Abstract
The genome sequence of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough was reanalyzed to design unique 70-mer oligonucleotide probes against 2,824 probable protein-coding regions. These included three genes not previously annotated, including one that encodes a c-type cytochrome. Using microarrays printed with these 70-mer probes, we analyzed the gene expression profile of wild-type D. vulgaris grown on cathodic hydrogen, generated at an iron electrode surface with an imposed negative potential of −1.1 V (cathodic protection conditions). The gene expression profile of cells grown on cathodic hydrogen was compared to that of cells grown with gaseous hydrogen bubbling through the culture. Relative to the latter, the electrode-grown cells overexpressed two hydrogenases, the hyn-1 genes for [NiFe] hydrogenase 1 and the hyd genes, encoding [Fe] hydrogenase. The hmc genes for the high-molecular-weight cytochrome complex, which allows electron flow from the hydrogenases across the cytoplasmic membrane, were also overexpressed. In contrast, cells grown on gaseous hydrogen overexpressed the hys genes for [NiFeSe] hydrogenase. Cells growing on the electrode also overexpressed genes encoding proteins which promote biofilm formation. Although the gene expression profiles for these two modes of growth were distinct, they were more closely related to each other than to that for cells grown in a lactate- and sulfate-containing medium. Electrochemically measured corrosion rates were lower for iron electrodes covered with hyn-1, hyd, and hmc mutant biofilms than for wild-type biofilms. This confirms the importance, suggested by the gene expression studies, of the corresponding gene products in D. vulgaris-mediated iron corrosion.
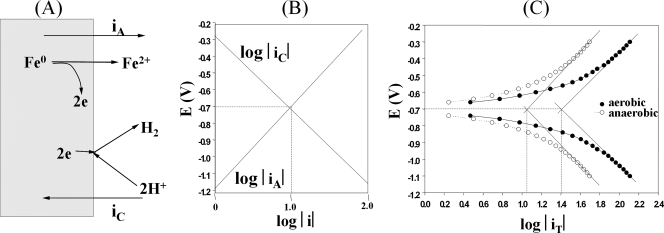
Metal corrosion involves the dissolution of metal ions at anodic sites, with the electrons remaining in the metal. In the case of metallic iron, ferrous ions are generated (Fe0 → Fe2+ + 2e). In addition, oxygen is reduced under aerobic conditions, while protons are reduced under anaerobic conditions at cathodic sites drawing electrons from the metal (7). These two processes, schematically depicted in Fig. 1A, give rise to an anodic and cathodic current, iA and iC, respectively. These currents are of opposite signs and depend on the metal's potential (E), as shown in Fig. 1B. Hence, the total current measured in an external wire connected to the electrode would be iT = iA + iC, and iT = 0 at the lines' point of intersection (Fig. 1B). The identical absolute values iA and iC represent the corrosion current, icorr, and the corresponding potential is the corrosion potential, Ecorr. The metabolic activity of microorganisms can contribute to the corrosion process, and sulfate-reducing bacteria (SRB) were implicated as early as 1934 (37). SRB can increase the icorr value by using cathodically produced hydrogen to reduce sulfate to sulfide or by using produced sulfide to precipitate anodically produced ferrous ions as FeS. However, produced FeS may also form a film on the iron surface, reducing the icorr value (7). As indicated in Fig. 1B, imposing a negative potential on the iron surface boosts formation of cathodic hydrogen (iC) and lowers the corrosive flux of ferrous ions (iA). This procedure, known as cathodic protection (CP), is commonly practiced to lower the corrosion rate of steel pipelines. Most hydrogenase-containing SRB, like the well-known Desulfovibrio spp., do not effectively use electrons derived from iron as the electron donor for sulfate reduction at potentials at or above the Ecorr value (E ≥ −0.7 V). Under these conditions, these SRB can only oxidize iron cometabolically in the presence of an organic electron donor for sulfate reduction, such as lactate (6). Desulfobacterium strains using iron efficiently as the sole electron donor for sulfate reduction (i.e., in the absence of an organic electron donor) have been isolated (12). It was suggested that these may be able to channel electrons directly from iron to the cytoplasmic sulfate reduction pathway through outer membrane electron transport complexes, as used by Fe(III)-reducing bacteria (12).
FIG. 1.
Anaerobic corrosion of iron. (A) Anodic and cathodic currents. (B) Dependence of log|iA| and log|iC|, where |iA| and |iC| are the absolute values of iA and iC, on electrode potential E. The intersection point has the coordinates (log|icorr|, Ecorr). (C) Tafel plot of actual experimental data in which E is plotted against log|iT|, the absolute value of the measured current: |iT| = ||iA| − |iC||.
In the present article, we focus on the mechanism by which Desulfovibrio vulgaris strain Hildenborough grows on cathodically protected iron electrodes. A point of interest is whether, in the absence of an organic electron donor like lactate, this requires H2 evolution from the electrode or whether effective growth at potentials only marginally below the Ecorr value is possible through direct transfer of electrons from the electrode to the cells. We used newly constructed microarrays to determine whether the gene products used for cathodic hydrogen oxidation were the same or different from those used for oxidation of gaseous hydrogen identified previously (3).
MATERIALS AND METHODS
Probe selection.
The MAGPIE software package (16) was used to reanalyze the published genome sequence of D. vulgaris (21), determined at The Institute of Genomic Research, now the J. Craig Venter Institute (JCVI), to contain 3,379 currently annotated coding sequences (CDSs). A large number of open reading frames (ORFs), called initially by MAGPIE, was reduced to a smaller number of probable CDSs by several criteria and through the use of bioinformatics programs. This included NCBI BLASTp (2), used to determine the amino acid sequence homology of ORFs with genes in the NCBI nonredundant sequence database, Emboss Hmoment (15), used to calculate the maximum mean hydrophobic moment of ORF-derived protein sequences, JCVI's rbs finder (11), used to determine the ribosome binding site (rbs) sequence and position relative to the ORF start codon, and WebLogo (8), used to determine the consensus rbs for 200 highly expressed genes (see Table S1 in the supplemental material). The sequences were also processed with the Emboss cusp software program (30) to derive a D. vulgaris codon usage table, which served in turn to determine the codon adaptation index (CAI) for each ORF using the CodonW software program (33).
Probe sequence creation and microarray manufacturing.
A total of 2,824 70-mer oligonucleotides were designed using the Osprey software program (18), one for each designated probable CDS (see Table S2 in the supplemental material). Their target melting temperature in 0.1 M NaCl was 87°C, matching that of oligonucleotides for a previously designed D. vulgaris microarray (5). The optimal design conditions for our oligonucleotides were as follows, in descending order of importance: (i) a melting temperature within 5°C of the target (i.e., from 82 to 92°C); (ii) no secondary binding of the oligonucleotide elsewhere in the genome with a melting temperature within 15°C of that for the target site; (iii) no formation of hairpins or dimers with a stability of more than 19 kcal/mol in 0.1 M NaCl; (iv) no long high-G+C stretches, to prevent “sticky” hybridization. Optimal oligonucleotide probes satisfying all four design conditions were found for 1,180 genes. A survey of design conditions violated by suboptimal probes is provided elsewhere (see Fig. S1F in the supplemental material). Additionally, eight oligonucleotides matching the optimal conditions but not targeting a CDS were included as negative hybridization controls.
The designed 70-mer oligonucleotides were custom synthesized by Invitrogen (Carlsbad, CA) on an average scale of 25 pmol/oligonucleotide. They were dissolved in 3× standard saline citrate (SSC) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) to a concentration of 40 pmol/μl and spotted in 0.7-nl quantities, using a ChipWriter Pro spotter (Bio-Rad, Hercules, CA), onto Corning UltraGAPS amino-silane 25-by-75-mm coated slides (Fisher Scientific). The spotting was done at the Southern Alberta Microarray Facility (SAMF). These arrays will be referred to as SAMF arrays. Spot quality was assessed by visual inspection of slides hybridized to cyanine-3-labeled random nanomers. In order to prevent localized hybridization problems from skewing the expression results, the CDS probes were laid out in a random pattern to separate probes for genes located in the same operon. All probes were spotted in duplicate with duplicate spots located in different halves of the array grid. In addition to spotting probes targeting D. vulgaris CDSs, several control probes were printed. Negative controls included blank spots, 3× SSC, and 70-mer oligonucleotide probes without homology to any D. vulgaris CDS. The positive-control spots consisted of 70-mer oligonucleotides probes to human and Arabidopsis genes for which RNA is available to add as spiked-in controls.
RNA and DNA extraction, labeling, hybridization, and data analysis.
The RNA was purified from D. vulgaris cultures with the Qiagen RNA Easy kit, and the RNA quality and quantity were assessed as described previously (3). Genomic DNA (gDNA) was isolated as described elsewhere (39). Amino-modified cDNA was generated from 10 μg of total RNA using the Invitrogen SuperScript indirect cDNA labeling system and random hexamers. Amino-modified gDNA was generated using 1 μg of gDNA with the Invitrogen BioPrime Plus Array CGH genomic labeling system. The amino-modified cDNA was then coupled with Alexa fluor 647 and the amino-modified gDNA with Alexa fluor 555. The dried cDNA and gDNA probes were mixed and diluted to 20 μl using hybridization solution (3). Following incubation, washing, and drying (3), the slides were scanned with a PerkinElmer Scanarray 5000 scanner (PerkinElmer, MA). Spot and background intensities were quantified using JCVI's Spotfinder software (32) with the gDNA channel being used to control for spot uniformity and labeling efficiency (5, 34), as described previously (3). Array analysis used RNA extracted from two independent biological samples, which were hybridized to a minimum of four arrays, each containing duplicate spots. Data were normalized (3), and differentially expressed genes were identified, using the Stanford Tools software package (32), as those with an expression change of at least twofold with a q value of less than 5%. Functional categories for global analysis were defined by clusters of orthologous genes (COGs) (35). A bootstrapped supported-hierarchical-clustering tree was created using JCVI's multiple experiment viewer from genes identified as significant by a one-way analysis of variance with adjusted Bonferroni correction at the 1% confidence level (32). The clusters were created using Euclidean distance and average linkage.
Strains, media, and culture conditions.
Desulfovibrio vulgaris subsp. vulgaris Hildenborough (NCIMB 8303), lacking the 202-kb megaplasmid (21), was the wild-type strain from which D. vulgaris Hyd100 (ΔhydAB Cmr), D. vulgaris Hyn100 (ΔhynAB), and D. vulgaris H801 (Δhmc) were derived by mutagenesis as described elsewhere (13, 17, 29). These were grown in Widdel-Pfennig medium with lactate and sulfate (WP-LS) and a gas phase of 10% (vol/vol) CO2 and 90% (vol/vol) N2 or in WP-HAS medium with 5 or 50% (vol/vol) hydrogen as the electron donor in 10% CO2 and balance N2 (3, 38, 40). The latter medium had, in addition to CO2, 3 mM acetate as the carbon source. In cases where the hydrogen was supplied electrochemically, this medium will be referred to as WP-EAS medium with the value of E indicated. The experimental setup for batchwise growth with a continuous gas flow of hydrogen (100 ml/min per bioreactor) was as described elsewhere (3). For electrochemical studies, a 5% (vol/vol) inoculum of a log-phase culture, grown in WP-LS or WP-HAS, was used as the inoculum.
Electrochemical analyses.
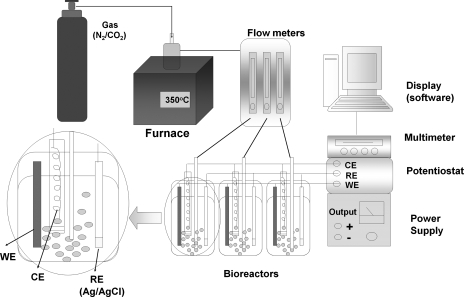
Mixed gas (10% [vol/vol] CO2, balance N2), which was passed through a heated copper reduction tube (350 °C) to remove traces of oxygen, was bubbled into a glass biocorrosion reactor with a working volume of 400 ml at a flow rate of 50 ml/min (Fig. 2). A conventional three-electrode system was used. A low-carbon steel coupon (ASTM A366; 8 by 1 by 0.1 cm) served as the working electrode (WE). The WE was polished with ethanol, dried in air, and sterilized (160°C, 2 h). A platinum helical wire was used as the counter electrode (CE), whereas an Ag/AgCl electrode (MF-2079; Bioanalytical Systems, Inc., West Lafayette, IN) was used as the reference electrode. A diagram of the experimental setup is shown in Fig. 2. Rates of corrosion (Fig. 1B, the anodic loss of metal represented by iA) are reported for Ecorr, the potential where iA = iC = icorr. Values for icorr and Ecorr are derived from the dependence of log |iT| on imposed potential, as represented in Tafel plots (Fig. 1C), or by fitting potentiodynamic curves to equations for iA and iC (28). Data were acquired with a model 410 potentiostat (Electrosynthesis Co, NY) coupled to a multimeter (Keithley 2700), which monitored the current using Testpoint software (Fig. 2). Corrosion rates (CR) in milli-inches per year (mpy) were calculated as follows: CR = (icorr × K × EW)/(d × A), where K is 1.288 × 105, EW is the equivalent weight of iron (55.8 g per 2 mol of electrons), d is the density of iron (7.8 g cm−3), and A is the electrode area in cm2. Alternatively, icorr was derived from electrochemical impedance spectra recorded with an SI1260 or SI1287 electrochemical interference/impedance gain-phase analysis spectroscope (Solartron, United Kingdom) in the frequency range of 0.05 to 100,000 Hz. For large-scale biomass production, D. vulgaris was grown on an iron electrode with a total surface area of 17.1 cm2 in WP-EAS medium (E = −1.1 V) for approximately 2 days, at which point half of the sulfate concentration in the bioreactor was reduced. The electrode-attached biomass was scraped off with a scalpel under anaerobic conditions, after which RNA was extracted as described before (3).
FIG. 2.
Schematic of the biocorrosion reactors. The reactors were gassed with 10% (vol/vol) CO2 and 90% N2 at a rate of 50 ml/min per bioreactor. Each bioreactor contained an iron WE, a platinum CE kept in a separate glass compartment sealed off with an agar plug, and an Ag/AgCl reference electrode (RE). The experimental setup allows the potential (EWE) to be changed while recording the current (iT).
Analytical measurements.
The concentration of ferrous ions was determined as previously described (24, 26). Lactate and acetate concentrations were determined by high-performance liquid chromatography, whereas sulfate and sulfide concentrations were determined spectrophotometrically (22).
Microarray data accession number.
The microarray data presented in this paper have been entered into NCBI GEO (14) under accession number GSE10388.
RESULTS
Probe selection.
The primary annotation of the D. vulgaris chromosome indicates 3,379 protein CDSs, identified using automated methods relying on interpolated hidden Markov models generated from a predefined training set of known genes (21). This includes a large number of genes for hypothetical proteins found only in D. vulgaris. In order to ensure that only ORFs with a high probability of being true CDSs were used for microarray construction, the genome sequence was reanalyzed. MAGPIE (16) identified 13,625 ORFs, based solely on the existence of in-frame start and stop codons. This number was reduced to 6,550 by excluding ORFs on the opposite strand of CDSs, confirmed by MAGPIE level 1 evidence (BLAST homology with an e-value below 10−25). Hand curation gave 2,659 CDSs, identified by strong homology with 1 or more known genes, whereas 67 were identified based on weaker homologies (see Table S3A, bottom, in the supplemental material). A total of 641 potential CDSs for hypothetical proteins occurring only in D. vulgaris (or in the related strain DP4, of which the sequence was not available at the time of microarray design) were analyzed in terms of protein length, protein hydrophobicity, rbs sequence, rbs position relative to the start codon, CAI, and GC codon bias. Relative to the average for all JCVI CDSs, these represented proteins or gene sequences with some or all of the following characteristics: a smaller-than-average protein sequence length (see Fig. S1A in the supplemental material), a lower protein hydrophobic moment (see Fig. S1C in the supplemental material), an rbs sequence different from the consensus rbs (AGGAG), an increased separation of the rbs and the putative start codon (see Fig. S1D in the supplemental material) compared to the consensus (10 nucleotides [nt]), a lower G+C content (see Fig. S1B in the supplemental material), and surprisingly a lower CAI (see Fig. S1E in the supplemental material). Hence, a significant fraction of these 641 may not be real CDSs. We included probes for 98 of these on the array with features considered closest to JCVI ORFs (see Table S3A in the supplemental material) and excluded 543 (see Table S3B in the supplemental material), based in part on the features outlined in Fig. S1 in the supplemental material. Of the 543 excluded ORFs, 139 had homologs in the recently completed genome sequence of Desulfovibrio vulgaris subsp. vulgaris strain DP4 (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi), as indicated in Table S3 in the supplemental material. The DP4 genome is virtually identical to the Hildenborough genome in both gene organization and sequence. The 139 ORFs are likely to represent genuine CDSs, and probes recognizing these will be included in future versions of the microarray. In addition, we did not include probes for 32 CDSs (see Table S3B, bottom, in the supplemental material), because these had virtually identical paralogs elsewhere in the genome.
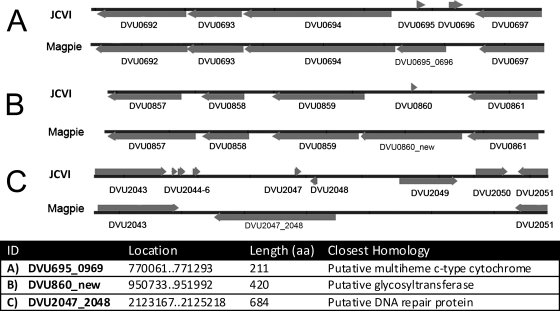
Reanalysis of the D. vulgaris Hildenborough genome indicated three probable genes not previously annotated (Fig. 3). DVU0695_0696 replaces two small ORFs (Fig. 3A) (DVU0695 and DVU0696) and encodes a 211-amino-acid (aa) multiheme cytochrome in a probable operon with DVU0694 to DVU0692 for molybdopterin oxidoreductase. It is conserved in strain DP4 (Dvul_2268; 211/211) and in D. desulfuricans G20 (Dde_2932; 147/211) in what appears to be the same operon. DVU0860_new encodes a predicted glycosyltransferase (420 aa) and replaces the 33-aa ORF DVU0860 (Fig. 3B). Dvul_2122 and Dde_1117 are its homologs in the other sequenced Desulfovibrio spp. DVU2047_2048 replaces DVU2047 and DVU2048 (Fig. 3C) and encodes a predicted 684-aa DNA repair protein with homology to a DP4 gene (Dvul_2616) but no G20 homology. Probes targeting these new D. vulgaris genes were spotted on the array, and both DVU0695_0696 and DVU0860_new, but not DVU2047_2048, were subsequently found to be expressed (see Table S4 in the supplemental material).
FIG. 3.
Survey of three newly annotated genes found by MAGPIE in the D. vulgaris genome, as explained in the text. DVU0695_696 replaces DVU0695 and DVU0696 (A), DVU0860_new replaces DVU0860 (B), and DVU2047_2048 replaces DVU2047 and DVU2048 (C). The new locus tags (ID), their location in the genome (bp), the length of the encoded protein (aa), and their closest homolog, as identified by BLAST search, are indicated.
Microarray quality assessment.
On average, 27 ± 18 spots out of a total of 5,678 (0.48%) were not detected when labeled gDNA was hybridized to the microarrays (see Fig. S2 in the supplemental material). The on-slide hybridization consistency was high (Pearson correlation of 0.980 for all expressed duplicate spots averaged for four arrays), as was the interslide hybridization consistency (Pearson correlation of 0.951 between all expressed spots on duplicate arrays, averaged for four arrays). Comparison of the SAMF arrays with the 70-mer oligonucleotide Virtual Institute for Microbial Stress and Survival (VIMSS) arrays constructed previously (5) in determining gene expression changes for cells grown with hydrogen or lactate, indicated 140 genes to be differentially expressed in the same direction on both. There was a Pearson correlation of 0.985 of the RC,i ratios determined with the two array platforms for these 140 genes (RC,i = Ri,H2/Ri,lactate). The average absolute value for the expression change for these 140 genes was 2.03 for the VIMSS arrays and 2.17 for the SAMF arrays. Both arrays indicated the same direction of change for expression of the periplasmic D. vulgaris hydrogenases DVU1769 and DVU1918. This expression change was also confirmed by reverse transcription-PCR (3). A set of 105 genes, believed to be required for growth on hydrogen (3), showed quantitatively similar gene expression changes on the two array platforms. Exceptions were genes for an alcohol dehydrogenase (DVU2545), a lactate dehydrogenase (DVU0600), the heterodisulfide reductase complex (DVU0848 to DVU0850), and the Hyn1 hydrogenase (DVU1921 and DVU1922). In all of these cases, expression changes for these genes were qualitatively in the same direction.
The quality of the SAMF microarrays was further tested by checking the correlation of differential expression ratios (RC,i) for genes in the same operon. This was computed as the Pearson product-moment correlation between the percentile rank of raw spot intensities (foreground minus background). Pearson's method is unaffected by downstream effects (reduced expression of 3′ genes compared to that of 5′ genes in many Desulfovibrio operons). Using the 639 predicted D. vulgaris operons (1), around 350 genes per replicate were considered “expressed” using a 1-mRNA/gDNA intensity as the threshold level. Random pairs of genes from these operons were tested for product-moment correlation (R, the tendency of genes to go up or down together linearly), and the average correlation for all operons was calculated. The random pair selection process was repeated 1,000 times to generate a confidence interval of R = 0.711 to 0.717, using a data set comprised of 5 hybridizations. The correlation implied that approximately 0.51% (R2 = 0.7142) of the expression change of a gene can be accounted for by other genes in the same operon. Performing the same analysis for data obtained with the VIMSS microarrays yielded data with a confidence interval of R = 0.714-0.718, suggesting data from both microarray platforms to be equally trustworthy and comparable to those published to date. These correlations are better than the similarly derived Spearman's confidence interval (R = 0.65 to 0.67) for the best analysis method for the Affymetrix Escherichia coli gene chips (19).
Global gene expression in electrode-grown D. vulgaris.
Growth of D. vulgaris on an iron electrode suspended in WP-EAS medium using the culturing system outlined in Fig. 2 required a low working electrode potential (EWE) of −1.1 V. Hydrogen evolves from the electrode, the anodic current (iA) is negligible (Fig. 1B), and the electrode is protected from corrosion (Fe0 → Fe2+ + 2e) under these CP conditions. Growth was not observed at higher electrode potentials, e.g., at an EWE value of −0.7 V, corresponding to Ecorr (Fig. 1C), when the bioreactor was filled with WP-EAS medium. RNA was extracted at the point where half of the sulfate of the medium was used. For wild-type cells grown under CP conditions, 85% of total RNA was extracted from the electrode, whereas for cells grown in WP-LS medium at an E value of −0.3 V, this was only 9% (see Table S5 in the supplemental material). Hence, under CP conditions, most cells grew as an electrode-associated biofilm.
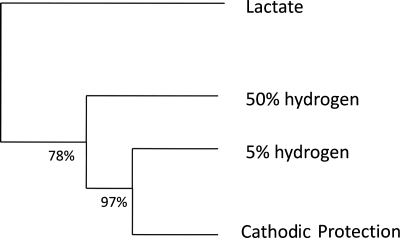
When the gene expression pattern of cells grown under CP conditions was compared with that of cells grown with gaseous hydrogen, a smaller shift in the gene expression pattern was observed (56 genes up-regulated and 40 genes down-regulated) than when that of cells grown under CP conditions was compared with that of cells grown with lactate (193 genes up- and 229 genes down-regulated) (Table 1). The bootstrap-supported hierarchical clustering tree (Fig. 4) also illustrates that the gene expression pattern in CP-grown cells is more similar to that of hydrogen-grown cells than to that of lactate-grown cells. CP-grown cells showed the greatest gene expression changes in the following COG categories: viii, cell envelope biogenesis, outer membrane; ix, cell motility and secretion; xiii, energy production and conversion; and xv, amino acid transport and metabolism.
TABLE 1.
Numbers of genes up- or downregulated in D. vulgaris wild-type cells grown under different conditionsa
| COG functional category | No. of genes up- or down-regulated for growth condition
|
|||||||
|---|---|---|---|---|---|---|---|---|
| CP vs 50%, up | CP vs 50%, down | CP vs 5%, up | CP vs 5%, down | CP vs H2, up | CP vs H2, down | CP vs lactate, up | CP vs lactate, down | |
| i, translation, ribosomal structure, and biogenesis | 4 | 21 | 3 | 1 | 0 | 1 | 15 | 2 |
| ii, RNA processing and modification | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| iii, transcription | 5 | 4 | 3 | 1 | 2 | 0 | 4 | 4 |
| iv, DNA replication, recombination, and repair | 1 | 2 | 0 | 0 | 0 | 0 | 5 | 2 |
| v, cell division and chromosome partitioning | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 1 |
| vi, defense mechanisms | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0 |
| vii, posttranslational modification, protein turnover, chaperones | 6 | 6 | 0 | 0 | 0 | 0 | 4 | 11 |
| viii, cell envelope biogenesis, outer membrane | 9 | 5 | 3 | 3 | 3 | 1 | 6 | 12 |
| ix, cell motility and secretion | 14 | 3 | 9 | 0 | 5 | 0 | 1 | 6 |
| x, signal transduction mechanisms | 13 | 10 | 3 | 7 | 4 | 2 | 16 | 16 |
| xi, extracellular structures | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| xii, intracellular trafficking, secretion, and vesicular transport | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 1 |
| xiii, energy production and conversion | 22 | 21 | 11 | 20 | 7 | 7 | 17 | 29 |
| xiv, carbohydrate transport and metabolism | 0 | 6 | 0 | 2 | 0 | 0 | 5 | 6 |
| xv, amino acid transport and metabolism | 19 | 22 | 7 | 6 | 6 | 3 | 10 | 15 |
| xvi, nucleotide transport and metabolism | 1 | 3 | 0 | 0 | 0 | 0 | 2 | 1 |
| xvii, coenzyme metabolism | 7 | 3 | 3 | 1 | 3 | 0 | 10 | 3 |
| xviii, lipid metabolism | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 |
| xix, inorganic ion transport and metabolism | 5 | 11 | 4 | 4 | 2 | 4 | 7 | 8 |
| xx, secondary metabolites biosynthesis, transport, and catabolism | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| xxi, general function prediction only | 13 | 12 | 5 | 4 | 5 | 2 | 12 | 20 |
| xxii, function unknown | 73 | 55 | 24 | 37 | 19 | 20 | 63 | 79 |
| Overallb | 198 | 188 | 78 | 89 | 56 | 40 | 193 | 229 |
CP, 50%, 5%, and lactate indicate cells grown in WP medium with cathodic hydrogen, 50% (vol/vol) hydrogen, 5% (vol/vol) hydrogen, or lactate as the electron donor for sulfate reduction, respectively. “Up” and “down” indicate up- and down-regulation, respectively.
The overall total does not count genes belonging to multiple categories more than once.
FIG. 4.
Bootstrap-supported hierarchical clustering tree for gene expression patterns for wild-type D. vulgaris grown in WP-LS (lactate), WP-HAS (50% [vol/vol] or 5% [vol/vol] hydrogen), or WP-EAS (EWE = −1.1 V; cathodic protection conditions, cells grown on cathodic hydrogen).
Specific gene expression in electrode-grown D. vulgaris.
Genes for GGDEF/EAL domain proteins were among the most highly up- and down-regulated (Table 2, DVU1959 and DVU3106) when CP-grown cells were compared with H2-grown cells. Such proteins have been implicated as signaling factors that promote the switch from the planktonic lifestyle to the biofilm lifestyle in many bacteria (9, 31). Relatively few genes for enzymes in carbohydrate synthesis, which could contribute to increased cell adhesion and biofilm formation (10), were up-regulated (Table 2, DVU1892 and DVU0117) in the CP condition. In contrast, many genes for flagellar components or for proteins involved in flagellar biosynthesis were up-regulated (Table 2, DVU0313 to DVU2444), suggesting that the electrode-associated D. vulgaris cells retained motility. Up- or down-regulation of several genes encoding proteins involved in periplasmic binding, transport, and excretion also occurred (Table 2, DVU0058 to DVU1343).
TABLE 2.
Selected genes that are up- or down-regulated for wild-type D. vulgaris cells grown under different conditionsa
| Gene IDb | Annotation | Log2RC value
|
|
|---|---|---|---|
| CP vs 50% H2 | CP vs 5% H2 | ||
| DVU1921 | [NiFe] hydrogenase isozyme 1, small subunit (hyn-1B) | 5.03 | 4.09 |
| DVU1922 | [NiFe] hydrogenase isozyme 1, large subunit (hyn-1A) | 5.36 | 4.05 |
| DVU1917 | [NiFeSe] hydrogenase, small subunit (hysB) | 2.40 | −1.01 |
| DVU1918 | [NiFeSe] hydrogenase, large subunit (hysA) | 2.57 | −1.48 |
| DVU1769 | [Fe] hydrogenase, large subunit (hydA) | 1.46 | 3.10 |
| DVU1770 | [Fe] hydrogenase, small subunit (hydB) | 0.54 | 2.37 |
| DVU0531 | HmcF | 2.41 | 2.03 |
| DVU0532 | HmcE | 3.60 | 1.96 |
| DVU0533 | HmcD | ND | 2.27 |
| DVU0534 | HmcC | 2.97 | 1.46 |
| DVU0535 | HmcB | 2.54 | 1.93 |
| DVU0536 | HmcA | ND | 2.65 |
| DVU1959 | EAL domain/GGDEF domain protein | 4.41 | 2.11 |
| DVU3106 | GGDEF domain protein | 3.97 | 4.14 |
| DVU0117 | Glycosyl transferase | 3.11 | 2.17 |
| DVU1892 | Glycosyl transferase | 3.24 | 2.45 |
| DVU0313 | Flagellar M-ring protein FliF | 2.19 | 1.00 |
| DVU0314 | Flagellar basal-body component FliE | 1.87 | 1.30 |
| DVU0315 | Flagellar basal-body rod protein FlgC | 2.50 | 1.95 |
| DVU0316 | Flagellar basal-body rod protein FlgB | 2.48 | 2.24 |
| DVU0512 | Flagellar basal-body rod protein, putative | 2.05 | 1.45 |
| DVU0513 | Flagellar basal-body rod protein FlgG | 1.31 | 0.59 |
| DVU0514 | FlgA family protein | 1.85 | 1.29 |
| DVU0515 | Flagellar L-ring protein FlgH | 1.59 | 0.84 |
| DVU0523 | Negative regulator of flagellin synthesis FlgM | 1.74 | 2.03 |
| DVU0863 | Flagellar hook-associated protein 2, putative | 2.41 | 1.90 |
| DVU1441 | Flagellin | 2.24 | 1.34 |
| DVU1442 | Flagellin FlaG, putative | 1.61 | 2.45 |
| DVU2082 | Flagellin, putative | 3.75 | 2.96 |
| DVU2444 | Flagellin | 5.09 | 3.69 |
| DVU2893 | Flagellar basal-body rod protein, putative | 1.47 | 1.28 |
| DVU0058 | Efflux transporter, RND family, MFP subunit | 2.31 | 2.11 |
| DVU0097 | Polyamine ABC transporter, permease protein | 1.47 | 1.23 |
| DVU0098 | Polyamine ABC transporter, ATP-binding protein | 1.40 | 1.06 |
| DVU1340 | Transcriptional regulator, FUR family | −2.27 | −1.14 |
| DVU1341 | Cation ABC transporter, permease | −2.85 | −1.44 |
| DVU1341 | Cation ABC transporter, ATP binding | −2.41 | −1.02 |
| DVU1343 | Cation ABC transporter, periplasmic binding | −2.41 | −1.37 |
| DVU2110 | l-Lactate permease | 3.52 | 3.00 |
| DVU2584 | Transporter, CorA family | 2.45 | 2.75 |
| DVU2661 | Twin-arginine translocation pathway signal sequence | 2.97 | 2.23 |
| DVU2667 | Phosphate ABC transporter, phosphate-binding protein | 2.99 | 2.12 |
Either 5% (vol/vol) hydrogen, 50% (vol/vol) hydrogen, or cathodically produced hydrogen (CP) was the sole electron donor for sulfate reduction. q values, indicating the statistical significance of the tabulated expression values, can be found in Table S4 in the supplemental material.
Gene identifier (ORF name).
With respect to energy and especially hydrogen metabolism, both hyn-1 genes (DVU1921 [hynB1] and DVU1922 [hynA1]) for [NiFe] hydrogenase 1 were among the most strongly up-regulated genes for the entire genome (Table 2; see also Table S4 in the supplemental material). All five hmc genes for the high-molecular-weight cytochrome (Hmc) complex, an electron-transferring complex located in the cytoplasmic membrane (13), as well as the hydAB genes (DVU1769 and DVU1770) for [Fe] hydrogenase were also up-regulated. The Hmc complex has been implicated in hydrogen oxidation by deletion analysis (13) but was not found to be strongly up-regulated in gaseous-hydrogen-grown cells compared to levels in lactate-grown cells (3).
Determination of corrosion rates.
The corrosion current (icorr) was determined without bacteria present under both anaerobic and aerobic conditions (Fig. 1C). The derived values, converted to corrosion rates (CRs), were 4.6 mpy under anaerobic conditions and 10.3 mpy under aerobic conditions. Under the latter conditions, oxygen reduction is part of the cathodic reaction (2H+ + O2 + 4e [electrode] → 2 OH−). The anaerobic abiotic rates were constant with time, as indicated in Fig. 5C and 6B.
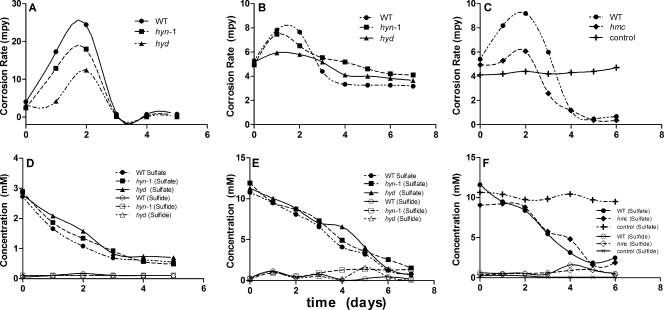
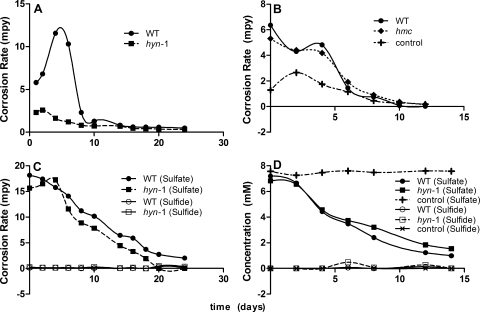
FIG. 5.
(A to C) Anaerobic corrosion rates in biocorrosion reactors filled with WP-LS medium as a function of time. The reactors were inoculated with D. vulgaris wild-type (WT) or mutant strains, as indicated. The iron coupon working electrodes had a measured EWE value of −0.3 V. Corrosion rates are reported for an EWE (= Ecorr) value of −0.7 V. (D to F) Concentrations of sulfate and sulfide as a function of time.
FIG. 6.
(A and B) Anaerobic corrosion rates in biocorrosion reactors filled with WP-EAS medium as a function of time. The reactors were inoculated with D. vulgaris wild-type (WT) or mutant strains, as indicated. The iron coupon working electrodes had a set EWE value of −1.1 V. Corrosion rates are reported for an EWE (Ecorr) value of −0.7 V. (C and D) Concentrations of sulfate and sulfide as a function of time.
Experiments in the presence of D. vulgaris wild-type or mutant strains were conducted under two different conditions, in which the three bioreactors (Fig. 2) were filled with either WP-LS medium with an average electrode potential of −0.3 V or WP-EAS medium with the electrode potential set at −1.1 V. The CRs were calculated from the icorr value at an Ecorr value of −0.7 V. Hence, iA, corresponding to the rate of ferrous ion production, was greater than the icorr value under the first set of conditions and smaller than the icorr value under the second set of conditions, as explained in Fig. 1B. Indeed, soluble ferrous ions were present under the former conditions (up to 2 mM) but not under the latter conditions (results not shown). No significant soluble sulfide was observed in the bulk phase under either of these conditions (Fig. 5D to F and Fig. 6C and D), because sulfide precipitated as FeS and because the bioreactors were continuously gassed with 10% (vol/vol) CO2 and 90% N2, removing H2S. This was to mimick the culture conditions of the earlier study (3), in which gaseous hydrogen (5% or 50% H2, 10% CO2, and balance N2) was provided at 100 ml/min and in which cultures in WP-LS (with lactate as the sole electron donor for sulfate reduction) were gassed with 10% CO2 and 90% N2.
The results of three experiments conducted in WP-LS medium, one conducted with low concentrations of lactate and sulfate (5 and 3 mM, respectively; Fig. 5A) and two conducted with high concentrations of lactate and sulfate (20 and 10 mM, respectively; Fig. 5B and C), are summarized in Fig. 5. In the case of all three experiments, the initial CR was close to the anaerobic abiotic CR of 4.6 mpy. The presence of wild-type or mutant D. vulgaris strains actively reducing sulfate transiently increased the CR (Fig. 5A to C). When sulfate reduction stopped, because the limiting concentration of lactate had been used (data not shown), the CRs settled to values at or below that of the abiotic control. Hence, the presence of actively growing Desulfovibrio strains in the bioreactors, which were in part present as an electrode-associated biofilm (see Table S5 in the supplemental material; 9% for the wild type), increased the CR. The reasons for the larger increase in the CR for the wild type at low lactate and sulfate concentration (Fig. 5A; ΔCR = 20 mpy) than at high lactate and sulfate concentrations (Fig. 5B and C; ΔCR = 3 to 5 mpy) are not known. The hyn1, hyd, and hmc mutant strains increased the CR less than the wild-type strain, suggesting that [NiFe] hydrogenase 1, [Fe] hydrogenase, and the Hmc complex contribute to the corrosion-enhancing metabolism of D. vulgaris.
The results of experiments conducted in WP-EAS medium under CP conditions are presented in Fig. 6. The wild type had a significantly increased CR, which was not observed for the hyn1 mutant (Fig. 6A), whereas the hmc mutant and the wild type increased the CR similarly (Fig. 6B). Sulfate reduction with associated growth could not be established for the hyd mutant, lacking [Fe] hydrogenase, suggesting this enzyme to be essential for growth under CP conditions.
DISCUSSION
The design and application of microarrays for transcript analysis of D. vulgaris Hildenborough have been described previously. Zhang et al. (41, 43) used NimbleGen System, Inc., microarrays, in which 13 unique 24-mer oligonucleotides were spotted for each of 3,548 CDSs indicated by the genome sequence. Chhabra et al. (5) and He et al. (20) used arrays in which single, unique 70-mer oligonucleotides for 3,482 CDSs were spotted. These VIMSS arrays were also used to profile the transcriptome of D. vulgaris when grown with different gas-phase hydrogen concentrations (3). It is of interest to compare the probe sequences designed for these arrays (20) with those designed by the Osprey software (18) in the current study. The SAMF probes are significantly closer to the 5′ start site of the CDSs than the VIMSS probes (see Table S6 in the supplemental material) (average distances, 179 and 559 nt, respectively). As a result, there is little probe overlap (see Table S6 in the supplemental material): of 2,595 CDSs, only 686 share between 1 and 70 nt (average, 46 nt). Nevertheless, arrays based on these two distinct sets of probes indicated very similar gene expression changes for D. vulgaris grown with hydrogen compared to results with lactate.
Because the hydrogen electron donor is exclusively generated at the electrode under CP conditions, the majority of the biomass is electrode associated (see Table S5 in the supplemental material). Bubbles emerging from the electrode surface did not effectively feed the planktonic population, since these were removed by the continuous gassing with N2-CO2, which also effectively stripped H2S (Fig. 5 and 6). Hence, gene expression changes of hydrogen-grown versus CP-grown cells will in part reflect the transition from the planktonic lifestyle to the biofilm lifestyle. This transition involves cells that are planktonic and motile (state A), surface associated with gliding motility through additional flagella (state B), and in a surface-attached, nonmotile, mature biofilm state (state C) (4). States A, B, and C can have increasing concentrations of the nucleotide signal cyclic di-GMP, as demonstrated for different bacteria (4, 23, 36). This signal is formed by GGDEF-domain proteins and removed by EAL-domain proteins, which synthesize and hydrolyze cyclic di-GMP, respectively. Bifunctional GGDEF/EAL-domain proteins also occur commonly. The fact that proteins in this class are among the most up-regulated (Table 1; DVU1959 and DVU3106) and the most down-regulated (see Table S4 in the supplemental material; DVU0763) of the entire genome, when CP cells and planktonically grown cells are compared, may reflect this lifestyle change.
The up-regulation of many genes for flagellar proteins in CP-grown cells (Table 2; see also Table S4 in the supplemental material), compared to cells grown in 50% (vol/vol) H2, indicates that the former are in the surface-associated, motile state (state B), not in the nonmotile, mature biofilm state (state C). The increased flagellar gene expression may allow effective gliding motility in the much more viscous substratum-associated biofilm (4, 23, 36). Relative to mature biofilm cells, cells in state B overexpress few genes for carbohydrate synthesis. In contrast, cells in state C have reduced expression of flagellum genes and increased expression of genes for enzymes involved in exopolysaccharide biosynthesis. Zhang et al. (42) have reported the gene expression pattern of D. vulgaris biofilms grown on iron coupons suspended in a continuous-culture bioreactor fed with lactate- and sulfate-containing medium for a period of 20 days. The biofilm that formed on these coupons was characterized by reduced expression of flagellum genes and increased expression of many genes involved in polysaccharide biosynthesis, indicating that these represent the mature biofilm state (state C). The striking difference between gene expression in the two biofilm types is presented in Table S7 in the supplemental material.
CP-grown biofilm and 50% (vol/vol) hydrogen-grown planktonic cells engage in similar energy metabolism with CP-grown cells experiencing a cell density that is 2 orders of magnitude higher. Perhaps due to the increased competition experienced by CP-grown cells, they overexpress virtually all known components of this energy metabolism pathway (27) relative to cells grown planktonically with 50% (vol/vol) H2, including genes for three of four hydrogenases, for the Hmc complex but not the other membrane-bound electron transfer complexes, for ATP synthase, and for the main enzymes of the sulfate reduction pathway (Table 2; see also Table S4 in the supplemental material). Of the three nickel-containing hydrogenases, genes for the large and small subunits of Hyn-1 are among the most up-regulated of the entire genome and those for Hys are also up-regulated, whereas those for Hyn-2 are down-regulated. The hyd genes for iron-only hydrogenase are also up-regulated under CP conditions, and the special enzymatic properties of this enzyme (high turnover, low affinity) may be important to explain why the hyd mutant could not be grown under CP conditions.
SRB may increase the cathodic and anodic currents (Fig. 1A). The lower CRs observed in D. vulgaris strains missing either Hyn1, Hyd, or the Hmc complex (Fig. 5 and 6) illustrate that these proteins facilitate metabolism that enhances iA, iC, or both. The gene expression results further demonstrate this connection, since the hyn1, hyd, and hmc genes are upregulated under CP conditions. The lower CRs observed when SRB metabolism has ceased (Fig. 5C) may be explained by formation of an iron sulfide film on the iron surface area, reducing the rate of the corrosion reaction (7, 25).
D. vulgaris does not appear to have an outer membrane electron transport complex (21). The Hmc complex and other membrane-bound electron transfer complexes are located in the inner membrane. Differently from Fe(III)-reducing Geobacter and Shewanella spp., which have outer membrane electron transport complexes, D. vulgaris cannot reduce extracellular Fe2O3 (26). The results obtained in this study similarly indicate that the organism does not grow by accepting electrons directly from metallic iron. Growth requires either an imposed low potential (EWE = −1.1 V), conditions under which hydrogen evolves from the metal surface, or, at higher potential (e.g., EWE = −0.3 V), the presence of the organic electron donor (lactate) to drive reduction of sulfate to sulfide. Corrosion (iron oxidation) is cometabolic with lactate oxidation under the latter conditions. SRB that use iron efficiently as an electron donor for sulfate reduction in the absence of an organic electron donor, like the Desulfobacterium strains identified by Dinh et al. (12), may be able to directly extract electrons from the iron surface, and it would be most interesting to elucidate the mechanism used.
Supplementary Material
Acknowledgments
This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant to G.V. and by an NSERC Strategic Grant to G.V. and C.W.S. with NOVA Research and Technology Corporation and the Dow Chemical Corporation as industrial partners. The latter was matched in part by the Alberta Science and Research Authority, Innovation and Science Research Investments Program (ASRA-ISRIP).
We thank Viola Birss, Department of Chemistry, University of Calgary, for electrochemistry support. Mayi Arcellana-Panlilio and Xiuling Wang of the Southern Alberta Microarray Facility are thanked for array printing and hybridization advice. Hong Tran is thanked for helping with the hand curation of the D. vulgaris genome.
Footnotes
Published ahead of print on 29 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alm, E. J., K. H. Huang, M. N. Price, R. P. Koche, K. Keller, I. L. Dubchak, and A. P. Arkin. 2005. The MicrobesOnline web site for comparative genomics. Genome Res. 15:1015-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caffrey, S. M., H.-S. Park, J. K. Voordouw, Z. He, J. Zhou, and G. Voordouw. 2007. Function of periplasmic hydrogenases in the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 189:6159-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caiazza, N. C., J. H. Merritt, K. M. Brothers, and G. A. O'Toole. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:3603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chhabra, S. R., Q. He, K. H. Huang, S. P. Gaucher, E. J. Alm, Z. He, M. Z. Hadi, T. C. Hazen, J. D. Wall, J. Zhou, A. P. Arkin, and A. K. Singh. 2006. Global analysis of heat shock response in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 188:1817-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cord-Ruwisch, R., and F. Widdel. 1986. Corroding iron as a hydrogen source for sulfate reduction in growing cultures of sulfate-reducing bacteria. Appl. Microbiol. Biotechnol. 25:169-174. [Google Scholar]
- 7.Crolet, J.-L. 2005. Microbial corrosion in the oil industry: a corrosionist's view, p. 143-169. In B. Ollivier and M. Magot (ed.), Petroleum microbiology. ASM Press, Washington, DC.
- 8.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Argenio, D. A., and S. I. Miller. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497-2502. [DOI] [PubMed] [Google Scholar]
- 10.Davey, M. E., and M. J. Duncan. 2006. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J. Bacteriol. 188:5510-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinh, H. T., J. Kuever, M. Mussmann, A. W. Hassel, M. Stratmann, and F. Widdel. 2004. Iron corrosion by novel anaerobic microorganisms. Nature 427:829-832. [DOI] [PubMed] [Google Scholar]
- 13.Dolla, A., B. K. Pohorelic, J. K. Voordouw, and G. Voordouw. 2000. Deletion of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough hampers hydrogen metabolism and low-redox-potential niche establishment. Arch. Microbiol. 174:143-151. [DOI] [PubMed] [Google Scholar]
- 14.Edgar, R. M., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenberg, D., R. M. Weiss, and T. C. Terwilliger. 1984. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. USA 81:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaasterland, T., and C. W. Sensen. 1996. Fully automated genome analysis that reflects user needs and preferences. A detailed introduction to the MAGPIE system architecture. Biochimie 78:302-310. [DOI] [PubMed] [Google Scholar]
- 17.Goenka, A., J. K. Voordouw, W. Lubitz, W. Gaertner, and G. Voordouw. 2005. Construction of a [NiFe]-hydrogenase deletion mutant of Desulfovibrio vulgaris Hildenborough. Biochem. Soc. Trans. 33:59-60. [DOI] [PubMed] [Google Scholar]
- 18.Gordon, P. M., and C. W. Sensen. 2004. Osprey: a comprehensive tool employing novel methods for the design of oligonucleotides for DNA sequencing and microarrays. Nucleic Acids Res. 32:e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harr, B., and C. Schlotterer. 2006. Comparison of algorithms for the analysis of Affymetrix microarray data as evaluated by co-expression of genes in known operons. Nucleic Acids Res. 34:e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, Q., K. H. Huang, Z. He, E. J. Alm, M. W. Fields, T. C. Hazen, A. P. Arkin, J. D. Wall, and J. Zhou. 2006. Energetic consequences of nitrite stress in Desulfovibrio vulgaris Hildenborough, inferred from global transcriptional analysis. Appl. Environ. Microbiol. 72:4370-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heidelberg, J. F., R. Seshadri, S. A. Haveman, C. L. Hemme, I. T. Paulsen, J. F. Kolonay, J. A. Eisen, N. Ward, B. Methe, L. M. Brinkac, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, D. Fouts, D. H. Haft, J. Selengut, J. D. Peterson, T. M. Davidsen, N. Zafar, L. Zhou, D. Radune, G. Dimitrov, M. Hance, K. Tran, H. Khouri, J. Gill, T. R. Utterback, T. V. Feldblyum, J. D. Wall, G. Voordouw, and C. M. Fraser. 2004. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22:554-559. [DOI] [PubMed] [Google Scholar]
- 22.Hubert, C., M. Nemati, G. Jenneman, and G. Voordouw. 2005. Corrosion risk associated with microbial souring control using nitrate or nitrite. Appl. Microbiol. Biotechnol. 68:272-282. [DOI] [PubMed] [Google Scholar]
- 23.Kim, Y. K., and L. L. McCarter. 2007. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J. Bacteriol. 189:4094-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovley, D. R., and E. J. P. Phillips. 1986. Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl. Environ. Microbiol. 51:683-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pankhania, I. P., A. N. Moosavi, and W. A. Hamilton. 1986. Utilization of cathodic hydrogen by Desulfovibrio vulgaris (Hildenborough). J. Gen. Microbiol. 132:3357-3365. [Google Scholar]
- 26.Park, H. S., S. Lin, and G. Voordouw. 2008. Ferric iron reduction by Desulfovibrio vulgaris Hildenborough wild type and energy metabolism mutants. Antonie van Leeuwenhoek 93:79-85. [DOI] [PubMed] [Google Scholar]
- 27.Pereira, I. A. C., S. A. Haveman, and G. Voordouw. 2007. Biochemical, genetic and genomic characterization of anaerobic electron transport pathways in sulphate-reducing delta-proteobacteria, p. 215-240. In L. L. Barton and W. A. Hamilton (ed.), Sulphate-reducing bacteria: environmental and engineered systems. Cambridge University Press, Cambridge, United Kingdom.
- 28.Piron, D. L. 1994. The electrochemistry of corrosion. National Association of Corrosion Engineeers, Houston, TX.
- 29.Pohorelic, B. K., J. K. Voordouw, E. Lojou, A. Dolla, J. Harder, and G. Voordouw. 2002. Effects of deletion of genes encoding Fe-only hydrogenase of Desulfovibrio vulgaris Hildenborough on hydrogen and lactate metabolism. J. Bacteriol. 184:679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 31.Ryan, R. P., Y. Fouhy, J. F. Lucey, and J. M. Dow. 2006. Cyclic Di-GMP signaling in bacteria: recent advances and new puzzles. J. Bacteriol. 188:8327-8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 33.Sharp, P. M., E. Bailes, R. J. Grocock, J. F. Peden, and R. E. Sockett. 2005. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 33:1141-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talaat, A. M., S. T. Howard, W. T. Hale, R. Lyons, H. Garner, and S. A. Johnston. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 36.Thormann, K. M., S. Duttler, R. M. Saville, M. Hyodo, S. Shukla, Y. Hayakawa, and A. M. Spormann. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 188:2681-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Wolzogen Kuehr, C. A. H., and I. S. van der Vlugt. 1934. The graphitization of cast iron as an electrobiochemical process in anaerobic soil. Water 18:147-165. [Google Scholar]
- 38.Voordouw, G. 2002. Carbon monoxide cycling by Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 184:5903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voordouw, G., V. Niviere, F. G. Ferris, P. M. Fedorak, and D. W. Westlake. 1990. Distribution of hydrogenase genes in Desulfovibrio spp. and their use in identification of species from the oil field environment. Appl. Environ. Microbiol. 56:3748-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 1. Springer-Verlag, New York, NY. [Google Scholar]
- 41.Zhang, W., D. E. Culley, M. Hogan, L. Vitiritti, and F. J. Brockman. 2006. Oxidative stress and heat-shock responses in Desulfovibrio vulgaris by genome-wide transcriptomic analysis. Antonie van Leeuwenhoek 90:41-55. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, W., D. E. Culley, L. Nie, and J. C. Scholten. 2007. Comparative transcriptome analysis of Desulfovibrio vulgaris grown in planktonic culture and mature biofilm. Appl. Microbiol. Biotechnol. 76:447-457. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, W., D. E. Culley, J. C. Scholten, M. Hogan, L. Vitiritti, and F. J. Brockman. 2006. Global transcriptomic analysis of Desulfovibrio vulgaris on different electron donors. Antonie van Leeuwenhoek 89:221-237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.