Abstract
PCR-restriction fragment length polymorphism (RFLP) analysis of a 960-bp fragment of the Campylobacter gyrB gene with either DdeI or XspI restriction enzymes generated unique digestion patterns for 12 different Campylobacter species. In addition, PCR assays using species-specific primer sets targeting gyrB were specific for the respective Campylobacter species. Therefore, PCR-RFLP analysis and species-specific PCR assays based on the gyrB gene provide valuable tools for rapid and unambiguous identification of the majority of Campylobacter species.
Campylobacter spp. are an important cause of bacterial gastrointestinal infections worldwide. Travel to developing countries, contact with household pets, and consumption of contaminated vegetables, shellfish, water, and especially poultry are risk factors for infection (2, 6, 11). The genus Campylobacter consists of 16 species and 6 subspecies (12), which may cause disease in humans and animals (10). An additional species has recently been described (3). C. jejuni accounts for the majority of human morbidity, although the other thermotolerant species, C. coli, C. lari, and C. upsaliensis, have also been isolated from clinical samples. Other species have been linked with diarrheal illness and periodontal disease (C. concisus, C. gracilis, C. rectus, and C. showae) as well as meningitis and septicemia (C. fetus) (10). C. lari, C. upsaliensis, and C. fetus subsp. fetus have been associated with food- and waterborne outbreaks of gastroenteritis (1, 4, 8). Due to the technical limitations of cultural and phenotypic methods employed for detection, isolation, and typing, the incidence of Campylobacter spp., especially non-jejuni species, is likely underreported.
The gyrB gene encodes the subunit B protein of DNA gyrase, a type II topoisomerase that catalyzes the negative supercoiling of bacterial DNA. Because the frequency of base substitutions in gyrB exceeded that of 16S rRNA within the species Pseudomonas putida, analysis based on gyrB was more discriminating than that based on 16S rRNA (19). Species identification and detection methods based on gyrB have been developed for Bacillus spp. and Vibrio spp. (15, 18). In this study, we identified the sequence polymorphisms in the Campylobacter gyrB gene and developed species-specific PCR assays and PCR-restriction fragment length polymorphism (RFLP) using the restriction enzymes DdeI and XspI to differentiate 12 Campylobacter species.
PCR amplification and sequencing and analysis of the Campylobacter gyrB gene.
Bacterial strains used to sequence the gyrB gene and to determine the specificity of the resultant gyrB-based PCR assays are listed in Table S1 in the supplemental material. The DNA was isolated as described previously (16) or was purified using PrepMan Ultra reagent (Applied Biosystems, Foster City, CA). PCR amplification of the gyrB gene and direct sequencing of the PCR products were performed using a GeneAmp 9700 thermal cycler (Applied Biosystems). The universal primer set for the PCR amplification of ca. 1,250 bp (1,253 or 1,256 bp) of the gyrB gene region from all strains was 5′-TAATACGACTCACTATAGGGGTCGACCAYGCNGGNGGNAARTTYGA-3′ (T7-FWD; the T7 promoter sequence attached to the 5′ end is underlined) and 5′-GATTTAGGTGACACTATAGCTCGAGCCRTCNACRTCNGCRTCNGTCAT-3′ (SP6-REV; the SP6 promoter sequence attached to the 5′ end is underlined). The DNA template (1 μl) was PCR amplified in a 100-μl reaction volume containing 1× PCR buffer, 4 mM MgCl2, 0.625 U rTaq DNA polymerase (Takara Bio Inc., Shiga, Japan), a 0.2 mM concentration (each) of the four deoxynucleoside triphosphates (dNTPs), and a 0.4 μM concentration of each primer. The amplification conditions were as follows: initial denaturation (95°C for 5 min), followed by 30 cycles each of denaturation (95°C for 1 min), annealing (60°C for 1 min), and extension (72°C for 1 min). The primers used for the DNA sequencing were 5′-TAATACGACTCACTATAGGGGTCGAC-3′ (T7kai) and 5′-GATTTAGGTGACACTATAGCTCGAG-3′ (SP6kai). DNA sequences were determined from both strands by extension from the attached promoter sequences (T7kai and SP6kai primers) and by primer walking using the ABI Prism dye terminator cycle sequencing kit (Applied Biosystems). Products were resolved on an ABI Prism 310 automated sequencer (Applied Biosystems). For phylogenetic analysis, the gyrB sequences of 12 species of Campylobacter were aligned using the DNASIS Pro program (version 2.0) (Hitachi, Tokyo, Japan). Distance matrices using the Kimura two-parameter correction and phylogenetic analysis using the neighbor-joining method (13) were performed with the CLUSTAL W program (14) on the DDBJ website (www.ddbj.nig.ac.jp/Welcome-e.html).
The major topology of the phylogenetic neighbor-joining tree constructed from the partial gyrB gene sequences derived in this study was similar to that previously reported for the 16S rRNA gene sequences (5, 7). However, gyrB provides higher resolution for Campylobacter species, with lower interspecies sequence similarities (ranging from 58.3 to 89.2% [see Table S2A in the supplemental material]) than those reported for the 16S rRNA gene (ranging from 89 to 99% [see Table S2B in the supplemental material]) (5). To illustrate, Gorkiewicz et al. (5) reported that the limitation of 16S rRNA analysis is its inability to differentiate C. jejuni, C. coli, and atypical C. lari strains, which shared identical 16S rRNA gene sequences and therefore were assigned to a common cluster. Earlier reports indicated that these thermotolerant strains exhibited a 98.1% homology based on partial 16S rRNA sequencing (9, 17). In the current study, gyrB gene sequence analyses discriminated these thermotolerant taxa. The C. jejuni isolates shared identical sequences and were clearly distinct from C. coli, though the two species had the highest similarity (89.2%) of the 12 Campylobacter species examined.
The gyrB sequences of C. fetus subsp. fetus ATCC 15296 from the American Type Culture Collection (ATCC), C. fetus subsp. fetus NADC 5513 from the National Animal Disease Center (NADC), C. fetus subsp. venerealis NADC 5519, and C. hyointestinalis ATCC 35217 had a unique 3-base insertion at positions 823 to 825 that is unlike the same region in the other Campylobacter species, which resulted in an additional amino acid in the protein sequence. C. fetus subsp. fetus and C. hyointestinalis are phylogenetically close, as inferred earlier when a 98% homology was calculated based on 16S rRNA sequence analysis (9). That gyrB offers higher resolution between C. fetus subsp. fetus and C. hyointestinalis (84% similarity [see Table S2A in the supplemental material]) than 16S rRNA should expedite the identification of these two species. C. fetus subsp. fetus and C. fetus subsp. venerealis strains shared identical gyrB gene sequences, however, suggesting that gyrB may not be a suitable marker for the identification of these subspecies. A similar conclusion was made following a comparison of rpoO and 16S rRNA sequences in the two C. fetus subspecies (9).
The primary objective of this study was to determine if the gyrB gene sequences were sufficiently unique to serve as suitable targets for Campylobacter species identification. Multiple alignments of the 12 Campylobacter gyrB sequences were performed, a matrix representing the sequence variations among the strains was calculated, and a dendrogram was constructed from these data (see Fig. S1 in the supplemental material). Analysis of the dendrogram showed that all 12 Campylobacter species were clearly differentiated in the constructed phylogenetic tree. The major topology of the tree based on the partial gyrB gene sequences was similar to that of one previously reported that was based on 16S rRNA gene sequence analyses (5).
PCR-RFLP for the differentiation of Campylobacter species.
A universal primer mix prepared using 12 primer sets complementary to the gyrB sequence of each species (Table 1) was used to amplify a 960-bp gyrB fragment from each Campylobacter strain. The DNA template (1 μl) was amplified in a 100-μl reaction volume containing 1× PCR buffer, 2 mM MgCl2, 0.625 U rTaq (Takara) DNA polymerase, a 0.4 mM concentration of each of the four dNTPs, and the universal primer mixture consisting of a 10 nM concentration of each primer. The cycling conditions consisted of an initial denaturation at 95°C for 10 min, followed by 50 cycles of denaturation (95°C for 15 s), annealing (65°C for 1 min), and extension (72°C for 1 min), with a final 7-min extension at 72°C.
TABLE 1.
Sequences used to prepare the universal primer mix for amplification of the 960-bp gyrB gene sequence for each Campylobacter species
| Strain | Universal mix forward primer | Universal mix reverse primer |
|---|---|---|
| C. jejuni NADC 5096 | CGTCAAGAATTTTCAGAAGGTAAAGTTATC | TTTTAAAATTTTATCTAGTCTTGCTTTTTC |
| C. coli NADC 5095 | CGCCAAGAATTTTCAGAAGGTAAAGTCATC | TTTTAAAATTTTATCTAATCTTGCTTTTTC |
| C. concisus ATCC 33237 | AGACAAGAATTTGCAAAAGGTATCCCTCAA | CTTTAAAATTTTATCCAGTCTTGCCTTTTC |
| C. curvus ATCC 35224 | AGGCAAGAATTTCAAAAAGGTATCCCGGTA | TTTTAAAATTTTATCGAGGCGCGATTTTTC |
| C. showae ATCC 51146 | AGACAAGAATTTTCAAAAGGTATCCCTCAA | TTTTAAAATTTTATCTAGTCTTGCTTTTTC |
| C. mucosalis ATCC 49352 | AGGCAAGAATTTGCAAAAGGAATTCCAGTA | TTTTAAAATTTTATCTAATCTTGATTTTTC |
| C. fetus subsp. fetus ATCC 15296 | CGTCAAGAGTTTTCAAAAGGAATACCCCAA | TTTTAAAATTTTATCAAGTCTACTTTTTTC |
| C. hyointestinalis ATCC 35217 | CGCCAAGAATTCGCCGAAGGCATACCTCAA | TTTAAGAATTTTATCAAGCCTACTTTTTTC |
| C. sputorum subsp. sputorum ATCC 33562 | AGACAAGAGTTTTCAAAAGGTGTTCCTACA | TTTTAAAATTTTTTCAAGACCTGCTTTTTC |
| C. helveticus ATCC 51210 | AGACAAGAATTTTCTAAAGGTCTAATTGCA | TTTTAAAATTTTATCCAGCCTTGCTTTTTC |
| C. upsaliensis ATCC 49816 | CGCCAAGAATTTGCTAAAGGGCAAATAGCT | TTTTAAAATTTTATCCAGTCTTGCTTTTTC |
| C. lari ATCC 35221 | AGACAAGAATTTTCAGAAGGAAAAGTAACA | TTTTAAAATTTTATCAAGTCTTGCTTTTTC |
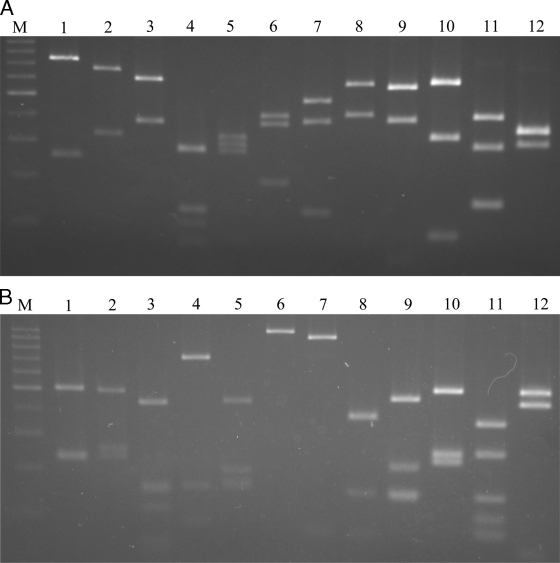
Computational restriction fragment length analyses of the 960-bp amplified region predicted that the DdeI and XspI enzymes would generate species-specific digestion patterns. For RFLP analysis, the purified PCR products were digested in a total volume of 20 μl with either 5 U of DdeI (Toyobo, Osaka, Japan) or 10 U of XspI (Takara). The resulting fragments were size separated using 3.0% agarose prepared in 1× Tris-acetate-EDTA buffer and stained with Sybr green I dye (Invitrogen, Carlsbad, CA). PCR-RFLP results using DdeI and XspI are shown in Fig. 1A and B, respectively. All Campylobacter species studied had species-specific DdeI and XspI digestion patterns. Furthermore, computer analysis of the sequences using the DNASIS program predicted the unambiguous identification of the 12 species of Campylobacter by double digestion of the 960-bp gyrB region with MboI and HindIII. This was also confirmed experimentally with the 960-bp PCR product (data not shown). Kärenlampi et al. (7) demonstrated that partial groEL sequencing and the resultant PCR-RFLP analyses were more discriminating than Campylobacter species identification based on 16S rRNA. A similar conclusion was made when the rpoO gene sequences of 16 Campylobacter species were compared with their 16S rRNA sequences (9).
FIG. 1.
Patterns from PCR-RFLP with DdeI (A) and XspI (B) for C. jejuni NADC 5096 (lane 1), C. coli NADC 5095 (lane 2), C. concisus ATCC 33237 (lane 3), C. curvus ATCC 35224 (lane 4), C. showae ATCC 51146 (lane 5), C. mucosalis ATCC 49352 (lane 6), C. fetus subsp. fetus ATCC 15296 (lane 7), C. hyointestinalis ATCC 35217 (lane 8), C. sputorum subsp. sputorum ATCC 33562 (lane 9), C. helveticus ATCC 51210 (lane 10), C. upsaliensis ATCC 49816 (lane 11), and C. lari ATCC 35221 (lane 12). Lane M, 100-bp molecular size markers.
PCR with Campylobacter species-specific primers and specificity testing.
Twelve different oligonucleotide primer sets were designed based on regions that were dissimilar among the different species (Table 2). Template DNA (2.5 μl) was amplified in a 25-μl reaction volume containing 1× GeneAmp PCR Gold buffer, 0.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems), a 200 μM concentration (each) of the four dNTPs, and a 0.2 μM concentration (each) of the species-specific primers. The cycling conditions for C. jejuni, C. coli, C. lari, C. concisus, C. showae, C. curvus, C. fetus, and C. helveticus were the following: initial denaturation at 95°C for 10 min and 30 cycles of 95°C for 20 s, 69°C for 20 s, and a final extension at 72°C for 7 min. For C. upsaliensis, C. mucosalis, and C. hyointestinalis, the annealing temperature and time were 68°C for 1 min, and for C. sputorum, they were 65°C for 20 s. A specific PCR product was generated for each of the respective target Campylobacter species (Fig. 2). No false-positive results were observed when DNA from the nontarget Campylobacter species was used, and furthermore, nonspecific bands were not observed with DNA from the non-Campylobacter strains tested (see Table S1 in the supplemental material). Thus, the species-specific primer sets based on gyrB sequences can be used for the rapid detection and identification of Campylobacter species.
TABLE 2.
PCR primer sequences targeting gyrB used for Campylobacter species-specific identification
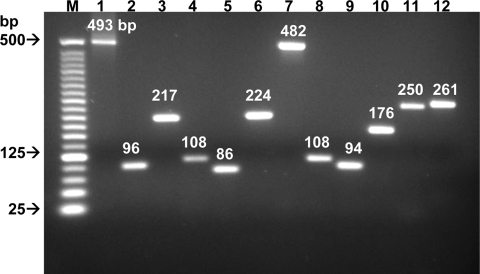
| Strain | Forward primer | Reverse primer | Amplicon length (bp) | Location |
|---|---|---|---|---|
| C. jejuni NADC 5096 | AGAATGGGTTTAACTCGTGTGATAAGT | TACCACGCAAAGGCAGTATAGCT | 493 | 84-576 |
| C. coli NADC 5095 | AAATGCTAGTGCTAGGGAAAAAGACTCT | TGAGGTTCAGGCACTTTTACACTTACTAC | 96 | 125-220 |
| C. concisus ATCC 33237 | AGCGGGCCTAACAAGAGTTATTACA | TGTAAGCACGTCAAAAACCATCTTT | 217 | 86-302 |
| C. curvus ATCC 35224 | CTGCCAAAGTAAGGACGCAAGTATA | GGCAAGATCGCCTGAAATACG | 108 | 458-565 |
| C. showae ATCC 51146 | AGGGTTTAAGCATAGGAACGCTG | CACCAGATAAAGCTCGCTGATCG | 86 | 415-500 |
| C. mucosalis ATCC 49352 | TGCGATTATGAACAAGGCCCTA | TCGCTTGAAACACACGGTCA | 224 | 335-558 |
| C. fetus subsp. fetus ATCC 15296 | AGAGCTGGGCTTACAAGAGCTATTACA | GGTAAAATCGCTTGAAACGCTCTAT | 482 | 84-565 |
| C. hyointestinalis ATCC 35217 | CGGTCAAAAGATGACTTTTGAAGTACTT | GCTTCCCTGCCACGAGCT | 108 | 272-379 |
| C. sputorum subsp. sputorum ATCC 33562 | AGCTTTACTTGCTGCAAGAGGAAGA | AGGAAGCGTTCCAACAGAAAAGTT | 94 | 350-443 |
| C. helveticus ATCC 51210 | CAATAACATACGCACACCAGATGGA | CAGGCACTTTAACGCTCACTATGG | 176 | 38-213 |
| C. upsaliensis ATCC 49816 | GCTTACGCGTGTAATTACAAACTATGTC | AATTGCCTTAGCCTCGATAGGG | 250 | 92-341 |
| C. lari ATCC 35221 | CTATGTTCGTCCTATAGTTTCTAAGGCTTC | CCAGCACTATCACCCTCAACTAAATAA | 261 | 257-517 |
FIG. 2.
Species-specific identification of Campylobacter species. Each lane represents the results of PCR assays using one set of primers and DNA from each of the following 12 Campylobacter spp.: C. jejuni primers (lane 1), C. coli (lane 2), C. concisus (lane 3), C. curvus (lane 4), C. showae (lane 5), C. mucosalis (lane 6), C. fetus subsp. fetus (lane 7), C. hyointestinalis (lane 8), C. sputorum (lane 9), C. helveticus (lane 10), C. upsaliensis (lane 11), and C. lari (lane 12). Lane M, 25-bp molecular size markers.
To determine if gyrB gene sequences were sufficiently unique to distinguish the Campylobacter species of public health significance, we sequenced a 1,020-bp region of the gyrB gene of 12 Campylobacter species and demonstrated that PCR-RFLP and direct PCR analyses with species-specific primer sets unambiguously distinguished the 12 species. DNA sequence analyses showed that the resultant PCR-RFLP and PCR assays were more discriminating for Campylobacter species identification than similar analyses based on the 16S rRNA gene. In addition to accelerating the identification of currently recognized species, gyrB gene sequence information will facilitate taxonomic studies of novel Campylobacter species. As new species of Campylobacter emerge, their gyrB gene can be sequenced, and high-fidelity PCR primers can be designed for the new taxa.
Nucleotide sequence accession numbers.
The gyrB gene sequences determined in this study and accession numbers have been deposited in the DDBJ (DNA Data Bank of Japan) nucleotide sequence database (see Table S1 in the supplemental material).
Supplementary Material
Acknowledgments
We thank Yanhong Liu, Lori Bagi, and Terence Strobaugh (USDA, ARS, Eastern Regional Research Center) and Hiroko Fukuda and Kumi Arai (National Food Research Institute, Japan) for technical assistance and helpful discussions. We acknowledge John Cherry (USDA, ARS, Eastern Regional Research Center) for facilitating this collaboration.
This work was supported by funding through the Integrated Research Program for Functionality and Safety of Food Toward an Establishment of a Healthy Diet from the Ministry of Agriculture, Forestry, and Fisheries of Japan to Susumu Kawasaki and Shinichi Kawamoto. This work was also supported by a fellowship to Pina Fratamico in 2004 from the Organization for Economic Co-operation and Development, Directorate for Food, Agriculture, and Fisheries, contract no. JA00024150.
Footnotes
Published ahead of print on 22 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Broczyk, A., S. Thompson, D. Smith, and H. Lior. 1987. Water-borne outbreak of Campylobacter laridis-associated gastroenteritis. Lancet i:164-165. [DOI] [PubMed] [Google Scholar]
- 2.Damborg, P., K. E. P. Olsen, E. Møller Nielsen, and L. Guardabassi. 2004. Occurrence of Campylobacter jejuni in pets living with human patients infected with C. jejuni. J. Clin. Microbiol. 42:1363-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foster, G., B. Holmes, A. G. Steigerwalt, P. A. Lawson, P. Thorne, D. E. Byrer, H. M. Ross, J. Xerry, P. M. Thompson, and M. D. Collins. 2004. Campylobacter insulaenigrae sp. nov., isolated from marine mammals. Int. J. Syst. Evol. Microbiol. 54:2369-2373. [DOI] [PubMed] [Google Scholar]
- 4.Goossens, H., B. A. J. Giesendorf, P. Vandamme, L. Vlaes, C. van den Borre, A. Koeken, W. G. Quint, W. Blomme, P. Hanicq, D. S. Koster, H. Hofstra, J.-P. Butzler, and J. van der Plas. 1995. Investigation of an outbreak of Campylobacter upsaliensis in day care centers in Brussels: analysis of relationships among isolates by phenotypic and genotypic typing methods. J. Infect. Dis. 172:1298-1305. [DOI] [PubMed] [Google Scholar]
- 5.Gorkiewicz, G., G. Feierl, C. Schober, F. Dieber, J. Köfer, R. Zechner, and E. L. Zechner. 2003. Species-specific identification of campylobacters by partial 16S rRNA gene sequencing. J. Clin. Microbiol. 41:2537-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobs-Reitsma, W. 2000. Campylobacter in the food supply, p. 467-481. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 7.Kärenlampi, R. I., T. P. Tolvanen, and M.-L. Hänninen. 2004. Phylogenetic analysis and PCR-restriction fragment length polymorphism identification of Campylobacter species based on partial groEL gene sequences. J. Clin. Microbiol. 42:5731-5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein, B. S., J. M. Vergeront, M. J. Blaser, P. Edmonds, D. J. Brenner, D. Janssen, and J. P. Davies. 1986. Campylobacter infection associated with raw milk. An outbreak of gastroenteritis due to Campylobacter jejuni and thermotolerant Campylobacter fetus subsp. fetus. JAMA 255:361-364. [DOI] [PubMed] [Google Scholar]
- 9.Korczak, B. M., R. Stieber, S. Emler, A. P. Burnens, J. Frey, and P. Kuhnert. 2006. Genetic relatedness within the genus Campylobacter inferred from the rpoB sequences. Int. J. Syst. Evol. Microbiol. 56:937-945. [DOI] [PubMed] [Google Scholar]
- 10.Lastovica, A. J., and M. B. Skirrow. 2000. Clinical significance of Campylobacter and related species other than Campylobacter jejuni and C. coli, p. 89-120. In I. Nachamkin and M. J. Blaser, Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 11.Manning, G., C. G. Dowson, M. C. Bagnall, I. H. Ahmed, M. West, and D. C. Newell. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.On, S. L. W. 2001. Taxonomy of Campylobacter, Arcobacter, Helicobacter and related bacteria: current status, future prospects and immediate concerns. J. Appl. Microbiol. 90:1S-15S. [DOI] [PubMed] [Google Scholar]
- 13.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 14.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkateswaran, K., N. Dohmoto, and S. Harayama. 1998. Cloning and nucleotide sequence of the gyrB gene of Vibrio parahaemolyticus and its application in detection of this pathogen in shrimp. Appl. Environ. Microbiol. 64:681-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wesley, I. V., and J. H. Bryner. 1989. Antigenic and restriction enzyme analysis of isolates of Campylobacter fetus subsp. venerealis recovered from persistently infected cattle. Am. J. Vet. Res. 50:807-813. [PubMed] [Google Scholar]
- 17.Wesley, I. V., R. D. Wesley, M. Cardella, F. E. Dewhirst, and B. J. Paster. 1991. Oligodeoxynucleotide probes for Campylobacter fetus and Campylobacter hyointestinalis based on 16S rRNA sequences. J. Clin. Microbiol. 29:1812-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada, S., E. Ohashi, N. Agata, and K. Venkateswaran. 1999. Cloning and nucleotide sequence analysis of gyrB of Bacillus cereus, B. thuringiensis, B. mycoides, and B. anthracis and their application to the detection of B. cereus in rice. Appl. Environ. Microbiol. 65:1483-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto, S., and S. Harayama. 1995. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl. Environ. Microbiol. 61:1104-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.