Abstract
The goal of this study was to determine the impact of physiological growth states (batch exponential and batch stationary growth) and growth modes (substrate-limited chemostat, substrate-sufficient exponential batch, and substrate-depleted stationary batch growth) on several measures of growth and responses to Cd(II)-mediated inhibition of Nitrosomonas europaea strain 19718. The specific oxygen uptake rate (sOUR) was the most sensitive indicator of inhibition among the different responses analyzed, including total cell abundance, membrane integrity, intracellular 16S rRNA/DNA ratio, and amoA expression. This observation remained true irrespective of the physiological state, the growth mode, or the mode of Cd(II) exposure. Based on the sOUR, a strong time-dependent exacerbation of inhibition (in terms of an inhibition coefficient [Ki]) in exponential batch cultures was observed. Long-term inhibition levels (based on Ki estimates) in metabolically active chemostat and exponential batch cultures were also especially severe and comparable. In contrast, the inhibition level in stationary-phase cultures was 10-fold lower and invariable with exposure time. Different strategies for surviving substrate limitation (a 10-fold increase in amoA expression) and starvation (the retention of 16S rRNA levels) in N. europaea cultures were observed. amoA expression was most negatively impacted by Cd(II) exposure in the chemostat cultures, was less impacted in exponential batch cultures, and was least impacted in stationary batch cultures. Although the amoA response was consistent with that of the sOUR, the amoA response was not as strong. The intracellular 16S rRNA/DNA ratio, as determined by fluorescence in situ hybridization, also did not uniformly correlate with the sOUR under conditions of inhibition or no inhibition. Finally, Cd(II)-mediated inhibition of N. europaea was attributed partially to oxidative stress.
Nitrification is often the rate-limiting step in engineered biological nitrogen removal systems. In addition to having inherently low specific growth rates and biomass yields, nitrifying bacteria are typically more sensitive to physical, chemical, and environmental perturbations than heterotrophic bacteria (22, 32, 33, 49). Therefore, it is important to understand and quantify the effects of such perturbations on nitrification. While the responses of nitrifying bacteria to inhibitors such as heavy metals have been widely quantified using different measurements, the reported inhibition coefficients vary by over an order of magnitude (2-4, 6, 18, 24, 28, 29, 39, 49). In this study, the factors that result in such variability in measured inhibition constants (Ki) were systematically examined using Nitrosomonas europaea 19718 as a model ammonia-oxidizing bacterium and Cd(II) as a model inhibitor. Recent evidence suggests that the degree of starvation significantly and negatively correlates with the expression of ammonia monooxygenase and ammonia oxidation activity in batch cultures of N. europaea (5, 52). Given this parallel link between activity measures of ammonia oxidation and substrate availability in different phases of batch growth, it was hypothesized that different modes of growth (batch and continuous culture), which are associated with different culture substrate concentrations, may also impact the rates of ammonia oxidation and result in various estimates of Ki. Cd(II), a potent nitrification inhibitor (26, 46) and a potential oxidative stressor (1, 8, 15, 50), was chosen since the oxidative stress and stationary-phase responses in N. europaea are not controlled by oxyR and rpoS genes, as they are in several other bacteria (9).
Thus, the primary objectives of this study were to (i) compare the inhibitory responses of and estimates of Ki for batch (exponential- and stationary-growth-phase) and chemostat N. europaea 19718 cultures exposed to discrete and pulse Cd(II) perturbations over time; (ii) identify the most sensitive measured response to Cd(II) exposure in N. europaea among total cell abundance, membrane integrity, respirometric activity, 16S rRNA content, and ammonia monooxygenase subunit A (amoA) expression; and (iii) evaluate the role of oxidative stress mechanisms in the inhibitory response of N. europaea to Cd(II).
MATERIALS AND METHODS
Cell cultivation.
N. europaea (ATCC 19718) was grown at room temperature in a medium containing 280 mg of ammonia-nitrogen/liter in batch cultures (volume, 4 liters) and chemostat reactors (volume, 1.75 liters; dilution rate, 0.39 day−1). The dilution rate was chosen to mimic the average substrate flux in batch cultures. Cultures were monitored by measuring ammonia (colorimetric phenate [11]), nitrite (ion chromatography [11]), total cell concentration, and membrane integrity (by using BacLight; Invitrogen, Carlsbad, CA). In experiments involving Cd(II), culture samples were acidified to a pH of 2.0 with nitric acid and stored at 4°C until being analyzed by atomic absorption spectrometry for total cadmium or by potentiometry for cationic cadmium.
Respirometry-based biokinetic monitoring.
The biokinetics of ammonia oxidation were determined using a previously described batch respirometric assay (10). The maximum ammonia oxidation activity per cell was expressed as the specific oxygen uptake rate (sOUR) and was calculated by dividing the slope of the respirograms (dissolved oxygen concentrations versus time) by the total cell concentration. The sOUR allows for discrimination between the effects of changing cell concentrations (for instance, during batch growth) and the effects of specific activities (for instance, during inhibition), which is not possible with volumetric oxygen uptake.
FISH and image processing.
Fluorescence in situ hybridization (FISH) was performed using probe Cy3-NSO1225 followed by DAPI (4′,6′-diamidino-2-phenylindole) staining (36). Image acquisition was performed with an epifluorescence microscope (Axioskop 2; Carl Zeiss, Thornwood, NY) and a Zeiss MRm camera. For each sample, 10 grayscale images of random fields were acquired using Axiovision 4.0. Images were summarized by the fluorescence ratio (with fluorescence calculated as the area times the intensity), defined as the ratio of the probe fluorescence signal or the autofluorescence signal (no probe) to the DAPI signal. The intracellular 16S rRNA/DNA ratio was approximated as the average difference in the fluorescence ratio between the probe and autofluorescence control images (42).
Quantitative reverse transcriptase PCR for determining amoA expression.
amoA expression was quantified using newly designed primers, amoA1f (5′-GGACTTCACGCTGTATCTG-3′) and amoA1r (5′-GTGCCTTCTACAACGATTGG-3′), and the results were normalized according to the 16S rRNA gene abundance, which was quantified using primers BACT1369f and PROK 1492R (48). An amoA gene fragment cloned with primers A189 (23) and amoA2R′ (41) was used as a standard. A quantitative reverse transcriptase PCR analysis was conducted with an iCycler (Bio-Rad Laboratories, Hercules, CA) using Sybr green (Applied Biosystems, Foster City, CA) chemistry.
Cd(II) exposure studies and Ki estimation.
The impact of Cd(II) was determined as a function of physiological growth states (substrate-sufficient exponential batch and substrate-starved stationary batch growth) as well as growth modes (substrate-limited chemostat growth and substrate-sufficient batch or starved batch growth) of N. europaea cultures. Inhibition studies were conducted by (i) exposing stationary- and exponential-phase N. europaea batch cultures to three discrete Cd(II) concentrations (0.1, 0.5, and 1 mM) over 1, 4, and 7 h (discrete batch exposure; design 1); (ii) exposing chemostat cultures to a pulse of Cd(II) at concentrations increased in 2 mM steps for 20 h, which resulted in a peak reactor Cd(II) concentration of 0.5 mM (chemostat pulse; design 2); and (iii) exposing batch stationary and exponential cultures to identical 20-h Cd(II) pulses administered directly into the batch vessels, which resulted in Cd(II) profiles similar to those in design 2 (fed-batch pulse; design 3). Control cultures that received no Cd(II) were maintained for each design. Designs 2 and 3 permitted a direct comparison between inhibition levels in substrate-limited chemostat and (substrate-sufficient or -starved) batch cultures at similar time-variable Cd(II) concentrations. Designs 1 and 3 permitted a comparison between the effects of short-term and long-term exposures on batch cultures.
Inhibition was measured as a reduction in the sOUR and described using a noncompetitive inhibition model (equation 1). Ki [expressed as the millimolar concentration of Cd(II)] was estimated via nonlinear regression, as follows:
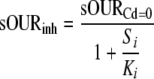 |
(1) |
where sOURinh is the inhibited sOUR (in milligrams of O2 per minute), sOURCd=0 is the uninhibited sOUR (in milligrams of O2 per minute), Si is the Cd(II) concentration [expressed as the millimolar Cd(II) concentration], and Ki is the inhibition coefficient being estimated [expressed as the millimolar Cd(II) concentration].
Evaluating oxidative stress as a mechanism for Cd(II) inhibition.
To evaluate oxidative stress mechanisms, exponential cultures of N. europaea were spiked with 0.01, 0.05, and 0.2 mM Cd(II) and amended with 5 mM N-acetyl-l-cysteine (NAC) 3 h after the initial Cd(II) spike. These lower Cd(II) concentrations were chosen to avoid the complete inactivation of the test cultures and were generally in the range of estimated Ki. Hydrogen peroxide (0.1, 0.5, and 1 mM H2O2) was used as a positive control for oxidative stress. The ameliorating impact of NAC on inhibited cells was determined by sOUR measurements. The potential for cationic cadmium to chelate NAC was tested by coincubating Cd(II) at doses of 1, 10, and 22.5 mM at pH 5 (to maximize the binding of cationic Cd2+ with NAC) and 5 mM NAC for 1, 2, and 3 h.
RESULTS
Batch growth of N. europaea.
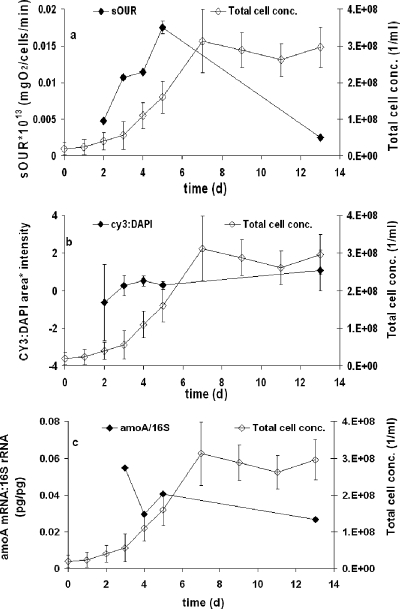
The sOUR was the most sensitive indicator of the change in the physiological growth state from substrate sufficiency (exponential batch culture) to starvation (stationary batch culture) (Fig. 1a). Though amoA expression decreased as N. europaea batch cultures transitioned from mid-exponential growth phase to stationary phase (Fig. 1c), the relative decrease in amoA expression (51%) was not as high as that in sOUR (85%). In contrast to the trends in sOUR and amoA expression, the intracellular 16S rRNA/DNA ratio determined via FISH signal intensity measures did not vary significantly over the period of batch growth and did not decrease upon the transition into stationary phase (Fig. 1b). These results highlight the sensitivity of direct activity-based measures such as the sOUR in describing changes in physiology. These results confirm results shown previously that the intracellular rRNA content may not necessarily track changes in physiology for all organisms (7, 42, 43).
FIG. 1.
Measurements during batch growth of N. europaea 19718 in the absence of Cd(II) inhibition. (a) Total cell concentrations (conc.) and sOURs; (b) FISH signal intensities; and (c) amoA expression relative to 16S rRNA gene abundance. d, day.
Short-term exposure of batch cultures to Cd(II) at discrete concentrations (design 1).
For both exponential and stationary growth at all three Cd(II) concentrations and for the three exposure times, the total cell concentrations and live-cell fractions were nearly constant in Cd(II)-exposed and control [no-Cd(II)] batch N. europaea cultures (data not shown). This result suggests that cell lysis and membrane disruption did not occur to a significant extent in response to Cd(II) exposure. However, sOUR was rapidly and strongly inhibited in both growth phases, even at the lowest Cd(II) concentration and the shortest exposure time tested. Estimates of Ki (equation 1) for exponential cultures decreased as the time of Cd(II) exposure increased (Table 1), indicating that cadmium became more inhibitory with increasing time of exposure. In contrast, the level of inhibition of stationary-phase cultures was relatively stable with time, as reflected by nearly constant Ki estimates (Table 1). These Ki values illustrate the sensitivity and increasing susceptibility of exponential batch cultures to Cd(II) with increasing exposure time.
TABLE 1.
Impact of physiological states and Cd(II) exposure times on measured inhibition coefficients
| Design no. and description and time point |
Ki [mM Cd(II)tot] duringa:
|
|
|---|---|---|
| Exponential growth | Stationary phase | |
| Design 1, discrete Cd(II) exposure for: | ||
| 1 h | 0.034 | 0.014 |
| 4 h | 0.012 | 0.013 |
| 7 h | 0.008 | 0.017 |
| Design 2, 20-h Cd(II) pulse to chemostat | 0.003 ± 0.003 | 0.003 ± 0.003 |
| Design 3, 20-h Cd(II) fed-batch pulse | 0.002 ± 0.000 | 0.033 ± 0.006 |
Values for designs 2 and 3 are averages ± standard deviations.
Cd(II) pulse to chemostat cultures (design 2).
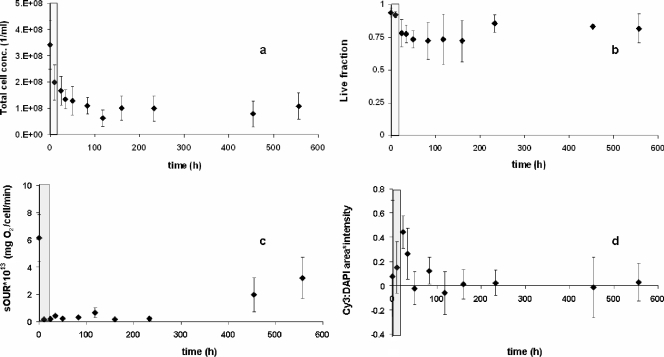
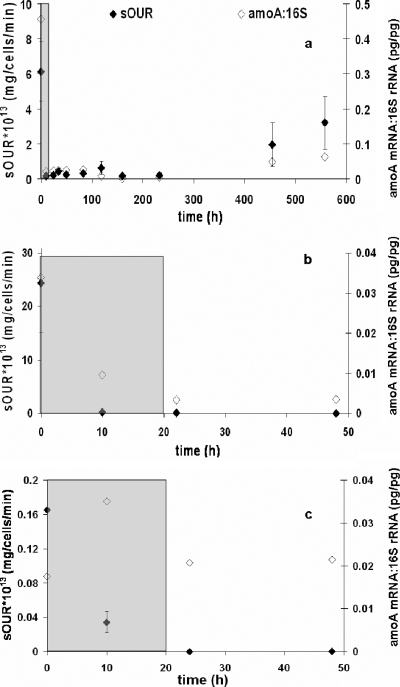
The peak total Cd(II) concentration reached 0.5 mM before declining at the end of the pulse. It took almost 600 h for near-complete washout of the Cd(II) from the chemostat, suggesting significant association of Cd(II) with N. europaea cellular material (data not shown). The sOUR was again the most rapid and sensitive indicator of inhibition (Fig. 2c). Reduction in total cell concentrations was observed and could be attributed to cell washout from a reduction in the specific growth rate or a reduction in cell yields (Fig. 2a). Membrane integrity-based measures of live-cell fractions decreased moderately in response to Cd(II) (Fig. 2b). FISH signal intensity measures remained elevated until the 50-h time point (Fig. 2d), by which near-complete inhibition of the sOUR had already occurred (Fig. 2c). Significantly, the reduction of amoA expression and that of the sOUR correlated strongly (Fig. 3a). Based on Cd(II) pulses imposed on two independent chemostat cultures, a Ki of 0.003 ± 0.003 mM total Cd(II) [Cd(II)tot; average ± standard deviation] was computed (Table 1). This result shows that of all inhibitory responses, those related to activity (sOUR and amoA expression) were by far the most rapid and sensitive.
FIG. 2.
Representative impacts of Cd(II) pulses to chemostat cultures on total cell concentrations (conc.) (a), the live-cell fraction (b), sOURs (c), and Cy3-DAPI relative signal intensities (d). The gray bar indicates the duration of the Cd(II) pulse.
FIG. 3.
Comparison between impacts of physiological state and growth mode on amoA expression (⋄) and sOUR responses (⧫) to Cd(II) pulse exposure in a chemostat (a), fed-batch pulse exposure in exponential phase (b), and fed-batch pulse exposure in stationary phase (c).
Fed-batch Cd(II) pulse to batch cultures (design 3).
The peak total Cd(II) concentrations reached 0.6 and 0.7 mM in the stationary and exponential batch cultures, respectively, and were nearly identical to those in the chemostat (data not shown). The rates of Cd(II) accumulation in the batch and chemostat cultures were also similar. Thus, the levels of inhibition by the Cd(II) pulses to all three cultures (chemostat, batch stationary, and batch exponential cultures) could be directly compared using computed Ki values. However, the response of the batch cultures to the fed-batch pulse was tracked for only 50 h to minimize changes in the target physiological state. The fed-batch pulses of Cd(II) yielded Ki estimates of 0.033 ± 0.006 and 0.002 ± 0.000 mM Cd(II)tot for stationary- and exponential-phase cultures, respectively (Table 1). Notably, the estimates of Ki obtained for Cd(II) pulsed into a chemostat [0.003 ± 0.003 mM Cd(II)tot] and the fed-batch pulse exposure of exponentially growing batch cultures [0.002 ± 0.000 mM Cd(II)tot] were not significantly different. In contrast, stationary-phase cultures subjected to 20 h of fed-batch Cd(II) pulse exposure were about 10 times less sensitive [corresponding to a 10-fold-higher Ki, 0.033 ± 0.006 mM Cd(II)tot]. A time-variable exacerbation of inhibition was also observed in exponential cultures exposed to short-term discrete Cd(II) pulses for either 1, 4, or 7 h, with the Ki decreasing from 0.034 mM (1 h) to 0.008 mM (7 h). The longer-term fed-batch pulse of Cd(II) resulted in a 10-fold reduction in the Ki for exponential cultures. For stationary-phase cultures, the estimated Ki was fairly constant and ranged from 0.014 mM (1 h) to 0.017 mM (7 h). Note that the 20-h fed-batch pulse to stationary-phase cultures corresponded to a higher Ki (less inhibition) of 0.033 ± 0.006 mM Cd(II)tot. These Ki estimates indicate that the fed-batch Cd(II) pulse to exponential batch cultures may be an attractive alternative to measure inhibition in chemostat cultures, which are more complicated and analytically intensive to maintain than batch cultures.
There was a finite but lower reduction in amoA expression than in the sOUR in exponential batch cultures exposed to Cd(II) through the 20-h fed-batch pulse (Fig. 3b). However, in stationary-phase cultures exposed to Cd(II) through the 20-h fed-batch pulse, there was little reduction in amoA expression (Fig. 3c). These results show that inhibited stationary-phase cultures retained both higher relative sOURs and higher-level amoA transcript abundance than inhibited exponential-phase cultures. Total cell counts, FISH measures, and live-cell fractions were consistently unresponsive to the Cd(II) pulse for both exponential and stationary cultures (data not shown).
Cd(II)-mediated inhibition is related to oxidative stress.
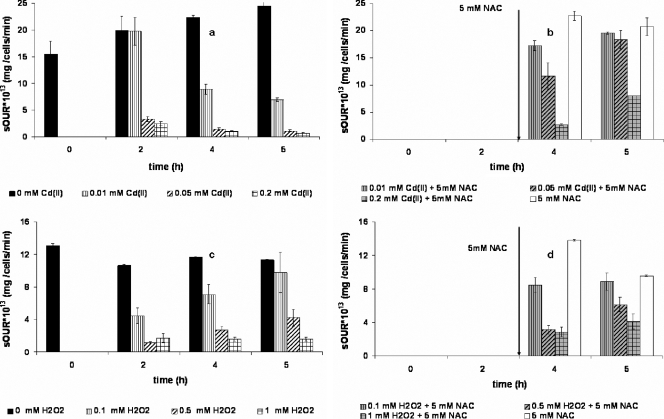
The addition of 5 mM NAC after 3 h of Cd(II) exposure significantly reduced inhibition in exponentially growing batch cultures to near control levels at Cd(II) concentrations of up to 0.2 mM (Fig. 4a and b). In the absence of NAC, N. europaea cultures were inhibited at all Cd(II) concentrations for the duration of the test (Fig. 4a). By itself, 5 mM NAC did not inhibit respiration (Fig. 4b and d). Inhibition by hydrogen peroxide, a positive control for oxidative stress, was moderately reversed even in the absence of NAC (Fig. 4c). The beneficial impact of NAC on H2O2-induced inhibition was evident only at the highest H2O2 concentration of 1 mM (Fig. 4d). These results show that Cd(II)-mediated inhibition was attributable to oxidative stress but was mechanistically distinct from H2O2-mediated inhibition.
FIG. 4.
Time-variable inhibition by Cd(II) (a) and H2O2 (c) and NAC-mediated recovery from inhibition (b and d) among exponential-phase N. europaea cells. Arrows in panels b and d indicate the time point at which 5 mM NAC was added.
DISCUSSION
Impact of physiological states, growth modes, and Cd(II) exposure on inhibition.
In keeping with our first hypothesis, the results of this study implicate the physiological state and growth mode as principal factors that contribute to variability in estimates of the Ki. Increased resistance to physical, chemical, and environmental stressors in response to starvation (which results in higher estimates of Ki) in several microorganisms has been documented previously (21). A recent study has shown that of the genes expressed at higher levels in N. europaea during substrate deprivation, most are involved in the mitigation of oxidative stress (52). Therefore, increased resistance in stationary phase to a potential oxidative stressor like Cd(II) may be expected.
The decreased susceptibility to Cd(II) during stationary phase relative to that during exponential growth may be due to reduced active uptake of Cd(II) under conditions of dwindling energy resources. In contrast, the increased susceptibility of chemostat cultures may be indirectly related to their increased competitive nutrient uptake capability, which may enable the internalization of toxicants such as Cd(II). Cd(II) internalization by bacterial cells is mediated by broad-specificity magnesium transporters (40). Indeed, one of the three gene clusters coding for cobalt, zinc, and cadmium transport in N. europaea (czc) is spatially close to an Mg2+ transporter gene (the Ne1633 gene [9]). The time-variable decrease in Ki for exponential batch cultures (design 1), leading ultimately to a 10-fold reduction in Ki under conditions of long-term exposure (design 3), points to time-variable intracellular accumulation of Cd(II), as shown previously (27).
Role of oxidative stress in Cd(II)-mediated inhibition.
Cd(II) causes direct or indirect oxidative stress in yeast and bacteria (1, 8, 15, 50). Cd(II) also preferentially displaces Zn(II) and Fe(II) from metalloproteins, causing protein inactivation and the release of cationic Fe3+, which may in turn cause the formation of reactive oxygen species (47). In this study, the distinct reversal of Cd(II)-mediated inhibition by NAC, a known scavenger of reactive oxygen species and a source of glutathione (14), indicated the possible contribution of Cd(II)-related oxidative stress in N. europaea. The chelation of Cd2+ by NAC was evaluated and rejected as a mechanism that ameliorated Cd(II) toxicity (data not shown). The observed NAC-free recovery from hydrogen peroxide inhibition in N. europaea cultures may be explained by the increased expression of innate responses to oxidative stress (38). The absence of NAC-free recovery from Cd(II) toxicity indicates modes of Cd(II) toxicity beyond oxidative stress in N. europaea.
Role of physiological state and growth mode on biokinetics and gene expression.
From an ecological perspective, the sOUR and amoA expression profiles highlight two contrasting strategies in N. europaea for adaptation to varying substrate fluxes in batch or continuous growth. On the one hand, by channeling energy resources away from ammonia oxidation during batch growth, N. europaea and other ammonia oxidizers may be better prepared to survive additional stressors, especially during stationary phase. On the other hand, increased amoA expression during chemostat growth may confer higher efficiency for substrate utilization in substrate-limited continuous-growth environments since ammonia is also the preferred assimilative nitrogen source for several microorganisms (35).
The results of this study compare favorably with previously documented reductions in specific ammonia oxidation activity and amoA expression in response to ammonia deprivation in the stationary phase (5, 30, 52). The reduction in amoA expression during stationary phase by a factor of approximately 2.1 (Fig. 1c) is consistent with a recent report that documented a threefold reduction in amoA expression in stationary phase (52).
Based on the entire experimental set, relative amoA expression during chemostat growth ranged from 7 to 27 times higher than expression during uninhibited batch growth. The increased expression of genes encoding periplasmic binding proteins and the increased metabolism of energy-yielding substrates in chemostats relative to those in batch cultures of Escherichia coli have indeed been recently shown (17). A similar general strategy may be employed by N. europaea as well.
Measurement of nitrification activity and inhibition in the environment.
The applicability of the sOUR as a rapid and sensitive indicator of inhibition compared to total cell concentrations or the membrane integrity was shown in this study. Although the sOUR is analytically facile to measure, it does not allow for discrimination between the activity and the inhibition of different ammonia-oxidizing communities in mixed populations. While the concentration and activity of nitrifying bacteria in the environment can be measured via an array of molecular tools targeting 16S rRNA (13, 19, 31, 37, 45, 51), 16S rRNA genes (20, 34), amoA DNA (25, 41), and amoA mRNA (5, 12), this study demonstrates that the relative expression of functional genes such as amoA may be a viable alternate to the sOUR for measuring ammonia oxidation activity attributed to specific communities in uninhibited and inhibited cultures. Though the amoA primer set used here targets a wide spectrum of cultured and noncultured betaproteobacterial ammonia oxidizers, a more general approach would be first to elucidate the overall ecology of ammonia-oxidizing bacteria and archaea in such communities. Subsequently, appropriate molecular tools could be developed to quantitatively track their abundance and activity.
16S rRNA-based FISH did not quantitatively track sOUR and amoA trends in the presence or absence of Cd(II) and may not be a conclusive descriptor of N. europaea specific activity. This lack of correlation between 16S rRNA abundance and activity in ammonia-oxidizing bacteria and other organisms that can maintain high 16S rRNA levels in excess of the protein-synthesizing requirement under starvation and stress conditions has been observed previously (5, 16, 30, 44). Thus, the correlation between 16S rRNA abundance and physiological activity shown previously (7, 42, 43) is not necessarily universal and needs to be evaluated on a case-specific basis.
Despite differences in respective uninhibited sOUR values and amoA expression, inhibition induced by fed-batch Cd(II) pulse exposure in exponentially growing N. europaea cultures effectively paralleled inhibition induced in chemostat cultures, as reflected in similar Ki values. Therefore, the fed-batch pulse modification (design 3) of the traditional discrete pulse (design 1) is a valuable tool for inferring inhibition in continuous reactors while saving the analytical effort to run such reactors.
Footnotes
Published ahead of print on 1 February 2008.
REFERENCES
- 1.Babai, R., and E. Z. Ron. 1998. An Escherichia coli gene responsive to heavy metals. FEMS Microbiol. Rev. 167:107-111. [DOI] [PubMed] [Google Scholar]
- 2.Bedard, C., and R. Knowles. 1989. Physiology, biochemistry and specific inhibitors of CH4, NH4+ and CO oxidation by methanotrophs and nitrifiers. Microbiol. Rev. 53:68-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beg, S. A., R. H. Siddiqi, and S. Ilias. 1982. Inhibition of nitrification by arsenic, chromium and fluoride. J. Water Pollut. Control Fed. 54:482-488. [Google Scholar]
- 4.Benmossa, H., G. Martin, Y. Richard, and A. Leprince. 1986. Inhibition of nitrification by heavy metal cations. Water Res. 20:1333-1339. [Google Scholar]
- 5.Bollmann, A., I. Schmidt, A. M. Saunders, and M. H. Nicolaisen. 2005. Influence of starvation on potential ammonia-oxidizing activity and amoA mRNA levels of Nitrosospira briensis. Appl. Environ. Microbiol. 71:1276-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braam, F., and A. Klapwijk. 1981. Effect of copper on nitrification in activated sludge. Water Res. 15:1093-1098. [Google Scholar]
- 7.Bremer, H., and P. P. Dennis. 1987. Modulation of chemical composition and other parameters of the cell by growth rate, p. 1527-1542. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 8.Brennan, R. J., and R. H. Schiestl. 1996. Cadmium is an inducer of oxidative stress in yeast. Mutat. Res. 356:171-178. [DOI] [PubMed] [Google Scholar]
- 9.Chain, P., J. Lamerdin, F. Larimer, W. Regala, V. Lao, M. Land, L. Hauser, A. Hooper, M. Klotz, J. Norton, L. Sayavedra-Soto, D. Arciero, N. Hommes, M. Whittaker, and D. Arp. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185:2759-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandran, K., and B. F. Smets. 2005. Optimizing experimental design to estimate ammonia and nitrite oxidation biokinetic parameters from batch respirograms. Water Res. 39:4969-4978. [DOI] [PubMed] [Google Scholar]
- 11.Eaton, A. D., L. S. Clesceri, and A. E. Greenberg (ed.). 2005. Standard methods for the examination of water and wastewater, 21st ed. American Public Health Association, Washington, DC.
- 12.Ebie, Y., N. Noda, H. Miura, M. Matsumura, S. Tsuneda, A. Hirata, and Y. Inamori. 2004. Comparative analysis of genetic diversity and expression of amoA in wastewater treatment processes. Appl. Microbiol. Biotechnol. 64:740-744. [DOI] [PubMed] [Google Scholar]
- 13.Egli, K., C. Langer, H.-R. Siegrist, A. J. B. Zehnder, M. Wagner, and J. R. van der Meer. 2003. Community analysis of ammonia and nitrite oxidizers during start-up of nitritation reactors. Appl. Environ. Microbiol. 69:3213-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ercal, N., H. Gurer-Orhan, and N. Aykin-Burns. 2001. Toxic metals and oxidative stress, part I: mechanisms involved in metal induced oxidative damage. Curr. Top. Med. Chem. 1:529-539. [DOI] [PubMed] [Google Scholar]
- 15.Ferianc, P., A. Farewell, and T. Nystrom. 1998. The cadmium-stress stimulon of Escherichia coli K-12. Microbiology 144:1045-1050. [DOI] [PubMed] [Google Scholar]
- 16.Flardh, K., P. S. Cohen, and S. Kjelleberg. 1992. Ribosomes exist in large excess over the apparent demand for protein synthesis during carbon starvation in marine Vibrio sp. strain CCUG 15956. J. Bacteriol. 174:6780-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franchini, A. G., and T. Egli. 2006. Global gene expression in Escherichia coli K-12 during short-term and long-term adaptation to glucose-limited continuous culture conditions. Microbiology 152:2111-2127. [DOI] [PubMed] [Google Scholar]
- 18.Gernaey, K., L. Verschuere, L. Lutyen, and W. Verstraete. 1997. Fast and sensitive acute toxicity detection with an enrichment nitrifying culture. Water Environ. Res. 69:1163-1169. [Google Scholar]
- 19.Gieseke, A., U. Purkhold, M. Wagner, R. Amann, and A. Schramm. 2001. Community structure and activity dynamics of nitrifying bacteria in a phosphate-removing biofilm. Appl. Environ. Microbiol. 67:1351-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harms, G., A. C. Layton, H. M. Dionisi, I. R. Gregory, V. M. Garrett, S. A. Hawkins, K. G. Robinson, and G. S. Sayler. 2003. Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ. Sci. Technol. 37:343-351. [DOI] [PubMed] [Google Scholar]
- 21.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, DC.
- 22.Hockenbury, M. R., and C. P. L. J. Grady. 1977. Inhibition of nitrification—effects of selected organic compounds. J. Water Pollut. Control Fed. 49:768-777. [Google Scholar]
- 23.Holmes, A. J., A. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 24.Hooper, A. B., and K. R. Terry. 1973. Specific inhibitors of ammonia oxidation in Nitrosomonas. J. Bacteriol. 115:480-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoshino, T., N. Noda, S. Tsuneda, A. Hirata, and Y. Inamori. 2001. Direct detection by in situ PCR of the amoA gene in biofilm resulting from a nitrogen removal process. Appl. Environ. Microbiol. 67:5261-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu, Z., K. Chandran, D. Grasso, and B. F. Smets. 2002. Effect of nickel and cadmium speciation on nitrification inhibition. Environ. Sci. Technol. 36:3074-3078. [DOI] [PubMed] [Google Scholar]
- 27.Hu, Z., K. Chandran, D. Grasso, and B. F. Smets. 2003. Impact of metal sorption and internalization on nitrification inhibition. Environ. Sci. Technol. 37:728-734. [DOI] [PubMed] [Google Scholar]
- 28.Hyman, M. R., C. Y. Kim, and D. J. Arp. 1990. Inhibition of ammonia monooxygenase in Nitrosomonas europaea by carbon disulfide. J. Bacteriol. 172:4775-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hynes, R. K., and R. Knowles. 1983. Inhibition of chemoautotrophic nitrification by sodium chlorate and sodium chlorite: a reexamination. Appl. Environ. Microbiol. 45:1178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnstone, B. H., and R. D. Jones. 1988. Physiological effects of long-term energy-source deprivation on the survival of a marine chemolithotrophic ammonium oxidizing bacterium. Mar. Ecol. Prog. Ser. 49:295-303. [Google Scholar]
- 31.Juretschko, S., G. Timmermann, M. Schmid, K.-H. Schleifer, A. Pommerening-Roser, H.-P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly, R. T., I. D. S. Henriques, and N. Love. 2004. Chemical inhibition of nitrification in activated sludge. Biotechnol. Bioeng. 85:683-694. [DOI] [PubMed] [Google Scholar]
- 33.Kong, Z., P. Vanrolleghem, and W. Verstraete. 1996. Simultaneous determination of inhibition kinetics of carbon oxidation and nitrification with a respirometer. Water Res. 30:825-836. [Google Scholar]
- 34.Kowalchuk, G. A., J. R. Stephen, W. DeBoer, J. I. Prosser, T. M. Embley, and J. W. Woldendopr. 1997. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madigan, M. T., and J. M. Martinko. 2006. Brock biology of microorganisms, 11th ed. Prentice Hall, Upper Saddle River, NJ.
- 36.Manz, W., U. Szewzyk, P. Ericsson, R. Amann, K. H. Schleifer, and T. A. Stenstrom. 1993. In situ identification of bacteria in drinking water and adjoining biofilms by hybridization with 16S and 23S rRNA-directed fluorescent oligonucleotide probes. Appl. Environ. Microbiol. 59:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mobarry, B., M. Wagner, V. Urbain, B. E. Rittmann, and D. A. Stahl. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mongkolsuk, S., and J. D. Helmann. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 45:9-15. [DOI] [PubMed] [Google Scholar]
- 39.Neufeld, R. D., A. J. Hill, and D. O. Adekoya. 1980. Phenol and free ammonia inhibition to Nitrosomonas activity. Water Res. 14:1695-1703. [Google Scholar]
- 40.Nies, D. H. 1992. Resistance to cadmium, cobalt, zinc, and nickel in microbes. Plasmid 27:17-28. [DOI] [PubMed] [Google Scholar]
- 41.Okano, Y., K. R. Hristova, C. M. Leutenegger, L. E. Jackson, R. F. Denison, B. Gebreyesus, D. Lebauer, and K. M. Scow. 2004. Application of real-time PCR to study effects of ammonium on population size of ammonia-oxidizing bacteria in soil. Appl. Environ. Microbiol. 70:1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poulsen, L. K., G. Ballard, and D. A. Stahl. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosset, R., J. Julien, and R. Monier. 1966. Ribonucleic acid composition of bacteria as a function of growth rate. J. Mol. Biol. 18:308-320. [DOI] [PubMed] [Google Scholar]
- 44.Schmid, M. C., B. Maas, A. Dapena, K. van de Pas-Schoonen, J. van de Vossenberg, B. Kartal, L. van Niftrik, I. Schmidt, I. Cirpus, J. G. Kuenen, M. Wagner, J. S. Sinninghe Damste, M. Kuypers, N. P. Revsbech, R. Mendez, M. S. M. Jetten, and M. Strous. 2005. Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl. Environ. Microbiol. 71:1677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schramm, A., D. de Beer, M. Wagner, and R. Amann. 1998. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl. Environ. Microbiol. 64:3480-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stephen, J. R., Y.-J. Chang, S. J. Macnaughton, G. A. Kowalchuk, K. T. Leung, C. A. Flemming, and D. C. White. 1999. Effect of toxic metals on indigenous soil β-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stohs, S. J., and D. Bagchi. 1995. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 18:321-336. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomlinson, T. G., A. G. Boon, and C. N. A. Trotman. 1966. Inhibition of nitrification in the activated sludge process of sewage disposal. J. Appl. Bacteriol. 29:266-291. [DOI] [PubMed] [Google Scholar]
- 50.VanBogelen, R. A., P. M. Kelley, and F. C. Neidhardt. 1987. Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. J. Bacteriol. 169:26-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner, M., D. R. Noguera, S. Juretschko, G. Rath, H. P. Koops, and K. H. Schleifer. 1998. Combining fluorescent in-situ hybridization (FISH) with cultivation and mathematical modeling to study population structure and function of ammonia-oxidizing bacteria in activated sludge. Water Sci. Technol. 37:441-449. [Google Scholar]
- 52.Wei, X., T. Yan, N. G. Hommes, X. Liu, L. Wu, C. McAlvin, M. G. Klotz, L. A. Sayavedra-Soto, J. Zhou, and D. J. Arp. 2006. Transcript profiles of Nitrosomonas europaea during growth and upon deprivation of ammonia and carbonate. FEMS Microbiol. Lett. 257:76-83. [DOI] [PubMed] [Google Scholar]