Abstract
Here we introduce a method for quantitative analysis of planktonic protists and microalgae from preserved field samples combining morphological and small-subunit (SSU) rRNA gene sequence analysis. We linked a microscopic screening with PCR of single cells using field samples preserved with Lugol's iodine solution. Cells possessing a rigid cell wall were incubated with Viscozyme and subsequently with proteinase K for cell disruption; this was unnecessary for fragile cells. The addition of sodium thiosulfate to the PCR tube considerably decreased the inhibiting effect of the fixative (iodine) on the PCR and thus allowed for successful single-cell PCR even of long DNA fragments (up to as many as 3,000 base pairs). We further applied the protocol to investigate the dominant SSU rRNA genotypes in distinct flagellate morphospecies originating from different samples. We hypothesized that despite the morphological similarity, protist morphospecies in different habitats or sampled during different seasons are represented by different genotypes. Our results indicate species-specific differences: the two species Ochromonas sp. and Dinobryon divergens were represented by several different genotypes each, and for the latter species, the dominating genotype differed with habitat. In contrast, Dinobryon pediforme, Dinobryon bavaricum, and Synura sphagnicola were exclusively represented by a single genotype each, and the respective genotype was the same in different samples. In summary, our results highlight the significance of molecular variation within protist morphospecies.
Linking a specific protist or microalgal small-subunit (SSU) rRNA gene sequence from environmental surveys to a specific morphotype is often problematic. Molecular surveys do not usually provide any information on the morphology of the organism (see references 19, 25, and 27 but compare with reference 10), whereas morphological surveys concentrate on preserved samples, which are usually not considered for molecular analyses (7). One main way to overcome these problems is to link sequence analysis with morphological investigations from preserved plankton samples on a per cell basis.
Successful sequence analysis has already been demonstrated for preserved specimens, but it has various shortcomings. Most methods either require relatively large amounts of template DNA (i.e., cultured material, preserved tissues, or environmental DNA collected on filters or by centrifugation [18]) or amplification is limited to short fragments or both (2, 4, 6). It is therefore no coincidence that attempts to analyze the DNA sequence from preserved microplankton samples focused mainly on alveolate taxa, i.e., organisms presumably with a high copy number of the SSU rRNA gene (dinoflagellates [5, 11, 13, 29]; ciliates [9]). Still, despite the presumably high gene copy number in the alveolates investigated so far, success with field samples is usually low.
Among the most common fixatives for microalgae and protists are formaldehyde and Lugol's iodine solution (12, 20, 32). Formaldehyde-preserved samples are generally problematic for molecular analyses, as formaldehyde may cause severe cell loss (e.g., reference 20 and references therein). Formaldehyde may further reduce the PCR efficiency in a storage time-dependent manner (17) and can alter the DNA structure and may thus cause sequencing errors, specifically C-T and G-A mutations during PCR (8, 26).
Lugol's iodine solution seems less problematic with respect to sequence analysis but still seems to require at least a 10-fold-higher cell concentration in the PCR compared to unpreserved PCR (5, 13, 30; see reference 6 for successful amplification of short fragments of around 200 base pairs).
We propose an optimized protocol combining microscopic screening with direct PCR of single protist and microalgal cells using field samples preserved with Lugol's iodine solution. We also successfully applied the protocol to investigate the dominant SSU rRNA genotypes in distinct flagellate taxa affiliated with the same morphospecies but originating from different samples. We hypothesized that despite the morphological similarity, protist morphospecies in different habitats or sampled during different seasons would be dominated by different genotypes.
MATERIALS AND METHODS
Media and stock solutions.
The following media, stock solutions, and chemicals were used in our study. NSY-IB medium is an inorganic basal medium for the maintenance of cultured strains. It contains the following substances: 75 mg of MgSO4·7H2O liter−1, 1.43 mg of Ca(NO3)2·4H2O liter−1, 16 mg of NaHCO3 liter−1, 5 mg of KCl liter−1, 2.8 mg of K2HPO4 liter−1, 4.4 mg of Na2EDTA liter−1, 3.2 mg of FeCl3·6H2O liter−1, 1.0 mg of H3BO3 liter−1, 0.2 mg of MnCl2·4H2O liter−1, 0.02 mg of ZnSO4·7H2O liter−1, 0.01 mg of CuSO4·6H2O liter−1, 0.01 mg of CoCl2·6H2O liter−1, 0.006 mg of Na2MoO4·2H2O liter−1, 0.1 mg of NiCl2·6H2O liter−1 (15); thiosulfate stock solution (62 g Na2S2O3·5H2O liter−1); thiosulfate working solution (50 μl of thiosulfate stock solution added to 1 ml of NSY-IB medium); Lugol's iodine stock solution (100 g KI liter−1 and 50 g I2 liter−1); Lugol's iodine working solution (2 ml of Lugol's iodine stock solution added to 98 ml of filtered NSY-IB medium (using 0.2-μm syringe filters).
Overcoming PCR inhibition caused by Lugol's iodine solution.
We tested the concentration-dependent inhibition of the PCR by Lugol's iodine solution and the effect of thiosulfate. Thiosulfate is commonly used to remove the dark stain from organisms preserved with Lugol's iodine solution for better identification of inner cell structures (21, 24, 31). We suspect that molecular iodine embedded in DNA is (partly) removed as the thiosulfate reduces the molecular iodine. We expected, therefore, that the fixation-related inhibition of the PCR would be partly neutralized.
(i) General setup.
Two axenic strains of chrysomonad flagellates were used as test organisms during the development of the method: Poterioochromonas malhamensis strain DS and Spumella sp. strain JBC07 (3). The flagellates were kept in axenic cultures. For the experiments, they were transferred to inorganic NSY-IB medium and fed with the bacterial strain Listonella pelagia CB5 (cf. reference 3). If not stated otherwise, 10 μl of dense flagellate cultures (corresponding to 100 to 200 cells) was used as the template for PCR without prior extraction of DNA.
In the first experiment, we tested for the effect of Lugol's iodine solution on the PCR at different thiosulfate concentrations: we investigated the effects of all combinations of 0, 0.09, 0.44, 0.88, 2.6, 5.3, 7.9, and 13.2 μl Lugol's iodine stock solution (corresponding to 0, 4.5, 22, 44, 130, 265, 395, and 660 μg I2 ml−1, respectively) and 0, 39, 195, 390, 975, and 1,950 μg sodium thiosulfate ml−1 on the PCR.
In a second experiment, the final concentrations of iodine in the PCR mix ranged from 0.0009 to 13.2 μl Lugol's iodine stock solution ml−1 (corresponding to 0.045 and 660 μg I2 ml−1, respectively); experiments were run in the absence and presence of 390 μg Na2S2O3 ml−1 (corresponding to the recommended concentration [see “Recommended protocol” below]). In addition, control treatments without fixative were tested.
(ii) DNA amplification and sequence handling.
Flagellate 18S rRNA was amplified with the following broad eukaryotic SSU rRNA targeting primers: the forward primer EK82f (5′-GAAACTGCGAATGGCTC-3′) and the reverse primer Proto5r (5′-GACGGGCGGTGTGTAC-3′).
Each PCR mixture contained 1.25 U of Taq polymerase (Qiagen), 5 μl of 10× PCR buffer, 200 nM of each primer, 200 μM of each deoxynucleoside triphosphate, 21.75 μl of water, and 20 μl of liquid containing the template DNA. The water for the PCR was distilled, then filtered using a 0.2-μm syringe filter, and finally autoclaved. Reactions were carried out in an Eppendorf Mastercycler gradient starting with a denaturation step of 3 min at 94°C, followed by 35 cycles, with 1 cycle consisting of denaturation (94°C for 1 min), annealing (52°C for 1 min), elongation (72°C for 2 min), and a final extension step of 5 min at 72°C. PCR products were checked on an agarose gel.
The PCR products were purified using the QIAquick PCR purification kit (Qiagen), following the instructions of the supplier. Subsequently, the products were quantified (Spectrophotometer Nano Drop ND-1000; program ND-1000 V3.3.0) and commercially sequenced (SMB, Berlin, Germany). The sequences were processed as previously described in reference 3, using the program BioEdit 5.0.9. The SSU rRNA gene sequences have been deposited in the National Center for Biotechnology Information (NCBI) database (for accession numbers, see Table 2 and see Table S1 in the supplemental material). Sequences were submitted to the BLAST search program at the NCBI.
TABLE 2.
Genotypes of individual cells affiliated with selected morphospecies originating from different habitats or seasonsa
| Species and strain | Genotypeb | GenBank accession no. | Lake | Sampling date (day.mo.yr) | Abundance (no. of cells ml−1) |
|---|---|---|---|---|---|
| Dinobryon bavaricum | |||||
| FU28-11 | Bavaricum 1 | EU024971 | Fuschlsee | 10.07.06 | 1.7 |
| FU28-13 | Bavaricum 1 | EU024972 | |||
| FU28-14 | Bavaricum 1 | EU024973 | |||
| FU44-4 | Bavaricum 1 | EU024979 | Fuschlsee | 30.10.06 | 5.9 |
| FU44-14 | Bavaricum 1 | EU024982 | |||
| FU44-61 | Bavaricum 1 | EU076735 | |||
| Dinobryon divergens | |||||
| FU28-24 | Divergens 1 | EU024976 | Fuschlsee | 10.07.06 | 9.3 |
| FU28-25 | Divergens 1 | EU024977 | |||
| FU28-27 | Divergens 1 | EU024978 | |||
| WA28-6 | Divergens 2 | EU025019 | Wallersee | 10.07.06 | 240 |
| WA28-7 | Divergens 3 | EU076736 | |||
| WA28-8 | Divergens 4 | EU025020 | |||
| WA28-34 | Divergens 4 | EU076737 | |||
| WA28-35 | Divergens 4 | EU076738 | |||
| WA28-37 | Divergens 4 | EU076739 | |||
| Dinobron pediforme | |||||
| LO128-14 | Pediforme 1 | EU005402 | Loibersbacher Teich 1 | 10.07.06 | 3,650 |
| LO128-16 | Pediforme 1 | EU024992 | |||
| LO128-17 | Pediforme 1 | EU024993 | |||
| LO134-9c | Pediforme 1 | EU024998 | Loibersbacher Teich 1 | 21.08.06 | 820 |
| LO134-19 | Pediforme 1 | EU025000 | |||
| LO134-20 | Pediforme 1 | EU025001 | |||
| LO228-51 | Pediforme 1 | EU025007 | Loibersbacher Teich 2 | 10.07.06 | 27 |
| LO228-76c | Pediforme 1 | EU025008 | |||
| LO228-77 | Pediforme 1 | EU025009 | |||
| LO234-1 | Pediforme 1 | EU025013 | Loibersbacher Teich 2 | 21.08.06 | 7,750 |
| LO234-4c | Pediforme 1 | EU025014 | |||
| LO234-5c | Pediforme 1 | EU025015 | |||
| Ochromonas sp. | |||||
| LO128-108 | Ochromonas 1 | EU024995 | Loibersbacher Teich 1 | 10.07.06 | 7 |
| LO128-155c | Ochromonas 1 | EU024996 | |||
| LO128-157 | Ochromonas 1 | EU076740 | |||
| LO128-158 | Ochromonas 2 | EU076741 | |||
| LO128-159 | Ochromonas 3 | EU076742 | |||
| LO128-160d | Ochromonas 1 | EU076743 | |||
| LO128-161c | Ochromonas 1 | EU076744 | |||
| LO128-162 | Ochromonas 1 | EU076745 | |||
| LO134-4 | Ochromonas 4 | EU024997 | Loibersbacher Teich 1 | 21.08.06 | 5,720 |
| LO134-15c | Ochromonas 1 | EU024999 | |||
| LO134-23 | Ochromonas 5 | EU025002 | |||
| LO134-25 | Ochromonas 1 | EU025003 | |||
| Synura sphagnicola | |||||
| LO228-6 | Sphagnicola 1 | EU025004 | Loibersbacher Teich 2 | 10.07.06 | 525 |
| LO228-7 | Sphagnicola 1 | EU025005 | |||
| LO228-33 | Sphagnicola 1 | EU025006 | |||
| LO234-7 | Sphagnicola 1 | EU025016 | Loibersbacher Teich 2 | 21.08.06 | 1,470 |
| LO234-8 | Sphagnicola 1 | EU025017 | |||
| LO234-9 | Sphagnicola 1 | EU025018 |
The strain number of the isolated cells, genotype, and GenBank accession number are shown for each investigated cell. The place of origin (lake) and the respective abundance is given for each morphospecies in the respective sample.
The same genotype number corresponds to 100% similarity between the investigated cells, different genotype numbers within a morphospecies indicate a difference of a single base pair (within the analyzed sequence) from the sequences of other cells of the same morphospecies. Only the sequences of Dinobryon divergens cells originating from Lake Fuschlsee differed in two positions from sequences of D. divergens cells originating from Lake Wallersee.
The sequences obtained from these strains have one mixed base site, but we assume that the base in question corresponds to that present in the other strains at the respective position.
This strain represents probably a different genotype, but the three base positions in question could not be unequivocally identified (i.e., K instead of A, R instead of T, and N instead of A).
Application to field samples. (i) Dealing with free DNA in plankton samples.
Preserved plankton samples often contain dissolved DNA due to disintegration of (some) cells during net sampling and sample processing (later on referred to as free DNA). We tested several protocols to prevent amplification of such free DNA in the PCR, specifically washing the whole plankton sample, washing individual cells, and combinations of these two approaches. For description of the washing steps, see “Recommended protocol” below. After each washing step, 10 μl of the fluid was taken and handled as described for the single cell up to the seminested PCR.
(ii) Quantitative analysis and applicability to different taxonomic groups.
An optimized protocol (see “Recommended protocol” in the Results and Discussion) was applied to organisms affiliated with different taxonomic groups in order to test for broad applicability of the proposed method. The main focus was on the Chrysophyceae and Synurophyceae (both stramenopiles) and the Dinozoa and Ciliophora (both alveolates) following the higher-level classification of Adl and coworkers (1). Field samples were taken from ponds and lakes in the Salzkammergut area in Austria, specifically from the ponds Loibersbacher Teich 1 and Loibersbacher Teich 2 and from the lakes Fuschlsee and Wallersee. Plankton samples were preserved with Lugol's iodine solution (20 μl Lugol's iodine stock solution ml−1) and stored at 4°C in the dark until further processing.
Primers and PCR protocols were chosen based on the morphological analysis of video recordings taken before picking the cells. In general, we used the primers and PCR conditions as stated above (see “DNA amplification and sequence handling”). For organisms that are not targeted by broad eukaryotic primers, a preliminary test was included, i.e., several cells were picked, and different primers and PCR protocols were tested until suitable protocols for all target organisms were discovered. In all cases, an additional negative control was taken from the remaining 400-μl drop after the last washing step and tested to check for contamination by free DNA. If there was too little PCR product for sequencing, a subsequent seminested PCR was conducted using 1 μl of the initial PCR product and appropriate primers, i.e., usually the forward primer Sogin 2f (5′-AGGGTTCGATTCCGGAG-3′) and the reverse primer Proto5r.
(iii) SSU rRNA genotype distribution within and among samples.
We further applied the proposed method to investigate the dominant SSU rRNA genotype within distinct microalgal morphospecies. We compared the genotype composition in samples originating from different locations and/or seasons (see Table 2). Specifically, we selected the chrysophyte taxa Dinobryon divergens and Dinobryon pediforme for spatial comparisons and D. pediforme, Dinobryon bavaricum, Synura sphagnicola, and Ochromonas sp. for seasonal analysis. For each of these species, we analyzed at least three individuals per sample (if present). This was done in order to determine the dominant genotype of the respective morphospecies.
RESULTS AND DISCUSSION
Overcoming PCR inhibition caused by Lugol's iodine solution.
Successful single-cell PCR from preserved plankton samples has been demonstrated for cultured strains using ethanol or methanol fixation (18), formalin or methanol fixation (13), osmium tetroxide fixation (29), and Lugol's iodine solution (14). All these methods are generally problematic for low concentrations of template DNA. The above studies therefore focused on alveolate taxa (dinoflagellates and ciliates), i.e., on taxa that presumably have a relatively high copy number of the SSU rRNA gene.
Accordingly, in our experiments, Lugol's iodine solution inhibited the PCR already at levels corresponding to less than 0.1 μl stock solution ml−1 (Table 1). Simple dilution, i.e., repeated washing in sterile media did not solve this problem for two reasons. First, the preserved cells still did not allow for successful PCR unless high concentrations of template DNA were used (i.e., many cells per PCR), and second, particularly fragile cells tended to burst in medium free of Lugol's iodine solution (data not shown).
TABLE 1.
PCR yield with different concentrations of Lugol's iodine solution and sodium thiosulfate in the PCR mix
| Lugol's iodine solution concn (μl of stock solution ml−1) | PCR efficiency with the following sodium thiosulfate concn (μg ml−1)a:
|
|||||
|---|---|---|---|---|---|---|
| 0 | 39 | 195 | 390 | 975 | 1,950 | |
| 0 | 44.8 (100) | 47.3 (105.6) | 45.1 (100.9) | 45.4 (101.5) | 16.8 (37.6) | 2.3 (5.2) |
| 0.088 | 1.5 (3.3) | 44.8 (100) | 32.4 (72.4) | 33 (73.7) | 27.7 (61.8) | 1.5 (3.4) |
| 0.44 | 0 (0) | 41.6 (92.9) | 26.1 (58.3) | 37.2 (83.2) | 24.1 (53.9) | 1.4 (3.1) |
| 0.88 | 0.2 (0.5) | 1.5 (3.3) | 42.8 (95.6) | 31.2 (69.6) | 20 (44.7) | 0.7 (1.5) |
| 2.64 | 0.1 (0.1) | 1.8 (4.1) | 37.1 (82.8) | 26 (58.1) | 15.3 (34.3) | 1.6 (3.6) |
| 5.28 | 0.5 (1.1) | 1.1 (2.5) | 1.3 (2.8) | 33.6 (75.1) | 21 (47) | 0.9 (2) |
| 7.92 | 0.6 (1.3) | 0.1 (0.3) | 0.3 (0.6) | 0.6 (1.4) | 14.8 (33) | 0.8 (1.8) |
| 13.2 | 0.7 (1.5) | 1.8 (3.9) | 0.4 (0.8) | 2.7 (6.1) | 0.7 (1.6) | 0.7 (1.5) |
The efficiency or yield of the PCR is shown as the amount of PCR product (ng of DNA μl−1) based on a constant amount of template DNA. The relative efficiency, i.e., the relative amount of DNA gained during PCR as a percentage of total DNA gained without the addition of the respective chemicals, is given in parentheses.
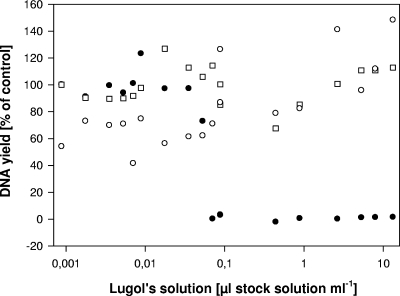
To solve this problem, we applied an alternative approach. By adding thiosulfate to the PCR master mix, the iodine fixation was (partly) reversed, allowing for successful PCR in the presence of much higher concentrations of Lugol's iodine solution (Table 1). A final concentration of 390 μg Na2S2O3 ml−1 in the PCR mix counterbalanced iodine concentrations of more than 600 μg I2 ml−1 (i.e., covering the usual range of iodine concentrations in preserved samples; Fig. 1). We did not observe any fixation artifacts during PCR, i.e., the SSU rRNA gene sequence using single cells of the flagellate strain JBC07 was identical for preserved and unpreserved cells and identical to the published sequence of that strain (GenBank accession number EF577165). The method was suitable for neutral, acidic, and alkaline Lugol's solution (data not shown).
FIG. 1.
Inhibition of the PCR by Lugol's iodine solution. The PCR yield is expressed as a percentage of the control treatment (DNA yield of the control without Lugol's iodine solution, 100%). The black circles indicate the PCR yield in the absence of thiosulfate, and the white symbols indicate the PCR yield in the presence of thiosulfate. Cultures of Poterioochromonas (circles) and Spumella (squares) were used as template in these experiments.
Recommended protocol.
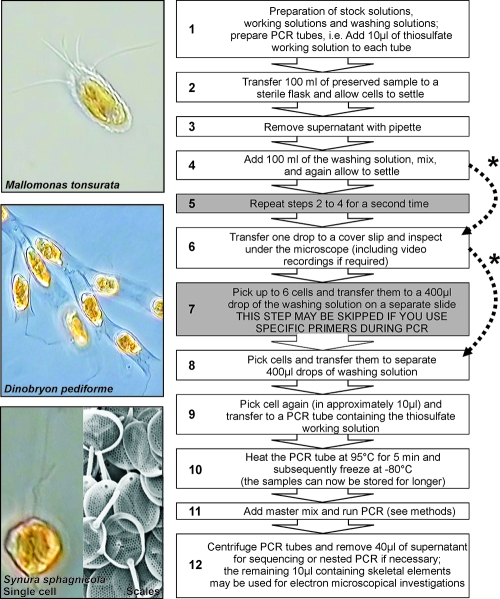
Prior to the analysis, the plankton samples preserved with Lugol's iodine solution were washed as follows (Fig. 2). First, we washed the whole sample, i.e., 100-ml portions of the preserved samples were transferred to fresh flasks, and the cells were allowed to settle. Subsequently, the liquid was gently removed with a pipette, leaving roughly 5 ml of residual material (0.5 cm of the water column at the bottom). Afterwards, 95 ml of the washing solution was added, and the sample was mixed. This procedure was repeated a second time.
FIG. 2.
Proposed protocol. The essential steps of the protocol are depicted with a white background, those with a gray background may be skipped depending on the level of contamination/cell concentrations in the original sample and the specificity of the primers used during PCR (indicated by the dotted arrows marked with an asterisk). For an explanation, see the text. Examples of the morphological analysis are shown on the left for three species. For Synura sphagnicola, a scanning electron microscopy image of the scales, which can be extracted from the PCR tubes after running the PCR, is also shown.
Afterwards, we washed individual target cells. One milliliter of the preserved sample was transferred to a cover slide and inspected for target cells at a total magnification of ×200 (Zeiss Axiovert 200). Target cells were then checked at a magnification of ×400 or ×630, and digital recordings were made for later morphological analysis (Panasonic KR222E with Dazzle Video Creator 150; program Pinnacle Studio Version 9.4.3.). After inspection and video recording, the single cells were picked at ×200 magnification with a glass pipette, and up to six cells (corresponding to 5 to 50 μl) were transferred to a 400-μl drop of Lugol's iodine working solution on a separate slide. These cells were picked again and separately transferred to one drop (400 μl) of Lugol's iodine working solution each. From this final drop, the cells were picked again and (in a drop of approximately 10 μl) transferred to a 200-μl PCR tube already containing 10 μl of the thiosulfate working solution (corresponding to 390 μg Na2S2O3 ml−1). For a negative control for free DNA, 10 μl of the remaining fluid was transferred to a second PCR tube and processed the same way. The PCR tubes were subsequently heated at 95°C for 5 min in an Eppendorf Mastercycler gradient and immediately afterwards shock frozen at −80°C in order to break cells and to denature proteins (Fig. 2).
Organisms possessing a rigid cell wall (e.g., dinoflagellates, cryptophytes, and diatoms) were treated in a slightly different way in order to break the cell wall. For the last washing step of the individual cells, we used NSY-IB medium instead of Lugol's iodine working solution. After the cells were picked, they were incubated in a 200-μl PCR tube with 1 μl of 1:100 diluted Viscozyme L solution (Sigma-Aldrich) in a total volume of ∼20 μl at 37°C for 2 h in an Eppendorf Thermomixer comfort. Afterwards, 1 μl of 1:10 diluted proteinase K solution (proteinase K [>600 × 10−3 absorbance units/ml]; Qiagen) was added. The tubes were incubated at 55°C for 50 min and subsequently heated at 95°C for 10 min (16). Afterwards, the samples were further processed as described above.
Application to field samples. (i) Dealing with free DNA in plankton samples.
As most eukaryotic cells contain many copies of the rRNA genes (specifically, ciliates and dinoflagellates may contain up to several hundred or thousand copies), even the disintegration of only a few cells may cause a serious contamination of the sample. The application of the two washing methods as applied in our protocol sufficiently excluded such background contamination even when broad eukaryotic primers were applied. Using more specific primers circumvented the amplification of free DNA even without washing single cells (data not shown). However, we recommend picking several cells from any morphospecies under investigation as an internal control and testing a negative control from the surrounding fluid of the cell.
(ii) Quantitative analysis and applicability to different taxonomic groups.
We recommend the proposed method for a quantitative analysis of protist and microalgae communities combining morphological and molecular investigations. Our protocol allowed for amplification of the nearly full-length SSU rRNA gene from single cells for any protist and algal taxon tested (see Table S1 in the supplemental material). We successfully and quantitatively analyzed DNA fragments of up to 3,000 (data not shown) even though we concentrated on a fragment of between 1,200 and 1,500 base pairs for analysis. All tested morphospecies, including several fragile taxa, were successfully investigated by the proposed method (GenBank accession numbers EU005402, EU024970, EU024971, EU024974, EU024975, EU024977, EU024980 to EU024984, EU024986 to EU024991, EU024994, EU025002, EU025010 to EU025012, EU025018, EU025021, and EU025022 [see Table S1 in the supplemental material]). The combination of washing protocols with the addition of thiosulfate in the PCR therefore significantly improved the single-cell PCR from preserved plankton samples.
Other molecular standard methods are limited in linking morphology and full sequence information; while providing sequence data, clone libraries provide mostly presence-absence information and are severely biased by primer specificity and taxon-specific differences in gene copy number (28). In contrast, fluorescence in situ hybridization targets only short oligonucleotide sequences and requires a detailed a priori knowledge of the sequence and probe specificity.
In contrast to conventional molecular surveys, the proposed method allows the linking of morphological and molecular screening approaches directly and quantitatively based on preserved plankton samples (see Table S1 in the supplemental material). Further, not only abundant taxa but also comparatively rare ones, taxa with a low gene copy number and taxa that are negatively selected by broad eukaryotic primers, can easily and quantitatively be analyzed by the proposed method (see Table S1 in the supplemental material).
(iii) SSU rRNA genotype distribution within and among samples.
The proposed method is particularly well suited to detect minute genotypic differences between individuals of the same morphospecies. Genotypic variation within nominal protist taxa is of special interest with respect to the debate on microbial distribution and biogeographies (3, 22, 23, 33). We applied our method to check for differences in the dominant genotypes in protist taxa affiliated with the same morphospecies but originating from different habitats or seasons. We tested specifically for spatial differences for Dinobryon pediforme and Dinobryon divergens (both Chrysophyceae) and, in addition, for seasonal differences for Ochromonas sp., D. pediforme, Dinobryon bavaricum (Chrysophyceae), and Synura sphagnicola (Synurophyceae). The results indicate considerable differences between the tested morphospecies (Table 2). For some species, i.e., Dinobryon bavaricum, Dinobryon pediforme, and Synura sphagnicola, we found no intraspecific variation in the SSU rRNA gene sequence between individuals. This was independent of the time and place of origin.
In contrast, for other species, e.g., Ochromonas sp. and Dinobryon divergens, the morphospecies were represented by different SSU rRNA genotypes. For these latter morphospecies, our data indicate a different genotype composition in the different samples. For instance, the dominant genotype of D. divergens from Lake Fuschlsee was not found in Lake Wallersee. A high molecular variation has also been demonstrated for other nominal nanoflagellate taxa (e.g., for Spumella sp. [K. Pfandl et al., submitted for publication], Paraphysomonas vestita [2], Neobodo designis [33], and Rhynchomonas nasuta [22]). A conclusive judgment would require in-depth investigation of the respective morphospecies, which was, however, not in the scope of this study. It remains a future challenge to identify those species in which the morphotype corresponds to a specific phylotype and to separate those in which this is not the case.
Supplementary Material
Acknowledgments
We thank G. Cronberg and H. Preisig for their help with the morphological analyses and L. Eisl and A. Wiedlroither for their technical assistance. We further thank G. Cronberg for providing the scanning electron microscopy image of S. sphagnicola.
The Austrian Science Fund (project 18315) and the Alpine Research Program of the Austrian Academy of Sciences (CLIME project) provided financial support for this work.
Footnotes
Published ahead of print on 22 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adl, S. M., A. G. B. Simpson, M. A. Farmer, R. A. Andersen, O. R. Anderson, J. R. Barta, S. S. Bowser, G. Brugerolle, R. A. Fensome, S. Frederico, T. Y. James, S. Karpov, P. Kugrens, J. Krug, C. E. Lane, L. A. Lewis, J. Lodge, D. H. Lynn, D. G. Mann, R. M. McCourt, L. Mendoza, Ø. Moestrup, S. E. Mozley-Standridge, T. A. Nerad, C. A. Shearer, A. V. Smirnov, F. W. Spiegel, and M. F. J. R. Taylor. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52:399-451. [DOI] [PubMed] [Google Scholar]
- 2.Bertozzini, E., A. Penna, E. Pierboni, I. Bruce, and M. Magnani. 2005. Development of new procedures for the isolation of phytoplankton DNA from fixed samples. J. Appl. Phycol. 17:223-229. [Google Scholar]
- 3.Boenigk, J., K. Pfandl, P. Stadler, and A. Chatzinotas. 2005. High diversity of the “Spumella-like” flagellates: an investigation based on the SSU rRNA gene sequences of isolates from habitats located in six different geographic regions. Environ. Microbiol. 7:685-697. [DOI] [PubMed] [Google Scholar]
- 4.Bonin, S., F. Petrera, J. Rosai, and G. Stanta. 2005. DNA and RNA obtained from Bouin's fixed tissues. J. Clin. Pathol. 58:313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowers, H. A., T. Tengs, H. B. Glasgow, J. M. Burkholder, P. A. Rublee, and D. W. Oldach. 2000. Development of real-time PCR assays for rapid detection of Pfisteria piscida and related dinoflagellates. Appl. Environ. Microbiol. 66:4641-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connell, L. 2002. Rapid identification of marine algae (Raphidophyceae) using three-primer PCR amplification of nuclear internal transcribed spacer (ITS) regions from fresh and archived material. Phycologia 41:15-21. [Google Scholar]
- 7.Cronberg, G., and R. Laugaste. 2005. New species of Uroglena and Ochromonas (Chromulinales, Chrysophyceae) from Estonia. Nova Hedwigia Beih. 128:43-63. [Google Scholar]
- 8.Douglas, M. P., and S. O. Rogers. 1998. DNA damage caused by common cytological fixatives. Mutat. Res. 401:77-88. [DOI] [PubMed] [Google Scholar]
- 9.Dyal, P. L., S. Hope, D. M. Roberts, and T. M. Embley. 1995. Use of the PCR and fluorescent-probes to recover SSU ribosomal-RNA gene-sequences from single cells of the ciliate protozoan Spathidium. Mol. Ecol. 4:499-503. [DOI] [PubMed] [Google Scholar]
- 10.Edvardsen, B., K. Shalchian-Tabrizi, K. S. Jakobsen, L. K. Medlin, E. Dahl, S. Brubak, and E. Paasche. 2003. Genetic variability and molecular phylogeny of Dinophysis species (Dinophyceae) from Norwegian waters inferred from single cell analyses of rDNA. J. Phycol. 39:295-408. [Google Scholar]
- 11.Galluzzi, L., A. Penna, E. Bertozzini, M. Vila, E. Garcés, and M. Magnani. 2004. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl. Environ. Microbiol. 70:1199-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gifford, D. J., and D. A. Caron. 2000. Sampling, preservation, enumeration and biomass of marine protozooplankton, p. 193-221. In R. P. Harris, P. H. Weibe, J. Lenz, H. R. Skjoldal, and M. Huntley (ed.), ICES zooplankton methodology manual. Academic Press, London, United Kingdom.
- 13.Godhe, A., D. M. Anderson, and A.-S. Rehnstam-Holm. 2002. PCR amplification of microalgal DNA for sequencing and species identification: studies on fixatives and algal growth stages. Harmful Algae 1:375-382. [Google Scholar]
- 14.Guillou, L., E. Nezan, V. Cueff, E. E. L. Denn, M. A. Cambon-Bonavita, P. Gentien, and G. Barbier. 2002. Genetic diversity and molecular detection of three toxic dinoflagellate genera (Alexandrium, Dinophysis, and Karenia) from French coasts. Protist 153:223-238. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, M. W., H. Lünsdorf, Q. L. Wu, M. Schauer, M. G. Höfle, J. Boenigk, and P. Stadler. 2003. Isolation of novel ultramicrobacteria classified as Actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ki, J. S., G. Y. Jang, and M. S. Han. 2004. Integrated method for single-cell DNA extraction, PCR amplification, and sequencing of the ribosomal DNA from harmful dinoflagellates Cochlodinium polykrikoides and Alexandrium catenella. Mar. Biotechnol. 6:587-593. [DOI] [PubMed] [Google Scholar]
- 17.Koppelstaetter, C., P. Jennings, K. Hochegger, P. Perco, R. Ischia, H. Karkoszka, and G. Mayer. 2005. Effect of tissue fixatives on telomere length determination by quantitative PCR. Mech. Ageing Dev. 126:1331-1333. [DOI] [PubMed] [Google Scholar]
- 18.Marin, I., A. Aguilera, B. Reguera, and J. Abad. 2001. Preparation of DNA suitable for PCR amplification from fresh and fixed single dinoflagellate cells. BioTechniques 30:88-93. [DOI] [PubMed] [Google Scholar]
- 19.Massana, R., J. Castresana, V. Balagué, L. Guillou, K. Romari, A. Groisillier, K. Valentin, and C. Pedrós-Alió. 2004. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl. Environ. Microbiol. 70:3528-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Modigh, M., and S. Castaldo. 2005. Effects of fixatives on ciliates as related to cell size. J. Plankton Res. 27:845-849. [Google Scholar]
- 21.Pomroy, A. J. 1984. Direct counting of bacteria preserved with Lugol iodine solution. Appl. Environ. Microbiol. 47:1191-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheckenbach, F., C. Wylezich, M. Weitere, K. Hausmann, and H. Arndt. 2005. Molecular identity of strains of heterotrophic flagellates isolated from surface waters and deep-sea sediments of the South Atlantic based on SSU rDNA. Aquat. Microb. Ecol. 38:239-247. [Google Scholar]
- 23.Shankle, A. M., X. Mayali, and J. S. Franks. 2004. Temporal patterns in population genetic diversity of Prorocentrum micans (Dinophyceae). J. Phycol. 40:239-247. [Google Scholar]
- 24.Sherr, E. B., and B. F. Sherr. 1993. Preservation and storage of samples for enumeration of heterotrophic protists, p. 207-212. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, London, United Kingdom.
- 25.Slapeta, J., D. Moreira, and P. Lopez-Garcia. 2005. The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proc. Biol. Sci. 272:2073-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srinivasan, M., D. Sedmak, and S. Jewell. 2002. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am. J. Pathol. 161:1961-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoeck, T., and S. Epstein. 2003. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl. Environ. Microbiol. 69:2657-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoeck, T., B. Hayward, G. T. Taylor, R. Varela, and S. S. Epstein. 2006. A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist 157:31-43. [DOI] [PubMed] [Google Scholar]
- 29.Takano, Y., and T. Horiguchi. 2005. Acquiring scanning electron microscopical, light microscopical and multiple gene sequence data from a single dinoflagellate cell. J. Phycol. 42:251-256. [Google Scholar]
- 30.Tengs, T., H. A. Bowers, A. P. Ziman, D. K. Stoecker, and D. W. Oldach. 2001. Genetic polymorphism in Gymnodinium galatheanum chloroplast DNA sequences and the development of a molecular detection assay. Mol. Ecol. 10:515-523. [DOI] [PubMed] [Google Scholar]
- 31.Throndsen, J. 1978. Preservation and storage, p. 69-74. In A. Sournia (ed.), Phytoplankton manual. UNESCO, Paris, France.
- 32.Utermöhl, H. 1958. Zur Vervollkommnung der quantitativen Phytoplankton-Methodik. Mitt. Int. Ver. Theor. Angew. Limnol. 9:1-38. [Google Scholar]
- 33.von der Heyden, S., and T. Cavalier-Smith. 2005. Culturing and environmental DNA sequencing uncover hidden kinetoplastid biodiversity and a major marine clade within ancestrally freshwater Neobodo designis. Int. J. Syst. Evol. Microbiol. 55:2605-2621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.