Abstract
The survival of Salmonella enterica was recently shown to increase when the bacteria were sequestered in expelled food vacuoles (vesicles) of Tetrahymena. Because fresh produce is increasingly linked to outbreaks of enteric illness, the present investigation aimed to determine the prevalence of protozoa on spinach and lettuce and to examine their interactions with S. enterica, Escherichia coli O157:H7, and Listeria monocytogenes. Glaucoma sp., Colpoda steinii, and Acanthamoeba palestinensis were cultured from store-bought spinach and lettuce and used in our study. A strain of Tetrahymena pyriformis previously isolated from spinach and a soil-borne Tetrahymena sp. were also used. Washed protozoa were allowed to graze on green fluorescent protein- or red fluorescent protein-labeled enteric pathogens. Significant differences in interactions among the various protist-enteric pathogen combinations were observed. Vesicles were produced by Glaucoma with all of the bacterial strains, although L. monocytogenes resulted in the smallest number per ciliate. Vesicle production was observed also during grazing of Tetrahymena on E. coli O157:H7 and S. enterica but not during grazing on L. monocytogenes, in vitro and on leaves. All vesicles contained intact fluorescing bacteria. In contrast, C. steinii and the amoeba did not produce vesicles from any of the enteric pathogens, nor were pathogens trapped within their cysts. Studies of the fate of E. coli O157:H7 in expelled vesicles revealed that by 4 h after addition of spinach extract, the bacteria multiplied and escaped the vesicles. The presence of protozoa on leafy vegetables and their sequestration of enteric bacteria in vesicles indicate that they may play an important role in the ecology of human pathogens on produce.
Outbreaks of food-borne illnesses caused by Escherichia coli O157:H7 and Salmonella enterica have recently received national attention. In less than 4 months in the fall of 2006, two outbreaks sickened nearly 400 people in at least 33 states, killing at least 3 people (11, 14). The outbreaks were traced to contaminated fresh spinach (E. coli) and tomatoes (Salmonella). Prior to these recent outbreaks, there have been at least 16 outbreaks of illness caused by E. coli O157:H7 associated with spinach or lettuce since 1995 (35). The Centers for Disease Control and Prevention recently compiled data that revealed that fresh produce was the most important vehicle of food-borne illness in 2005 (T. Ayers, personal communication). One of the recurrent and most crucial questions that emerge from these outbreaks is how these human pathogens survived the harsh environmental conditions on produce in the field and the sanitizer treatments during processing. Although localization in hidden microsites on produce has been suggested as a possible stress avoidance mechanism (7), the role of microbial communities in the persistence of human pathogens on produce has been relatively unexplored.
Protozoa are common members of the natural microflora of plants. Several species of amoebae have been found in association with fresh salad vegetables (31), and the commonly studied ciliated protozoan strain Tetrahymena pyriformis ATCC 30202 was isolated from spinach. The role of protozoa in the protection and survival of the food-borne pathogen S. enterica was studied recently by Brandl et al. (9), who observed enhanced survival of S. enterica in food vacuoles (vesicles) released by a Tetrahymena sp. isolated from moist soil. The vesicles were also shown to protect the bacteria from low concentrations of calcium hypochlorite (9). The objective of the present study was to determine whether protozoa isolated from fresh produce can also expel vesicles or trap pathogens in their cysts when fed food-borne pathogens such as E. coli O157:H7, S. enterica, and L. monocytogenes. In addition, protozoan population sizes were monitored following grazing to determine whether the protozoa could utilize the pathogen as a food source.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli O157:H7 strain MB269 is strain EDL933, an isolate from an outbreak linked to hamburgers (30). It was transformed with plasmid pKT-KAN (1) to impart green fluorescence by expression of the green fluorescent protein (GFP). S. enterica serovar Thompson strains MB108 (8) and MB156 (10) are derived from clinical isolate RM1987, which was isolated in an outbreak linked to cilantro; these strains were transformed with pWM1032 to impart red fluorescence by expression of red fluorescent protein (DsRed) and with pGT-KAN to impart green fluorescence via GFP. L. monocytogenes strain 2387, an isolate from fresh mint leaves, was transformed with gfp via pNF8 and kindly provided by Lisa Gorski (USDA, ARS, Albany, CA); this strain was described previously (15). All of the above plasmids are stably maintained in their host strains. pKT-KAN and pWM1032 encode kanamycin resistance, pGT-KAN encodes gentamicin resistance, and pNF8 encodes lyncomycin resistance. E. coli O157:H7 MB269 and S. enterica MB108 were grown in nutrient broth containing 40 μg/ml kanamycin, S. enterica MB156 was grown in nutrient broth containing 15 μg/ml gentamicin, and L. monocytogenes RM2387 was grown on Trypticase soy broth with 25 μg/ml lyncomycin. All human pathogens were grown at 37°C. Erwinia chrysanthemi strain AC4150 (12), the plant pathogen that was used in the plant experiments, was grown at 28°C in nutrient broth with 50 μg/ml nalidixic acid.
Prevalence of protozoan groups on supermarket produce.
Spinach and romaine lettuce were purchased from various grocery stores, and the water present on the produce was allowed to drain into the plastic bags provided at the stores. Each sample of lettuce and spinach weighed approximately 500 to 550 g and 250 to 300 g, respectively. Water alone from the misting devices was also collected in separate bags. Aliquots (50 μl) of the drainage from produce and from the misting water were examined directly for the presence and number of various protozoan and metazoan taxa that included ciliates, flagellates, amoebae, and nematodes. Subsamples for enumeration of bacteria and flagellates were diluted and then stained with acridine orange prior to observation via epifluorescence microscopy on 0.22-μm-pore-size Millipore black 25-mm-diameter filters, similar to the methods of Hobbie et al. (17). Other subsamples were examined by phase-contrast microscopy under an inverted microscope. All data were converted to numbers of organisms per milliliter of the drainage from produce.
Isolation of protozoa from fresh produce.
Romaine lettuce heads and bundled spinach, unbagged, were purchased from two supermarkets, placed in plastic bags from the produce section of the stores, and immediately returned to the laboratory for isolation of protozoa. The fresh lettuce or spinach leaves were washed with sterile Tris-buffered saline solution (TBSS; 2 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2, 1 mM Tris [pH 6.8 to 7.2]) while they were still in the closed, clean plastic bags. This was similar to the method followed by Rude et al. (31), in which they rinsed spinach with 300 ml of water in a closed bag. The leaf washings were then transferred aseptically into sterile petri dishes. Autoclaved wheat grains were added to the petri dishes to enrich for protozoa in the produce wash water. Enrichment would permit easier isolation because of higher concentrations of a given organism and possibly reveal the presence of more species by inducing excystment of certain cyst-forming species. The dishes were then incubated at 25°C for 24 to 48 h. After incubation, the suspensions in the dishes were observed under an inverted microscope for the presence of protozoa.
Washed GFP- and DsRed-labeled pathogens were added to the mixed protozoan populations resulting from the enrichment cultures to determine whether the protozoa would ingest the bacteria. All groups of protozoa ingested the bacteria; however, the amoebae and ciliates appeared to ingest more cells more rapidly than the flagellates did; therefore, amoebae and ciliates were targeted for isolation and experimentation.
Isolation of amoebae was done by transferring aliquots from petri dishes onto nonnutrient agar (NNA) plates seeded with nonpathogenic E. coli (ATCC 33153) and incubating the plates at 30°C for 24 h. Amoebae were picked with sterile glass micropipettes from agar plates and tested for the ability to ingest pathogens.
Ciliates were isolated by serial dilution in Cereal Leaves Medium (Sigma, St. Louis, MO) prepared by boiling 1.0 g in distilled water for 5 min and filtering it through Whatman no. 40 filter paper. The filtered broth was then distributed in 125-ml glass flasks and autoclaved for 20 min.
During the study, two species of ciliates and one species of amoeba were isolated from produce. The amoeba species isolated from lettuce was identified as Acanthamoeba palestinensis on the basis of the Illustrated Guide to the Protozoa (25) and a key to the freshwater free-living amoebae (28), which includes the morphology of trophozoites and cysts, as well as growth temperature limits. One ciliate was identified morphologically as Colpoda steinii (from spinach) because of its unique morphological characteristics by using species descriptions by Kudo (24) and in the Illustrated Guide to the Protozoa (25). The other ciliate (from romaine lettuce) was identified molecularly by its rRNA gene sequence as a novel strain of Glaucoma most closely related to Glaucoma scintillans.
For sequencing of the rRNA gene, Glaucoma sp. cells were grown as described below and fixed in ethanol added to a final concentration of 70%. Several hundred alcohol-fixed cells were allowed to dry in the bottom of a 0.5-ml Microfuge tube and then digested in 50 μl of a proteinase K solution (1% Triton X-100, 50 mM Tris-HCl [pH 7.8], 5 mM CaCl2, 100 μg/ml proteinase K) held at 50°C for 2 h. The proteinase K was then inactivated by heating the tube to 95°C for 5 min. An aliquot of 0.5 μl of the digested ciliate solution was then added to a 100-μl PCR mixture containing 5 U of Taq DNA polymerase (USB Corporation), the commercial buffer provided with the enzyme, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, and 0.2 μM each forward (GGGGACAAGTTTGTACAAAAAAGCAGGCTAACCYGGTYGATCCTGCCA) and reverse (GGGGACCACTTTGTACAAGAAAGCTGGGTCYGCAGGTTCACCTAC) primer. The primers contain sequence elements required for cloning with the Gateway system (Invitrogen Corp.), as well as sequence elements complementary to target regions at the 5′ and 3′ ends of eukaryotic small-subunit rRNA coding regions (although the primers contained sequences useful for cloning, the amplification products were sequenced directly). After an initial 2 min of denaturation at 95°C, the material was sent through 35 amplification cycles (95°C for 30 s, 50°C for 1 min, 72°C for 4 min). The amplified DNA was then sent to GenHunter, Inc. (Nashville, TN), and sequenced in both directions with primers complementary to conserved regions in eukaryotic small-subunit rRNAs. The completed sequence was then used to perform a BLAST search of GenBank files to identify similar sequences.
Glaucoma sp. was rendered axenic by being cultured at 25°C in 5 ml of two-thirds-strength Plate Count Broth (PCB; Difco) supplemented with 200 U/ml penicillin and 200 μg/ml streptomycin. After several passages with antibiotics, the ciliates grew well in two-thirds-strength PCB without antibiotics. The other ciliate, C. steinii, could not be rendered axenic and was maintained in Cereal Leaves Medium with nonpathogenic E. coli. The amoeba species was maintained on NNA plates seeded with a lawn of nonpathogenic E. coli.
Other protozoa used for interaction experiments.
In addition to the protozoa isolated from lettuce and spinach in our study, two species of Tetrahymena from other sources were also tested. One was isolated from moist soil by Brandl et al. (9) for their study with S. enterica (strain MB125; formerly SSU); the other species was T. pyriformis (ATCC 30202), originally isolated from spinach. Both ciliate species were maintained axenically in two-thirds-strength PCB at 25°C.
Protozoan and bacterial interaction experiments.
For grazing experiments, 2-week-old cultures of Glaucoma sp. were used because of the slow growth of these ciliates, whereas 3-day-old cultures of C. steinii and 5-day-old cultures of the two Tetrahymena species were used. Ciliates were washed with TBSS by centrifugation at 500 × g and resuspended in TBSS. Amoebae were washed from NNA plates and suspended in TBSS. The concentration of protozoa in stock suspensions was determined by differential interference contrast (DIC) microscopy. For grazing tests, 24-h-old broth cultures of S. enterica MB108, E. coli O157:H7 MB269, and L. monocytogenes RM2387 were washed twice by centrifugation and the final pellet was resuspended in TBSS. The concentration of bacteria in the stock suspension was determined by epifluorescence microscopy.
Cocultures of the protozoa and bacteria were established at a ratio of bacteria to protozoa of 10,000:1. Controls consisted of suspensions of washed protozoa only. Cocultures were incubated at 25°C for 24 h. All coculture experiments with bacteria and protozoa were performed twice with two replicate suspensions.
Vesicle production and change in concentrations of protozoa.
After 24 h, 25 μl was sampled from each replicate coculture and the released vesicles containing pathogens were counted with a hemacytometer under a Nikon Microphot light microscope. A Leica TS confocal laser scanning microscope (CLSM) was used to view the fluorescent bacterial cells in optical sections of vacuoles inside the protozoa and vesicles expelled into the suspension. Concentrations of protozoa were determined also after 24 h by counts under the light microscope with DIC.
Production of vesicles on produce surfaces.
Broth cultures of S. enterica MB156 and E. chrysanthemi AC4150 were washed twice in TBSS and coinoculated onto young potted cilantro plants at 104 cells of each strain per leaf as described previously (8). The cilantro leaves were gently wounded with tweezers prior to inoculation to promote soft-rot production by the plant pathogen and thus increase the S. enterica cell density on the leaves. This helped the visualization of vesicles containing GFP-labeled S. enterica under the CLSM. Twenty-four hours after inoculation with the bacterial strains, the plants were inoculated with cells of Tetrahymena sp. strain MB125 that were grown and washed as described above. During the entire experiment, the plants were incubated at 28°C in a chamber allowing for the presence of free water on the plant surface. Five leaves were sampled from each of three replicate cilantro plants 24 h after inoculation with the protozoa, and leaf discs were mounted in water for observation under the CLSM and detection of vesicles containing S. enterica.
Fate of E. coli O157:H7 in vesicles after addition of spinach extract.
Spinach extract was used to simulate the type of nutrients that protozoa and bacteria on cut or damaged spinach leaves may encounter. The extract was made by grinding leaves of packaged spinach and sterilizing the liquid extract by passage through a 0.22-μm-pore-size filter. The filtrate was diluted 1:4 with sterile distilled water.
Vesicles were produced in cocultures of E. coli O157:H7 MB269 and the T. pyriformis isolate from spinach (ATCC 30202) incubated for 24 h. Spinach extract was added to the coculture, and the ability of the bacteria to grow within the vesicles was assessed by a modified direct viable count method (20) based on the Kogure assay of cell viability (23). In this assay, nalidixic acid, which prevents bacterial cells from dividing, causes the elongation of cells that are actively growing. Nalidixic acid was added to a final concentration of 20 μg/ml of the vesicle suspension in TBSS containing diluted spinach extract. The suspensions of vesicles were then incubated at 37°C and examined at 0, 2, 3, and 4 h for the presence and localization of elongated cells.
Entrapment of food-borne pathogens in cysts of protozoa.
A. palestinensis and C. steinii formed cysts, which were tested for entrapment of food-borne pathogens by adding 100 μl of encystment medium (0.1 M KCl, 0.02 M 2-amino-2-methyl-1,3-propanediol, 0.008 M MgSO4, 0.0004 M CaCl2, [pH 8.8]) to the coculture after 24 h of grazing. The cysts were observed by epifluorescence microscopy and by optical sectioning with the CLSM for the presence of fluorescent bacterial cells trapped inside them.
Data analysis.
To determine whether there were significant differences in the numbers of vesicles produced by each protozoan species when cocultured with each of the three different bacterial species, one-way analysis of variance, along with Tukey's pairwise comparison, was performed with Statistical Analysis Software 9.1 for Windows. This statistical test was also used to determine whether the difference in the concentrations of protozoan cells after feeding on the three pathogens for 24 h was significant. Significant differences were reported at P < 0.05.
RESULTS
Prevalence of protozoa on grocery store produce.
Examination of water drained from produce revealed that the composition of the protist flora was heterogeneous among all of the samples of produce purchased at the store (Table 1). The number of individuals within each group of protists also varied greatly between samples and stores. Spinach appeared to have the highest diversity and numbers of microorganisms. Flagellates were the most prevalent in all of the samples, ranging approximately from 1 × 105 to 8 × 106/ml. Some samples contained more than 100 ciliates/ml of drained water, which could account for as much as 3,500 ciliates, in total, per spinach bundle. Although we detected only flagellates in the subsamples of drained water among all of the six lettuce heads examined, various types of protists were present in the enrichment cultures from leaf washings. No protozoa were present in water from the misting devices.
TABLE 1.
Concentration of microorganisms in a subsample of water drained from one head of lettuce or one bundle of spinacha
| Product and sample no. | No. of cells/ml of drained water
|
||||
|---|---|---|---|---|---|
| Bacteria | Ciliates | Flagellates | Amoebae | Nematodes | |
| Spinach | |||||
| 1 | 1.5 × 108 | —b | 5.0 × 105 | — | — |
| 2 | 2.2 × 109 | — | 3.0 × 106 | — | — |
| 3 | 4.2 × 108 | 1.0 × 102 | 8.2 × 106 | 3.2 × 101 | — |
| 4 | 5.9 × 108 | 1.6 × 102 | 3.6 × 106 | 2.7 × 101 | 5.0 × 101 |
| 5 | 1.6 × 107 | — | 7.4 × 105 | — | — |
| 6 | 4.9 × 107 | — | 2.6 × 105 | — | — |
| 7 | 5.2 × 107 | — | 2.6 × 105 | — | — |
| 8 | 8.6 × 107 | — | — | — | — |
| Romaine lettuce | |||||
| 1 | 12.0 × 106 | — | — | — | — |
| 2 | 2.8 × 107 | — | 1.5 × 106 | — | — |
| 3 | 9.5 × 106 | — | 1.8 × 105 | — | — |
| 4 | 4.0 × 106 | — | 1.1 × 105 | — | — |
| 5 | 1.0 × 107 | — | 3.7 × 105 | — | — |
| 6 | 1.6 × 106 | — | — | — | — |
Samples of 1.5 to 35 ml were tested. Samples were purchased on different days.
—, none observed in two subsamples.
Organisms isolated from romaine lettuce and spinach.
Two species of ciliates were isolated from leafy produce in the present study and were identified as Glaucoma sp. (romaine lettuce) and C. steinii (spinach) (Fig. 1). Both are bacterivorous and ingested the GFP- and DsRed-labeled enteric pathogens in our initial screen. An amoeba strain, identified as A. palestinensis, was isolated from lettuce. Other types of protozoa, including flagellates, were detected in the enrichment cultures but were not isolated either because they did not ingest high numbers of bacteria or because they were not easily culturable.
FIG. 1.
DIC images. (Left) Glaucoma sp., with oral aperture evident at right; (right) C. steinii, focused on a section through the center of the cell.
Vesicle production and change in the number of protozoa.
The release of vesicles containing viable pathogens was observed with Glaucoma sp. and the two Tetrahymena species. C. steinii and the amoeba species did not expel vesicles with the pathogens. No vesicles were observed in the unfed controls for any protozoan species. After 24 h of feeding, Glaucoma sp. released vesicles with all three pathogens; however, the protozoan produced the fewest vesicles with L. monocytogenes. The vesicles contained intact fluorescent pathogens. Figure 2 shows the vacuoles inside a Glaucoma sp. cell that contain GFP-labeled E. coli O157:H7 cells, and Fig. 3 shows vesicles released by the same ciliate and also containing the pathogen. The number of vesicles produced per cell by Glaucoma sp. after grazing on E. coli O157:H7, S. enterica, and L. monocytogenes ranged from 88 to 220, 81 to 124, and 12 to 28, respectively. The average number of vesicles produced by Glaucoma sp. grazing on L. monocytogenes was significantly lower than the number produced by this ciliate grazing on E. coli O157:H7 or S. enterica (P < 0.05) (Fig. 4). There was no difference in the number of vesicles produced by Glaucoma sp. grazing on E. coli O157:H7 compared with grazing on S. enterica.
FIG. 2.
Optical section through Glaucoma sp. containing GFP-labeled E. coli O157:H7 cells. (Left) DIC image; (right) corresponding fluorescent confocal image showing food vacuoles containing intact bacteria.
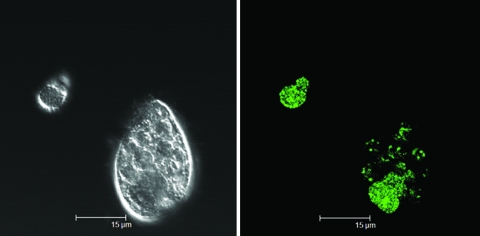
FIG. 3.
(Left) DIC image of a cluster of vesicles released from Glaucoma sp. that fed on GFP-labeled E. coli O157:H7. (Right) Corresponding confocal image of the same cluster showing green-fluorescing E. coli O157:H7 within vesicles.
FIG. 4.
Comparison of the numbers of vesicles produced by Glaucoma sp. after feeding on three pathogens at 25°C.
T. pyriformis also expelled vesicles with E. coli O157:H7 and S. enterica but not with L. monocytogenes. Observation of aliquots of the cocultures of T. pyriformis and L. monocytogenes failed to reveal the presence of such vesicles by epifluorescence, as well as DIC, microscopy. The number of vesicles expelled per T. pyriformis cell ranged from 34 to 79 with E. coli O157:H7 and 83 to 158 with S. enterica. A significant difference was observed between the numbers of vesicles produced by this ciliate with the two latter pathogens (P < 0.05) (Fig. 5).
FIG. 5.
Comparison of the numbers of vesicles produced by T. pyriformis after feeding on three pathogens at 25°C.
The other ciliate, a Tetrahymena sp. isolated from a field in California, also made vesicles containing E. coli O157:H7 cells (Fig. 6). This ciliate had previously been shown to produce vesicles with S. enterica but rarely with L. monocytogenes (9); therefore, these pathogens were not tested with this Tetrahymena sp. in the present study. The number of vesicles released by this ciliate when grazing on E. coli O157:H7 ranged from 30 to 150 per protist cell.
FIG. 6.
Cell of Tetrahymena strain MB1125 containing E. coli O157:H7 and an expelled vesicle outside the cell. (Left) DIC image; (right) fluorescent confocal image.
Protozoa increased in numbers after they fed on pathogens for 24 h. T. pyriformis, the soil-borne Tetrahymena sp., and C. steinii increased to a greater extent than did Glaucoma sp. or the amoebae. Because of differences in the growth rates of the different species, statistical comparisons of increases were not appropriate. The average increase in each protozoan species is shown in Table 2. No change in protist cell concentration was observed in the control in which Glaucoma sp. cells were cultured alone. Control suspensions of C. steinii and A. palestinensis decreased slightly, whereas the control suspensions of both Tetrahymena species increased slightly.
TABLE 2.
Growth of protozoa in cocultures with food-borne pathogenic bacteria and changes in numbers of cells over 24 h
| Protozoan | Food-borne pathogen | No. of protozoan cells/ml
|
% Increase in 24 h | |
|---|---|---|---|---|
| Initial | Final | |||
| G. scintillans | E. coli O157:H7 | 2.5 × 103 | 8.6 × 103 | 244 |
| S. enterica | 2.6 × 103 | 5.5 × 103 | 111 | |
| Listeria | 2.0 × 103 | 4.4 × 103 | 120 | |
| Control | 2.5 × 103 | 2.5 × 103 | 0 | |
| C. steinii | E. coli O157:H7 | 4.3 × 103 | 3.0 × 104 | 579 |
| S. enterica | 4.3 × 103 | 3.6 × 104 | 837 | |
| Listeria | 4.3 × 103 | 3.5 × 104 | 713 | |
| Control | 4.3 × 103 | 5.7 × 102 | −86 | |
| A. palestinensis | E. coli O157:H7 | 4.14 × 103 | 4.2 × 104 | 914 |
| S. enterica | 4.14 × 103 | 4.6 × 104 | 1,011 | |
| Listeria | 4.14 × 103 | 5.2 × 104 | 1,156 | |
| Control | 4.14 × 103 | 2.0 ×102 | −95 | |
| T. pyriformis | E. coli O157:H7 | 2.9 × 103 | 2.7 × 104 | 1,025 |
| S. enterica | 2.9 × 103 | 3.7 × 104 | 1,175 | |
| Listeria | 2.9 × 103 | 5.9 × 104 | 1,934 | |
| Control | 2.9 × 103 | 3.2 × 103 | 10 | |
| Tetrahymena sp. strain MB125 | E. coli O157:H7 | 4.9 × 103 | 3.9 × 104 | 1,256 |
| Control | 4.9 × 103 | 6.0 × 103 | 22 | |
Fate of E. coli O157:H7 in vesicles.
E. coli O157:H7 cells increased four to five times in length 3 h after the addition of spinach extract and nalidixic acid to vesicle suspensions, and many cells appeared to be free outside the vesicles. By 4 h, only a few intact vesicles were visible in the samples and most bacterial cells were free in the suspension. Empty vesicle material appeared to be left in the samples. Figure 7 shows vesicles with large E. coli O157:H7 cells, some of which appear to be exiting a vesicle.
FIG. 7.
(Left) DIC image of E. coli O157:H7 in an expelled vesicle of T. pyriformis without added nutrients; (right) E. coli O157:H7 in vesicles after 4 h of incubation with spinach extract and nalidixic acid. Note the elongated cells exiting the large vesicle.
Entrapment in cysts.
Cyst forms were observed with C. steinii and A. palestinensis. The cysts started appearing within 24 h after the addition of encystment medium. Approximately 50 cysts of C. steinii were observed for the presence of entrapped pathogens, but no viable fluorescent pathogen was observed inside any cysts. Likewise, no viable pathogens were observed inside cysts of the amoeba.
Vesicle production on plants.
Confocal microscopy of cilantro leaves revealed the presence of vesicles containing S. enterica cells (Fig. 8). The vesicles were located mostly in regions of the leaves where plant damage or rot was visible. These sites were also heavily colonized by the plant and human pathogens and by Tetrahymena. The detectable presence of vesicles was variable from leaf to leaf, as several leaves from different plants did not appear to have detectable vesicles despite large populations of the human pathogen and the protist.
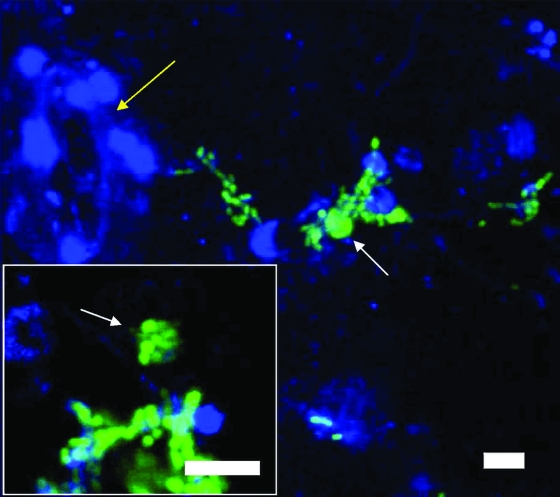
FIG. 8.
Confocal micrographs of vesicles (white arrows) containing cells of GFP-labeled S. enterica MB156. The vesicles were expelled by Tetrahymena sp. during grazing on the leaves of cilantro plants inoculated with the enteric pathogen and 24 h later with the protist. Note the presence of distinct green fluorescent bacterial cells in the vesicle shown in the insert. The autofluorescence of the plant tissue was assigned the pseudocolor blue. The yellow arrow indicates the presence of a stomate. Scale bars, 10 μm.
DISCUSSION
Protozoa that were detected in the water drained from spinach bundles purchased at the supermarket included flagellates, amoebae, and ciliates. The romaine lettuce samples examined in this study did not harbor a great diversity of protozoa, since only flagellates were detected directly from the leaf washings without enrichment. This apparent lack of diversity may be explained by the fact that the outermost leaves are trimmed off the lettuce heads at harvest, thus leaving the most-colonized leaves in the field. However, the protozoan populations resulting from enrichment procedures yielded more species, including ciliates and amoebae. This indicates that some ciliates may have been encysted and others may have been present in numbers below our detection level for direct enumeration in the aliquots of drained water. Because amoebae tend to adhere strongly to surfaces, they were most likely underrepresented in the water drained from the produce.
Flagellates appeared to be present in high numbers in all of the samples. These small protists ingest only a few bacteria at a time, and their role in the survival of bacteria on plant surfaces remains to be investigated. While flagellates predominated on spinach, a significant number of ciliates and amoebae were additionally detected in some samples. Although our detection method did not enable us to recover all of the protozoa present on the produce samples that were tested, it is clear that the distribution of types of protozoa among produce samples is heterogeneous.
In a separate experiment aimed at isolating protozoa from produce and investigating their interaction with human pathogens, we identified two ciliated protozoa associated with bundled spinach and whole lettuce, namely, C. steinii and Glaucoma sp., respectively. In addition, a strain of A. palestinensis was isolated from a head of romaine lettuce. We used also in these studies a strain of T. pyriformis from the ATCC collection which was isolated from spinach and Tetrahymena sp. strain MB125 (in cocultures with E. coli O157:H7 only), as they allowed us to comparatively test a wider range of protozoan species.
Of these five protozoan species, three expelled vesicles filled with intact E. coli O157:H7 and S. enterica. Only Glaucoma sp. produced vesicles while grazing on L. monocytogenes, but they were very few. In a previous study by Brandl et al. (9), Tetrahymena sp. strain MB125 also rarely expelled L. monocytogenes-containing vesicles, compared to the abundance of vesicles produced during grazing on S. enterica and E. coli O157:H7. The authors suggested that this difference may be due to the ability of S. enterica to arrest digestion in the Tetrahymena vacuole. According to another report, Listeria may escape from the food vacuole and replicate in the cytoplasm of the protozoa (29). However, L. monocytogenes replication in the cytoplasm was not apparent under the CLSM in the previous (9) and present studies.
E. coli O157:H7 and S. enterica resisted digestion by Glaucoma sp. and T. pyriformis, as was shown with other protozoan species (2, 9). Although C. steinii and A. palestinensis ingested all three pathogens readily, none of them extensively survived digestion by these protozoa, neither by being expelled in vesicles nor by being within their cysts. Cysts have been shown to be a potential source of pathogens (21, 32), and it is possible that protozoan species other than the two tested in this study could trap enteric pathogens in their cysts. The lack of vesicle production in cocultures of C. steinii and A. palestinensis with any of the three pathogens tested, and also in cocultures of Tetrahymena sp. strain MB125 and Glaucoma sp. with L. monocytogenes, suggests a certain degree of specificity in the interaction between protozoa and enteric pathogens.
The degree of population increase varied with each protozoan species fed a given bacterial pathogen. The greatest increase in protozoan concentration in cocultures was observed with C. steinii and T. pyriformis, whereas Glaucoma sp. and the amoeba species increased only slightly with the pathogens. An increase was observed also with Tetrahymena sp. strain MB125. These differences may be caused in part by different rates of feeding when bacterial cells are the food source, as previously observed for various protozoa (27) and by Taylor and Berger (33). Biomass differences among the protozoa may be a factor affecting their various growth rates. Protozoa that released undigested bacteria in vesicles nevertheless derived nutrients from some of the bacterial cells in their vacuoles or from bacterial by-products in the coculture.
It remains unclear whether the bacterial cells are sequestered in the vesicles by a membrane or if some material holds the cells together. However, expelled vesicles appear to protect bacterial pathogens from harsh conditions, as shown by Berk et al. (5) and Brandl et al. (9). Moreover, the vesicles released by Tetrahymena cocultured with Legionella pneumophila resisted desiccation (3) and UV light (4). A recent study of L. pneumophila cocultured with amoebae showed that the long-term survival of the legionellae may have been due to their presence in released vesicles (6).
On the other hand, protozoa that host pathogens might simply enhance the pathogens' survival by physically protecting the undigested bacteria inside their cells from disinfectants such as chlorine, as was observed by King et al. (22) for protozoa feeding on coliforms. Furthermore, bacteria such as E. coli O157:H7 (2) and S. enterica (34) are able to replicate inside the food vacuoles of protozoa. Similar results were reported with Pasteurella multocida (19), Mycobacterium avium (13), and L. pneumophila (18). This implies a complex role for protozoa in the ecology of human pathogens.
Observations by confocal microscopy revealed that vesicles containing S. enterica were produced by Tetrahymena on wet leaf surfaces. The GFP-labeled S. enterica cells were brightly fluorescent, indicating that they were most likely viable cells (9). Some of the observed vesicles contained more than 20 cells (Fig. 3). This number is sufficiently high to suggest that at least a portion of the cells in such aggregates would be shielded from various physical and chemical stresses that prevail on the plant surface (16), as well as from subsequent grazing by other protozoan cells. We have previously provided evidence that S. enterica cells located in Tetrahymena vesicles were more resistant to low chlorine concentrations than cells remaining free in suspension (9). Thus, vesicles may protect human pathogens on contaminated leaves from the sanitizers used during fresh produce processing.
It is clear, however, that such vesicles would only be produced when the surface of produce is wet in order to enable Tetrahymena to graze by filter feeding on bacteria that are free in the water film on the plant surface. These conditions may be met in the preharvest environment during dew, rain, or overhead irrigation and in the postharvest environment when water is misted onto produce at the market and remains in the bagged product. It may be amplified when soft rot is present and induces higher populations of the human pathogen, in addition to creating an aqueous environment for grazing by protozoa, as was demonstrated in this study.
We observed in our study that many E. coli O157:H7 cells in vesicles were not only intact and brightly fluorescing but also able to grow and escape the vesicles upon addition of spinach extract, as determined by the modified Kogure protocol for bacterial viability (23). Manasherob et al. (26) demonstrated that spores of Bacillus thuringiensis can germinate, grow, and sporulate in vesicles of T. pyriformis, as well as exit the vesicles.
The present study revealed that certain protozoa isolated directly from produce can sequester S. enterica and E. coli O157:H7 in expelled vesicles. It also provides the first evidence that viable human pathogens can multiply in, and exit from, these protozoan vesicles. Most important for the microbial safety of fresh produce is the demonstration that the process of vesicle formation and expulsion can occur directly on leaves of wet produce. Thus, protozoa may interact with enteric pathogens on produce surfaces in a manner that may have significant implications for food safety and public health.
Acknowledgments
This work was supported in part by USDA grant 2005-35201-15323 (to S.G.B.); by the Center for the Management, Utilization, and Protection of Water Resources at Tennessee Technological University; and by U.S. Department of Agriculture Agricultural Research Service CRIS project 201-5325-210-44.
We thank Lisa Gorski of the USDA/ARS, Albany, CA, for the gift of L. monocytogenes RM2387.
Footnotes
Published ahead of print on 29 February 2008.
REFERENCES
- 1.Barak, J. D., L. C. Whitehand, and A. O. Charkowski. 2002. Differences in attachment of Salmonella enterica serovars and Escherichia coli O157:H7 to alfalfa sprouts. Appl. Environ. Microbiol. 68:4758-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker, J., T. J. Humphrey, and M. W. R. Brown. 1999. Survival of Escherichia coli O157:H7 in a soil protozoan: implications for disease. FEMS Microbiol. Lett. 173:291-295. [DOI] [PubMed] [Google Scholar]
- 3.Berk, S. G., J. S. Thomas, and R. S. Ting. 2003. Protozoan phagosomes enhance survival of Legionella pneumophila in a low-humidity environment, abstr. Q-195, p. 549. Abstr. 103rd Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 4.Berk, S. G., K. S. Redding, J. S. Thomas, and E. L. Williams. 2004. Ciliate phagosomes enhance survival of Legionella pneumophila exposed to UV irradiation, abstr. Q-057, p. 82. Abstr. 104th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 5.Berk, S. G., R. S. Ting, G. W. Turner, and R. J. Ashburn. 1998. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl. Environ. Microbiol. 64:279-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouyer, S., C. Imbert, M. H. Rodier, and Y. Hechard. 2007. Long-term survival of Legionella pneumophila associated with Acanthamoeba castellanii vesicles. Environ. Microbiol. 9:1341-1344. [DOI] [PubMed] [Google Scholar]
- 7.Brandl, M. T. 2006. Human pathogens and the health threat of the phyllosphere, p. 269-285. In M. J. Bailey, A. K. Lilley, T. M. Timms-Wilson, and P. T. N. Spencer-Philips (ed.), Microbial ecology of aerial plant surfaces. CABI Publishing, Wallingford, United Kingdom.
- 8.Brandl, M. T., and R. E. Mandrell. 2002. Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl. Environ. Microbiol. 68:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandl, M. T., B. M. Rosenthal, A. F. Haxo, and S. G. Berk. 2005. Enhanced survival of Salmonella enterica in vesicles released by a soil-borne Tetrahymena species. Appl. Environ. Microbiol. 71:1562-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandl, M. T., W. G. Miller, A. H. Bates, and R. E. Mandrell. 2005. Production of autoinducer 2 in Salmonella enterica serovar Thompson contributes to its fitness in chickens but not on cilantro leaf surfaces. Appl. Environ. Microbiol. 71:2653-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2007. Multistate outbreaks of Salmonella infections associated with raw tomatoes eaten in restaurants—United States, 2005-2006. Morb. Mortal. Wkly. Rep. 56:909-911. http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5635a3.htm. [PubMed] [Google Scholar]
- 12.Chatterjee, A. K., K. K. Thurn, and D. A. Feese. 1983. Tn5-induced mutations in the enterobacterial phytopathogen Erwinia chrysanthemi. Appl. Environ. Microbiol. 45:644-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirillo, J. D., S. Falkow, L. S. Tompkins, and L. E. Bermudez. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Food and Drug Administration. 2007. FDA finalizes report on 2006 spinach outbreak. Food and Drug Administration, Washington, DC. http://www.fda.gov/bbs/topics/NEWS/2007/NEW01593.html.
- 15.Gorski, L., J. D. Palumbo, and K. D. Nguyen. 2004. Strain-specific differences in the attachment of Listeria monocytogenes to alfalfa sprouts. J. Food Prot. 67:2488-2495. [DOI] [PubMed] [Google Scholar]
- 16.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobbie, J. E., R J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holden, E. P., H. H. Winkler, D. O. Wood, and E. D. Leinbach. 1984. Intracellular growth of Legionella pneumophila within Acanthamoeba castellanii Neff. Infect. Immun. 45:18-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hundt, M. J., and C. G. Ruffolo. 2005. Interaction of Pasteurella multocida with free-living amoebae. Appl. Environ. Microbiol. 71:5458-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joux, F., and P. Lebaron. 1997. Ecological implication of an improved direct viable count method for aquatic bacteria. Appl. Environ. Microbiol. 63:3643-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilvington, S., and J. Price. 1990. Survival of L. pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure. J. Appl. Bacteriol. 68:519-525. [DOI] [PubMed] [Google Scholar]
- 22.King, C. H., E. B. Shotts, Jr., R. E. Wooley, and K. G. Porter. 1988. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl. Environ. Microbiol. 54:3023-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kogure, K., U. Simidu, and N. Taga. 1984. An improved direct viable count method for aquatic for aquatic bacteria. Arch. Hydrobiol. 102:117-122. [Google Scholar]
- 24.Kudo, R. R. 1966. Protozoology, fifth ed., p. 869-870. Charles C Thomas, Publisher, Springfield, IL.
- 25.Lee, J. J., H. Hunter, and E. C. Bovee. 1985. Illustrated guide to the protozoa. Society of Protozoologists, Lawrence, KS.
- 26.Manasherob, R., E. Ben-Dov, A. Zaritsky, and Z. E Barak. 1998. Germination, growth, and sporulation of Bacillus thuringiensis subsp. israelensis in excreted food vacuoles of the protozoan Tetrahymena pyriformis. Appl. Environ. Microbiol. 64:1750-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell, G. C., J. H. Baker, and H. A. Sleigh. 1998. Feeding of freshwater flagellate Bodo saltans, on diverse bacteria. J. Protozool. 35:219-222. [Google Scholar]
- 28.Page, F. C. 1988. A new key to freshwater and soil gymnamoebae. Freshwater Biological Association, Ambleside, Cumbria, United Kingdom.
- 29.Parry, J. D. February 2005. Sugar-coated bacteria: wolves in sheep's clothing? Microbiol. Today 2005:18-21. http://www.socgenmicrobiol.org.uk/pubs/micro_today/pdf/020504.pdf. [Google Scholar]
- 30.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 31.Rude, R. A., G. J. Jackson, J. W. Bier, T. K. Sawyer, and N. G. Risty. 1984. Survey of fresh vegetables for nematodes, amoebae, and Salmonella. J. Assoc. Off. Anal. Chem. 67:613-615. [PubMed] [Google Scholar]
- 32.Steinert, M., K. Birkness, E. White, B. Fields, and F. Quinn. 1998. Mycobacterium avium bacilli grown saprozoically in coculture with Acanthamoeba polyphaga and survive within cysts walls. Appl. Environ. Microbiol. 64:2256-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor, W. D., and J. Berger. 1976. Growth responses of cohabiting ciliate protozoa to various prey bacteria. Can. J. Zool. 54:1111-1114. [Google Scholar]
- 34.Tezcan-Merdol, D., M. Ljungström, J. Winiecka-Krusnell, E. Linder, L. Engstrand, and M. Rhen. 2004. Uptake and replication of Salmonella enterica in Acanthamoeba rhysodes. Appl. Environ. Microbiol. 70:3706-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.U.S. Department of Agriculture/Agricultural Research Service. 2006. USDA funds research on prevention and control of E. coli O157:H7 in fresh produce. U.S. Department of Agriculture/Agricultural Research Service, Washington, DC. http://www.ars.usda.gov/News/News.htm?modecode=53-25-21-00.