Qu, M. A. Singer, T. A. Richmond, Y. Wu, R. D. Green, and B. Ren. 2005. A high-resolution map of active promoters in the human genome. Nature 436876-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinzler, K. W., and B. Vogelstein. 1996. Lessons from hereditary colorectal cancer. Cell 87159-170. [DOI] [PubMed] [Google Scholar]
- 26.Korinek, V., N. Barker, P. J. Morin, D. van Wichen, R. de Weger, K. W. Kinzler, B. Vogelstein, and H. Clevers. 1997. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science 2751784-1787. [DOI] [PubMed] [Google Scholar]
- 27.Li, Q., S. Harju, and K. R. Peterson. 1999. Locus control regions: coming of age at a decade plus. Trends Genet. 15403-408. [DOI] [PubMed] [Google Scholar]
- 28.Li, W., C. A. Meyer, and X. S. Liu. 2005. A hidden Markov model for analyzing ChIP-chip experiments on genome tiling arrays and its application to p53 binding sequences. Bioinformatics 21(Suppl. 1)i274-i282. [DOI] [PubMed] [Google Scholar]
- 29.Loh, Y. H., Q. Wu, J. L. Chew, V. B. Vega, W. Zhang, X. Chen, G. Bourque, J. George, B. Leong, J. Liu, K. Y. Wong, K. W. Sung, C. W. Lee, X. D. Zhao, K. P. Chiu, L. Lipovich, V. A. Kuznetsov, P. Robson, L. W. Stanton, C. L. Wei, Y. Ruan, B. Lim, and H. H. Ng. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38431-440. [DOI] [PubMed] [Google Scholar]
- 30.Lomvardas, S., G. Barnea, D. J. Pisapia, M. Mendelsohn, J. Kirkland, and R. Axel. 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126403-413. [DOI] [PubMed] [Google Scholar]
- 31.Lustig, B., B. Jerchow, M. Sachs, S. Weiler, T. Pietsch, U. Karsten, M. van de Wetering, H. Clevers, P. M. Schlag, W. Birchmeier, and J. Behrens. 2002. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol. 221184-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polakis, P. 2000. Wnt signaling and cancer. Genes Dev. 141837-1851. [PubMed] [Google Scholar]
- 33.Radtke, F., and H. Clevers. 2005. Self-renewal and cancer of the gut: two sides of a coin. Science 3071904-1909. [DOI] [PubMed] [Google Scholar]
- 34.Reimand, J., M. Kull, H. Peterson, J. Hansen, and J. Vilo. 2007. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35W193-W200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 2902306-2309. [DOI] [PubMed] [Google Scholar]
- 36.Sabates-Bellver, J., L. G. Van der Flier, M. de Palo, E. Cattaneo, C. Maake, H. Rehrauer, E. Laczko, M. A. Kurowski, J. M. Bujnicki, M. Menigatti, J. Luz, T. V. Ranalli, V. Gomes, A. Pastorelli, R. Faggiani, M. Anti, J. Jiricny, H. Clevers, and G. Marra. 2007. Transcriptome profile of human colorectal adenomas. Mol. Cancer Res. 51263-1275. [DOI] [PubMed] [Google Scholar]
- 37.Schones, D. E., A. D. Smith, and M. Q. Zhang. 2007. Statistical significance of cis-regulatory modules. BMC Bioinform. 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sierra, J., T. Yoshida, C. A. Joazeiro, and K. A. Jones. 2006. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 20586-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spilianakis, C. G., and R. A. Flavell. 2004. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 51017-1027. [DOI] [PubMed] [Google Scholar]
- 40.Spilianakis, C. G., M. D. Lalioti, T. Town, G. R. Lee, and R. A. Flavell. 2005. Interchromosomal associations between alternatively expressed loci. Nature 435637-645. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi, M., Y. Nakamura, K. Obama, and Y. Furukawa. 2005. Identification of SP5 as a downstream gene of the beta-catenin/Tcf pathway and its enhanced expression in human colon cancer. Int. J. Oncol 271483-1487. [PubMed] [Google Scholar]
- 42.Takemaru, K. I., and R. T. Moon. 2000. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J. Cell Biol. 149249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thorpe, C. J., G. Weidinger, and R. T. Moon. 2005. Wnt/beta-catenin regulation of the Sp1-related transcription factor sp5l promotes tail development in zebrafish. Development 1321763-1772. [DOI] [PubMed] [Google Scholar]
- 44.van Beest, M., D. Dooijes, M. van De Wetering, S. Kjaerulff, A. Bonvin, O. Nielsen, and H. Clevers. 2000. Sequence-specific high mobility group box factors recognize 10-12-base pair minor groove motifs. J. Biol. Chem. 27527266-27273. [DOI] [PubMed] [Google Scholar]
- 45.Van der Flier, L. G., J. Sabates-Bellver, I. Oving, A. Haegebarth, M. De Palo, M. Anti, M. E. Van Gijn, S. Suijkerbuijk, M. Van de Wetering, G. Marra, and H. Clevers. 2007. The intestinal Wnt/TCF signature. Gastroenterology 132628-632. [DOI] [PubMed] [Google Scholar]
- 46.van de Wetering, M., R. Cavallo, D. Dooijes, M. van Beest, J. van Es, J. Loureiro, A. Ypma, D. Hursh, T. Jones, A. Bejsovec, M. Peifer, M. Mortin, and H. Clevers. 1997. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88789-799. [DOI] [PubMed] [Google Scholar]
- 47.van de Wetering, M., M. Oosterwegel, K. van Norren, and H. Clevers. 1993. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J. 123847-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van de Wetering, M., I. Oving, V. Muncan, M. T. Pon Fong, H. Brantjes, D. van Leenen, F. C. Holstege, T. R. Brummelkamp, R. Agami, and H. Clevers. 2003. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 4609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van de Wetering, M., E. Sancho, C. Verweij, W. de Lau, I. Oving, A. Hurlstone, K. van der Horn, E. Batlle, D. Coudreuse, A. P. Haramis, M. Tjon-Pon-Fong, P. Moerer, M. van den Born, G. Soete, S. Pals, M. Eilers, R. Medema, and H. Clevers. 2002. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111241-250. [DOI] [PubMed] [Google Scholar]
- 50.van Houte, L., A. van Oers, M. van de Wetering, D. Dooijes, R. Kaptein, and H. Clevers. 1993. The sequence-specific high mobility group 1 box of TCF-1 adopts a predominantly alpha-helical conformation in solution. J. Biol. Chem. 26818083-18087. [PubMed] [Google Scholar]
- 51.Wei, C. L., Q. Wu, V. B. Vega, K. P. Chiu, P. Ng, T. Zhang, A. Shahab, H. C. Yong, Y. Fu, Z. Weng, J. Liu, X. D. Zhao, J. L. Chew, Y. L. Lee, V. A. Kuznetsov, W. K. Sung, L. D. Miller, B. Lim, E. T. Liu, Q. Yu, H. H. Ng, and Y. Ruan. 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell 124207-219. [DOI] [PubMed] [Google Scholar]
- 52.Yang, A., Z. Zhu, P. Kapranov, F. McKeon, G. M. Church, T. R. Gingeras, and K. Struhl. 2006. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell 24593-602. [DOI] [PubMed] [Google Scholar]
- 53.Yochum, G. S., S. McWeeney, V. Rajaraman, R. Cleland, S. Peters, and R. H. Goodman. 2007. Serial analysis of chromatin occupancy identifies beta-catenin target genes in colorectal carcinoma cells. Proc. Natl. Acad. Sci. USA 1043324-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng, Y., S. Z. Josefowicz, A. Kas, T. T. Chu, M. A. Gavin, and A. Y. Rudensky. 2007. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445936-940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.