Abstract
The latex-clearing protein (LcpK30) from the rubber-degrading bacterium Streptomyces sp. strain K30 is involved in the cleavage of poly(cis-1,4-isoprene), yielding isoprenoid aldehydes and ketones. Lcp homologues have so far been detected in all investigated clearing-zone-forming rubber-degrading bacteria. Internal degenerated oligonucleotides derived from lcp genes of Streptomyces sp. strain K30 (lcpK30), Streptomyces coelicolor strain A3(2), and Nocardia farcinica strains IFM10152 and E1 were applied in PCR to investigate whether lcp homologues occur also in the non-clearing-zone-forming rubber-utilizing bacteria Gordonia polyisoprenivorans strains VH2 and Y2K, Gordonia alkanivorans strain 44187, and Gordonia westfalica strain Kb1, which grow adhesively on rubber. The 1,230- and 1,224-bp lcp-homologous genes from G. polyisoprenivorans strain VH2 (lcpVH2) and G. westfalica strain Kb1 (lcpKb1) were obtained after screening genomic libraries by degenerated PCR amplification, and their translational products exhibited 50 and 52% amino acid identity, respectively, to LcpK30. Recombinant lcpVH2 and lcpKb1 harboring cells of the non-rubber-degrading Streptomyces lividans strain TK23 were able to form clearing zones and aldehydes on latex overlay-agar plates, thus indicating that lcpVH2 and lcpKb1 encode functionally active proteins. Analysis by gel permeation chromatography demonstrated lower polymer concentrations and molecular weights of the remaining polyisoprenoid molecules after incubation with these recombinant S. lividans strains. Reverse transcription-PCR analysis demonstrated that lcpVH2 was transcribed in cells of G. polyisoprenivorans strain VH2 cultivated in the presence of poly(cis-1,4-isoprene) but not in the presence of sodium acetate. Anti-LcpK30 immunoglobulin Gs, which were raised in this study, were rather specific for LcpK30 and did not cross-react with LcpVH2 and LcpKb1. A lcpVH2 disruption mutant was still able to grow with poly(cis-1,4-isoprene) as sole carbon source; therefore, lcpVH2 seems not to be essential for rubber degradation in G. polyisoprenivorans.
Investigations of bacterial degradation of natural rubber (NR) revealed two groups of NR-degrading bacteria according to their strategy for substrate utilization. (i) Members of the first group form clearing zones on latex overlay-agar plates, indicating an extracellular enzyme activity. Representatives belong to the genera Actinomadura, Actinoplanes, Dactylosporangium, Micromonospora, Microtetraspora, Nocardia, and Streptomyces (22, 23). (ii) Members of the second group exhibit adhesive growth with direct contact of the cells with the NR material and extensive disintegration of the substrate. Representatives belong to the genera Gordonia, Mycobacterium, and Nocardia (2, 28, 29, 30, 41).
Lcp (latex-clearing protein) has been considered a key enzyme in NR degradation by clearing-zone-forming gram-positive bacteria (33), and RoxA (rubber oxygenase) has been considered a key enzyme in NR degradation by the gram-negative bacterium Xanthomonas sp. strain 35Y (24). RoxA is an extracellular diheme protein secreted by this strain during growth on NR (24). Purified RoxA degraded poly(cis-1,4-isoprene) by oxidative cleavage at the double bonds, yielding 12-oxo-4,8-dimethyltrideca-4,8-diene-1-al as the main cleavage product; other minor cleavage products differed only in the number of repetitive isoprene units (9). In vitro experiments also revealed occurrence of two 18O atoms in the reduced degradation product 12-hydroxy-4,8-dimethyltrideca-4,8-diene-1-ol, thereby disclosing a dioxygenase mechanism (10).
The lcp gene was identified in the gram-positive Streptomyces sp. strain K30, which belongs to the first group of NR-degrading bacteria, by Rose et al. (33). UV mutagenesis produced mutants with a clearing-zone-negative phenotype on latex overlay-agar plates and the inability to mineralize NR. A genomic DNA fragment from Streptomyces sp. strain K30, which restored the latex-positive phenotype in the mutants, comprised three open reading frames possibly involved in NR degradation. The translational product of one gene exhibited similarities to a putatively secreted protein of Streptomyces coelicolor strain A3(2) and was designated Lcp (latex-clearing protein). The translational products of the two other open reading frames exhibited strong similarities to putative heterodimeric molybdenum hydroxylases (OxiAB) representing some isochinoline oxidoreductases and aldehyde dehydrogenases. Heterologous expression of lcp in Streptomyces lividans strain TK23, which is not able to utilize rubber, enabled this strain to form clearing zones on latex overlay-agar plates and to degrade NR as demonstrated by gel permeation chromatography (GPC) analysis and by staining with Schiff's reagent indicating the presence of compounds with aldehyde groups among the degradation products of NR. According to a hypothetical degradation pathway, the rubber molecules are cleaved by Lcp to aldehydes and ketones with low molecular weights which are then possibly further oxidized by OxiAB to the corresponding acids and activated and metabolized via the β-oxidation pathway in Streptomyces sp. strain K30 (34).
Very little is known about the biochemical mechanism of rubber degradation in adhesively growing bacteria. Members of the genus Gordonia serve as model organisms to investigate this aspect (2, 5, 12, 28, 29, 30). Due to the importance of Lcp for biodegradation of NR in Streptomyces sp. strain K30, the occurrence and diversity of Lcp homologues in members of the genus Gordonia were investigated in this study. This study should unravel whether Lcp homologues occur only in clearing-zone-forming bacteria or in any rubber-degrading bacteria. One Lcp homologue was recently identified in a thermophilic adhesively growing strain of Nocardia farcinica (20). To identify Lcp homologues in Gordonia sp., degenerate PCR primers specific for genes coding for Lcp were designed based on DNA sequences from Streptomyces sp. strain K30, S. coelicolor strain A3(2), and the N. farcinica strains IFM10152 and E1. Two identified lcp-homologous genes were then further characterized.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cultivation conditions.
Bacteria and plasmids used in this study are listed in Table 1. All strains of the genera Gordonia, Nocardia, and Streptomyces were grown at 30°C on either standard I complex nutrient broth (St-I; Merck, Darmstadt, Germany) or mineral salts medium (MSM) (38). Carbon sources were added to liquid MSM as indicated in the text. Liquid cultures in Erlenmeyer flasks were incubated on a horizontal rotary shaker. Solid media were prepared by addition of 1.5% (wt/vol) agar-agar. For preparation of latex overlay-agar plates, MSM agar plates, containing 1% (wt/vol) glucose, were covered with an overlay of MSM agar containing 0.2% (vol/vol) latex concentrate (Neotex Latz; Weber & Schaer, Hamburg, Germany) plus 1% (wt/vol) glucose. Escherichia coli was cultivated at 37°C in Luria-Bertani broth (LB) (36). Antibiotics were applied according to the method of Sambrook et al. (36) and as indicated in the text. Protoplasts of S. lividans strain TK23 were prepared from cells grown in modified YEME (3% [wt/vol] yeast extract, 5% [wt/vol] Bacto peptone, 3% [wt/vol] malt extract, 34% [wt/vol] sucrose) medium (19). R5 agar plates were used for protoplast regeneration (25).
TABLE 1.
Bacterial strains and plasmids and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Relevant marker(s) or sequence (5′→3′) | Reference, source, or function |
|---|---|---|
| Strains | ||
| Gordonia species | ||
| G. polyisoprenivorans strain VH2 | Poly(cis-1,4-isoprene)-degrading wild type | 2 |
| G. polyisoprenivorans strain Y2K | Poly(cis-1,4-isoprene)-degrading wild type | 2 |
| G. polyisoprenivorans mutant A12 | lcpVH2 disruption mutant of G. polyisoprenivorans strain VH2; Kmr | This study |
| G. polyisoprenivorans mutant A17 | lcpVH2 disruption mutant of G. polyisoprenivorans strain VH2; Kmr | This study |
| G. polyisoprenivorans mutant A29 | lcpVH2 disruption mutant of G. polyisoprenivorans strain VH2; Kmr | This study |
| G. polyisoprenivorans mutant A34 | lcpVH2 disruption mutant of G. polyisoprenivorans strain VH2; Kmr | This study |
| G. alkanivorans strain 44187 | Poly(cis-1,4-isoprene)-degrading wild type | 26 |
| G. westfalica strain Kb1 | Poly(cis-1,4-isoprene)-degrading wild type | 30 |
| Nocardia farcinica strain IFM10152 | Poly(cis-1,4-isoprene)-degrading wild type | 21 |
| Streptomyces species | ||
| Streptomyces sp. strain K30 | Poly(cis-1,4-isoprene)-degrading and clearing-zone-forming wild type | 33 |
| S. lividans strain TK23 | Non-poly(cis-1,4-isoprene)-degrading and non-clearing-zone-forming wild type | 19 |
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 (rk− mk+) supE44 relA1 λ−lac [F′ proAB lacIqlacZΔM15 Tn10(Tcr)] | 13 |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rk− mk+) phoA supE44 λ−thi-1 gyrA96 relA1 | Roche Applied Science, Penzberg, Germany |
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm lacY1(DE3) | Novagen, Madison, WI |
| Plasmids | ||
| pBluescript SK− | AprlacPOZ′ | Stratagene, San Diego, CA |
| pHC79 | Cosmid (Apr Tcr) | 18 |
| pGEM-T Easy | E. coli cloning vector (Apr T-tailing) | Promega, Madison, WI |
| pGEM-T Easy::lcpVH2::aph | E. coli cloning vector (Apr), containing the cloned gene lcpVH2 from G. polyisoprenivorans strain VH2 with an inserted aph (Kmr) | This study |
| pET-23a | E. coli expression vector (Apr T7 promoter) | Novagen, Madison, WI |
| pET-23a::lcpK30his | E. coli expression vector (Apr T7 promoter), containing the cloned gene lcpK30 from Streptomyces sp. strain K30 | This study |
| pIJSK::lcpK30 | E. coli-Streptomyces shuttle vector, containing the cloned gene lcpK30 from Streptomyces sp. strain K30 (Apr TsrrmelC1 melC2) | Henrike Wernsmann, unpublished data |
| pIJSK | E. coli-Streptomyces shuttle vector (Apr TsrrmelC1 melC2) | This study |
| pIJSK::lcpVH2 | E. coli-Streptomyces shuttle vector, containing the cloned gene lcpVH2 from G. polyisoprenivorans strain VH2 (Apr TsrrmelC1 melC2) | This study |
| pIJSK::lcpVH2::aph | E. coli-Streptomyces shuttle vector, containing the cloned gene lcpVH2::aph from G. polyisoprenivorans mutant A17 (Apr Tsrr KmrmelC1 melC2) | This study |
| pIJSK::lcpKb1 | E. coli-Streptomyces shuttle vector, containing the cloned gene lcpKb1 from G. westfalica strain Kb1 (Apr TsrrmelC1 melC2) | This study |
| Oligonucleotides | ||
| P265f | CGCTGCCCG(AG)CGGA(CT)T(GC)CC(GC) | Degenerated PCR primer based on lcp-homologous sequences, with P701r amplification of a 436-bp PCR product |
| P701r | CTGTG(GC)(AC)AGGTGACCA(AGT)GCATGTCG | Degenerated PCR primer based on lcp-homologous sequences, with P265f amplification of a 436-bp PCR product |
| PVH2_117fBglII | ATATAGATCTGCGAGGTGCGTTGAATACCG | With PVH2_1752rBglII, 1,635-bp lcpVH2 PCR product of G. polyisoprenivorans strain VH2 (BglII restriction sites used for cloning are underlined) |
| PVH2_1752rBglII | ATATAGATCTAGGACCTCGGTTCCGGTGAAC | With PVH2_117fBglII, 1,635-bp lcpVH2 PCR product of G. polyisoprenivorans strain VH2 (BglII restriction sites used for cloning are underlined) |
| PVH2_360f(RT) | TTCAATCAGCGACGGGGCAC | With PVH2_911r(RT), 551-bp PCR product of G. polyisoprenivorans strain VH2 used for RT-PCR |
| PVH2_911r(RT) | AGCATGTCGGCGAGTTTCTGG | With PVH2_360f(RT), 551-bp PCR product of G. polyisoprenivorans strain VH2 used for RT-PCR |
| PVH2_449f | TGACGAAGGCCAGCAGCAGG | With PVH2_2392r, 1,944-bp lcpVH2 PCR product of G. polyisoprenivorans strain VH2 |
| PVH2_2392r | AGGAAGTGCAGTTGCGCGGTC | With PVH2_449f, 1,944-bp lcpVH2 PCR product of G. polyisoprenivorans strain VH2 |
| PKb1_57fBglII | ATATAGATCTCTGGCGTTGATTGGATTCGGG | With PKb1_1789rBglII, 1,732-bp lcpKb1 PCR product of G. westfalica strain Kb1 (BglII restriction sites used for cloning are underlined) |
| PKb1_1789rBglII | ATATAGATCTGCGTCTCCCGTTCAGCAAATGG | With PKb1_57fBglII, 1,732-bp lcpKb1 PCR product of G. westfalica strain Kb1 (BglII restriction sites used for cloning are underlined) |
| PLcp_NtermNdeI | GGCATATGGACGGTTCAGCAG | With PLcp_CtermBamHI, 1,235-bp lcpK30 PCR product of Streptomyces sp. strain K30 (NdeI restriction sites used for cloning are underlined) |
| PLcp_CtermBamHI | AAAGGATCCGGACGGGCGGTTGACGTCCGG | With PLcp_NtermNdeI, 1,235-bp lcpK30 PCR product of Streptomyces sp. strain K30 (BamHI restriction sites used for cloning are underlined) |
Isolation, analysis, and manipulation of DNA.
Plasmid DNA was prepared from crude lysates by the alkaline extraction method (8). Total DNA of Streptomyces, Gordonia, and Nocardia was prepared as described by Ausubel et al. (4) with modifications as recently described (12). Recombinant DNA techniques for S. lividans strain TK23 were performed as described previously (25). DNA was transferred to Gordonia polyisoprenivorans strain VH2 by electroporation (3). DNA was restricted with restriction endonucleases (Gibco/BRL, Gaithersburg, MD) under conditions recommended by the manufacturer. All other genetic procedures and manipulations were conducted as described by Sambrook et al. (36).
GPC.
Cleavage of poly(cis-1,4-isoprene) by recombinant strains of S. lividans strain TK23 was verified by GPC. After a cultivation period of 8 weeks, samples were prepared in chloroform and analyzed as previously described (20) employing a Waters-GPC system (Waters, Milford, CT) consisting of a 515 high-pressure liquid chromatography pump, a 410 differential refractometer, a 717plus autosampler, and four in-series-connected Styragel columns (HR3, HR4, HR5, and HR6 with pore sizes of 103, 104, 105, and 106 Å, respectively). Molecular weights of poly(cis-1,4-isoprene) and of cleavage products were calculated from retention times of defined poly(cis-1,4-isoprene) standards (PSS Polymer Standards Service GmbH, Mainz, Germany).
Cosmid cloning and sequencing.
A cosmid library of Gordonia westfalica strain Kb1 was prepared from partially restricted (EcoRI) genomic DNA yielding preferentially fragments in the 30- to 40-kbp size range. The latter were ligated to pHC79 (18) and packaged into λ particles (17), and ampicillin-plus-tetracycline-resistant transductants of E. coli strain DH5α were selected. Clones containing hybrid cosmids of this cosmid library of G. westfalica strain Kb1 and of another cosmid library of G. polyisoprenivorans strain VH2 (Quyen Banh, unpublished data) were selected on LB-ampicillin-tetracycline agar plates. All the colonies from several plates (around 50 on each plate) were pooled, plasmid DNA was prepared from each pool, and putative fragments containing the lcp genes were amplified by PCR at an annealing temperature of 61°C using the ImmoMix ready-to-use PCR mixture (BioLine, Randolph, MA) and the degenerated primers P265f and P701r (Table 1). The number of colonies from the master plate of the identified pool was decreased and used for further screening by the described PCR amplification. Using this method, 35 and 25 pools comprising 700 and 500 clones of G. westfalica strain Kb1 or G. polyisoprenivorans strain VH2, respectively, were screened for the presence of lcp-containing hybrid plasmids. Hybrid cosmids pHC79::535, pHC79::549, and pHC79::632 from G. westfalica strain Kb1 and pHC79::290 from G. polyisoprenivorans strain VH2 contained lcp-homologous sequences. These hybrid cosmids were used as template DNA for sequencing reactions by primer walking. DNA was sequenced in a 48-capillary 3730 DNA Analyzer electrophoresis system (Applied Biosystems, Foster City, CA). Oligonucleotides were purchased from MWG Biotech (Ebersberg, Germany). For sequencing of inserts in the pGEM-T Easy vector (Promega, Madison, WI), the universal M13 forward primer and M13 reverse primer were used.
Nested PCR.
The 3′ region of lcp from G. westfalica strain Kb1 (lcpKb1) was amplified by nested PCR employing one biotinylated oligonucleotide, which binds closely to the 3′ end of the known lcpKb1 sequence, plus four degenerated walker primers, which were expected to anneal in the 3′-flanking region of known lcp sequences according to the work of Mishra et al. (32). A set of four PCRs was carried out using genomic DNA of G. westfalica strain Kb1 as template and each single biotinylated primer in combination with each of the four walking primers under conditions described by Mishra et al. to amplify the flanking region (32). Streptavidin-coupled magnetic beads (Roche, Switzerland) were then applied according to the manufacturer's protocol to isolate biotinylated PCR products which were subsequently used as template for a second set of PCRs. For this a specific nested primer was designed, binding even more closely to the 3′ region of the known sequence than the corresponding biotinylated oligonucleotide. The specific nested primer was used together with the general nested primer, also taken from the work of Mishra et al. (32), for PCR, and the obtained amplification products were then cloned into the vector pGEM-T Easy (Promega, Madison, WI) and sequenced (see “Cosmid cloning and sequencing”). The designed biotinylated and nested oligonucleotides were purchased from MWG Biotech (Ebersberg, Germany).
Sequence analyses.
Database searches of the predicted protein sequences were performed employing the BLAST program provided by EMBL/Heidelberg (1). Multiple sequence alignments and the generation of sequence contigs were carried out using the program ClustalX (39). Phylogenetic trees were constructed using the program TREE 1.6.5. The SignalP 3.0 server was used for prediction of the presence and location of signal peptide cleavage sites in amino acid sequences (15).
Construction of plasmids.
The coding regions of lcpVH2 from G. polyisoprenivorans strain VH2 and lcpKb1 from G. westfalica strain Kb1, including regions approximately 400 bp upstream of the start codons and approximately 100 bp downstream of the stop codons, were amplified by PCR using oligonucleotides PVH2_117fBglII plus PVH2_1752rBglII for lcpVH2 and PKb1_57fBglII plus PKb1_1789rBglII for lcpKb1 (Table 1) and Pfx DNA polymerase (Gibco BRL) according to the manufacturer's instructions. For complementation experiments in S. lividans strain TK23, the E. coli-Streptomyces shuttle vector pIJSK::lcpK30 was used as a positive control and also for cloning of lcpVH2 and lcpKb1. The vector pIJSK::lcpK30 conferred ampicillin resistance for selection in E. coli and thiostrepton resistance for selection in Streptomyces (Henrike Wernsmann, unpublished data). To obtain pIJSK from pIJSK::lcpK30, lcpK30 was excised by restriction with BglII. It was then also ligated to lcpVH2 or lcpKb1, yielding pIJSK::lcpVH2 or pIJSK::lcpKb1, respectively (see Fig. S1 in the supplemental material). All plasmids were transferred to S. lividans strain TK23 by protoplast transformation (25).
The coding region of lcpK30 from Streptomyces sp. strain K30 was amplified by PCR using oligonucleotides Lcp_NtermNdeI and Lcp_CtermBamHI (Table 1) and Pfx DNA polymerase (Gibco BRL) according to the manufacturer's instructions. The PCR product was then cloned into the NdeI-BamHI-linearized plasmid pET-23a, yielding plasmid pET-23a::lcpK30his, which was subsequently transferred to E. coli BL21(DE3).
To study the loss of functionality of the disrupted lcpVH2 gene, it was amplified from the disruption mutant A17 by PCR using primers PVH2_117fBglII and PVH2_1752bBglII (Table 1). The resulting 2,635-bp lcpVH2::aph fragment was cloned into the BglII-linearized pIJSK vector. The hybrid plasmid pIJSK::lcpVH2::aph was subsequently transferred to S. lividans strain TK23 by protoplast transformation (25).
Expression of six-His-tagged LcpK30 in E. coli strain BL21(DE3), isolation of IBs, and generation of anti-LcpK30 antibodies.
E. coli strain BL21(DE3) harboring plasmid pET-23a::lcpK30his was cultivated in LB medium at 37°C to an optical density at 600 nm of 0.5, and then expression was induced by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM for 3 h yielding cells with inclusion bodies (IBs). For isolation of IBs, the cells of a 100-ml culture were harvested, resuspended in 4 ml 20 mM Tris-HCl (pH 8.0) buffer, and disrupted by a twofold French press passage at 1,000 MPa. The disrupted cells were centrifuged at 25,000 × g for 15 min at 4°C. The obtained pellet was resuspended in 3 ml cold IB wash buffer (2 M urea, 20 mM Tris-HCl, 0.5 M NaCl, 2% Triton X-100, pH 8.0) by sonication (1 min/ml with an amplitude of 40 μm) with a Bandelin Sonopuls GM200 ultrasonic disintegrator. After 15 min of centrifugation at 4°C and 25,000 × g, treatment with IB wash buffer, resuspension by sonication, and centrifugation were repeated three times. The purified IBs were dissolved in sodium dodecyl sulfate (SDS) denaturation buffer (27). A sample, consisting of the dissolved IBs containing the extracted Lcp protein, was separated by SDS-polyacrylamide gel electrophoresis and excised from the gel, and its identity was confirmed by matrix-assisted laser desorption ionization-time of flight analysis, before it was used for generation of custom polyclonal antibodies in rabbits by Eurogentec (Seraing, Belgium). Purified polyclonal rabbit anti-LcpK30 immunoglobulin G (IgG) antibodies were obtained from the serum by chromatography on protein A-Sepharose (16).
Preparation of crude cell extracts and extracellular protein fractions.
For preparative purposes, cells of G. polyisoprenivorans strain VH2, G. westfalica strain Kb1, Streptomyces sp. strain K30, and recombinant strains of S. lividans TK23 were grown in MSM (38) containing the carbon sources and for the periods described in the text. Cells were sedimented from the aqueous phase by 30 min of ultracentrifugation at 100,000 × g at 4°C to remove cells and as many particles as possible from the supernatant. Cells were used for preparation of crude cell extracts by a threefold passage through a French pressure cell (1,000 MPa). The clear supernatant was filtrated using a 0.22-μm membrane filter (Roth, Karlsruhe, Germany); the proteins were subsequently concentrated by precipitation with 0.015% (vol/vol) sodium deoxycholate plus 10% (wt/vol) trichloric acid as previously described (31) and then resuspended in an appropriate volume of distilled water and 2% (vol/vol) 1 M Tris-HCl (pH 9.0). Small amounts of extracellular proteins were also concentrated from these supernatants by applying Vivaspin 500 centrifugal filter units with a 10-kDa-cutoff polyethersulfone membrane (Sartorius, Göttingen, Germany).
SDS-polyacrylamide gel electrophoresis, Western blot analysis, and other immunological analyses.
Protein contents of crude cell extracts and extracellular protein fractions were estimated by the dye binding principle method using bovine serum albumin as standard (11). Samples of crude cell extracts and extracellular protein fractions representing a quantity of 50 μg protein were separated in 11.5% (wt/vol) SDS-polyacrylamide gels (27). Proteins were visualized with Coomassie brilliant blue R250 (42). For immunological detection of Lcp, the proteins were transferred from gels onto polyvinylidene difluoride (PVDF) membranes, according to the work of Towbin et al. (40) and the manufacturer (GE Healthcare, Buckinghamshire, United Kingdom). Proteins on the membrane were stained with Ponceau S and analyzed immunologically employing 400 μl of the polyclonal rabbit anti-LcpK30 IgG solution. IgG antibodies were visualized on immunoblots using anti-rabbit IgG-alkaline phosphatase conjugates (Sigma-Aldrich), converting 5-bromo-4-chloro-3-indolyl-phosphate dipotassium/nitrotetrazolium blue chloride (Sigma-Aldrich) into an insoluble dark product. Dot blotting experiments were done as described by the manufacturer of the PVDF membrane (GE Healthcare, Buckinghamshire, United Kingdom).
RT-PCR analysis of total RNA from G. polyisoprenivorans strain VH2.
DNA-free total RNA of G. polyisoprenivorans strain VH2 was prepared by DNase I treatment of an RNA sample kindly provided by Quyen Banh in our laboratory. For identification of lcpVH2-derived mRNA, reverse transcription-PCR (RT-PCR) was applied using oligonucleotides PVH2_360f(RT) and PVH2_911r(RT) (Table 1). RT-PCR was carried out using a commercial kit (One Step RT-PCR kit; Qiagen, Hilden, Germany) according to the manufacturer's protocol and 0.5 ng RNA as template. To exclude any DNA contamination that could serve as template for PCR, template RNA was added in a control experiment, after inactivation of reverse transcriptase for 15 min at 95°C in the presence of Taq polymerase: the absence of PCR products indicated that the RT-PCR products were not derived from contaminating DNA.
Nucleotide sequence accession numbers.
The nucleotide sequences of lcpVH2 and lcpKb1 investigated in this study have been deposited in the GenBank database under accession numbers EU013941 and EU013942, respectively.
RESULTS
Identification of lcp homologues in rubber-degrading species of the genus Gordonia.
The occurrence of lcp-homologous genes in the genomes of rubber-utilizing species belonging to the genus Gordonia was investigated. Two degenerated PCR primers, P265f and P701r (Table 1), were designed based on conserved sequences within the known lcp genes from Streptomyces sp. strain K30, S. coelicolor strain A3(2), and N. farcinica strains IFM10152 and E1. Total DNA of the rubber-utilizing bacteria G. polyisoprenivorans strains VH2 and Y2K, Gordonia alkanivorans strain 44187, and G. westfalica strain Kb1 was isolated and used as template DNA in a gradient PCR (annealing temperature of 49 to 61°C) applying the ImmoMix ready-to-use PCR mixture (BioLine, Randolph, MA) and primers P265f and P701r. Specific PCR products were obtained at an annealing temperature of 61°C from all four templates as well as from N. farcinica strain IFM10152 DNA, which was used as a positive control. The PCR products were then cloned into the pGEM-T Easy vector (Promega, Madison, WI) and sequenced. Comparison of the sequences obtained to the NCBI protein database showed significant similarities to the already-known lcp genes. The translational products of the 433-bp sequences from G. polyisoprenivorans strains VH2 and Y2K exhibited 60% amino acid identity and those of the 425- and 431-bp sequences from G. alkanivorans strain 44187 and G. westfalica strain Kb1 showed 63 and 65% amino acid identity, respectively, to the corresponding region of Lcp from Streptomyces sp. strain K30. This provided for the first time strong evidence that Lcp homologues occur also in rubber-degrading bacteria belonging to the genus Gordonia that grow adhesively on the polymer (see Fig. S2 in the supplemental material).
Identification of the complete nucleotide sequences of lcpVH2 from G. polyisoprenivorans strain VH2 and lcpKb1 from G. westfalica strain Kb1.
To obtain the complete nucleotide sequences of lcp homologues from two Gordonia species, which have been studied in most detail in the past (2, 5, 12, 29, 30), cosmid libraries of G. westfalica strain Kb1 or G. polyisoprenivorans strain VH2 were screened for occurrence of recombinant E. coli clones harboring lcp homologues by clones harboring a degenerated PCR amplification. Hybrid cosmids containing lcp-homologous sequences were then used as template DNA for sequencing reactions by primer walking. Using cosmid pHC79::290, a 2,987-bp sequence was determined containing the complete lcpVH2 from G. polyisoprenivorans strain VH2. Cosmids pHC79::535, pHC79::549, and pHC79::632, which were all identified in the library of G. westfalica strain Kb1, did not yield the complete lcpKb1 sequence due to an internal EcoRI restriction site at position 1130 of lcpKb1. Therefore, the missing nucleotides encoding the C-terminal region of LcpKb1 were amplified by nested PCR as described in Materials and Methods (32). The obtained PCR products were cloned into the pGEM-T Easy vector (Promega, Madison, WI) and sequenced. Two steps of nested PCRs were performed to obtain a 2,505-bp sequence containing the complete lcpKb1.
Phylogenetic analysis of the amino acid sequence derived from the lcp homologues of G. polyisoprenivorans strain VH2 (lcpVH2), G. westfalica strain Kb1 (lcpKb1), the N. farcinica strains E1 (lcpE1) and IFM10152 (lcpIFM10152), Streptomyces sp. strain K30 (lcpK30), and S. coelicolor strain A3(2) [lcpA3(2)] revealed the closest relationship between LcpVH2 and LcpKb1 (see Fig. S3 in the supplemental material); this is not surprising since their hosts belong to the same genus. Both are also more closely related to the Nocardia Lcps than to the Lcp of Streptomyces. LcpVH2 and LcpKb1 exhibited 50 and 52% amino acid identity, respectively, to LcpK30.
Cloning of lcpVH2 and lcpKb1 and complementation experiments in S. lividans strain TK23.
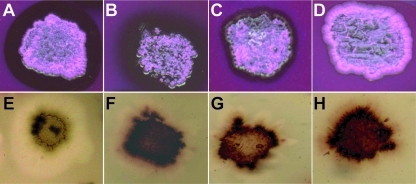
To test the functionality of the two Gordonia lcp genes, fragments containing the 1,635-bp lcpVH2 and the 1,732-bp lcpKb1 sequences were amplified by PCR. Both fragments comprised beside the entire lcp genes about 400 bp of the region upstream of the respective start codons to include their own promoter regions and about 100 bp of the region downstream of the putative stop codon. Both PCR products were cloned via the pGEM-T Easy vector into the newly constructed E. coli-Streptomyces shuttle vector pIJSK, yielding plasmids pIJSK::lcpVH2 and pIJSK::lcpKb1. Plasmid pIJSK::lcpK30, containing the lcp gene of Streptomyces sp. strain K30 including 300 bp upstream and 100 bp downstream of the start and stop codon, was used as positive control. All plasmids were transferred to S. lividans strain TK23, and the resulting recombinant strains were incubated on latex overlay-agar plates containing 1% (wt/vol) glucose for 10 days and stained with Schiff's reagent, to detect aldehydes. Aldehydes occurred around colonies of the positive control (S. lividans strain TK23 containing pIJSK::lcpK30 [Fig. 1A ]), and in addition clearing zones became visible after an incubation of 30 days (Fig. 1E) as observed previously in our laboratory (Henrike Wernsmann, unpublished data), whereas the negative control, S. lividans strain TK23 harboring only the vector pIJSK, did not form aldehydes (Fig. 1D) or clearing zones (Fig. 1H) in latex overlay-agar plates. Since the recombinant strains of S. lividans strain TK23 harboring pIJSK::lcpVH2 (Fig. 1B and 1F) or pIJSK::lcpKb1 (Fig. 1C and 1G) formed aldehydes after 10 days of incubation and clearing zones after 30 days, the homologues lcpVH2 and lcpKb1 from both Gordonia species code for active enzymes.
FIG. 1.
Formation of aldehydes and clearing zones on latex overlay-agar plates. Recombinant strains of S. lividans strain TK23 harboring plasmid pIJSK::lcpK30 (A), pIJSK::lcpVH2 (B), pIJSK::lcpKb1 (C), or pIJSK (D) were incubated for 10 days at 30°C on latex overlay-agar plates containing 0.5% (wt/vol) glucose and stained with Schiff's reagent to detect aldehydes; results are shown in the upper part of the figure. Clearing-zone formation was documented for the same recombinant strains of S. lividans strain TK23 in the corresponding panels at the bottom of the figure after 30 days of incubation at 30°C on latex overlay-agar plates containing 0.5% (wt/vol) glucose. Thiostrepton (25 μg/ml) was added to the medium for plasmid maintenance.
The region with stained aldehydes appeared diffuse without a clearly visible border on latex overlay-agar plates incubated with S. lividans strain TK23 harboring pIJSK::lcpVH2 (Fig. 1B), as with latex overlay-agar plates incubated with S. lividans strain TK23 pIJSK::lcpK30 (Fig. 1A). Furthermore, the stained regions were bigger with recombinant strains of S. lividans strain TK23 harboring pIJSK::lcpK30 or pIJSK::lcpVH2 than with S. lividans strain TK23 harboring pIJSK::lcpKb1, where the stained region was much more closely restricted to the cells (Fig. 1C).
It was also tested if the wild-type G. polyisoprenivorans strain VH2 and G. westfalica strain Kb1 were able to form aldehydes on latex overlay-agar plates. For this, both strains were incubated on latex overlay-agar plates containing 1% (wt/vol) glucose for 10 days and stained with Schiff's reagent, to detect the formation of aldehydes. Little aldehyde formation very close to the cells could be detected on latex overlay-agar plates incubated with G. polyisoprenivorans strain VH2. With G. westfalica strain Kb1 only very weak aldehyde formation occurred. Both strains did not form clearing zones even after 30 days of incubation (data not shown).
GPC analysis of poly(cis-1,4-isoprene) after incubation with recombinant strains of S. lividans strain TK23.
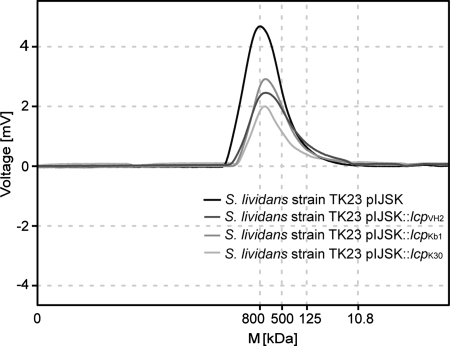
Cleavage of poly(cis-1,4-isoprene) by recombinant strains of S. lividans strain TK23 harboring pIJSK::lcpVH2 or pIJSK::lcpKb1 was verified by GPC. S. lividans strain TK23 containing plasmid pIJSK::lcpK30 or pIJSK was used as a positive or negative control, respectively. Cells of these recombinant strains were cultivated in 50 ml MSM in the presence of 0.25% (wt/vol) poly(cis-1,4-isoprene) with an average molecular mass of 800 kDa (catalog no. 182141; Sigma-Aldrich, Steinheim, Germany) and 0.5% (wt/vol) glucose. GPC analysis of the residual poly(cis-1,4-isoprene) after 8 weeks of incubation was performed as described in Materials and Methods. Poly(cis-1,4-isoprene) incubated with S. lividans strain TK23 harboring pIJSK, which was used as a negative control, showed no change in peak height and area and thus in molecular mass of the 800-kDa polymer (Fig. 2). In contrast, a decrease of 43.3% in height and 27.1% in the peak area (total weight loss of 33.88 mg) occurred after incubation of poly(cis-1,4-isoprene) with strain TK23 harboring pIJSK::lcpKb1, and a decrease of 43.6% in height and 53.8% in the peak area (total weight loss of 67.25 mg) occurred with strain TK23 containing pIJSK::lcpVH2. With the positive control, i.e., strain TK23 harboring pIJSK::lcpK30, the poly(cis-1,4-isoprene) peak showed a decrease in height of 54.6% and in area of 56.7% (total weight loss of 70.88 mg) in comparison to the reference. This clearly indicated that these strains were able to cleave the polyisoprene polymer (Fig. 2). These results agree with observations made for poly(cis-1,4-isoprene) incubated with recombinant strains of S. lividans TK23 strains harboring lcpK30 (33) or lcpE1 (20).
FIG. 2.
Changes of molecular masses of poly(cis-1,4-isoprene) after incubation with recombinant strains of S. lividans strain TK23. The diagram represents GPC elution profiles for residual chloroform-soluble polymers after incubation of synthetic poly(cis-1,4-isoprene) with an average molecular mass (M) of 800 kDa (catalog no. 182141; Sigma-Aldrich, Steinheim, Germany) with recombinant strains of S. lividans strain TK23 harboring plasmid pIJSK::lcpK30, pIJSK::lcpVH2, pIJSK::lcpKb1, or pIJSK for 8 weeks.
Transcription analysis of lcpVH2 in G. polyisoprenivorans strain VH2.
The occurrence of an lcp-homologous gene in G. polyisoprenivorans strain VH2 and other species of Gordonia and the finding that this lcpVH2 gene was functional in S. lividans strain TK23 harboring pIJSK::lcpVH2 raised the question whether lcpVH2 is also expressed in its natural host. Therefore, RT-PCR was employed to investigate transcription of lcpVH2 in G. polyisoprenivorans strain VH2 qualitatively. RT-PCR was done with DNA-free total RNA obtained after DNase I treatment and with primers PVH2_360f(RT) and PVH2_911r(RT) (Table 1). This yielded a PCR product of the expected size of 551 bp representing the central region of lcpVH2 which was not obtained if RNA isolated from acetate-grown cells of strain VH2 was analyzed. This clearly demonstrated that lcpVH2 was transcribed in cells of G. polyisoprenivorans strain VH2 cultivated in the presence of poly (cis-1,4-isoprene) and not in the presence of acetate. The absence of PCR products in the control indicated that the RT-PCR product was not derived from contaminating DNA (Fig. 3).
FIG. 3.
Transcription analysis of lcpVH2 in G. polyisoprenivorans strain VH2. Expression of lcpVH2 was analyzed by RT-PCR of samples containing total RNA isolated from cells of G. polyisoprenivorans strain VH2 in the logarithmic growth phase. The resulting PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide, and a negative image is presented. Cells were grown in MSM with either 0.2% (wt/vol) sodium acetate (lanes 1 and 2) or 0.25% (wt/vol) poly(cis-1,4-isoprene) (lanes 3 and 4) as sole carbon source. Lanes 1 and 3 represent the controls to detect DNA contamination, whereas lanes 2 and 4 represent the RT-PCR assay. A 100-bp DNA ladder (lanes M; MBI Fermentas, Germany) was used for size comparison.
Application of antibodies raised against Lcp of Streptomyces sp. strain K30 for detection of Lcp proteins.
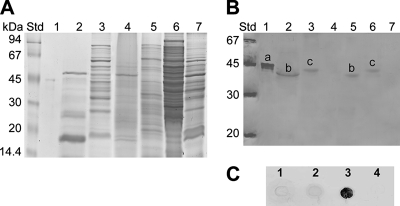
Because the genomes of G. polyisoprenivorans strain VH2 and G. westfalica strain Kb1 encode definitely a functional lcp gene that is transcribed, it should be investigated if detectable Lcp protein occurs in the intra- and extracellular protein fractions of these bacteria. Cells of both strains were grown in MSM, either for 8 weeks in medium containing 0.25% (wt/vol) poly(cis-1,4-isoprene) or for 3 days in medium containing 0.2% (wt/vol) sodium acetate as sole carbon source. To test the functionality of the available antibodies raised against the Streptomyces sp. strain K30 LcpK30, cells of this bacterium were grown in MSM, either for 8 weeks in medium containing 0.2% (wt/vol) latex or for 4 days in medium containing 0.5% (wt/vol) glucose as sole carbon source. To determine heterologous expression of lcp in the recombinants of S. lividans strain TK23 harboring pIJSK::lcpVH2, pIJSK::lcpKb1, or pIJSK::lcpK30, these strains and the negative control, S. lividans strain TK23 harboring pIJSK, were cultivated in MSM with 0.5% (wt/vol) glucose plus 0.2% (wt/vol) latex for 4 days. Crude cell extracts and extracellular protein fractions were prepared and analyzed by SDS-polyacrylamide gel electrophoresis (Fig. 4A) and Western blotting (Fig. 4B). The Western blot prepared with antibodies raised against LcpK30 gave an intensive immunoreaction signal with a protein exhibiting a size of approximately 46 kDa with the purified six-His-tagged LcpK30 protein. This molecular mass corresponded well to that calculated for the six-His-tagged LcpK30. Applying anti-LcpK30 IgG antibodies to the extracellular protein fraction of S. lividans strain TK23 harboring pIJSK::lcpK30, a signal from an approximately 42-kDa protein occurred as was expected for the extracellular LcpK30. Considering cleavage of the signal peptide of LcpK30 between amino acid positions 30 and 31, the latter represented the mature protein as calculated with the SignalP program (15). Western blots of the crude cell extract prepared from this strain revealed a signal representing a protein of approximately 45 kDa. The anti-LcpK30 IgG antibodies also recognized the approximately 42- and 45-kDa proteins in Western blots obtained from the extracellular fraction or from the crude cell extracts of the wild-type Streptomyces sp. strain K30 grown on latex. Crude cell extracts of the negative controls, i.e., S. lividans strain TK23 harboring pIJSK and Streptomyces sp. strain K30 grown on 0.5% (wt/vol) glucose, revealed no immunoreaction signal (Fig. 4B), thus demonstrating that the anti-LcpK30 IgG antibodies were useful and specific against LcpK30.
FIG. 4.
Immunological detection of Lcp from Gordonia species. (A) Electropherogram of an SDS-polyacrylamide gel after separation of proteins from crude cell extracts and extracellular protein fractions. Proteins in the gel were stained with Coomassie brilliant blue R250. (B) Western blot employing anti-LcpK30 IgG antibodies prepared from an SDS-polyacrylamide gel. Std, molecular mass standard; lanes 1, six-His-tagged LcpK30 protein; lanes 2, extracellular protein fraction of S. lividans strain TK23 harboring pIJSK::lcpK30; lanes 3, crude cell extract of S. lividans strain TK23 harboring pIJSK::lcpK30; lanes 4, crude cell extract of S. lividans strain TK23 harboring pIJSK; lanes 5, extracellular protein fraction of Streptomyces sp. strain K30 wild type grown on latex; lanes 6, crude cell extract of Streptomyces sp. strain K30 wild type grown on latex; lanes 7, crude cell extract of Streptomyces sp. strain K30 wild type grown on glucose. Streptomyces sp. strain K30 was grown in MSM either with 0.2% (wt/vol) latex or with 0.5% glucose. The recombinant strain of S. lividans strain TK23 containing plasmid pIJSK::lcpK30 was cultivated in MSM with 0.5% (wt/vol) glucose plus 0.2% (wt/vol) latex as carbon sources. In the Western blot the anti-LcpK30 IgG antibodies recognized the approximately 46-kDa six-His-tagged LcpK30 protein (a), the approximately 45-kDa LcpK30 with signal peptide (b), and the approximately 42-kDa LcpK30 after signal peptide cleavage (c). Furthermore, Lcp present in crude cell extracts was detected in dot blot assays. (C) Crude cell extracts of the recombinant strains of S. lividans strain TK23 containing plasmids pIJSK::lcpVH2 (lane 1), pIJSK::lcpKb1 (lane 2), pIJSK::lcpK30 (lane 3), or pIJSK (lane 4), cultivated for 1 week at 30°C in MSM containing 0.5% (wt/vol) glucose plus 0.2% (wt/vol) latex as carbon sources, were applied to a PVDF membrane, and the immunological analysis was done as described in the manual from the manufacturer (GE Healthcare, Buckinghamshire, United Kingdom).
No immunoreactions occurred with crude cell extracts or extracellular protein fractions of G. polyisoprenivorans strain VH2 and G. westfalica strain Kb1 or of the recombinant S. lividans TK23 strain harboring pIJSK::lcpVH2 or pIJSK::lcpKb1 (data not shown). Thus, dot blot experiments should demonstrate if the generated antibodies raised against the Streptomyces sp. strain K30 LcpK30 were suitable for detection of the Lcp homologues of G. polyisoprenivorans strain VH2 and G. westfalica strain Kb1. Due to possible proteolysis in extracellular fractions, only crude cell extracts were analyzed in order to obtain clear results. Crude cell extracts were obtained from the recombinant S. lividans TK23 strains harboring pIJSK::lcpVH2, pIJSK::lcpKb1, pIJSK::lcpK30, or pIJSK after cultivation in MSM with 0.5% (wt/vol) glucose plus 0.2% (wt/vol) latex for 1 week and were analyzed in a dot blot assay. We applied 5 μl of each sample to a PVDF membrane and used anti-LcpK30 IgG antibodies as described in the manual from the manufacturer (GE Healthcare, Buckinghamshire, United Kingdom). No immunoreactions occurred with crude cell extracts obtained from the recombinant S. lividans TK23 strains harboring pIJSK::lcpVH2, pIJSK::lcpKb1, or pIJSK, whereas the crude cell extract of S. lividans strain TK23 harboring pIJSK::lcpK30 gave an intensive signal (Fig. 4C), thus demonstrating that the anti-LcpK30 IgG antibodies were specific against LcpK30 and did not cross-react with the Lcp homologues.
Construction of a lcpVH2 disruption mutant.
To investigate the relevance of lcpVH2 for degradation of poly(cis-1,4-isoprene) by G. polyisoprenivorans strain VH2, this gene was disrupted. A 1,635-bp sequence, comprising the complete coding region of lcpVH2 (positions 313 to 1590) and adjacent regions, was amplified by PCR using oligonucleotides PVH2_117fBglII and PVH2_1752rBglII (Table 1). As lcpVH2 exhibited a unique restriction site for XhoI 719 nucleotides downstream of the putative start codon, whereas the pGEM-T Easy vector (Promega, Madison, WI) did not contain a cleavage site for XhoI, the 1,635-bp lcpVH2 PCR fragment was cloned into pGEM-T Easy after addition of 3′-A overhangs (A-tailing). The resulting hybrid plasmid containing lcpVH2 was then linearized with XhoI, the 3′-protruding ends were blunted with T4 DNA polymerase, and an approximately 1,000-bp SmaI-SmaI kanamycin resistance cassette (aph) was subsequently inserted at position 719 of lcpVH2. Primers PVH2_117fBglII and PVH2_1752rBglII were then used to amplify the resulting 2,635-bp lcpVH2::aph DNA fragment by PCR, and the fragment was then transferred to G. polyisoprenivorans strain VH2 by electroporation. Recombinant clones with integration of the 2,635-bp lcpVH2::aph fragment into the chromosome were selected on St-I medium agar plates containing kanamycin (50 μg/ml). About 200 kanamycin-resistant colonies were obtained and suspended in 50 μl TE buffer (1 mM EDTA, 10 mM Tris-HCl, pH 8.0), boiled for 15 min, and then used as template for colony PCR. Use of primers PVH2_449f and PVH2_2392r (Table 1), which bind 180 bp upstream of PVH2_117fBglII and 130 bp downstream of PVH2_1752rBglII, yielded four clones (A12, A17, A29, and A34) possessing the 2,944-bp lcpVH2::aph knockout PCR product as expected for the desired lcpVH2 disruption mutants. PCRs employing total DNA of the wild-type strain VH2 and of the four disruption mutants as templates confirmed these results (see Fig. S4A in the supplemental material). Total KpnI-digested DNA isolated from the wild type and the four mutants was separated in an agarose gel, stained with ethidium bromide, and transferred to a nylon membrane and then hybridized with digoxigenin-labeled lcpVH2. DNA also confirmed disruption of lcpVH2 in the mutants since the hybridizing KpnI fragments of these mutants were approximately 1 kbp larger than that of the wild type due to the inserted kanamycin resistance cassette (see Fig. S4B and C in the supplemental material).
Complementation experiments in S. lividans strain TK23 using the 2,635-bp lcpVH2::aph knockout DNA fragment.
The hybrid plasmid pIJSK::lcpVH2::aph was transferred to S. lividans strain TK23. The obtained recombinant strains were incubated on latex overlay-agar plates containing 1% (wt/vol) glucose for 10 days and were stained with Schiff's reagent; no aldehyde formation was detected. Also no clearing-zone formation was observed after 30 days of incubation (data not shown). Therefore, the disrupted lcpVH2::aph gene could, in contrast to lcpVH2, not confer aldehyde or clearing-zone formation on S. lividans strain TK23, thereby clearly demonstrating the inactivation of lcpVH2 by the disruption performed with aph in mutant A17.
Phenotypic analysis of the mutants.
To investigate the effect of lcp disruption on the utilization of poly(cis-1,4-isoprene), mutants A12, A17, A29, and A34 and the wild-type strain VH2 were cultivated in MSM containing 0.25% (wt/vol) poly(cis-1,4-isoprene) as sole carbon and energy source. However, after 30 days of incubation no effect of the inactivation of lcpVH2 on growth with poly(cis-1,4-isoprene) was observed. Both the mutants and the wild type showed similar adhesive growth on the rubber substrate (data not shown). Mutants A12, A17, A29, and A34 were also able to utilize acetonylacetone and methyl-branched isoprenoid compounds such as geranylacetone, farnesol, and squalene like the wild-type strain VH2. These volatile carbon sources were added to MSM agar plates which were sealed with Parafilm.
DISCUSSION
Recently, the lcp gene from the clearing-zone-forming bacterium Streptomyces sp. strain K30 was identified, and its involvement in degradation of poly(cis-1,4-isoprene) was unequivocally demonstrated (33). Lcp was considered a key protein occurring solely for clearing-zone-forming, rubber-degrading gram-positive bacteria, whereas polyisoprenoid degradation by gram-positive bacteria growing adhesively on rubber was considered to rely on a different type of protein. The present study was carried out to investigate whether Lcp homologues occur also in rubber-degrading bacteria belonging to the genus Gordonia, which serve as model organisms to study rubber degradation in adhesively growing bacteria, and whether Lcp is essential for rubber degradation in these bacteria. G. polyisoprenivorans strain VH2 is for example a much better rubber degrader, growing about six times faster in MSM containing poly(cis-1,4-isoprene) as sole carbon source than the clearing-zone-forming Streptomyces sp. strain K30. Recently, the occurrence of lcp-homologous genes was also described for thermophilic adhesively growing strains of N. farcinica (20). Rubber degradation in both clearing-zone-forming and adhesively growing bacteria is still only slightly understood (34).
As G. polyisprenivorans strains VH2 and Y2K, G. alkanivorans strain 44187, and G. westfalica strain Kb1 contain lcp-homologous genes, it must be concluded that Lcp is widespread in gram-positive, rubber-utilizing bacteria exhibiting an adhesive growth on rubber and that Lcp is not restricted to clearing-zone-forming actinobacteria. It is noticeable that so far no rubber-degrading gram-positive bacterium without Lcp has been found (20, 33; this study) and that Lcp comprises obviously a highly conserved type of a novel protein.
The functionality of the lcp homologues from Gordonia sp. was unequivocally demonstrated by clearing-zone and aldehyde formation. The specific characteristics of aldehyde formation regarding size and shape of the stained aldehyde regions in the recombinant strains of S. lividans strain TK23 harboring the lcp homologues from Gordonia sp. in comparison to strain TK23 containing lcpK30 from Streptomyces sp. strain K30 indicate a lower rate of protein secretion in the recombinant lcpVH2- and lcpKb1-containing strains than in the recombinant lcpK30-harboring strain. Due to the present signal sequence there are differences in native protein secretion and especially for secretion of heterologous proteins in S. lividans, as previously described (6, 14, 35, 37). These differences in heterologous protein secretion might also explain the different GPC profiles of poly(cis-1,4-isoprene) molecules after incubation with the recombinant lcpVH2-, lcpKb1-, or lcpK30-harboring strains of S. lividans TK23 (Fig. 2). However, this might be also due to different enzyme activities of the various Lcp homologues.
Furthermore, induction of lcpVH2 transcription in cells of G. polyisoprenivorans strain VH2 during growth on poly(cis-1,4-isoprene) but not on acetate (Fig. 3) confirmed again that Lcp is related to rubber degradation.
Although the Lcp homologues from Gordonia sp. exhibited significant amino acid identity to LcpK30, anti-LcpK30 IgG antibodies raised against Lcp from Streptomyces sp. strain K30 did not cross-react with these Lcp homologues. However, this was the first time that antibodies to an Lcp protein became available, and these antibodies were obviously rather specific for LcpK30 (Fig. 4B).
Disruption of lcpVH2 demonstrated that LcpVH2 has no essential role for rubber or isoprenoid degradation in G. polyisoprenivorans strain VH2. It cannot be excluded that through gene duplication at least one further lcp-homologous gene is present in this bacterium. However, completely sequenced genomes of N. farcinica strain IFM10152 (21) and S. coelicolor strain A3(2) (7) contain only a single lcp gene. Another explanation is the involvement of a different protein type in parallel to Lcp during rubber degradation in Gordonia. The latter could upon inactivation of the lcpVH2 compensate for its rubber-degrading capacity to allow full growth on natural rubber. This assumption is supported by a previously described transposon mutagenesis of G. polyisoprenivorans strain VH2 (5). Interestingly, none of 25,000 characterized mutants were defective in genes whose translational products were homologous to the two known enzymes, LcpK30 and RoxA, catalyzing the primary poly(cis-1,4-isoprene)-cleaving reaction identified in Streptomyces sp. strain K30 (33) and in Xanthomonas sp. strain 35Y (9), respectively. Therefore, it was suggested that the well-characterized rubber-cleaving enzyme RoxA (10) exclusively occurs in gram-negative bacteria (5). Further studies will be necessary to unravel rubber degradation in the gram-positive Gordonia sp. strains.
Supplementary Material
Acknowledgments
Financial support by a grant of the Deutsche Forschungsgemeinschaft (STE 386/10-1) is gratefully acknowledged.
We are also most grateful to Henrike Wernsmann and Quyen Banh for providing unpublished information as well as RNA and plasmid samples. Proteomic mass spectrometry was performed by S. König and A.-M. Mehlich (technical assistance) at the Integrated Functional Genomics core facility of the Interdisciplinary Center for Clinical Research at the Medical Faculty of the University of Münster (Germany) and is gratefully acknowledged.
Footnotes
Published ahead of print on 22 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arenskötter, M., D. Baumeister, M. M. Berekaa, G. Pötter, R. M. Kroppenstedt, A. Linos, and A. Steinbüchel. 2001. Taxonomic characterization of two rubber-degrading bacteria belonging to the species Gordonia polyisoprenivorans and analysis of hypervariable regions of 16S rDNA sequences. FEMS Microbiol. Lett. 205:277-282. [DOI] [PubMed] [Google Scholar]
- 3.Arenskötter, M., D. Baumeister, R. Kalscheuer, and A. Steinbüchel. 2003. Identification and application of plasmids suitable for transfer of foreign DNA to members of the genus Gordonia. Appl. Environ. Microbiol. 69:4971-4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology, vol. 1, 1st ed. John Wiley & Sons, Inc., New York, NY.
- 5.Banh, Q., M. Arenskötter, and A. Steinbüchel. 2005. Establishment of Tn5096-based transposon mutagenesis in Gordonia polyisoprenivorans. Appl. Environ. Microbiol. 71:5077-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender, E., K. P. Koller, and J. W. Engels. 1990. Secretory synthesis of human interleukin-2 by Streptomyces lividans. Gene 86:227-232. [DOI] [PubMed] [Google Scholar]
- 7.Bentley, S. D., K. F. Chater, A. M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 8.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braaz, R., P. Fischer, and D. Jendrossek. 2004. Novel type of heme-dependent oxygenase catalyses oxidative cleavage of rubber poly(cis-1,4-isoprene). Appl. Environ. Microbiol. 70:7388-7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braaz, R., W. Armbruster, and D. Jendrossek. 2005. Heme-dependent rubber oxygenase RoxA of Xanthomonas sp. cleaves the carbon backbone of poly(cis-1,4-isoprene) by a dioxygenase mechanism. Appl. Environ. Microbiol. 71:2473-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 12.Bröker, D., M. Arenskötter, A. Legatzki, D. H. Nies, and A. Steinbüchel. 2004. Characterization of the 101-kilobase-pair megaplasmid pKB1 isolated from the rubber-degrading bacterium Gordonia westfalica Kb1. J. Bacteriol. 186:212-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bullock, W. O., J. M. Fernandez, and J. M. Stuart. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques 5:376-379. [Google Scholar]
- 14.Eckhardt, T., J. Strickler, L. Gorniak, W. V. Burnett, and L. R. Fare. 1987. Characterization of the promoter, signal sequence, and amino terminus of a secreted β-galactosidase from “Streptomyces lividans.” J. Bacteriol. 169:4249-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emanuelsson, O., S. Brunak, G. von Heijne, and H. Nielsen. 2007. Locating proteins in the cell using TargetP, SignalP, and related tools. Nat. Protoc. 2:953-971. [DOI] [PubMed] [Google Scholar]
- 16.Hjelm, H., K. Hjelm, and J. Sjöquist. 1972. Protein A from Staphylococcus aureus. Its isolation by affinity chromatography and its use as an immunosorbent for isolation of immunoglobulins. FEBS Lett. 28:73-76. [DOI] [PubMed] [Google Scholar]
- 17.Hohn, B. 1979. In vitro packaging of lambda and cosmid DNA. Methods Enzymol. 68:299-309. [DOI] [PubMed] [Google Scholar]
- 18.Hohn, B., and J. Collins. 1980. A small cosmid for efficient cloning of large DNA fragments. Gene 11:291-298. [DOI] [PubMed] [Google Scholar]
- 19.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces. A laboratory manual. John Innes Foundation, Norwich, United Kingdom.
- 20.Ibrahim, E. M. A., M. Arenskötter, H. Luftmann, and A. Steinbüchel. 2006. Identification of poly(cis-1,4-isoprene) degradation intermediates during growth of moderately thermophilic actinomycetes on rubber and cloning of a functional lcp homologue from Nocardia farcinica strain E1. Appl. Environ. Microbiol. 72:3375-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa, J., A. Yamashita, Y. Mikami, Y. Hoshino, H. Kurita, K. Hotta, T. Shiba, and M. Hattori. 2004. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. USA 101:14925-14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jendrossek, D., G. Tomasi, and R. M. Kroppenstedt. 1997. Bacterial degradation of natural rubber: a privilege of actinomycetes? FEMS Microbiol. Lett. 150:179-188. [DOI] [PubMed] [Google Scholar]
- 23.Jendrossek, D., G. Tomasi, and H. G. Schlegel. 1997. Mikrobiologischer Abbau von Kautschuk. Nachrichten der Akademie der Wissenschaften in Göttingen. II mathematisch-physikalische Klasse. Nr 1. Vandenhoeck and Ruprecht, Göttingen, Germany.
- 24.Jendrossek, D., and S. Reinhardt. 2003. Sequence analysis of a gene product synthesized by Xanthomonas sp. during growth on natural rubber latex. FEMS Microbiol. Lett. 224:61-65. [DOI] [PubMed] [Google Scholar]
- 25.Kieser, T., M. J. Bipp, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical streptomyces genetics. John Innes Foundation. Norwich, United Kingdom.
- 26.Kummer, C., P. Schumann, and E. Stackebrandt. 1999. Gordonia alkanivorans sp. nov., isolated from tar-contaminated soil. Int. J. Syst. Bacteriol. 49:1513-1522. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Linos, A., and A. Steinbüchel. 1998. Microbial degradation of natural and synthetic rubbers by novel bacteria belonging to the genus Gordona. Kautsch. Gummi. Kunstst. 51:496-499. [Google Scholar]
- 29.Linos, A., A. Steinbüchel, C. Spröer, and R. M. Kroppenstedt. 1999. Gordonia polyisoprenivorans sp. nov., a rubber-degrading actinomycete isolated from automobile tire. Int. J. Syst. Bacteriol. 49:1785-1791. [DOI] [PubMed] [Google Scholar]
- 30.Linos, A., M. M. Berekaa, A. Steinbüchel, K. K. Kim, C. Spröer, and R. M. Kroppenstedt. 2002. Gordonia westfalica sp. nov., a novel rubber-degrading actinomycete. Int. J. Syst. Evol. Microbiol. 52:1133-1139. [DOI] [PubMed] [Google Scholar]
- 31.Mattow, J., U. E. Schaible, F. Schmidt, K. Hagens, F. Siejak, G. Brestrich, G. Haeselbarth, E.-C. Müller, P. R. Jungblut, and S. H. E. Kaufmann. 2003. Comparative proteome analysis of culture supernatant proteins from virulent Mycobacterium tuberculosis H37Rv and attenuated M. bovis BCG Copenhagen. Electrophoresis 24:3405-3420. [DOI] [PubMed] [Google Scholar]
- 32.Mishra, R. N., S. L. Singla-Pareek, S. Nair, S. K. Sopory, and M. K. Reddy. 2002. Directional genome walking using PCR. BioTechniques 33:830-832. [DOI] [PubMed] [Google Scholar]
- 33.Rose, K., K. B. Tenberge, and A. Steinbüchel. 2005. Identification and characterization of genes from Streptomyces sp. strain K30 responsible for clear zone formation on natural rubber latex and poly(cis-1,4-isoprene) rubber degradation. Biomacromolecules 6:180-188. [DOI] [PubMed] [Google Scholar]
- 34.Rose, K., and A. Steinbüchel. 2005. Biodegradation of natural rubber and related compounds: recent insights into a hardly understood catabolic capability of microorganisms. Appl. Environ. Microbiol. 71:2803-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowland, S. S., J. J. Zulty, M. Sathyamoorthy, B. M. Pogell, and M. K. Speedie. 1992. The effect of signal sequences on the efficiency of secretion of a heterologous phosphotriesterase by Streptomyces lividans. Appl. Microbiol. Biotechnol. 38:94-100. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Sathyamoorthy, M., D. Stemke, and M. K. Speedie. 1996. Native and heterologous protein secretion by Streptomyces lividans. Appl. Microbiol. Biotechnol. 46:347-352. [PubMed] [Google Scholar]
- 38.Schlegel, H. G., H. Kaltwasser, and G. Gottschalk. 1961. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch. Mikrobiol. 38:209-222.13747777 [Google Scholar]
- 39.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuchii, A., K. Takeda, and Y. Tokiwa. 1996. Colonization and degradation of rubber pieces by Nocardia sp. Biodegradation 7:41-48. [DOI] [PubMed] [Google Scholar]
- 42.Weber, K., and M. Osborn. 1969. The reliability of molecular weight determinations by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406-4412. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.