Abstract
rRNA-based studies, which have become the most common method for assessing microbial communities, rely upon faithful amplification of the corresponding genes from the original DNA sample. We report here an analysis and reevaluation of commonly used primers for amplifying the DNA between positions 27 and 1492 of bacterial 16S rRNA genes (numbered according to the Escherichia coli rRNA). We propose a formulation for a forward primer (27f) that includes three sequences not usually present. We compare our proposed formulation to two common alternatives by using linear amplification—providing an assessment that is independent of a reverse primer—and in combination with the 1492 reverse primer (1492r) under the PCR conditions appropriate for making community rRNA gene clone libraries. For analyses of DNA from human vaginal samples, our formulation was better at maintaining the original rRNA gene ratio of Lactobacillus spp. to Gardnerella spp., particularly under stringent amplification conditions. Because our 27f formulation remains relatively simple, having seven distinct primer sequences, there is minimal loss of overall amplification efficiency and specificity.
The study of microbial communities is important on multiple levels, from describing nutrient cycling and elucidating novel metabolisms to understanding how ecosystems are maintained and how mixtures of microbes can promote and/or upset the health status of their harboring host. Our ability to evaluate aspects of microbial ecology depends to a large extent on correctly identifying community members and their relative contributions to the overall makeup of the ecosystem.
The analysis of genes found in an environment as proxies for the organisms themselves has revolutionized our understanding of microbial communities (16). Studies of universal genes, especially the small-subunit rRNA (SSU rRNA), provide phylogenetic portraits of the communities, including organisms that have not yet been cultivated (10, 16, 32). These data increase in value with time, as newly cultivated species provide more anchor points that relate organismal phylogeny and physiology. Furthermore, communities are easily compared between locations and over time.
An essential contribution to the utility of this approach is the interspersion of more- and less-conserved sequences within the rRNA genes. The more varied portions distinguish the phylogenetic groups, while the conserved portions provide universal (or nearly universal) sequences for PCR primer binding. This allows specific amplification of the genes of interest out of total community genome DNA (the metagenome). Nearly all studies of bacterial SSU rRNA genes rely on primers designed over 15 years ago (32, 35). Although several groups have warned of the limitations of these primers, this has had little impact on common practice (reviewed in reference 30). This might be of little consequence in studies that seek only a qualitative portrait of community diversity, but with the increasing application of rRNA gene-based methods to analyze medically important samples (see, e.g., references 4, 5, 6, 9, 12, 13, 25, 29, and 36), overlooking some of the community components due to inefficient primer binding could have great practical implications.
Motivated by analyses of vaginal microbial communities (5, 12, 13, 36), we have reexamined two of the most commonly used primers for bacterial 16S rRNA genes, 27f (spanning positions 8 to 27 in Escherichia coli rRNA coordinates) and 1492r (commonly spanning positions 1492 to 1507, though longer versions are sometimes used) (2, 11, 16, 21, 28, 32, 35), which amplify nearly the entire length of the gene. We assessed the sequence variability at the corresponding primer-binding sites in the nearly 200,000 sequences from the Ribosomal Database Project II (RDP) (8, 17) and in the Sargasso Sea metagenomic data set (27). In doing so, we developed tools for extracting the relevant sequence regions and a heuristic algorithm for screening out sequences that are contributed by the inclusion of amplification primers in published sequences rather than by the original genomic DNA. Based on the observed sequence variation, we propose a new formulation of the 27f primer. We compare our formulation to two that are in current use for their influence on the relative abundances of Gardnerella and Lactobacillus rRNA genes in linear (unidirectional) and exponential (bidirectional) amplifications.
MATERIALS AND METHODS
Sequence data.
Sequences were from the RDP (release 9.36) (8; http://rdp.cme.msu.edu/) and the Sargasso Sea metagenome (27) (GenBank accession numbers AACY01000001 to AACY01811372).
Construction of a representative set of bacterial SSU rRNA gene sequences.
The identification of rRNA gene sequences in metagenomic data and the locating of primer-binding sites in the rRNA gene sequences were based on the similarity to sequences in a phylogenetically representative set of full-length bacterial SSU rRNA sequences. In the interests of completeness and accuracy, to as great an extent as feasible, these representative sequences were extracted from completed (or nearly completed) genome sequences available through the NCBI Entrez system (34) and The SEED (22). An initial set of sequences was handpicked for phylogenetic diversity and the familiarity of the taxonomic name. Other named sequences with less than 85% identity to those already chosen (as measured by BLASTN [3]) were added with the aid of Perl scripts to cover phylogenetic groups without complete genome data. Several other sequences (particularly from actinomycetes) were added to encompass more of the variations in structure observed in the first 200 nucleotides of the 16S rRNA.
In order to project primer-binding site coordinates from the E. coli 16S rRNA sequence to each of the other representative sequences, a multiple sequence alignment was produced using ClustalW (7). The initial alignment demonstrated that the endpoints of many of the 16S rRNA genes were misannotated. Where necessary, endpoints were manually adjusted. The resulting alignment was manually checked for accuracy at the primer locations.
Locating bacterial 16S rRNA genes in metagenomic data.
The Sargasso Sea metagenome sequence data from The SEED (22) were formatted as a nucleotide BLAST database. To locate bacterial rRNA genes in the metagenomic data, each of the representative rRNA gene sequences (described above) was used as a BLASTN (3) query with the following search parameters: a maximum expectation value of 10−12, an identity score of 1, a nonidentity score of −1, 10,000 maximum alignments, and no low-complexity filtering. The matching DNA regions were extracted, oriented in the rRNA-like direction, and placed in a database of metagenomic rRNA genes. The resulting collection comprised 1,137 rRNA gene sequence fragments. Because the start points and endpoints of the original clones are random, the sequence coverage of the SSU rRNA genes is relatively uniform from start to end, with somewhat over 400 sequences covering any given portion of the gene.
Identification of primer-binding sites in rRNA gene sequences.
The first task was to reliably identify and extract the rRNA gene sequences found at the primer-binding sites in a manner that is resistant to idiosyncratic variations, since characterizing these variations was our goal. We reasoned that the most reliable approach is to align each sequence with a related, full-length “representative” rRNA gene and use that alignment to project the known locations of the primer sites in the representatives onto the database sequences. The representative rRNA gene sequences (described above) were formatted as a nucleotide BLAST database. To identify a primer-binding site sequence in a given rRNA gene, the rRNA gene was used as a BLASTN (3) query with the following search parameters: a maximum expectation value of 10−8, an identity score of 1, a nonidentity score of −1, a maximum of five alignments, and no low-complexity filtering. The search results were scanned for matches overlapping the desired primer-binding site in one of the representative sequences, and the corresponding portion of the query was extracted. For consistency of data handling, all analyses were carried out in the RNA-like sequence orientation.
PCR primers.
Lyophilized oligonucleotides (Integrated DNA Technologies) were resuspended in 1 mM EDTA containing 0.001% Triton X-100. Nearly full length 16S rRNA genes were amplified using the 1492r primer (5′-TACCTTGTTACGACTT) and one of the following three 27f primer formulations: twofold-degenerate primer 27f-CM (5′-AGAGTTTGATCMTGGCTCAG, where M is A or C), fourfold-degenerate primer 27f-YM (5′-AGAGTTTGATYMTGGCTCAG, where Y is C or T), or sevenfold-degenerate primer 27f-YM+3. The sevenfold-degenerate primer 27f-YM+3 is four parts 27f-YM, plus one part each of primers specific for the amplification of Bifidobacteriaceae (27f-Bif, 5′-AGGGTTCGATTCTGGCTCAG), Borrelia (27f-Bor, 5′-AGAGTTTGATCCTGGCTTAG), and Chlamydiales (27f-Chl, 5′-AGAATTTGATCTTGGTTCAG) sequences.
For the quantitative PCR (qPCR) of Lactobacillus 16S rRNA gene sequences, the primers used were LactoF (5′-TGGAAACAGRTGCTAATACCG, where R is A or G) and LactoR (5′-GYCCATTGTGGAAGATTCCC); these primers amplify a fragment of about 200 bp in length, approximately between positions 200 and 400 of Lactobacillus 16S rRNA genes. They match all Lactobacillus 16S rRNA gene clones isolated from the human vaginal microbial DNA samples that we have analyzed and no other taxonomic groups in the samples. For the qPCR of Gardnerella 16S rRNA gene sequences, the primers used were GardnF (5′-GACTGAGATACGGCCCAGAC) and GardnR (5′-ATTCGAAAGGTACACTCACC); these primers amplify a fragment of about 180 bp in length, approximately between positions 180 and 360 of the Gardnerella vaginalis 16S rRNA gene.
Sample collection and genomic DNA extraction.
Human vaginal samples were collected from eight healthy, premenopausal women between the ages of 20 and 45 years through a study approved by the Institutional Review Boards of the University of Illinois at Urbana-Champaign and the Carle Foundation Hospital. Genomic DNA was isolated from 0.5-ml aliquots of the vaginal samples. To prepare DNA from each sample, 125 μl of 0.5 M Na-EDTA, pH 8.0, containing 75 mg/ml lysozyme was added and samples were incubated at 37°C for 30 min. Following the addition of 70 μl of 10% sodium dodecyl sulfate and 5 μl of 10-mg/ml proteinase K, the samples were subjected to three freeze-thaw cycles consisting of 5 min in a dry ice-ethanol bath followed by incubation at 37°C for 5 min, and finally they were held at 55°C for an additional 30 min. Following the addition of 70 μl of 5 M NaCl to the mixture and incubation on ice for 30 min, the samples were centrifuged at 16,000 × g for 20 min and the supernatants were extracted with phenol and phenol-chloroform-isoamyl alcohol, followed by ethanol precipitation. The genomic DNA was resuspended in 100 μl of 10 mM Tris-Cl, pH 8.0, containing 1 mM EDTA and stored at −80°C until used.
Linear amplification of vaginal microbial 16S rRNA genes.
Human vaginal microbial DNA samples were used as the substrate for the linear amplification of 16S rRNA genes using the following three 27f primer variants: 27f-CM, 27f-YM, and 27f-YM+3. Each primer variant was tested at three levels of stringency (the annealing temperatures were 48°C, 54°C, and 60°C). Reaction mixtures contained 0.1 ng of sample DNA in 25 μl PCR buffer (Invitrogen) containing 2 mM MgCl2, a 0.2 mM concentration of a deoxynucleoside triphosphate mix, a 200 nM concentration of the appropriate 27f primer variant, and 0.25 units of Platinum Taq DNA polymerase (Invitrogen). Reaction mixtures were incubated for 4 min at 94°C, followed by cycles of denaturation for 1 min at 94°C, annealing for 30 s at the appropriate temperature, and extension for 2 min at 72°C. A 10-μl aliquot was removed before amplification (zero-cycle sample). After five cycles, the amplification program was paused and 10-μl aliquots were removed from each reaction mixture (five-cycle sample). After the amplification program was resumed, reaction mixtures were incubated for a further five cycles, at the end of which another 10-μl aliquot was removed (10-cycle sample). Before being used as the substrate for qPCR, linearly amplified samples were diluted eightfold in 1 mM EDTA containing 0.001% Triton X-100, making them 0.0005 ng/μl in the original sample DNA.
PCR amplification of vaginal microbial 16S rRNA genes.
16S rRNA genes present in the human vaginal microbial DNA samples were amplified using the 1492r primer and one of the three formulations of the 27f primer. Amplification conditions were the same as those for linear amplification except for the addition of a 200 nM concentration of the reverse primer; all annealings were at 48°C, and the amplification was for 23 cycles. Based on our empirical estimates of the amplification efficiency, we calculated that 23 cycles of PCR increased the number of rRNA gene sequences by as much as 770,000-fold. To accurately compare the ratios of Lactobacillus to Gardnerella sequences present in samples before and after amplification, the 23-cycle PCR products were diluted 25,000-fold in 1 mM EDTA containing 0.001% Triton X-100. When 1 μl of the diluted PCR product was added to a 25-μl qPCR mixture, the overall dilution of amplified rRNA gene sequences was 625,000-fold.
qPCR.
Each sample to be analyzed was used as a template in two 25-μl qPCR mixtures, one for Lactobacillus and one for Gardnerella. For the analysis of linear amplification products, 2 μl of the diluted product (0.01 ng of original-sample DNA) was used for each reaction. For the analysis of PCR-amplified samples, the unamplified control contained 0.1 ng of original-sample DNA, while 1 μl of the 25,000-fold dilution was used for the exponentially amplified products. Each reaction mixture contained 1× iQ SYBR green supermix (Bio-Rad) and 100 nM concentrations of the forward and reverse primers. The reaction mixtures were incubated at 94°C for 4 min, followed by 33 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 15 s, and extension at 72°C for 45 s. Fluorescence was quantified using a Bio-Rad iCycler during the extension step of each PCR cycle. Each qPCR experiment was conducted in triplicate for a given DNA sample.
RESULTS
Primer-binding sites in database sequences.
As described in more detail in Materials and Methods, we devised and implemented a method to extract the bacterial SSU rRNA 27f and 1492r primer-binding site sequences from the data in the RDP and the Sargasso Sea metagenomic data. A key point of the method is that it assumes only a ≥50% sequence identity between the region containing the primer-binding site in the sequence being analyzed and at least one member of a diverse set of “reference” sequences. The method is general and can be used to extract any portion of a sequence that is sufficiently conserved or at least is flanked by conserved sequences.
Removing primer sequence contamination from rRNA gene data.
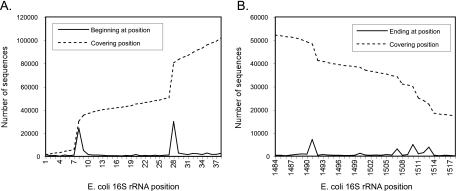
The bacterial SSU rRNA gene sequence data collected in the RDP were trimmed of nonribosomal sequences and oriented in the rRNA-like direction. However, it became clear during our extraction of primer-binding sites that many were PCR products that had not been trimmed to just the amplified DNA (i.e., they still included PCR primer sequences). In locating primer-binding sites, we used each RDP sequence as a BLASTN query against the representative sequences. For each query, we recorded the regions of similarity to the rRNA gene (mapped to E. coli coordinates). In Fig. 1A, we plot the total number of RDP sequences whose match covers a given nucleotide and the number of sequences whose similarity to rRNA starts at a given nucleotide (beginning point). The distribution is revealing. Out of 30,258 sequences with similarity extending to rRNA position 8, 24,340 start precisely there. Given that the 27f primer is most commonly a 20-mer spanning positions 8 to 27, this is the situation expected if the rRNA genes were amplified with a 27f primer and not trimmed of their primer sequence before database submission. The spot-checking of publications in which some of these sequences were reported supports this interpretation. Overall, out of 80,428 sequences that include position 28, 29,922 appear to be PCR products trimmed of the primer, 15,081 appear to be PCR products reported with part of the primer sequence, 24,340 appear to be PCR products reported with the full primer sequence, 5,918 retain similarity to rRNA at least 1 nucleotide past the primer, and 4,317 sequences (5.4% of the total) include similarity to rRNA at least 3 nucleotides past the primer.
FIG. 1.
Beginning and ending points of RDP bacterial 16S rRNA sequences in E. coli coordinates. The graphs show the number of sequences beginning or ending at a given point and the total number of sequences that include (cover) the position. (A) Region corresponding to E. coli positions 1 to 36, which includes the 27f primer-binding site; (B) region corresponding to E. coli positions 1484 to 1519, which includes the 1492r primer-binding site.
The same analysis was carried out in the region of the 1492r primer-binding site (Fig. 1B). The histogram of rRNA gene similarity endpoints clearly shows the 16-, 19-, and 22-nt-long versions of the primer. Overall, 7,139 sequences appear to be PCR products properly trimmed of the 1492r primer, 18,889 sequences appear to be PCR products reported with some or all of the 1492r primer sequence, and 18,522 sequences extend beyond the primer site. The larger number of sequences that continue beyond this primer site is due primarily to the use of reverse-amplification primers downstream of the 1492r-binding site (e.g., 1522r, 1525r, or a primer in the 23S rRNA gene, which commonly follows the 16S rRNA gene).
To maximize the chance that the sequences analyzed below contain bona fide primer-binding site sequences (not PCR primer sequences), we demanded that the sequences included in our analysis extend at least 3 nucleotides beyond the primer-binding site being analyzed and that the extension showed similarity to rRNA sequences. At the 27f primer-binding site, this excluded 80% of the sequences that might otherwise have been included. Requiring only 2 nucleotides beyond the primer-binding site did not significantly increase the data available for analysis but decreased our confidence in their validity. The application of these criteria resulted in the extraction of 4,315 (4,152 after the removal of sequences containing ambiguous nucleotides) 27f-binding site sequences and 17,940 (17,696 after the removal of sequences containing ambiguous nucleotides) 1492r-binding site sequences from the RDP database. Spot checks confirmed these to be largely devoid of PCR primer contamination.
Analysis of primer-binding site sequences.
The number of occurrences of the most commonly observed sequence variants at the 27f-binding site are shown in Table 1, as are the dominant phylogenetic groups in which each is observed. We also indicate which of several published 27f primer formulations precisely match the sequence. The two most common binding site variants cover most of the bacterial phyla. It is interesting that the sequence observed most often in Sargasso Sea rRNA genes (Table 1, second row) is not precisely matched by the common nondegenerate form of the primer, 27f-CC (AGAGTTTGATCCTGGCTCAG) (see, e.g., references 1, 4, 6, 9, and 25). The third most frequently observed binding site sequence is found in Actinobacteria and some Proteobacteria. It is accommodated by the 27f-YM primer but requires a C-A mispairing when the more common 27f-CC and 27f-CM formulations are used.
TABLE 1.
Occurrences of the most commonly observed sequences in 27f primer-binding sites, their phylogenetic distribution, and the primer formulations that they match precisely
| Primer binding site sequencea | No. of occurrences in:
|
Phylogenetic group(s) containing the binding site sequence | Exactly matching primer(s)b | |
|---|---|---|---|---|
| RDP v9.36 | Sargasso Sea data | |||
| AGAGTTTGATCCTGGCTCAG | 2,825 | 132 | Most Bacteria | 1, 2, 3, 4, 5, 6 |
| ...........A........ | 905 | 252 | Many Bacteria, especially enteric bacteria | 2, 3, 4, 5, 6 |
| ..........T......... | 59 | 2 | Actinobacteria, some Proteobacteria | 3, 4, 6 |
| ...A.......T...T.... | 57 | 0 | Chlamydiales | 6 |
| ......C............. | 54 | 0 | Atopobium and chloroplasts | |
| .................T.. | 35 | 0 | Borrelia spp. | 6 |
| ..........TA........ | 7 | 21 | Campylobacterales and Sphingomonadales | 3, 4, 6 |
| ..G...C...T......... | 21 | 0 | Bifidobacteriales | 6 |
| ..G................. | 17 | 0 | Thermotogales and Planctomycetales | 5 |
Sequence variations are shown as differences from the first (most common) sequence.
Primer formulations, with differences from the most common sequence in bold and examples of recent usage in parentheses, were as follows: 1 was 27f-CC (AGAGTTTGATCCTGGCTCAG [1, 4]); 2 was 27f-CM (AGAGTTTGATCMTGGCTCAG [11, 13]); 3 was 27f-YM (AGAGTTTGATYMTGGCTCAG [21]); 4 was AGAGTTTGATIITGGCTCAG (19, 31); 5 was AGRGTTTGATCMTGGCTCAG (15); and 6 was 27f-YM+3 (AGAGTTTGATYMTGGCTCAG plus AGAATTTGATCTTGGTTCAG plus AGAGTTTGATCCTGGCTTAG plus AGGGTTCGATTCTGGCTCAG [this paper]).
The fourth-most-observed binding site sequence is found in Chlamydiales. As noted by previous authors (32), this sequence differs at three positions from all of the common primer formulations.
The fifth-most-observed sequence is found primarily in Atopobium spp. and chloroplasts (but, interestingly, not in Cyanobacteria, which include the chloroplast ancestor). Given that this variant requires only a single T-G mispairing to bind the 27f-CC primer (and that component of 27f-CM and 27f-YM) and the mispairing is far from the 3′ end of the primer, it is unlikely to have a substantial effect on efficiency.
The sixth-most-observed sequence is that previously reported for Borrelia spp. (32). Although it requires only a single C-A mispairing with the 27f-CC primer (and the corresponding component of all other common formulations), the mismatch is in the third pair from the 3′ end, sufficiently close enough to raise concerns about efficiency in detecting members of this clinically important genus.
The 27f primer-binding site observed in Campylobacterales (which are clinically relevant) and Sphingomonadales (which are a major component of the Sargasso Sea data) precisely matches a component of the 27f-YM primer but requires one or two (consecutive) mispairings with the more common 27f-CM and 27f-CC primers.
The eighth-most-observed binding site variant in these data is of particular interest for our studies of vaginal microbiology. This sequence is observed throughout the Bifidobacteriales, including the genus Gardnerella (18) (GenBank accession numbers M58729 to M58744). This sequence requires three mispairings to bind the 27f-CC and 27f-CM primer formulations and two mispairings to bind the best-matching sequence in the 27f-YM formulation. Although all of the mispaired positions are 9 or more nucleotides from the 3′ end of the primer and this primer is routinely used with annealing temperatures far below its computed melting temperature, we were concerned that these mismatches still might introduce significant bias in detecting these organisms. This is analyzed in detail below.
The next-most-common binding site sequence has also been previously noted (28) and is found in Thermotogales and Planctomycetales. Interestingly, this sequence requires only a single A-C mispairing 17 nucleotides from the 3′ end of the 27f-CC, 27f-CM, and 27f-YM primer formulations, yet it has been incorporated into a primer used by Kuske et al. (15).
Continuing down our list of observed 27f primer-binding site sequences (data not shown), we could not assign additional sequences to phylogenetic groups or confidently conclude that they represented bona fide variants, as opposed to sequencing errors or other artifacts. Given this, we limited our present analysis to the sequences shown in Table 1. As genome projects and metagenomic studies improve the phylogenetic sampling of full-length sequences, we expect that additional variants will be found to be important.
In the case of the 1492r primer-binding site, the picture is entirely different. Restricting our analysis to a 16-nt-long version, 17,029 of 17,696 available binding sites have the sequence AAGTCGTAACAAGGTA. The most common alternative at this site, AAGTCGTAACAAGGAG, was observed 34 times. The examination of the surrounding sequences indicated that the last T in the canonical sequence is missing rather than that this represents two base changes. The only phylogenetically clustered subset of this variant was found in several Lactobacillus sequences reported in a single paper (23). Other sequences for rRNA genes from the same species include the missing T, suggesting that the variant is sometimes a sequencing error (the site is expected to suffer from “band compression,” making sequencing particularly difficult). The next-most-observed sequences were AAATCGTAACAAGGTA and AAGTCCTAACAAGGTA, each of which was seen 28 times. Except for the recurrence of the latter sequence in a group of the Comamonadaceae (33), there is no obvious phylogenetic clustering of these sequences. With this caveat regarding the latter sequence, we conclude that the current data support the use of TACCTTGTTACGACTT as a universal bacterial 1492r primer. This is generally in keeping with the published 16-nt version of 1492r and the corresponding regions of longer versions.
27f primer design.
When using the primer-binding site information gathered from the sequence databases to design primers for rRNA gene amplification, it is necessary to distinguish the actual sequence variation that we wish to accommodate from those due to errors or artifacts in the data (e.g., the contamination of the data with amplification primers, as discussed above). To decide whether a variant is real or not, there are two useful criteria: the first is to require that a given variant occur a minimum number of times, and the second is to require the phylogenetic congruency of a variant (we expect rare, but real, variants to be phylogenetically clustered). Each of the nine binding site sequence variants discussed above satisfies both of these criteria. Four of the nine 27f primer site variants in Table 1 precisely match components of the 27f-YM primer. The bifidobacterial and chlamydial sequences each have two and three differences from the best-matching component of the 27f-YM primer, suggesting a need to modify the primer design to better accommodate them. The Borrelia sequence is only a single base change from a 27f-YM primer component, but the mismatch is sufficiently close to the 3′ end (the third base) to potentially decrease priming efficiency. The remaining two sequence variants require only a single base mismatch (suggesting a relatively small destabilization) and are farther from the 3′ end of the primer, minimizing their deleterious influence on the priming efficiency of a bound oligonucleotide. For the current study, we have chosen not to make any adjustments for these last two sequences. Therefore, our goal is a primer design that will precisely match seven different sequences.
Given the analysis of the 27f primer-binding sites tabulated above, we decided that the best way to accommodate sequence variants was to make an equimolar mixture of the seven desired sequences. In this case, that meant synthesizing primers specific for Chlamydiales, Borrelia, and Bifidobacteriales separately and mixing one part of each with four parts of the fourfold-degenerate 27f-YM primer. We refer to this mixture as 27f-YM+3. This primer pool, containing only seven sequence variants, is able to accommodate the vast majority of the observed natural variation at the 27f primer-binding site. Since each individual sequence is diluted only sevenfold in the mix and amplifications are carried out with an excess of primer, we predicted comparable levels of amplification across all potential templates.
Amplification efficiency using the 27f-YM+3 primer.
We evaluated the amplification efficiency of our 27f-YM+3 primer mixture and compared it to the commonly used 27f-CM and 27f-YM primers. As a test system, we chose human vaginal microbiota DNA samples from clinically healthy women. Our analyses of these samples (data not shown), as well as other studies of vaginal microbiota (5, 12, 13, 36), had determined that Lactobacillus and Gardnerella are two of the most frequently observed genera. Lactobacillus rRNA genes are predicted to perfectly match and to be amplified efficiently with all three of the 27f primer formulations examined in this study (as well as with 27f-CC). However, whereas Gardnerella rRNA genes should precisely match a component of our proposed 27f-YM+3 primer, it would have three and two mismatches with the 27f-CM and 27f-YM primers, respectively, and might be less efficiently amplified in these last two cases. Therefore, we chose to evaluate the relative merits of primer formulations by comparing the amplification efficiencies of Lactobacillus and Gardnerella in these clinical samples.
We used real-time qPCR with primer pairs specifically targeted to Lactobacillus and Gardnerella to measure the change in the levels of these rRNA gene sequences following amplification with each of the three 27f primer formulations. In considering an experimental design, we were concerned about possible bias in any reverse primer that we might choose. Therefore, we performed an analysis with linear amplification using only a 27f primer, rather than the more common PCR. An additional benefit of the linear amplification was that it allowed us to explore the behavior of the 27f formulations at various temperatures without concern for the length and stability of a reverse primer.
Using qPCR, we measured the relative amounts of target DNA (Lactobacillus or Gardnerella rRNA genes) before amplification, after 5 linear amplification cycles, and after 10 linear amplification cycles. The primer used for the linear amplification was no primer, 27f-CM, 27f-YM, or 27f-YM+3. Our logic was that if linear amplification were perfect, for every 2 strands of target in the input DNA (i.e., each double-stranded gene), 5 cycles of linear amplification would yield 7 strands of target DNA (the original 2 plus 5 copies of the RNA-like strand) and 10 cycles would yield 12 strands of target DNA (the original 2 plus 10 copies). Less-efficient amplification should result in lesser increases in the target DNA.
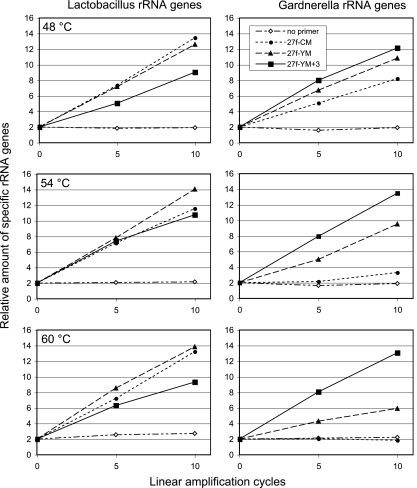
Figure 2 shows the influence of the three 27f primer formulations on the yield of Lactobacillus and Gardnerella sequences at three different annealing stringencies (temperatures). In the case of Lactobacillus (left panels), all primers at all temperatures tested amplified the starting material efficiently, though there was slightly less amplification with our 27f-YM+3 primer, which we attribute to the slightly higher dilution of exactly matching sequences in the primer population. In contrast, the results for the amplification of Gardnerella sequences are striking (right panels). At the highest stringency tested (60°C), the 27f-CM primer completely failed to amplify Gardnerella sequences and the 27f-YM primer was only marginally proficient, while our reformulated primer was able to amplify these sequences with high efficiency. As the annealing stringency decreased, there was an improvement in the ability of the 27f-CM and 27f-YM primers to amplify Gardnerella sequences. It is important to note that even at a low stringency (48°C, a commonly used temperature in microbial ecology rRNA gene-amplifying protocols), there were still discrepancies in the efficiencies with which the different primer formulations were able to amplify the target sequences.
FIG. 2.
Change in the abundances of Lactobacillus and Gardnerella rRNA genes during linear amplification with three different 27f primer formulations. Amplification was carried out with a human vaginal DNA sample at the following three different annealing temperatures: 48°C, 54°C, and 60°C. Each sample was analyzed by qPCR using primers specific for Lactobacillus (left panels) and Gardnerella (right panels). Data are relative fluorescence units of DNA in the 26th cycle of qPCR. The fluorescence units are normalized to “two strands” at zero cycles of linear amplification. Each data point represents the average of results from three independent experiments.
27f-YM+3 primer and clonal representation in rRNA gene libraries.
Having demonstrated that our reformulated 27f-YM+3 primer more efficiently amplified Gardnerella sequences than the usual 27f primers, we turned our attention to the effect of its use in rRNA gene library construction from clinical samples. The question is how does each of the 27f primer formulations alter the composition of the rRNA genes used in library construction relative to the makeup of the original material?
For PCR amplification, each of the three 27f primer formulations in this study was paired with a 16-nt-long version of the 1492r primer. Since this short version of the 1492r primer requires a relatively low annealing temperature for optimal amplification (48°C under our conditions), it was possible that little or no bias would be observed due to mispairings of the 27f primer. DNAs from three different human vaginal microbial samples were amplified with each of the three 27f primer formulations and the 1492r primer. We then used qPCR to evaluate the relative amounts of Lactobacillus and Gardnerella sequences before and after amplification. To eliminate the need for an absolute calibration of the qPCR efficiency and thereby simplify analysis, before performing qPCR, we diluted the amplified samples to approximately the rRNA gene levels in the unamplified samples. Control PCR amplifications without the 27f and 1492r primers generated no detectable product (data not shown). Critical threshold cycles (CT; the number of qPCR cycles needed to generate detectable product) were used as a measure of the amount of taxon-specific rRNA genes. To compare the ratios of sequences, we calculated the difference in critical thresholds for Lactobacillus and Gardnerella (i.e., ΔCT = CT Lactobacillus − CT Gardnerella). No difference in critical thresholds implies that the sequences are present in the sample in approximately equal amounts; a ΔCT of +1 connotes that there are roughly twice as many Gardnerella sequences as there are Lactobacillus sequences (a higher CT value means that more cycles were required for the rRNA gene to become detectable due to a lower starting concentration). Although absolute calibration of the Lactobacillus and Gardnerella qPCR amplification efficiencies is required to determine the actual ratio, our interest in the present study is the extent to which amplification with the 27f and 1492r primers alters the ratio and hence the ΔCT; that is, we are interested in the ΔΔCT due to the PCR amplification step.
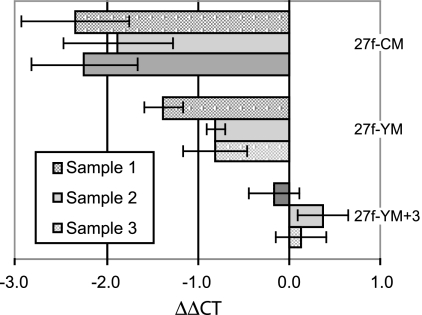
For each of the three DNA samples, Table 2 presents the Lactobacillus and Gardnerella ΔCT values (the average and standard error from three replicates) for the sample DNA and the amplification products using alternative 27f primer formulations. Before amplification (the “Sample DNA” row of Table 2), the different ΔCT values indicated that the clinical samples have different bacterial compositions (they were chosen for this property based on clone sequencing [data not shown]). For samples 1, 2, and 3, the Gardnerella/Lactobacillus rRNA gene sequence ratios are 1.54:1, 3.81:1, and 0.54:1, assuming that they have equal qPCR efficiencies. A ΔCT of +1 corresponds to a 1:2 ratio. However, what matters in this experiment is the change in the ΔCT that occurs with PCR amplification (ΔΔCT). The shift is particularly dramatic with the 27f-CM formulation, less so with the 27f-YM formulation, and least with the 27f-YM+3 formulation. This is most easily seen graphically (Fig. 3). For the 27f-CM primer, the ΔΔCT is ca. −2, reflecting an approximately fourfold underrepresentation of Gardnerella rRNA in the PCR product. For the 27f-YM primer, the ΔΔCT is ca. −1, reflecting an approximately twofold underrepresentation of Gardnerella rRNA in the PCR product. In contrast, for the 27f-YM+3 primer, the ΔΔCT is ca. 0.1, clearly showing a much more faithful representation of the Gardnerella/Lactobacillus rRNA gene ratio in the original samples.
TABLE 2.
Differences between Lactobacillus and Gardnerella critical ΔCTs for three samples of human vaginal microbial DNA
| 27f primer formulation | ΔCT for samplea:
|
||
|---|---|---|---|
| 1 | 2 | 3 | |
| Sample DNA | 0.63 ± 0.32 | 1.92 ± 0.23 | −0.93 ± 0.19 |
| 27f-CM | −1.70 ± 0.67 | 0.07 ± 0.38 | −3.17 ± 0.44 |
| 27f-YM | −0.73 ± 0.12 | 1.13 ± 0.30 | −1.73 ± 0.23 |
| 27f-YM+3 | 0.47 ± 0.20 | 2.30 ± 0.50 | −0.80 ± 0.31 |
Values are means ± the standard errors of the means from three separate experiments and were taken before and after PCR amplification with alternative 27f primer formulations.
FIG. 3.
Change in Lactobacillus and Gardnerella sequence ratios after PCR amplification using each of three 27f primer formulations and the 1492r primer. The values plotted are ΔΔCTs, the change in the Lactobacillus CT minus the Gardnerella CT resulting from 23 cycles of PCR amplification of the DNA samples. Values are the means and the corresponding standard errors from three experiments based on the same data as are shown in Table 2.
DISCUSSION
The introduction of rRNA gene-based methods for community analysis has revolutionized our view of the microbial world. Overcoming the limitations of culture-based protocols for environmental census, they have revealed a largely unanticipated diversity in all communities probed. These studies vastly expanded our knowledge of the extant microbial types, identifying entire new groups, sometimes even at the highest taxonomic levels. This explosion in the discovery of microbial novelty, while reshaping our thinking about the complexity of natural environments, left many questions unanswered.
In order to understand microbial communities, their structure, and their fluctuations, surveys need to be comprehensive, i.e., provide as vast a coverage of the present diversity as possible. Are our current methods up to the task? Also, surveys ideally need to provide quantitative measures of the microbial composition. How well can the relative abundance of different taxa in a community be assessed with our current methods?
Our motivation for the present studies was therefore twofold: to assess both the quantitative scope and comprehensiveness of the most commonly used method of rRNA gene amplification to characterize microbial communities. To address the issue of the quantitative analysis of individual components in microbial communities, we designed and tested a protocol for rRNA gene amplification that more faithfully maintained the relative abundances of the microbial types present in the clinical samples. To address the issue of comprehensiveness, we tested how well the most commonly used rRNA gene amplification primers represent the vastly expanded database of rRNA gene sequences and the subsequent effects on the surveys using them. We reformulated the commonly used primers and tested them for their coverage in the analysis of clinical vaginal microbial samples.
Our analyses of the 27f and 1492r primer-binding sites in SSU rRNA gene databases revealed that the termini of many (perhaps most) rRNA gene sequences in the databases are contaminated with sequences that do not originate from the organism that they are asserted to represent. This suggests a need for greater vigilance in sequence reporting and curation. We have proposed and implemented a criterion for the automated detection of, and thereby removal of, much of the contamination, with a possible loss of some valid sequences. Improvements to our procedure are possible by integrating the analyses of both the 5′ and 3′ ends of sequences (with few exceptions, if there is primer contamination at one end, then it is present at the other end as well). Similarly, all sequences first described in any particular publication tend to have or not have primer contamination.
Our analysis of 27f primer-binding sites revealed several sequence variations representing cohesive phylogenetic groups that are not accommodated by the commonly used 27f primer formulations, even though these sequences have been known for many years (e.g., see reference 32). This leads to the question of whether the mismatched nucleotides cause a systematic underrepresentation of the corresponding phylogenetic groups in rRNA gene libraries. Implicit in much of the microbial ecology literature is the assumption that a low primer annealing temperature (at least for the first several PCR cycles) provides a good and expedient method for accommodating primer-binding site sequence diversity, while avoiding the pitfalls of highly degenerate mixtures. We combined the 20-nucleotide-long 27f primer with a shorter (16 nucleotide) 1492r primer, necessitating a PCR annealing temperature well below that required for the longer 27f. Even under these low-stringency conditions, we found that the 27f-CM and 27f-YM primers discriminated against Gardnerella rRNA genes, with 27f-CM being less discriminatory than 27f-YM. Thus, we found that even low-stringency amplification did not fully tolerate the mismatches.
These results emphasize the importance of primers that exactly match their template. There are multiple ways to accommodate sequence variation when designing amplification primers. The most common way is to synthesize primers with two or more nucleotides at selected positions (degenerate primers). While this strategy allows the stringency of amplification to remain high, hence reducing spurious pairing, the sequences that are exact matches to each variant are diluted in the pool, leading to lower overall efficiency of binding. As the number of variable sites increases, the complexity of the pool increases exponentially, and some of the sequences in the pool may be superfluous, worsening the problems associated with the dilution of relevant sequences. In the present case, introducing degenerate positions to include the seven desired sequences would require the sequence AGRRTTYGATYHTGGYTYAG (where H is A or C or T), which is a mixture of 192 distinct sequences.
An alternative approach is to incorporate the noncanonical nucleotide inosine at variable positions (19, 31). Inosine forms stable pairs with all four nucleotides, although the strength of the interaction varies (pairings with pyrimidines are more stable) (20). Unlike the use of degenerate primers, an increase in the number of positions replaced with inosine can lead to promiscuous pairings and the amplification of spurious products. In the present case, introducing inosine at seven sites would create a primer that matches 16,384 sequences. We instead chose to maintain high specificity and low dilution by formulating a 27f primer mixture consisting only of the seven unique sequence variants, 27f-YM+3.
Although we have added three sequences to the 27f-YM primer, we have not been comprehensive in testing all components of our formulation or their effects on all phylogenetic groups for which the primer is intended to improve results. Instead, we focused on the behavior of two phylogenetic groups relevant to our current work on vaginal microbiota, though our results should be applicable to other groups. The literature suggests that most investigators either are unaware of the issue or expect that the effects will be insignificant. Our survey of the literature suggests that 27f-CC and 27f-CM are the two most common 27f primer formulations, with 27f-YM being the third most common. Even in studies of vaginal microbiota, the 27f-CC (6, 9, 25) and 27f-CM (13) primers are routinely used, each with three mismatches to the clinically relevant Gardnerella rRNA genes. One vaginal study used an even shorter version of 27f-CC (positions 10 to 27 in E. coli coordinates), resulting in three mismatches out of 18 base pairs with the Gardnerella rRNA genes, a fact acknowledged by the authors (29). Our results suggest that a minor modification of the primer design could yield substantially more representative results.
Even if we are willing to accept the quantitative bias introduced by mismatches when we rely on a lower stringency to tolerate them, there is a second reason to explicitly accommodate known sequence diversity in the primer. Relying on a low stringency leaves little or no buffer for additional, undiscovered diversity. Incorporating presently known diversity in the primer design will provide a more solid basis for sampling the unknown.
Overall experimental design.
Although the focus of our work is the comparison of primer formulations, we have attempted to critically analyze other aspects of the protocols. Our survey of the literature revealed a wide range of template DNA used in the PCR (in those cases where it is specified). Similarly, there is a wide range in the number of PCR cycles used. Only rarely do we find discussion of how published protocols were designed. When we do, the most important theme seems to be avoiding PCR saturation (e.g., references 14, 24, and 26), the point at which abundant rRNA gene types stop amplifying because they reanneal before binding the primer and DNA polymerase. This still leaves a trade off between the amount of template and the number of cycles. Here, we chose to minimize the consumption of the (often limited) template by computing the number of distinct rRNA genes expected in a given amount of DNA and using the smallest amount that will give few if any duplicates in the subsequent sequencing of random clones. Given that PCR amplification only increases the number of copies of the sequences originally present, the sequencing of random clones of the amplified DNA amounts to drawing random genes from this original gene pool, with replacement. If the pool has N rRNA genes and n clones are sequenced, then the probability that a copy of a specific one of the N genes has not been sequenced is e−μ, where μ is −n/N. The expected number of genes without a copy sequenced is described in the equation Ne−μ. The expected number of distinct genes that have been sequenced is described in the equation N − Ne−μ = N(1 − e−μ). The number of instances of resequencing copies of the same gene is the difference between the clones sequenced and the genes sequenced, which is n − N(1 − e−μ). When n is ≪N (which is true when there are many more genes in the sample than there are clones sequenced), this is approximately Nμ2/2. If n is equal to N, the expected number of instances of resequencing a gene is approximately one-half. If we take this as our goal, then it would require ∼105 rRNA genes to sequence 300 clones with minimal duplicates. If we estimate an average of 1 rRNA gene per 106 bp of bacterial DNA, this gives 1 rRNA gene per 1 fg of DNA, or 105 rRNA genes per 0.1 ng of bacterial DNA. This is 1,000-fold less DNA than that used in some studies. An important caveat is that all of the input DNA must be bacterial. If 99% of the DNA in a sample were from a eukaryal host, then 100-fold-more input DNA would be needed to produce the same PCR amplification product. To maintain less than one duplicate when sequencing more clones, say 1,000 instead of 300, it would require about 10 times the input DNA. However, even if the input DNA were maintained at 0.1 ng, the predicted number of duplicates in 1,000 clone sequences would be only five, so little sequencing effort would be wasted.
Given a specified amount of input DNA, our next goal was to determine the number of PCR cycles that would not saturate the amplification of the most-abundant sequences. Depending on the template, the primers, and the DNA polymerase, with 0.1 ng of template in a 25-μl reaction mixture, we observed the first evidence of saturation at 24 to 26 cycles (data not shown). Again, this value is low relative to the number of cycles of many (but not all) published protocols. It is impossible to say why so much DNA and so many cycles are common in the literature, but unless the reaction conditions are tremendously inefficient, we suspect that many investigators have driven their amplifications well into saturation, introducing additional bias into the resulting rRNA gene composition.
Carrying an earlier argument one step further, it might be argued that there are so many problems with using clone sequences to characterize a microbial community that there is no point in trying at all. Our first response is that it is the clone-based analyses that reveal the taxa that require more-accurate analysis. Microarrays, qPCR, and many other methods require a predefined list of what is being sought, and this generally comes from clone-based analyses. Missing taxa at this early step can lead to their exclusion from more-quantitative studies. Our second response to “why minimize bias in clone-based studies?” is that some analyses require a degree of resolution for which a complete (or nearly complete) sequence is necessary. For example, microbiologists have yet to understand the origins and implications of the pervasive heterogeneity of rRNA gene types in natural communities and how this diversity reflects the ecology and evolution of the communities. Microarrays and qPCR are poor methods for observing and measuring this diversity because one or two short fragments of the gene define their specificity. Although denaturing gradient gel electrophoresis and terminal restriction fragment length polymorphism fingerprinting can sample more of the rRNA genes, they still have much less resolving power than sequencing.
During the 20 years since the introduction of rRNA gene-based community analyses, it has been relatively easy to make new discoveries, explore new territories, sample new environments, and recognize novel microbial types. Future progress in microbial ecology hinges on our ability to perform comprehensive and quantitative studies. If in no other context, the increasing use of these methods in medical studies deserves this attention. Toward this end, it is vital that we regularly reevaluate the analytical methods that we use to study these systems.
Acknowledgments
This work was supported in part by the Research Board of the University of Illinois at Urbana-Champaign and by the Carle Foundation Hospital.
We thank Michelle Hughes and Barbara Hall for their clinical and clerical assistance with the sample collection. We thank Mengfei Ho, Angel Rivera, Noriko Nakamura, Abigail Salyers, Rex Gaskins, Lois Hoyer, and James Slauch for their helpful discussions. We also thank the reviewers for their suggestions.
Footnotes
Published ahead of print on 22 February 2008.
REFERENCES
- 1.Abulencia, C. B., D. L. Wyborski, J. A. Garcia, M. Podar, W. Chen, S. H. Chang, H. W. Chang, D. Watson, E. L. Brodie, T. C. Hazen, and M. Keller. 2006. Environmental whole-genome amplification to access microbial populations in contaminated sediments. Appl. Environ. Microbiol. 72:3291-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Bik, E. M., P. B. Eckburg, S. R. Gill, K. E. Nelson, E. A. Purdom, F. Francois, G. Perez-Perez, M. J. Blaser, and D. A. Relman. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA 103:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton, J. P., and G. Reid. 2002. Evaluation of the bacterial vaginal flora of 20 postmenopausal women by direct (Nugent score) and molecular (polymerase chain reaction and denaturing gradient gel electrophoresis) techniques. J. Infect. Dis. 186:1770-1780. [DOI] [PubMed] [Google Scholar]
- 6.Burton, J. P., E. Devillard, P. A. Cadieux, J.-A. Hammond, and G. Reid. 2004. Detection of Atopobium vaginae in postmenopausal women by cultivation-independent methods warrants further investigation. J. Clin. Microbiol. 42:1829-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coolen, M. J. L., E. Post, C. C. Davis, and L. J. Forney. 2005. Characterization of microbial communities found in the human vagina by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Appl. Environ. Microbiol. 71:8729-8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLong, E. F. 2004. Microbial population genomics and ecology: the road ahead. Environ. Microbiol. 6:875-878. [DOI] [PubMed] [Google Scholar]
- 11.DeLong, E. F., C. M. Preston, T. Mincer, V. Rich, S. J. Hallam, N. U. Frigaard, A. Martinez, M. B. Sullivan, R. Edwards, B. R. Brito, S. W. Chisholm, and D. M. Karl. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496-503. [DOI] [PubMed] [Google Scholar]
- 12.Fredricks, D. N., T. L. Fiedler, and J. M. Marrazzo. 2005. Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med. 353:1899-1911. [DOI] [PubMed] [Google Scholar]
- 13.Hyman, R. W., M. Fukushima, L. Diamond, J. Kumm, L. C. Giudice, and R. W. Davis. 2005. Microbes on the human vaginal epithelium. Proc. Natl. Acad. Sci. USA 102:7952-7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kainz, P. 2000. The PCR plateau phase—towards an understanding of its limitations. Biochim. Biophys. Acta 1494:23-27. [DOI] [PubMed] [Google Scholar]
- 15.Kuske, C. R., S. M. Barns, C. C. Grow, L. Merrill, and J. Dunbar. 2006. Environmental survey for four pathogenic bacteria and closely related species using phylogenetic and functional genes. J. Forensic Sci. 51:548-558. [DOI] [PubMed] [Google Scholar]
- 16.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsen, N., G. J. Olsen, B. L. Maidak, M. J. McCaughey, R. Overbeek, T. J. Macke, T. L. Marsh, and C. R. Woese. 1993. The Ribosomal Database Project. Nucleic Acids Res. 21:3021-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leblond-Bourget, N., H. Philippe, I. Mangin, and B. Decaris. 1996. 16S rRNA and 16S to 23S internal transcribed spacer sequence analyses reveal inter- and intraspecific Bifidobacterium phylogeny. Int. J. Syst. Bacteriol. 46:102-111. [DOI] [PubMed] [Google Scholar]
- 19.Lindh, J. M., O. Terenius, and I. Faye. 2005. 16S rRNA gene-based identification of midgut bacteria from field-caught Anopheles gambiae sensu lato and A. funestus mosquitoes reveals new species related to known insect symbionts. Appl. Environ. Microbiol. 71:7217-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin, F. H., and M. M. Castro. 1985. Base pairing involving deoxyinosine: implications for probe design. Nucleic Acids Res. 13:8927-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nercessian, O., Y. Fouquet, C. Pierre, D. Prieur, and C. Jeanthon. 2005. Diversity of Bacteria and Archaea associated with a carbonate-rich metalliferous sediment sample from the Rainbow vent field on the Mid-Atlantic Ridge. Environ. Microbiol. 7:698-714. [DOI] [PubMed] [Google Scholar]
- 22.Overbeek, R., T. Begley, R. M. Butler, J. V. Choudhuri, H. Y. Chuang, M. Cohoon, V. de Crecy-Lagard, N. Diaz, T. Disz, R. Edwards, M. Fonstein, E. D. Frank, S. Gerdes, E. M. Glass, A. Goesmann, A. Hanson, D. Iwata-Reuyl, R. Jensen, N. Jamshidi, L. Krause, M. Kubal, N. Larsen, B. Linke, A. C. McHardy, F. Meyer, H. Neuweger, G. Olsen, R. Olson, A. Osterman, V. Portnoy, G. D. Pusch, D. A. Rodionov, C. Ruckert, J. Steiner, R. Stevens, I. Thiele, O. Vassieva, Y. Ye, O. Zagnitko, and V. Vonstein. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavlova, S. I., A. O. Kilic, S. S. Kilic, J. S. So, M. E. Nader-Macias, J. A. Simoes, and L. Tao. 2002. Genetic diversity of vaginal lactobacilli from women in different countries based on 16S rRNA gene sequences. J. Appl. Microbiol. 92:451-459. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thies, F. L., W. König, and B. König. 2007. Rapid characterization of the normal and disturbed vaginal microbiota by application of 16S rRNA gene terminal RFLP fingerprinting. J. Med. Microbiol. 56:755-761. [DOI] [PubMed] [Google Scholar]
- 26.Thompson, J. R., L. A. Marcelino, and M. F. Polz. 2002. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res. 30:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 28.Vergin, K. L., E. Urbach, J. L. Stein, E. F. DeLong, B. D. Lanoil, and S. J. Giovannoni. 1998. Screening of a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl. Environ. Microbiol. 64:3075-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verhelst, R., H. Verstraelen, G. Claeys, G. Verschraegen, J. Delanghe, L. Van Simaey, C. De Ganck, M. Temmerman, and M. Vaneechoutte. 2004. Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae, Gardnerella vaginalis and bacterial vaginosis. BMC Microbiol. 21:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe, K., Y. Kodama, and S. Harayama. 2001. Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J. Microbiol. Methods 44:253-262. [DOI] [PubMed] [Google Scholar]
- 32.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen, A., M. Fegan, C. Hayward, S. Chakraborty, and L. I. Sly. 1999. Phylogenetic relationships among members of the Comamonadaceae, and description of Delftia acidovorans (den Dooren de Jong 1926 and Tamaoka et al. 1987) gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49:567-576. [DOI] [PubMed] [Google Scholar]
- 34.Wheeler, D. L., T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, V. Chetvernin, D. M. Church, M. DiCuccio, R. Edgar, S. Federhen, L. Y. Geer, Y. Kapustin, O. Khovayko, D. Landsman, D. J. Lipman, T. L. Madden, D. R. Maglott, J. Ostell, V. Miller, K. D. Pruitt, G. D. Schuler, E. Sequeira, S. T. Sherry, K. Sirotkin, A. Souvorov, G. Starchenko, R. L. Tatusov, T. A. Tatusova, L. Wagner, and E. Yaschenko. 2007. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 35(Database issue):D5-D12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson, K. H., R. B. Blitchington, and R. C. Greene. 1990. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J. Clin. Microbiol. 28:1942-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou, X., S. J. Bent, M. G. Schneider, C. C. Davis, M. R. Islam, and L. J. Forney. 2004. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 150:2565-2573. [DOI] [PubMed] [Google Scholar]