Abstract
Between March and May 2006, a Texas hospital identified five Mycobacterium mucogenicum bloodstream infections among hospitalized oncology patients using fluorescence high-performance liquid chromatography analysis of mycolic acids. Isolates from blood cultures were compared to 16 isolates from environmental sites or water associated with this ward. These isolates were further characterized by hsp65, 16S rRNA, and rpoB gene sequencing, hsp65 PCR restriction analysis, and molecular typing methods, including repetitive element PCR, random amplified polymorphic DNA PCR, and pulsed-field gel electrophoresis (PFGE) of large restriction fragments. Three of five patient isolates were confirmed as M. mucogenicum and were in a single cluster as determined by all identification and typing methods. The remaining two patient isolates were identified as different strains of Mycobacterium phocaicum by rpoB sequence analysis. One of these matched an environmental isolate from a swab of a hand shower in the patient's room, while none of the clinical isolates of M. mucogenicum matched environmental strains. Among the other 15 environmental isolates, 11 were identified as M. mucogenicum and 4 as M. phocaicum strains, all of which were unrelated by typing methods. Although the 16S rRNA gene sequences matched for all 14 M. mucogenicum isolates, there were two each of the hsp65 and rpoB sequevars, seven PCR typing patterns, and 12 PFGE patterns. Among the seven M. phocaicum isolates were three 16S rRNA sequevars, two hsp65 sequevars, two rpoB sequevars, six PCR typing patterns, and six PFGE patterns. This outbreak represents the first case of catheter-associated bacteremia caused by M. phocaicum and the first report of clinical isolates from a U.S. hospital. The investigation highlights important differences in the available typing methods for mycobacteria and demonstrates the genetic diversity of these organisms even within narrow confines of time and space.
Among the 125 species of Mycobacterium that are currently recognized officially (http://www.bacterio.cict.fr/), approximately 60 species are considered to be rapidly growing mycobacteria (RGM), defined by their ability to form visible colonies on solid agar within 7 days (23). Most of them have been associated with human disease (5, 9, 32, 33), and many are well described as causes of outbreaks or pseudo-outbreaks in health care settings, with bloodstream infections being one of the most commonly reported (9). RGM species are ubiquitous in both public and hospital water systems, where their persistence in biofilms and amoeba is enabled by their ability to tolerate disinfectants, chlorination, and temperature extremes (2, 29, 33). Mycobacterium mucogenicum was originally described as an organism which exhibited features of both Mycobacterium fortuitum and Mycobacterium chelonae and was designated in 1981 as an M. chelonae-like organism (24). It was confirmed to be a distinct species by 16S rRNA gene sequence analysis in 1995 (25). As a pathogen, M. mucogenicum has been found to be most often associated with posttraumatic wound infections and central venous catheter (CVC)-associated infections (34). Contamination of CVCs during bathing was found to be the route of infection by M. mucogenicum in an outbreak of bacteremias among five bone marrow transplant patients and one oncology patient at a tertiary-care hospital in Minnesota (18). A cord blood transplant recipient who developed a CVC infection after the patient herself had flushed her catheter with tap water was also reported (12).
Although conventional phenotypic testing parameters, such as carbon source utilization or growth characteristics on various media or in the presence of inhibitors, continue to distinguish Mycobacterium species, most novel species are currently recognized by unusual molecular parameters, including patterns of mycolic acid in the cell wall as determined by high-performance liquid chromatography (HPLC) and genomic sequence polymorphisms, most notably within signature regions of the gene coding for 16S rRNA. Criteria that defined distinct species having at least 1% divergence in the 16S rRNA sequence were published in 2003 (31). This definition was the basis for establishing three major taxonomic groups of pathogenic RGM: the Mycobacterium fortuitum group, the Mycobacterium chelonae-Mycobacterium abscessus group, and the Mycobacterium smegmatis group (4). It has since been proposed that the concatenation of multiple gene sequences offers the most robust phylogeny of mycobacteria (10), and to this end, a subsequent analysis of five genomic regions, including 16S rRNA, rpoB, hsp65, recA, and sodA, refined these RGM groups into separate groups: the M. mucogenicum group, the Mycobacterium mageritense group, and the Mycobacterium wolinskyi group. The M. mucogenicum group was found to be closely related to the M. chelonae-M. abscessus group, and one of three M. mucogenicum strains analyzed had substantial sequence differences from two others (3). Although it is now accepted that complete 16S rRNA sequence homology may be shared among some mycobacterial species, a polymorphic region of rpoB (nucleotides 2533 to 3255) with at least 3% sequence divergence among species was used as the principal criterion to report three novel RGM species in 2006: M. bolletii, M. phocaicum, and M. aubagnense (1).
Restriction fragment length polymorphic analysis of large genomic restriction fragments using pulsed-field gel electrophoresis (PFGE) has become the standard method to evaluate the relatedness of strains of several clinically important nontuberculous mycobacterium (NTM) species (33). More recently, PCR-based methods, such as repetitive element (rep) PCR and random amplified polymorphic DNA (RAPD) electrophoresis, have been applied to the task of comparing NTM isolates. Although these methods are considerably easier and faster than PFGE, their results are typically interpreted only presumptively pending final interpretation of PFGE patterns. The RAPD method utilizes a single primer in a low-annealing-temperature PCR, and the sites of primer annealing on opposite strands are affected by secondary structures of the target DNA template. Among RGM species, this method was first used at the CDC to evaluate collections of M. abscessus, M. chelonae, and M. mucogenicum for epidemiologic investigations, and the typing results in these studies were found to correlate with conventional typing results (8, 18, 19). The rep PCR typing method also utilizes one primer, but annealing is based upon DNA sequences that are present in most bacterial genera, including Mycobacterium. Following the demonstration that PCR-based typing of Mycobacterium tuberculosis and M. abscessus could be performed using an automated microfluidic lab chip instrument (8), a commercial kit for rep PCR using this automated platform (DiversiLab; Bacterial Barcodes, Inc., Athens, GA) was introduced and found to correlate with the results of conventional insertion sequence restriction fragment length polymorphic typing methods (7, 14).
On 3 May 2006, the Texas Department of State Health Services (TDSHS) notified the CDC of five oncology patients at hospital A who had developed bloodstream infections caused by M. mucogenicum. The ensuing environmental sampling study was undertaken to evaluate the extent of the genetic diversity of isolates of this species in water distribution systems affecting these patients. We used a multiphasic approach to evaluate the relationship among environmental and patient isolates in this outbreak.
MATERIALS AND METHODS
Outbreak investigation.
To evaluate risk factors for M. mucogenicum infection, a case-control study was performed. Case patients were patients in hospital A's oncology unit from 1 March to 10 May 2006 (>72 h admission) with positive blood cultures identified as M. mucogenicum by fluorescence HPLC mycolate analyses (16). Three controls were randomly selected for each case from patients who had at least one inpatient stay in hospital A's oncology unit >72 h between 1 March and 10 May 2006. Controls were not required to have blood cultures negative for M. mucogenicum. For controls with multiple admissions, the first hospitalization from 1 March to 10 May 2006 was used. Clinical and demographic data were abstracted from patients' medical records. Statistical analyses were performed using SPSS version 13.0 (Chicago, IL).
Environmental isolates.
Environmental samples were obtained from hospital sinks, showers, and two municipal water plant tanks. Samples of biofilms were obtained from the inside of faucets with transport swabs (Fisher Healthcare, Houston, TX) moistened with 10 ml Butterfield buffer (0.00425% monopotassium phosphate). Biofilm samples of the inside of two municipal water tank faucets were obtained with culture swabs (Becton Dickinson & Co. [BD], Sparks, MD). One-liter water samples were collected in sterile bottles (Nalge Nunc International, Rochester, NY), and sodium thiosulfate was added to a final concentration of 0.01% (wt/vol). The outside of faucets or showers was disinfected with 70% isopropanol before sampling. Water was collected immediately after turning on the tap. One ice machine, providing ice for patient consumption, was sampled by scooping ice into the sample bottle and allowing it to thaw. A handheld shower unit (head and tubing) from a seventh-floor case patient room and a section of copper plumbing removed from an eighth floor bathroom during renovations were shipped to the laboratory with the ends wrapped in Parafilm to prevent drying.
Biofilm swabs were decontaminated with cetyl pyridinium chloride (CPC) (Sigma-Aldrich, St. Louis, MO), 0.005% (wt/vol), for 30 min at room temperature, vortexed, and centrifuged at 4,000 × g for 20 min (17). The swabs were removed and streaked directly onto Middlebrook 7H10 agar plates (BD). The pellets were resuspended in 250 to 500 μl of water, and the suspension was streaked on R2A (BD) and Middlebrook 7H10 agar plates. Culture swabs were removed from the original tube, placed in 15-ml tubes containing 2 ml of 0.005% CPC, and processed as described above.
Water samples were decontaminated with 0.005% CPC, incubated at room temperature for 10 min, and centrifuged 20 min at 4,000 × g. Pellets were resuspended in 1 ml of complete Middlebrook 7H9 broth (Remel Co., Lenexa, KS). Aliquots were spread plated on R2A and 7H10 agar and incubated in 7H9 broth. All cultures were incubated at 30°C.
The outside of the handheld shower tubing and the copper pipe were disinfected with 10% bleach for 30 s, rinsed with 0.01% sodium thiosulfate, and wiped with 70% isopropanol. Surface swabs were streaked on R2A agar to confirm sterility. The metal casing of the shower tubing was cut and spread apart to expose the inner rubber tube. The outside of the rubber tube was disinfected in the same manner, clamped, and cut into 1-cm lengths. Rubber tubing sections were rinsed in phosphate-buffered saline and placed in 10 ml of phosphate-buffered saline containing 0.1% Tween 80. Biofilm was removed by three cycles of sonication/vortexing for 1 min/30 seconds each. Suspended biofilm was exposed to 0.005% CPC for 30 min, diluted in Butterfield buffer, and streaked on R2A and 7H10 agar plates. The copper pipe was aseptically cut into 1-cm lengths, each of which was then cut in half longitudinally and processed as described above.
Broth cultures and nonmotile cream or white colonies from agar plates were subcultured on 7H10 agar plates and tested for acid fastness using the Kinyoun method (17).
Chemical analyses.
Mycolic acids from patient isolates were initially prepared, esterified, and then subjected to fluorescence detection HPLC as previously described (16). The mycolate patterns of environmental and patient isolates were subsequently tested using a standard UV HPLC method (6).
Molecular analyses.
Crude DNA preps for PCR amplification methods were prepared using a bead agitation method (8). PCR restriction analysis (PRA) of a 441-bp region of hsp65 was performed as previously described using primer pair Tb11 and Tb12 (8, 21, 27). Regions of polymorphic genes that were sequenced included a hypervariable region of rpoB (nucleotides 2900 to 3328, M. mucogenicum numbering; GenBank accession no. AY147174) (1), hsp65 (nucleotides 14 to 1622, Mycobacterium tuberculosis numbering; GenBank accession no. AE000516), and 16S rRNA (nucleotides 16 to 1459 excluding primer binding sites, M. mucogenicum numbering; accession no. AY457075). The rpoB and hsp65 amplification and sequencing primers designed for this study are summarized in Table 1. The 16S rRNA gene was amplified as previously described (22). Sequence data were edited and compiled using the Wisconsin Sequence Analysis 11 package (Genetics Computer Group, Madison, WI). The multiple sequence alignments of 16S rRNA, hsp65, and rpoB were performed using the Clustal X program, v.1.81 from the PHYLIP software package (30). A phylogenetic tree was obtained from 16S rRNA, hsp65, and rpoB by using the neighbor-joining method with Kimura's two-parameter distance correlation model with 1,000 bootstrap replication in MEGA version 3.1 (20).
TABLE 1.
Oligonucleotide primers
| Gene or assay | Nucleotide positions or sequence (length) | Primer (usea) | GenBank accession no./reference |
|---|---|---|---|
| hsp65 | 14-33 (20) | 14f (S) | AE000516 |
| 1622-1603 (20) | 1622r (S) | ||
| 517-534 (18) | 517f (S) | ||
| 559-542 (18) | 559r (S) | ||
| 1102-1119 (18) | 1102f (S) | ||
| 1175-1158 (18) | 1175r (S) | ||
| 396-416 (21) | Tb11 (P) | 27 | |
| 836-817 (20) | Tb12 (P) | ||
| rep PCR | 5′-CTACGGCAAGGCGACGCTGACG (22) | Box A1R (T) | 11 |
| RAPD PCR | 5′-TGGTCGCGGC (10) | RAPD1 (T) | 35 |
| rpoB | 2900-2919 (20) | 2900f (S) | AY147174; 1 |
| 3328-3308 (20) | 3328r (S) | ||
| 16S rRNA gene | 16-32 (17) | f38 (S) | AY457075; 3 |
| 1459-1443 (17) | r1464 (S) |
S, sequencing primer; P, PCR amplification; T, PCR typing.
Analysis of large restriction fragments using PFGE was performed as previously described (15). Following digestion with AseI, chromosomal DNA fragments were subjected to electrophoresis using a Chef-DR III pulsed-field gel apparatus (Bio-Rad Laboratories, Hercules, CA) and ramped pulse times from 4 to 20 seconds for 20 h at 200V. Amplification for RAPD typing was performed using primer RAPD1 and conditions that were previously described (8). Primer BOXA1R and previously described conditions were used for rep PCR amplifications (11), except that HotStar Taq polymerase master mix (Qiagen, Valencia, CA) and an initial incubation for 15 min at 95°C during thermocycling were used (7). Portions of rep PCR or RAPD samples (5 μl) were electrophoresed in 1.5% agarose-Tris-borate-EDTA gels for 3 h at 100 V. After electrophoresis, gel lanes for PFGE, rep PCR, or RAPD procedures were compared with one another and isolates for which banding patterns differed from all other isolates by at least three DNA bands were interpreted as unrelated strains (28).
Nucleotide sequence accession numbers.
DNA sequences have been deposited in the GenBank database with the accession numbers EF551386 to EF551408 (16S rRNA), EF551409 toEF551431 (hsp65), and EF551432 to EF551457 (rpoB).
RESULTS
Outbreak investigation.
Five case patients were identified at hospital A (Table 2). The mean duration between CVC placement and admission date for four of five case patients was 32 days (one case patient was excluded from this analysis due to the unreliability of reported information), significantly less than 492 days for nine controls with CVCs (P = 0.025). All cases and controls reported bathing, and no statistically significant association was identified between case status and exposure to any hospital water source. Case patients had more days of neutropenia (defined as an absolute neutrophil count of <500) while in the oncology unit than the controls, but this difference was not significant (P = 0.197). Patient isolates from blood cultures were initially identified as M. mucogenicum by computer software analysis of fluorescence detection HPLC mycolate patterns.
TABLE 2.
Patient data
| Characteristic | Value
|
P valuea | |
|---|---|---|---|
| Case (n = 5) | Control (n = 15) | ||
| Male (%) | 2 (40) | 5 (33) | 0.787 |
| Median age | 67 | 68 | 0.887 |
| Mean no. of days in hospital during admission | 17.8 | 15.5 | 0.595 |
| Mean no. of showers taken during admission | 10.4 | 8.4 | 0.548 |
| No. with CVC in place at time of admission (%) | 5 (100) | 9 (60) | 0.091 |
| Mean no. of days CVC in place before admissionb | 32 | 492 | 0.025 |
| Mean no. of days of neutropeniac during admission | 3.8 | 1.1 | 0.197 |
| No. with skin breakdown documented in chart (%) | 2 (40) | 4 (27) | 0.573 |
| No. in unit following reported plumbing failure (13 March 2006) (%) | 3 (60) | 7 (35) | 0.176 |
Chi-square test for categorical variables; Student's t test for continuous variables.
Excludes value for one case patient for whom documentation was unreliable.
Absolute neutrophil count of <500.
A generator failure occurred during the outbreak time period, which reportedly dropped water pressure throughout the hospital, allowing gravity to direct water flow from plumbing on floors 8, 9, and 10 into areas of regular use by patients on floor 7, where the oncology unit was located. Floors 8 through 10 were undergoing renovations at the time, and hence the water in pipes on that floor had been stagnant. This event occurred 5 days prior to the first case patient's infection and 44 days prior to the last. An epidemiological investigation was subsequently undertaken.
Twelve swab samples, including two from municipal tanks, were obtained. Seven of these were positive for nonpigmented RGM organisms: six from inside the hospital and one from a municipal tank. Fifteen water samples (2 from municipal tanks, 1 ice sample from the hospital, and the rest from within the hospital) were obtained, and 12 were positive for NTM, all within the hospital. Eleven of the positive water samples contained RGM organisms, including the ice; two sink samples also contained M. gordonae. One sample of hot water from a sink did not contain RGM species but did contain a slowly growing Mycobacterium species presumptively assigned to the M. simiae/M. avium group (13). The showerhead tubing from patient room 3 contained RGM organisms. The copper plumbing piece, although nearly dry when sampled, contained an isolate of M. gordonae. All samples that were positive for NTM, except from the copper plumbing piece, also contained a variety of heterotrophic bacteria and/or fungi, as evidenced by growth on R2A.
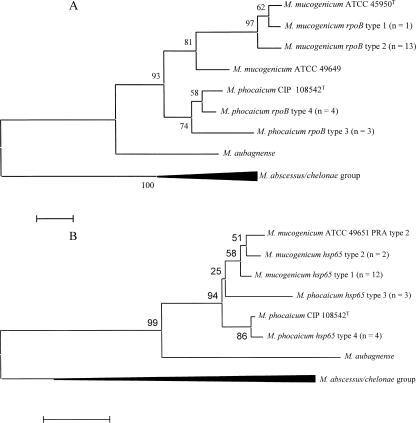
Among the RGM isolates, mycolic acid patterns determined by UV HPLC matched for 16 environmental and 5 patient isolates. This pattern was typical for M. mucogenicum and distinguished the 21 isolates by the appearance of two sets of mycolic acid peaks, one of which extracted earlier than major peaks observed with other nonpigmented RGM species (34). This subset was then evaluated by molecular methods (see Fig. 1 to 3). The rpoB sequences (429 bp) of the 21 isolates were compared with those of M. mucogenicum strains ATCC 49649 (accession no. AY147171) and ATCC 49650T (accession no. AY147170) and M. phocaicum strain CIP 108542T. Fourteen isolates matched M. mucogenicum, and seven matched M. phocaicum, according to previously defined criteria (1). Thirteen of 14 M. mucogenicum isolates (93%), including the 3 patient isolates, had rpoB type 2 and the fourteenth had type 1, both of which were more closely related to strain ATCC 49650T than to strain ATCC 49649 (Fig. 1A). There were two additional sequevars among the seven M. phocaicum isolates, neither of which matched the control strain (CIP 108542T). rpoB sequences for the two patient isolates of M. phocaicum did not match one another, but one shared an identical rpoB sequence with two environmental M. phocaicum isolates (type 3) and the other (isolate no. 6) with three environmental isolates (type 4), two of which also shared the same 16S rRNA gene sequence (16S rRNA type 2). The predominant 16S rRNA gene sequence (type 1) was found in 17 isolates, including all 14 M. mucogenicum isolates and 3 environmental M. phocaicum isolates (Table 3). Two additional sequevars were observed in three and one M. phocaicum isolate, respectively, and one of these (type 2) was shared among one patient isolate (isolate no. 6) and two environmental isolates (Table 3). In all, six polymorphisms were observed among the three 16S rRNA types, all within the first 500 bp. Sequence variation within hsp65 was observed only within 402 nucleotides of the PRA region (nucleotides 396 to 836). Two hsp65 sequevars were each observed among the 14 M. mucogenicum and 7 M. phocaicum isolates (Fig. 1B; Table 3). The type 1 hsp65 sequevar predominated among patient and environmental M. mucogenicum isolates, and the type 4 sequence was observed in two patient and two environmental M. phocaicum isolates. However, only one of the patient M. phocaicum isolates (isolate no. 6) also matched the rpoB and 16S rRNA gene sequences of these two environmental isolates.
FIG. 1.
Phylogenetic analyses of rpoB (429 nucleotides) (A) and hsp65 (402 nucleotides) (B) among 14 isolates of M. mucogenicum and 7 isolates of M. phocaicum from hospital A. Phylogenetic trees were prepared using the neighbor-joining method, and bootstrap percentages are shown at nodes. Bars represent 2.0% of the total rpoB sequence (approximately nine nucleotides) and 1% of the total hsp65 sequence (approximately 5 nucleotides). Type designations and isolates are shown in Table 3.
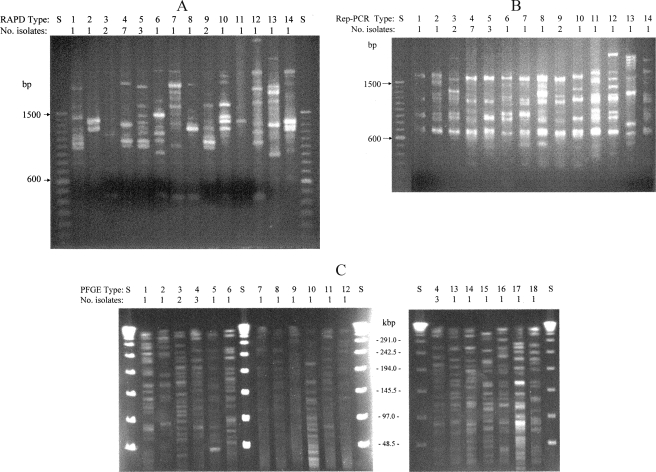
FIG. 3.
Typing patterns found among 14 M. mucogenicum and 7 M. phocaicum isolates from a Texas hospital. RAPD patterns (A) and rep PCR patterns (B). Patterns 1, 4, 5, 7, 8, and 10 among M. mucogenicum isolates and patterns 2, 3, 6, 9, and 11 among M. phocaicum isolates are included. Patterns 12 to 14 included M. mucogenicum control strain ATCC 49651 (pattern 12) and strains from an unrelated outbreak study (18). S, 100-bp ladder. (C) Large restriction fragment PFGE patterns. Lanes show patterns and number of isolates with each pattern. Patterns were found among M. mucogenicum isolates except for M. phocaicum patterns 2, 3, 5, 6, 8 and 13. S, 48-kb ladder.
TABLE 3.
Genotypes of 5 patient and 16 environmental isolates from hospital A in Texas, 2006
| Isolate no. | Sourceb | Species | Typea
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| PRA | hsp65c | rpoBc | 16S rRNA | PFGE | RAPD PCR | rep PCR | |||
| 3 | Blood | M. mucogenicum | 1 | 1 | 2 | 1 | 4 | 4 | 4 |
| 4 | Blood | M. mucogenicum | 1 | 1 | 2 | 1 | 4 | 4 | 4 |
| 5 | Blood | M. mucogenicum | 1 | 1 | 2 | 1 | 4 | 4 | 4 |
| 9 | Room 1 SCW | M. mucogenicum | 1 | 1 | 2 | 1 | 7 | 4 | 4 |
| 13 | Room 2 SCW | M. mucogenicum | 1 | 1 | 2 | 1 | 11 | 9 | 9 |
| 16 | Room 2 SHW | M. mucogenicum | 1 | 1 | 2 | 1 | 14 | 4 | 4 |
| 24 | 10th-floor DEP | M. mucogenicum | 1 | 1 | 2 | 1 | 18 | 4 | 4 |
| 21 | Nurse station faucet | M. mucogenicum | 3 | 2 | 2 | 1 | 15 | 10 | 10 |
| 11 | Room 1 ShH | M. mucogenicum | 3 | 2 | 1 | 1 | 9 | 8 | 8 |
| 12 | Room 1 HS | M. mucogenicum | 1 | 1 | 2 | 1 | 10 | 5 | 5 |
| 1 | Room 3 ShH | M. mucogenicum | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| 14 | Room 2 ShS | M. mucogenicum | 1 | 1 | 2 | 1 | 12 | 5 | 5 |
| 22 | Room 2 ShH | M. mucogenicum | 1 | 1 | 2 | 1 | 16 | 5 | 5 |
| 23 | Room 2 faucet | M. mucogenicum | 1 | 1 | 2 | 1 | 17 | 7 | 7 |
| 10 | Room 1 HS | M. phocaicum | 1 | 3 | 3 | 1 | 8 | 4 | 4 |
| 15 | Room 2 HS | M. phocaicum | 1 | 3 | 3 | 1 | 13 | 9 | 9 |
| 2 | Blood | M. phocaicum | 2 | 4 | 3 | 1 | 2 | 2 | 2 |
| 6 | Blood | M. phocaicum | 2 | 4 | 4 | 2 | 3 | 3 | 3 |
| 19 | Room 3 HS | M. phocaicum | 2 | 4 | 4 | 2 | 3 | 3 | 3 |
| 25 | Nurse station water | M. phocaicum | 2 | 4 | 4 | 2 | 6 | 11 | 11 |
| 7 | Municipal WTS | M. phocaicum | 1 | 3 | 4 | 3 | 5 | 6 | 6 |
Type designations are arbitrarily assigned. PRA patterns are shown in Fig. 2.
SCW, sink cold water; SHW, sink hot water; DEP, dead end pipe; ShH, showerhead; HS, hand shower; ShS, shower swab; WTS, water tank swab. Seventh-floor patient room numbers are coded.
Designations are sequevars shown in Fig. 1.
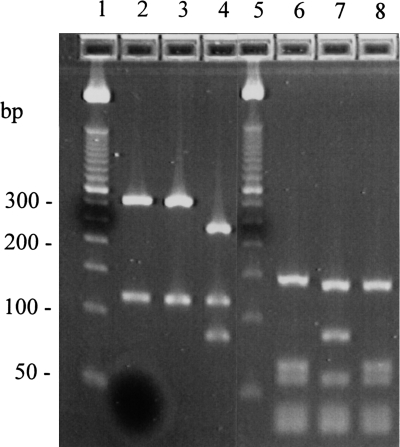
Three hsp65 PRA patterns (PRA types) were observed (Fig. 2), and the most common of these was PRA type 1 for which major restriction fragment sizes were 320 bp and 115 bp for BstEII and 140 bp, 65 bp, and 55 bp for HaeIII. This pattern was observed in 12 of 14 M. mucogenicum isolates (85.7%), including all 3 patient isolates and 3 environmental M. phocaicum isolates (42.9%). The PRA type 2 pattern which was found in two patient and two environmental M. phocaicum isolates had major BstEII fragments that were 320 bp and 115 bp and HaeIII fragments that were 140 bp, 85 bp, and 55 bp. These two patterns, PRA type 1 and 2, were previously reported for M. mucogenicum ATCC 49649 and ATCC 49651, respectively (26). An additional PRA type was found in two environmental M. mucogenicum isolates. (Fig. 2).
FIG. 2.
hsp65 PRA patterns of M. mucogenicum and M. phocaicum isolates from a Texas hospital. BstEII digests are in lanes 2 to 4 and HaeIII digests are in lanes 6 to 8. Patterns and numbers of isolates are as follows: type 1 (lanes 2 and 6), 320 + 115 bp for BstEII and 140 + 65 + 55 bp for HaeIII, 12 M. mucogenicum and three M. phocaicum; type 2 (lanes 3 and 7), 320 + 115 bp for BstEII and 140 + 85 + 55 bp for HaeIII, four M. phocaicum; type 3 (lanes 4 and 8), 240 + 115 + 85 bp for BstEII and 145 + 65 + 55 bp for HaeIII, two M. mucogenicum. Size standards (50-bp ladder) are in lanes 1 and 5.
The 21 isolates were represented by 11 distinct gel banding patterns (designated arbitrarily) as determined using either rep PCR or RAPD electrophoresis, all of which were different from the control strain ATCC 49651 (Fig. 3). The most common pattern (type 4) was found in three patient and four environmental isolates. One patient isolate of M. phocaicum matched an environmental isolate, not only by rpoB, 16S rRNA gene, and hsp65 sequence analysis but also by rep PCR, RAPD, and PFGE (type 3) (Table 3). The fifth patient isolate had unique patterns by the three typing methods. The most diversity among the isolates was observed using PFGE (Fig. 3C) for which 18 patterns and two small clusters were observed among the 21 isolates (Fig. 3C; Table 3).
DISCUSSION
This investigation demonstrates the widespread environmental prevalence and significant genetic diversity of NTM that were present in the water supplies of hospital A. It also underscores the need for careful analyses of results of the complex myriad of molecular methods that are currently available for identification and typing of NTM isolates, considering that all of the isolates were initially identified as M. mucogenicum but that two patient isolates were later found to be M. phocaicum. It seems likely that in previous outbreaks or samplings of M. mucogenicum, closely related species such as M. phocaicum may have been present, since identification has most often been based upon mycolic acid patterns which are indistinguishable for these two species.
As in most CVC infections caused by RGM organisms, the recovery of patients was achieved primarily by removal of the catheters without the need for specific antimicrobial therapy (32). From a therapeutic perspective, both species are susceptible to amikacin, cefoxitin, clarithromycin, imipenem, fluoroquinolones, and minocycline, and in contrast, only M. phocaicum is resistant to amoxicillin and trimethoprim-sulfamethoxazole (1).
Genetically similar strains of M. mucogenicum in clinical and environmental isolates suggest exposure to water was an infection risk in these oncology patients with CVCs. Due to a hospital construction project, the three floors directly above the oncology unit had been closed for several months prior to the outbreak, which would have reduced water flow through all pipes in these areas. As a result, the flow of water and possible sloughing of biofilm from floors 8 through 10 to floor 7 during the generator failure possibly created a very high load of NTM in the water on floor 7. Although this increased organism burden may have contributed to the infections among oncology patients who had recently placed CVCs, the exact source of the infections remains unknown, since only one of the patient isolates was genetically matched to an environmental isolate. However, our findings are consistent with environmental contamination, given that patients with recently placed CVCs may have been less experienced with proper covering during bathing and at greater risk to contaminate lines. Likewise, recently placed lines might not have had sufficient time for the formation of a fibrinous cuff around the insertion site, which would also facilitate the ingress of organisms. The molecular typing information supports this hypothesis, since one common strain of M. phocaicum was found in a patient and the water. Our findings underscore the importance of careful infection control training for all patients with CVCs.
The absence of clusters of any particular strain suggests that there was little evidence for cross-contamination of the various environmental sites sampled. There is further evidence that RGM species may persist for long periods of time, i.e., within aqueous biofilms, and the tendency for polymorphisms within the genes and sequences studied likely increases with time (10). There are no time course data, however, to prove that polymorphisms arise within a single clone in any given environmental M. mucogenicum clone. We concluded that only isolates that matched in every parameter should be considered common strain matches. When the extreme diversity of NTM organisms in aqueous environments is considered, however, it is nonetheless difficult to exclude water systems as likely sources of infections.
We found matches among three patient isolates and between a fourth patient isolate and an environmental isolate on the basis of all six methods used (Table 3). Strain clusters identified by rep PCR and RAPD electrophoresis were concordant, which indicates that either of these typing methods is suitable for these two species. Although PCR-based typing methods are considerably more convenient than PFGE, they do not offer the same degree of strain resolution. The fewest number of clusters was observed using PFGE, for which 18 distinct PFGE patterns among the 21 isolates support an equally large number of strains from the various sources sampled. The six PFGE patterns found among the M. phocaicum isolates likely extend the number of strains of this novel species to seven, including the type strain CIP 108542T (1).
The region of rpoB chosen for analysis was essentially a hypervariable region consisting of 429 nucleotides within a 723-bp region defined by Adékambi, whose sequence was ≥3% variable among 20 type strains of 15 RGM species and was ≤1.7% variable among strains within any of these species (1).
It has been noted that even full-length sequencing of 16S rRNA genes will not distinguish between the type strains of M. mucogenicum and recently defined M. phocaicum (1). Since we found extensive conservation throughout the 16S rRNA gene among RGM species that have been associated with outbreaks in health care institutions, the need for multiple gene analyses to confirm the identification of species is underscored. It was recently determined that analysis of hsp65, 16S rRNA genes, rpoB, sod, and recA (in a concatenated fashion) enabled the most robust phylogenetic distinction of Mycobacterium species (3, 10).
Although more sequevars were found among hsp65 and rpoB sequences than among 16S rRNA gene sequences, there was less diversity than was observed for PCR typing or PFGE results. We found that an hsp65 PRA pattern that was previously reported only in M. mucogenicum (21, 26) was present in some isolates of both species. Although we found sequence variation in the PRA region (nucleotides 396 to 836), we also found differences throughout hsp65. Among the three genes we sequenced, it is likely that molecular diversity among groups of organisms, such as those under epidemiologic investigation, will increase in a fashion that is parallel to the number and sophistication of tests available for such investigations.
Acknowledgments
We thank Allison Abell, Eric Amster, Anne Groda, Vivienne Heines, Eric Miller, Neil Pascoe, and Linda Simmons for assistance with the outbreak investigation.
Footnotes
Published ahead of print on 29 February 2008.
REFERENCES
- 1.Adékambi, T., P. Berger, D. Raoult, and M. Drancourt. 2006. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56:133-143. [DOI] [PubMed] [Google Scholar]
- 2.Adékambi, T., S. B. Salah, M. Khlif, D. Raoult, and M. Drancourt. 2006. Survival of environmental mycobacteria in Acanthamoeba polyphaga. Appl. Environ. Microbiol. 72:5974-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adékambi, T., and M. Drancourt. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 54:2095-2105. [DOI] [PubMed] [Google Scholar]
- 4.Adékambi, T., P. Colson, and M. Drancourt. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41:5699-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown-Elliott, B. A., and R. J. Wallace, Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15:716-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler, W. R., M. M. Floyd, V. Silcox, G. Cage, E. Desmond, P. S. Duffey, L. S. Guthertz, W. M. Gross, K. C. Jost, Jr., S. L. Ramos, L. Thibert, and N. Warren. 1996. Standardized method for HPLC identification of mycolic acids of Mycobacteria. Centers for Disease Control and Prevention, Atlanta, GA.
- 7.Cangelosi, G. A., R. J. Freeman, K. N. Lewis, D. Livingston-Rosanoff, K. S. Shah, S. J. Milan, and S. V. Goldberg. 2004. Evaluation of a high-throughput repetitive-sequence-based PCR system for DNA fingerprinting of Mycobacterium tuberculosis and Mycobacterium avium complex strains. J. Clin. Microbiol. 42:2685-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooksey, R. C., J. Limor, G. P. Morlock, and J. T. Crawford. 2003. Identifying Mycobacterium species and strain typing using a microfluidic labchip instrument. BioTechniques 35:786-794. [DOI] [PubMed] [Google Scholar]
- 9.De Groote, M. A., and G. Huitt. 2006. Infections due to rapidly growing mycobacteria. Clin. Infect. Dis. 42:1756-1763. [DOI] [PubMed] [Google Scholar]
- 10.Devulder, G., M. Perouse de Montclos, and J. P. Flandrois. 2005. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 55:293-302. [DOI] [PubMed] [Google Scholar]
- 11.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleming, G. A., H. Frangoul, T. S. Dermody, and N. Halasa. 2006. A cord blood transplant recipient with Mycobacterium mucogenicum central venous catheter infection after infusion of tap water. Pediatr. Infect. Dis. J. 25:567-569. [DOI] [PubMed] [Google Scholar]
- 13.Floyd, M. A., L. S. Guthertz, V. A. Silcox, P. S. Duffy, Y. Jang, E. P. Desmond, J. T. Crawford, and W. R. Butler. 1996. Characterization of an SAV organism and proposal of Mycobacterium triplex sp. nov. J. Clin. Microbiol. 34:2963-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Healy, M., J. Huong, T. Bittner, M. Lising, S. Frye, S. Raza, R. Schrock, J. Manry, A. Renwick, R. Nieto, C. Woods, J. Versalovic, and J. R. Lupski. 2005. Microbial DNA typing by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 43:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hector J. S., Y. Pang, G. H. Mazurek, Y. Zhang, B. A. Brown, and R. J. Wallace, Jr. 1992. Large restriction patterns of genomic Mycobacterium fortuitum DNA as strain-specific markers and their use in epidemiologic investigation of four nosocomial outbreaks. J. Clin. Microbiol. 30:1250-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jost, K. C., Jr., D. F. Dunbar, S. S. Barth, V. L. Headley, and L. B. Elliott. 1995. Identification of Mycobacterium tuberculosis and M. avium complex directly from smear-positive sputum specimens and BACTEC 12B cultures by high-performance liquid chromatography with fluorescence detection and computer-driven pattern recognition models. J. Clin. Microbiol. 33:1270-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent, P. T., and G. P. Kubica. 1985. Public health laboratory. A guide for the level III laboratory. U.S. Department of Health and Human Services publication no. (CDC) 86-8230. Centers for Disease Control, Atlanta, GA.
- 18.Kline, S., S. Cameron, A. Streifel, M. A. Yakrus, F. Kairis, K. Peacock, J. Besser, and R. C. Cooksey. 2004. An outbreak of bacteremias associated with Mycobacterium mucogenicum in a hospital water supply. Infect. Control Hosp. Epidemiol. 25:1042-1049. [DOI] [PubMed] [Google Scholar]
- 19.Knackmuhs, G., M. Gerwel, C. Patterson, N. Cipollone, M. Navitski, R. Lawler, A. Monaco, M. Dillon, E. Bresnitz, C. Tan, R. Cooksey, and C. A. Robertson. 2004. Mycobacterium chelonae infections associated with face lifts—New Jersey, 2002-2003. MMWR Morb. Mortal. Wkly. Rep. 53:192-194. [PubMed] [Google Scholar]
- 20.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 21.Leão, S. C., A. Martin, G. I. Mejia, J. C. Palomino, J. Robledo, M. A. da Silva Telles, and F. Portaels (ed.). 2004. Practical handbook for the phenotypic and genotypic identification of mycobacteria, p. 113-125. Vanden Broele, Brugge, Belgium.
- 22.Morey, R. E., R. L. Galloway, S. L. Bragg, A. G. Steigerwalt, L. W. Mayer, and P. N. Levett. 2006. Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J. Clin. Microbiol. 44:3510-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Runyon, E. H. 1970. Identification of mycobacterial pathogens using colony characteristics. Am. J. Clin. Pathol. 54:578-586. [DOI] [PubMed] [Google Scholar]
- 24.Silcox, V. A., R. C. Good, and M. M. Floyd. 1981. Identification of clinically significant Mycobacterium fortuitum complex isolates. J. Clin. Microbiol. 14:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Springer, B., E. C. Bottger, P. Kirschner, and R. J. Wallace, Jr. 1995. Phylogeny of the Mycobacterium chelonae-like organism based on partial sequencing of the 16S rRNA gene and proposal of Mycobacterium mucogenicum sp. nov. Int. J. Syst. Bacteriol. 45:262-267. [DOI] [PubMed] [Google Scholar]
- 26.Steingrube, V. A., J. L. Gibson, B. A. Brown, Y. Zhang, R. W. Wilson, M. Rajagopalan, and R. J. Wallace, Jr. 1995. PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria. J. Clin. Microbiol. 33:149-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas, V., K. Herrera-Rimann, D. S. Blanc, and G. Greub. 2006. Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network. Appl. Environ. Microbiol. 72:2428-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson, J. D., T. J. Gibson, F. Plewiak, F. Jeanmougin, and G. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tortoli, E. 2003. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin. Microbiol. Rev. 16:319-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner, D., and L. S. Young. 2003. Nontuberculous mycobacterial infections: a clinical review. Infection 32:257-270. [DOI] [PubMed] [Google Scholar]
- 33.Wallace, R. J., Jr., B. A. Brown, and D. E. Griffith. 1998. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu. Rev. Microbiol. 52:453-490. [DOI] [PubMed] [Google Scholar]
- 34.Wallace, R. J., Jr., V. A. Silcox, M. Tsukamura, B. A. Brown, J. O. Kilburn, W. R. Butler, and G. Onyi. 1993. Clinical significance, biochemical features, and susceptibility patterns of sporadic isolates of the Mycobacterium chelonae-like organism. J. Clin. Microbiol. 31:3231-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, Q., R. Kennon, M. A. Koza, K. Hulten, and J. E. Clarridge III. 2002. Pseudoepidemic due to a unique strain of Mycobacterium szulgai: genotypic, phenotypic, and epidemiological analysis. J. Clin. Microbiol. 40:1134-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]