Abstract
The presence and activities of urease genes were investigated in 49 clinical, food, and environmental Bacillus cereus isolates. Ten strains were shown to have urease genes, with eight of these strains showing growth on urea as the sole nitrogen source. Two of the urease-positive strains, including the sequenced strain ATCC 10987, could not use urea for growth, despite their capacities to produce active urease. These observations can be explained by the inability of the two strains to use ammonium as a nitrogen source. The impact of urea hydrolysis on acid stress resistance was subsequently assessed among the ureolytic B. cereus strains. However, none of the strains displayed increased fitness under acidic conditions or showed enhanced acid shock survival in the presence of urea. Expression analysis of urease genes in B. cereus ATCC 10987 revealed a low level of expression of these genes and a lack of pH-, nitrogen-, urea-, oxygen-, and growth phase-dependent modulation of mRNA transcription. This is in agreement with the low urease activity observed in strain ATCC 10987 and the other nine strains tested. Although a role for B. cereus ureolytic activity in acid survival cannot be excluded, its main role appears to be in nitrogen metabolism, where ammonium may be provided to the cells in nitrogen-limited, urea-containing environments.
Bacillus cereus is a gram-positive, spore-forming rod known to be present in various environments. Vegetative cells and/or spores can be found in soil (46), rhizospheres (5), air (17), insect guts (26), and foods (23, 36) and in the human gastrointestinal tract (18). B. cereus can cause a diarrheal and an emetic type of food-borne disease. The emetic type of disease occurs when B. cereus cells grow and produce the heat-stable emetic toxin cereulide in food. The emetic symptoms arise after ingestion of intoxicated food (1, 43). The diarrheal type of disease is caused upon ingestion of B. cereus vegetative cells and/or spores and their survival of passage through the stomach, followed by germination and/or growth in the human small intestine, where vegetative cells produce several enterotoxins (20, 43, 47).
B. cereus can encounter suboptimal conditions for growth in various environments. The presence and activity of alternative ways to metabolize nitrogen sources, such as urease-mediated degradation of urea, can give bacteria a growth advantage under nitrogen-limited conditions. In addition, the bacteria may be exposed to stress conditions and display adaptive stress responses to cope with suboptimal conditions, such as low pH. For instance, gram-positive bacteria may activate decarboxylases, deaminases, proton pumps, and urease to cope with acid stress (11).
Urease (EC 3.5.1.5) catalyzes the hydrolysis of urea, generating two molecules of ammonia and one molecule of carbon dioxide, and it was the first enzyme to be crystallized (41). Urease is present in various organisms, including plants, fungi, and bacteria. Urease activity has been associated with various diseases, such as infection-induced urinary stones and peptic ulceration (29). The role of urease in the pathogenesis of Helicobacter pylori, which causes peptic ulceration (25), is evident. Ammonia molecules, produced upon hydrolysis of urea, can bind protons and consequently elevate the pH. This mechanism has been shown to be essential for the colonization of the human stomach by H. pylori (42). Another role of urea hydrolysis is to provide ammonium for nitrogen metabolism, as has been reported for a range of bacterial species (22). A dual role of urease, in acid resistance and in nitrogen metabolism, for example, has been reported for Streptococcus salivarius (8), H. pylori (48), and Yersinia enterocolitica (50).
Rasko et al. (34) identified a urease utilization cluster composed of nine genes (BCE3657 to BCE3666) in the genome of B. cereus strain ATCC 10987 that is not present in other sequenced strains belonging to the B. cereus group. This cluster harbors three genes, ureA, ureB, and ureC, encoding the structural enzyme that, respectively, show similarities of 62%, 44%, and 65% to the structural urease genes of Bacillus subtilis (12). Besides the structural genes, the cluster harbors genes encoding accessory proteins (ureE, ureF, ureG, and ureD), which are required to incorporate nickel ions into the enzyme and to activate the enzyme (38). Furthermore, the urease cluster of B. cereus ATCC 10987 contains two additional genes for a putative urea (acetamide) transporter (ureI) and a nickel transporter (nikT).
Urea is present in various environments in which B. cereus can be found, including soil, food, and the human host, where urea is present in all body fluids and is finally excreted in the urine (0.4 M to 0.5 M urea) as a detoxification product (29). B. cereus encounters urea upon its interaction with the human host: human saliva contains 2.3 mM to 4.1 mM (13), the human stomach contains approximately 4.8 mM (32), and the urea concentration in human blood varies from 1.7 mM to 8.3 mM (27). Urea can also be present in various foods. Foods of animal origin generally contain urea; for example, milk contains 4.4 mM to 6.4 mM urea (7). Although urease may play an important role in the life cycle of B. cereus, information is lacking about its role in nitrogen metabolism and acid survival in this human pathogen.
In this study, we investigated the prevalence of urease genes and urease activity among 49 environmental, food, and clinical B. cereus isolates. Furthermore, urease gene expression and the roles of ureolytic activity in nitrogen metabolism and in acid resistance were assessed.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. cereus strains from various sources, including clinical, environmental, and food isolates, together with the type strain (ATCC 14579) and other sequenced strains, such as ATCC 10987 and PAL25 (also known as AH187 [35] and F4810/72 [21]), were used in this study (see Table 2). A number of strains were kindly provided by P. E. Granum (Norwegian School of Veterinary Science, Oslo, Norway), NIZO Food Research (Ede, The Netherlands), Institut National de la Recherche Agronomique (Avignon, France), and the National Institute for Public Health and the Environment (Bilthoven, The Netherlands). Stock cultures grown in brain heart infusion (BHI) (Becton Dickinson, France) broth were stored at −80°C in 50% glycerol. To prepare aerobic working cultures, 10 ml BHI in a 100-ml Erlenmeyer flask was inoculated with a droplet from the glycerol stock and incubated overnight at 30°C with shaking at 200 rpm. Oxygen-limited working cultures were prepared in 50 ml BHI in a completely filled 50-ml tube (Greiner Bio-one, Germany) and incubated overnight at 30°C.
TABLE 2.
B. cereus strains tested for the presence of ureABC genes; growth in minimal medium with glutamate, ammonium, and urea as sole nitrogen sources; and urease activity
| B. cereus strain | Origin (reference) | ureABC PCRa | Growth in minimal medium witha:
|
Urease activitya | ||
|---|---|---|---|---|---|---|
| Glutamate | Ammonium | Urea | ||||
| B434 | Pasteurized milk | − | + | + | − | − |
| B436 | Raw milk | − | + | + | − | − |
| B437 | Pasteurized milk | − | + | + | − | − |
| B439 | Pasteurized milk | − | + | + | − | − |
| B443 | Pasteurized milk | − | + | + | − | − |
| ATCC 14579 | Air (17) | − | + | + | − | − |
| PAL2 | Human fecesd | − | + | − | − | − |
| PAL3 | Human fecesd | + | + | + | + | + |
| PAL5b | Patient fecesd | + | + | + | + | + |
| PAL7 | Human fecesd | − | + | + | − | − |
| PAL17 | Human fecesd | − | − | + | − | − |
| PAL18b | Patient fecesd | − | + | + | − | − |
| PAL20 | Chilled food (9) | + | + | + | + | + |
| PAL22 | Chilled food (9) | − | + | + | − | − |
| PAL25c | Human vomit (2) | − | + | + | − | − |
| PAL26b | Meat loaf (28) | − | + | + | − | − |
| PAL27b | Pea soup (39) | + | + | + | + | + |
| PAL28b | Food poisoning (19) | − | + | + | − | − |
| 17 | Cream (40) | − | − | + | − | − |
| 55 | Cream (40) | + | + | + | + | + |
| 59 | Cream (40) | + | + | + | + | + |
| 61 | Cream (40) | + | + | + | + | + |
| 72 | Semiskim Milk (40) | − | + | + | − | − |
| 132 | Milk (40) | − | + | + | − | − |
| 43-92 | Milk (40) | + | + | − | − | + |
| 401-92 | Scrambled eggs (40) | − | + | + | − | − |
| 67-498 | Scrambled eggs (40) | − | + | − | − | − |
| 1230-88 | Stew (40) | − | + | + | − | − |
| F450183 | Clinical (PHLS) (40) | − | + | + | − | − |
| DSM 11821T | Pasteurized Milk (24) | − | − | + | − | − |
| KW85 | Unknown | − | + | + | − | − |
| F5581 | Unknown | − | + | + | − | − |
| TZ415 | Chilled food (9) | − | + | + | − | − |
| TZ426 | Unknown | − | + | − | − | − |
| TZ427 | Unknown | − | + | + | − | − |
| TZ428 | Unknown | − | + | − | − | − |
| Z4234 | Unknown | − | + | + | − | − |
| P21S | Unknown | + | + | + | + | − |
| Z421 | Unknown | − | + | + | − | − |
| F3752A/86c | Emetic outbreak (15) | − | + | + | − | − |
| B6/Ac | Unknown | − | + | + | − | − |
| F2797/87 | Unknown | − | + | − | − | − |
| F3351/87c | Emetic outbreak (15) | − | + | + | − | − |
| F3748/75 | Human feces (16) | − | + | + | − | − |
| F4635A/90 | Unknown | − | + | − | − | − |
| F4628/90 | Paella | − | + | − | − | − |
| F4626/90 | Milk (4) | − | + | + | − | − |
| F4623/90 | Unknown | − | + | − | − | − |
| ATCC 10987 | Spoiled cheese (34) | + | + | − | − | + |
+, positive result, PCR product with correct size, growth observed (final OD > 0.05), urease activity observed with cells grown in BHI; −, negative result, no PCR product, no growth observed (final OD < 0.05), no urease activity observed with cells grown in BHI. The total numbers of positive strains were as follows: ureABC PCR, 10; glutamate, 46; ammonium, 39; urea, 8; and urease activity, 9.
Diarrhea associated strain.
Emetic strain.
Strain obtained from an RIVM project on the incidence and pathogenesis of gastroenteritis.
PCR detection of urease genes.
The primers used in the PCR (listed in Table 1) were designed from the ureA (BCE_3662) and ureC (BCE_3664) genes from B. cereus ATCC 10987 (accession number AE017194) (34). Taq DNA polymerase (MBI Fermentas, Germany) was used to amplify the target genes. The PCR was performed in an MJ Research PTC-200 thermal cycler preset to 30 cycles of 15 seconds at 94°C, 15 seconds at 45°C, and 1 min at 72°C. Template DNA was obtained from 1 ml aerobic working culture, which was boiled two times for 1 min each time and put on ice after each boiling step.
TABLE 1.
PCR and RT−RT−PCR primers used in this study
| Primer name | Gene(s) | Gene ID(s) | Sequence |
|---|---|---|---|
| PCR | |||
| UreaseForw | ureABC | BCE3662−BCE3664 | ATCAGATATTCAAGTCGAGG |
| UreaseRev | ureABC | BCE3662−BCE3664 | CCAGGTGTTATTGTAGTTGC |
| RT−RT−PCR | |||
| QPCR_ureA_Forw | ureA | BCE3664 | TTCCGCCACATTTTTACCATC |
| QPCR_ureA_Rev | ureA | BCE3664 | ACGAAAGGAGAGGGGGCTTA |
| QPCR_ureG_Forw | ureG | BCE3659 | GCCCCTACATAAGGTGCCAAA |
| QPCR_ureG_Rev | ureG | BCE3659 | GCGCAGGGAGAAAAGATTCC |
| QPCR_ureI_Forw | ureI | BCE3657 | GGAATGCCCAAAAGAACCAA |
| QPCR_ureI_Rev | ureI | BCE3657 | CATTGCAACGGTTATGGGAAT |
| QPCR_nikT_Forw | nikT | BCE3656 | CCATGTTTTGCCGTTTTTCC |
| QPCR_nikT_Rev | nikT | BCE3656 | ACTCACAGGGGGTGGGATTT |
| QPCR_rpoA_Forw | rpoA | BCE0137 | GCCCAGGTCACGCTGACTAT |
| QPCR_rpoA_Rev | rpoA | BCE0137 | TCACGTGTTTGAGGCATTGG |
| QPCR_tufA_Forw | tufA | BCE0108 | ACCGCTTGAGCGTGTGGATATG |
| QPCR_tufA_Rev | tufA | BCE0108 | TAGCAGTAACAGCGGCACCA |
Assessment of nutrient utilization.
The utilization of glutamate, ammonium, and urea as sole nitrogen sources was tested in minimal medium. This medium was derived from GGGS medium (6) and contained 3.0 mM K2HPO4, 3.5 mM KH2PO4, 0.8 mM MgSO4, 0.04 mM MnCl2, 0.2 mM NaCl, 0.2 mM CaCl2, 0.05 mM ZnCl2, 0.04 mM FeCl3, 20 mM maltose, and 2 mM glutamate, ammonium, or urea (Merck, Germany, and Sigma, St. Louis, MO). One milliliter of aerobic working culture was spun down and washed with minimal medium without nitrogen sources. Subsequently, 200 μl of minimal medium supplemented with the desired nitrogen source was inoculated (1:100; initial optical density at 600 nm [OD600], 0.005 ± 0.003) with the washed culture in a microtiter plate (Greiner Bio-one, Germany). The plate was incubated for up to 6 days at 30°C. The OD600 was measured each day (uQuant; Bio-Tek Instruments). The result was scored positive if the final OD was 0.05 or higher.
Urease activity assay.
The urease activities of the strains that scored positive in the PCR test (Table 2) were tested as follows. One milliliter of oxygen-limited working culture in BHI was spun down and resuspended in 200 μl physiological salt solution (0.85% [wt/vol] NaCl in water), and a urease activity diagnostic tablet (Rosco Diagnostica, Denmark) containing urea and a pH indicator was added to the suspension. The suspension was incubated for 24 h at 30°C. The mixture changed color, from yellow (low pH) to purple (high pH), upon the increase of pH caused by the urease activity. The urease activities of B. cereus strains ATCC 10987 and ATCC 14579 (negative control) were also monitored in MES [2-(N-morpholino)ethanesulfonic acid] (Sigma, Germany) and MOPS [3-(N-morpholino)propanesulfonic acid] (Sigma, Germany) buffers. Twenty milliliters of oxygen-limited working culture was spun down and resuspended in 4 ml 10 mM MES or 10 mM MOPS buffer with 10 mM urea at pH 5.2 and pH 6.5, respectively. The pH was measured over time with a PHM 240 pH/ION meter (Radiometer, Denmark). To assess the amount of ammonia formed in the urease assay, MES, pH 5.0, including ATCC 10987 cells was titrated to pH 9.0 with 100 mM NH3 solution. The protein concentration of the aerobic working culture was determined by using the bicinchoninic acid assay to determine the urease enzyme activity of ATCC 10987 cells.
Acid growth assay and urea concentration measurements.
To assess the growth performance of the B. cereus strains under acid conditions, 200 μl BHI, BHI supplemented with 10 mM urea, and BHI supplemented with 10 mM urea and 10 μM the urease inhibitor flurofamide (Tocris Biosciences, United Kingdom) at pH 7 and at pH 5 were inoculated with 2 μl aerobic B. cereus working cultures. The growth performance was assessed in microtiter plates (Greiner Bio-one, Germany) and with incubation for 10 h at 30°C. Every hour, the absorbance (600 nm) was measured (uQuant; Bio-Tek Instruments). The growth of strains ATCC 10987 and ATCC 14579 was also tested in nutrient broth (NB) (Oxoid, England) at pH 7 and pH 5 with and without 10 mM added urea. The media used were acidified with HCl (37%; Merck, Germany) to pH 5.0. The pH was monitored with a PHM 240 pH/ION meter (Radiometer, Denmark). To quantify the endogenous urea concentration in BHI and NB, the Quantichrom urea assay kit (Bioassay Systems) was used according to the provided protocol.
Acid shock survival assay.
The impact of urea on the acid shock survival capacities of the B. cereus strains was assessed with stationary-phase cells in 20 ml BHI, in 20 ml BHI with 10 mM added urea, and in 20 ml BHI with 10 mM added urea and 10 μM flurofamide. The cultures were inoculated with 100 μl aerobic working cultures and were subsequently incubated at 30°C with aeration (200 rpm). After overnight incubation, a preset amount of HCl was added to reach the desired acidic pH values. At designated time points (0, 5, 10, 20, and 30 min), dilutions (10−1, 10−3, and 10−5) were made in peptone physiological salt solution (1 g/liter neutralized bacteriological peptone [Oxoid, England] and 8.5 g/liter NaCl in water). The survival of B. cereus was determined by droplet plating (31), with 5 μl of the cultures and the corresponding dilutions put on BHI agar plates (15 g/liter agar bacteriological; Oxoid, England) and incubated overnight at 30°C.
RNA isolation and RT-RT-PCR.
B. cereus strain ATCC 10987, with the highest ureolytic activity in the urease activity assay, was used in the real-time reverse-transcriptase (RT-RT) PCR analysis. To obtain RNA samples, 50 ml of oxygen-limited working culture was spun down and resuspended in 10 ml 10 mM MES or 10 mM MOPS buffer with 10 mM urea at pH 5.2 and pH 6.5, respectively. RNA isolation was performed by transferring 10 ml of the cultures at 0, 30, 90, and 180 min into a 50-ml Falcon tube, and the cultures were spun down at 13,000 × g for 30 seconds. After the supernatant was decanted, the cell pellets were snap-frozen in liquid nitrogen. Within 20 min after the cell pellets were frozen, TRI-reagent (Ambion, United Kingdom) was added to the pellets and RNA was extracted as described previously (44). The expression ratios of four genes from the urease cluster (ureA, ureG, ureI, and nikT) were determined using the oligonucleotides listed in Table 1. cDNA synthesis and RT-RT-PCR were performed as described previously, with tufA and rpoA as reference genes (45). All RT-RT-PCRs were performed in duplicate. Expression ratios between the time points were determined with the REST tool (33).
RESULTS
Occurrence of urease genes and urease activity.
The occurrence of urease genes in B. cereus isolates was tested by PCR, using primers designed based on the ureABC genes of B. cereus ATCC 10987 (UreaseForw and UreaseRev, listed in Table 1). If the ureABC genes are present, a 958-bp DNA fragment will be amplified in the PCR. This fragment was indeed found in 10 out of 49 PCRs (∼20%), and the corresponding strains were thus considered to harbor the ureABC cluster in their genomes (Table 2). The 10 ureABC PCR-positive strains and the ureABC-negative strains were subsequently tested for urease activity. In this test, the hydrolysis of urea and the concomitant production of ammonia resulted in an increase of the pH, which turned the pH indicator from yellow (low pH) to purple (high pH). This color change was observed in 9 of the 49 tested strains, with the ureABC-negative strains and ureABC-positive strain P21S scoring negative after 24 h of incubation (Table 2). ATCC 10987 showed the highest urease activity, scoring positive within 6 hours of incubation. Notably, in all cases, the pH increase was inhibited by the addition of the urease inhibitor flurofamide, which indicated that alkalinization was indeed dependent on urease activity.
Growth on urea as a nitrogen source.
The utilization of urea, as well as that of glutamate and ammonium, as sole nitrogen sources was tested in minimal medium for the 49 B. cereus isolates (Table 2). All 49 strains were able to grow on glutamate and/or ammonium chloride as a nitrogen source with maltose as the sole carbon source. As expected, none of the ureABC-negative strains showed growth on urea as a nitrogen source. Of the 10 urease-positive isolates, 8 showed growth on urea and ammonium as sole nitrogen sources, whereas 2 isolates did not, including the sequenced B. cereus strain ATCC 10987. One urease PCR-positive strain (P21S) that did not display urease activity (see above) was able to utilize urea for growth. This was likely due to a low urease activity that was sufficient for growth but insufficient to elevate the pH in the urease activity assay. In summary, out of the 10 ureABC-positive strains identified in the collection of 49 environmental, food, and clinical B. cereus isolates, 8 were shown to utilize urea for growth.
Urease activity under acidic conditions.
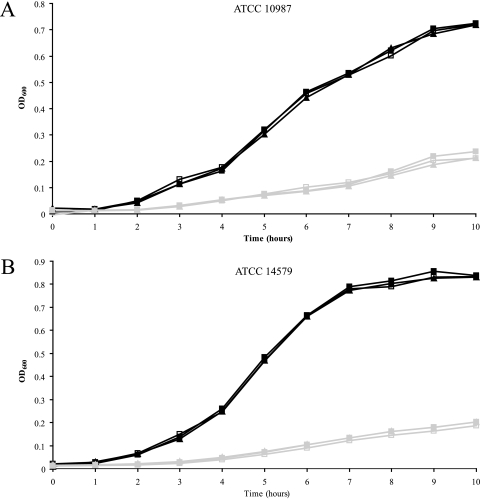
To elucidate a possible role for urease in the acid resistance of B. cereus, the ureABC-positive strains were subjected to two types of acid stress, i.e., growth at low pH and acid shock. The 10 urease-positive strains and the urease-negative type strain (ATCC 14579) were grown in BHI at pH 7 and pH 5 without and with extra urea (10 mM urea added) and with additional urea and flurofamide. Strains ATCC 14579 and ATCC 10987 were additionally tested in NB and NB with 10 mM urea added. Assessment of urea concentrations revealed BHI and NB to contain 1.1 mM and 0.5 mM urea, respectively. Neither the urease-positive strain ATCC 10987 nor the urease-negative strain ATCC 14579 showed differences in growth performance when the cells were cultured in BHI without additional urea, with extra urea, and with extra urea and flurofamide (Fig. 1). The other ureolytic strains showed similar growth profiles, and no urea-induced differences were detected (data not shown). Moreover, all growth performances appeared to be the same in the presence of flurofamide, indicating that urease activity did not contribute to the results obtained.
FIG. 1.
Impacts of urea and urease inhibitor on the growth (OD600) of B. cereus strains ATCC 10987 (A) and ATCC 14579 (B) in BHI at pH 7 (black lines) and pH 5 (gray lines). Cultures without supplements are depicted by open squares; the closed squares represent cultures with the addition of urea, and the closed triangles represent cultures supplemented with urea and flurofamide.
The role of urease activity in acid shock survival was also examined. For this, the cells were grown overnight in BHI, and with the addition of hydrogen chloride, the pH was set at different acidic pH values, ranging from 2.5 to 4.8. Data obtained with stationary-phase cells of strain ATCC 10987 exposed to pH 4.5 are shown in Fig. 2. In the 30 min of exposure at pH 4.5, without and with extra urea present and with extra urea and flurofamide present, a similar decrease in the number of viable cells was observed. With mid-exponential-phase cells of ATCC 10987 exposed to pH 4.8, similar results were obtained, i.e., urease activity did not contribute to survival capacity under the conditions tested (data not shown). Notably, similar observations were made for the nine other ureABC-positive strains (data not shown). These experiments revealed that urease activity, independent of the strains' capacities to metabolize the ammonia generated, did not contribute to B. cereus survival capacity under acid shock conditions.
FIG. 2.
Impacts of urea and urease inhibitor on survival of BHI-grown stationary-phase cells of B. cereus ATCC 10987 exposed to pH 4.5. Three exposed cultures, without added urea, with 10 mM added urea, and with added urea and flurofamide, as indicated at the right, were diluted and droplet plated. The time of low-pH exposure is depicted at the top of each plate, and the dilution factors are indicated below the plates. This experiment was performed in triplicate, and all replicates showed similar results; a typical result is shown here.
Urease activity and expression of urease genes.
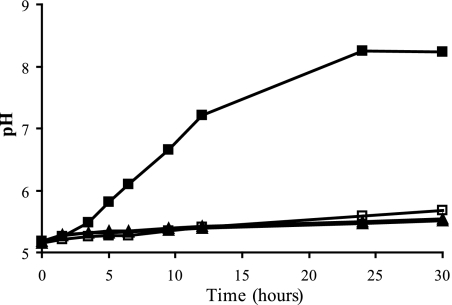
The urease activities of B. cereus ATCC 10987 and ATCC 14579 (negative control) were assessed by measuring the pH increase of the medium due to the formation of ammonium. Cells from overnight cultures were resuspended in MES buffer supplemented with urea without and with the urease inhibitor flurofamide present. The pH was monitored for 30 h, during which the pH increased from 5.2 to 8.2 in the presence of ATCC 10987 cells, whereas with flurofamide present, such a pH increase was not observed (Fig. 3). This indicates that addition of 10 μM (final concentration) flurofamide is sufficient to inhibit the urease activity, and similar results were obtained with all other ureolytic B. cereus strains tested (data not shown). Based on these results, the urease enzyme activity of strain ATCC 10987 was determined at approximately 1 nmol min−1 mg protein−1, indicating a very low activity level. The type strain ATCC 14579, which lacks the urease genes, did not provoke a pH increase under the conditions tested (Fig. 3).
FIG. 3.
Urease activity assay of B. cereus ATCC 10987 and ATCC 14579 in MES buffer. The ATCC 10987 cells in MES buffer supplemented with urea or with urea and flurofamide are depicted by closed squares and open squares, respectively. Closed triangles and open triangles represent the ATCC 14579 cells in MES buffer with urea or with urea and urease inhibitor, respectively.
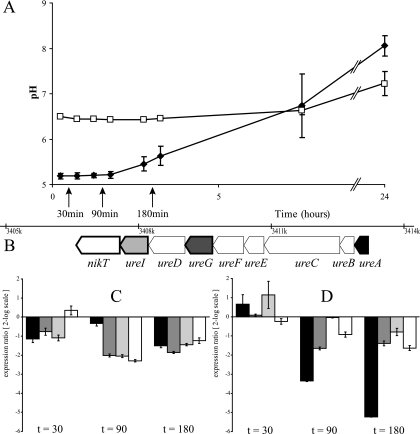
At the indicated time points in the urease activity assay (Fig. 4A), expression of urease genes (Fig. 4B) was quantified by RT-RT-PCR (Fig. 4C and D). Total-RNA samples were taken from ATCC 10987 cells resuspended in MES buffer with 10 mM urea at pH 5.2 and MOPS buffer with 10 mM urea at pH 6.5. The general expression of the four tested urease cluster genes was very low compared to the expression of the reference genes tufA and rpoA. The RNA levels of ureA, ureG, ureI, and nikT were approximately 32 times lower than the RNA levels of tufA and rpoA (the average threshold cycles of time zero samples were as follows: rpoA, 16.71; tufA, 15.34; ureA, 22.54; ureG, 20.53; ureI, 19.64; and nikT, 20.47). Significant changes in expression, according to the REST tool, could be obtained despite the low mRNA levels of the urease genes. However, no increase of relative expression could be observed when cells were exposed to a lower pH. Figure 4C and D indicates that the urease cluster genes were down-regulated compared to rpoA and tufA, even though the cells displayed ureolytic activity. In addition, the relative expression ratios of the urease genes were also determined at different phases of growth under aerobic conditions, and samples were taken from BHI cultures upon reaching ODs of 0.2, 0.5, and 5 and after overnight incubation. Under anaerobic growth conditions, samples were taken from BHI cultures upon reaching ODs of 0.2, 0.5, and 1 and after overnight incubation. Again, low levels of mRNAs of the urease genes were observed with no significant changes in the relative expression ratios of the urease genes during different growth stages in BHI under the conditions tested (data not shown). In conclusion, under a wide range of conditions tested, the expression of urease cluster genes was low, and no urea-, pH-, oxygen-, or nitrogen-dependent induction of expression could be observed.
FIG. 4.
Impacts of pH and urease activity on the relative expression ratios of urease cluster genes of B. cereus ATCC 10987. (A) pH increase over time of ATCC 10987 cells in MES buffer with 10 mM urea at pH 5.2 (closed diamonds) and of cells in MOPS buffer with 10 mM urea at pH 6.5 (open squares). The error bars represent the standard deviations between duplicate experiments, and the time points at which RNA samples were taken are indicated by arrows. (B) Positions of the genes investigated with RT-RT-PCR within the urease cluster: ureA (black), ureG (dark gray), ureI (light gray), and nikT (white). The other genes (ureB, ureC, ureE, ureF, and ureD) are indicated in white, and the gene aliases are indicated at the bottom. (C and D) Relative expression ratios compared to time zero of ureA, ureG, ureI, and nikT (the colors correspond to those in panel B) of ATCC 10987 cells exposed to pH 6.5 in MOPS buffer with 10 mM urea (C) and to pH 5.2 in MES buffer with 10 mM urea (D) at 30, 90, and 180 min (left, middle, and right, respectively). The expression ratios are expressed in log2 scale, and the error bars represent the standard errors between the duplicate experiments.
DISCUSSION
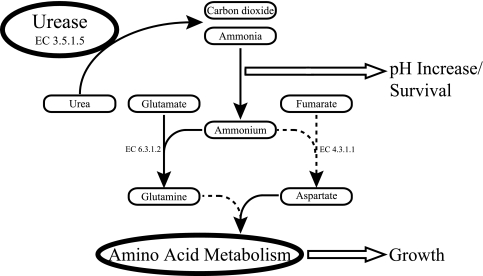
This study shows that 10 of the 49 (∼20%) tested clinical, food, and environmental B. cereus isolates harbor ureABC genes. Notably, only eight of these were able to use urea as a sole nitrogen source, whereas two of the strains, including the sequenced B. cereus strain ATCC 10987, could not. Remarkably, these two strains displayed ureolytic activity, and their failure to use urea as a nitrogen source can be explained by the inability to utilize ammonium for growth. The ammonium-negative phenotype of ATCC 10987 is possibly due to the lack of aspartate ammonia-lyase (EC 4.3.1.1) and the inability to utilize glutamine as a sole nitrogen source (30). The urease-positive strains that are capable of utilizing ammonium for growth conceivably incorporate ammonium into nitrogen metabolism via glutamine or aspartate (Fig. 5). Among the 49 isolates tested, eight strains have been associated with outbreaks (five diarrheal and three emetic strains), and only strains PAL5 and PAL27 (both diarrheal) were shown to be ureolytic. However, the limited number of disease-associated (diarrheal) B. cereus strains used in our study does not allow conclusions to be drawn as to the correlation (if any) between the presence of urease genes (and ureolytic activity) and the capability to cause disease.
FIG. 5.
Schematic representation of the functions of urease in B. cereus. Urea is hydrolyzed by urease (EC 3.5.1.5), forming carbon dioxide and ammonia. Subsequently, ammonia is converted into ammonium, which causes the pH to increase and may serve low-pH survival. Ammonium can be used as a nitrogen source and may be included in nitrogen metabolism via two possible routes supporting bacterial growth. The involved enzymes, glutamine synthetase and aspartate-ammonia lyase, are indicated by enzyme numbers (6.3.1.2 and 4.3.1.1, respectively). Note that the sequenced ATCC 10987 strain cannot use ammonium as a sole nitrogen source. The routes and/or enzymes missing in ATCC 10987 are indicated by dashed lines.
The formation of ammonium out of ammonia results in an elevation of the pH (Fig. 5), which may increase the fitness of ureolytic strains in acidic environments. It has been proposed (34) that the acquisition of the urease genes may have an impact on the survival of B. cereus strain ATCC 10987 under acidic conditions, e.g., the human stomach, as observed for H. pylori (42). However, growth of the ureolytic strains in urea-containing media with and without urease inhibitor at a low pH revealed no differences in fitness, i.e., none of the strains showed significant differences in the growth rate or final OD reached under the conditions tested. Furthermore, no differences in survival capacity were observed between cells of the ureolytic strains exposed to an acid shock in the presence of extra urea and cells exposed to an acid shock while urease activity was blocked by flurofamide. The results obtained can be explained by the low ureolytic activity displayed by the B. cereus strains tested. This is in contrast to the results obtained for S. salivarius (8) and H. pylori (42), where high ureolytic activity enabled the bacterial cells to cope with acid stress and, in the case of H. pylori, to colonize the human stomach.
The expression of urease genes can be constitutive, regulated by the presence of urea, pH, and/or nitrogen availability (10). The expression of the urease cluster in B. cereus ATCC 10987 appeared to be constitutive, because no up-regulation of the urease genes was observed after exposure to urea or acid downshift and under conditions of nitrogen limitation. Furthermore, the expression of the urease genes in B. cereus is generally very low compared to that of the housekeeping genes rpoA and tufA. The constitutive expression of the urease genes of B. cereus is in contrast with the regulation of the urease genes found in Proteus mirabilis (10), S. salivarius (37), and B. subtilis (10). The urease genes of P. mirabilis are expressed only when urea is present in the growth medium. The UreR regulator as present in the genome of P. mirabilis is involved in the urea-dependent induction of the urease cluster (10). Such a UreR regulator is absent in the genome of B. cereus ATCC 10987, as previously noted by Rasko et al. (34). In S. salivarius and other bacteria, urease expression is regulated by the pH; for example, at pH 5.5, urease activity increased 100-fold compared to that at pH 7.0 in S. salivarius (37). In B. subtilis, the expression of the urease operon is induced by nitrogen limitation (3) and regulated by a key transition phase transcription regulator, Spo0H (sigma factor σH) (49). This results in elevated expression of the urease genes in B. subtilis during stationary phase, which has also been shown for Y. enterocolitica (14). Screening of the genome of ATCC 10987 with a B. cereus-specific σH promoter consensus revealed that this strain does not harbor a σH promoter in front of the urease cluster (M. Tempelaars and T. Abee, unpublished results), and this corresponds to the absence of up-regulation of the urease genes in transition and stationary phase in B. cereus.
Even though a dual role of ureolytic activity in nitrogen metabolism and in acid stress survival has been described for Y. enterocolitica (50), S. salivarius (8), and H. pylori (48), we can conclude that none of the ureolytic B. cereus strains displayed increased fitness under acidic conditions or showed increased acid shock survival in the presence of urea. This is most likely linked to the low level of expression of the urease genes, the lack of modulation of their expression, and the resultant low level of ureolytic activity. Therefore, we conclude that the main role of B. cereus urease is in nitrogen metabolism, so that ammonia may be provided to the cells in nitrogen-limited environments.
Acknowledgments
We thank Marcel H. Zwietering (Laboratory of Food Microbiology, Wageningen University, Wageningen, The Netherlands) and Roy Moezelaar (Food Technology Centre, Wageningen University and Research Centre, Wageningen, The Netherlands) for fruitful discussions and critical reading of the manuscript.
Footnotes
Published ahead of print on 22 February 2008.
REFERENCES
- 1.Agata, N., M. Ohta, and K. Yokoyama. 2002. Production of Bacillus cereus emetic toxin (cereulide) in various foods. Int. J. Food Microbiol. 73:23-27. [DOI] [PubMed] [Google Scholar]
- 2.Ash, C., and M. D. Collins. 1992. Comparative analysis of 23S ribosomal RNA gene sequences of Bacillus anthracis and emetic Bacillus cereus determined by PCR-direct sequencing. FEMS Microbiol. Lett. 73:75-80. [DOI] [PubMed] [Google Scholar]
- 3.Atkinson, M. R., and S. H. Fisher. 1991. Identification of genes and gene products whose expression is activated during nitrogen-limited growth in Bacillus subtilis. J. Bacteriol. 173:23-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennik, M. H. J., E. J. Smid, F. M. Rombouts, and L. G. M. Gorris. 1995. Growth of psychrotrophic foodborne pathogens in a solid surface model system under the influence of carbon dioxide and oxygen. Food Microbiol. 12:509-519. [Google Scholar]
- 5.Berg, G., L. Eberl, and A. Hartmann. 2005. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7:1673-1685. [DOI] [PubMed] [Google Scholar]
- 6.Buono, F., R. Testa, and D. G. Lundgren. 1966. Physiology of growth and sporulation in Bacillus cereus. I. Effect of glutamic and other amino acids. J. Bacteriol. 91:2291-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlsson, J., J. Bergstrom, and B. Pehrson. 1995. Variations with breed, age, season, yield, stage of lactation and herd in the concentration of urea in bulk milk and individual cow's milk. Acta Vet. Scand. 36:245-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y. Y., C. A. Weaver, and R. A. Burne. 2000. Dual functions of Streptococcus salivarius urease. J. Bacteriol. 182:4667-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choma, C., M. H. Guinebretiere, F. Carlin, P. Schmitt, P. Velge, P. E. Granum, and C. Nguyen-The. 2000. Prevalence, characterization and growth of Bacillus cereus in commercial cooked chilled foods containing vegetables. J. Appl. Microbiol. 88:617-625. [DOI] [PubMed] [Google Scholar]
- 10.Collins, C. M., and S. E. D'Orazio. 1993. Bacterial ureases: structure, regulation of expression and role in pathogenesis. Mol. Microbiol. 9:907-913. [DOI] [PubMed] [Google Scholar]
- 11.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz-Ramos, H., P. Glaser, L. V. Wray, Jr., and S. H. Fisher. 1997. The Bacillus subtilis ureABC operon. J. Bacteriol. 179:3371-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawes, C., and G. H. Dibdin. 2001. Salivary concentrations of urea released from a chewing gum containing urea and how these affect the urea content of gel-stabilized plaques and their pH after exposure to sucrose. Caries Res. 35:344-353. [DOI] [PubMed] [Google Scholar]
- 14.de Koning-Ward, T. F., and R. M. Robins-Browne. 1997. A novel mechanism of urease regulation in Yersinia enterocolitica. FEMS Microbiol. Lett. 147:221-226. [DOI] [PubMed] [Google Scholar]
- 15.Ehling-Schulz, M., B. Svensson, M. H. Guinebretiere, T. Lindback, M. Andersson, A. Schulz, M. Fricker, A. Christiansson, P. E. Granum, E. Martlbauer, C. Nguyen-The, M. Salkinoja-Salonen, and S. Scherer. 2005. Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology 151:183-197. [DOI] [PubMed] [Google Scholar]
- 16.Finlay, W. J., N. A. Logan, and A. D. Sutherland. 2002. Bacillus cereus emetic toxin production in cooked rice. Food Microbiol. 19:431-439. [Google Scholar]
- 17.Frankland, G. C., and P. F. Frankland. 1887. Studies on some new micro-organisms obtained from air. Phil. Trans. R. Soc. Lond. B 178:257-287. [Google Scholar]
- 18.Ghosh, A. C. 1978. Prevalence of Bacillus cereus in the faeces of healthy adults. J. Hyg. 80:233-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granum, P. E., S. Brynestad, and J. M. Kramer. 1993. Analysis of enterotoxin production by Bacillus cereus from dairy products, food poisoning incidents and non-gastrointestinal infections. Int. J. Food Microbiol. 17:269-279. [DOI] [PubMed] [Google Scholar]
- 20.Granum, P. E., and T. Lund. 1997. Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 157:223-228. [DOI] [PubMed] [Google Scholar]
- 21.Haggblom, M. M., C. Apetroaie, M. A. Andersson, and M. S. Salkinoja-Salonen. 2002. Quantitative analysis of cereulide, the emetic toxin of Bacillus cereus, produced under various conditions. Appl. Environ. Microbiol. 68:2479-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jahns, T., A. Zobel, D. Kleiner, and H. Kaltwasser. 1988. Evidence for carrier-mediated, energy-dependent uptake of urea in some bacteria. Arch. Microbiol. 149:377-383. [Google Scholar]
- 23.Kotiranta, A., K. Lounatmaa, and M. Haapasalo. 2000. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2:189-198. [DOI] [PubMed] [Google Scholar]
- 24.Lechner, S., R. Mayr, K. P. Francis, B. M. Pruss, T. Kaplan, E. Wiessner-Gunkel, G. S. Stewart, and S. Scherer. 1998. Bacillus weihenstephanensis sp. nov. is a new psychrotolerant species of the Bacillus cereus group. Int. J. Syst. Bacteriol. 48:1373-1382. [DOI] [PubMed] [Google Scholar]
- 25.Lee, A., J. Fox, and S. Hazell. 1993. Pathogenicity of Helicobacter pylori: a perspective. Infect. Immun. 61:1601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luxananil, P., H. Atomi, S. Panyim, and T. Imanaka. 2001. Isolation of bacterial strains colonizable in mosquito larval guts as novel host cells for mosquito control. J. Biosci. Bioeng. 92:342-345. [DOI] [PubMed] [Google Scholar]
- 27.Mackay, E. M., and L. L. Mackay. 1927. The concentration of urea in the blood of normal individuals. J. Clin. Investig. 4:295-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Midura, T., M. Gerber, R. Wood, and A. R. Leonard. 1970. Outbreak of food poisoning caused by Bacillus cereus. Public Health Rep. 85:45-48. [PMC free article] [PubMed] [Google Scholar]
- 29.Mobley, H. L., and R. P. Hausinger. 1989. Microbial ureases: significance, regulation, and molecular characterization. Microbiol. Rev. 53:85-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mols, M., M. de Been, M. H. Zwietering, R. Moezelaar, and T. Abee. 2007. Metabolic capacity of Bacillus cereus strains ATCC 14579 and ATCC 10987 interlinked with comparative genomics. Environ. Microbiol. 9:2933-2944. [DOI] [PubMed] [Google Scholar]
- 31.Neblett, T. R. 1976. Use of droplet plating method and cystine-lactose-lactng electrolyte-deficient medium in routine quantitative urine culturing procedure. J. Clin. Microbiol. 4:296-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neithercut, W. D., A. M. el Nujumi, and K. E. McColl. 1993. Measurement of urea and ammonium concentrations in gastric juice. J. Clin. Pathol. 46:462-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasko, D. A., J. Ravel, O. A. Okstad, E. Helgason, R. Z. Cer, L. Jiang, K. A. Shores, D. E. Fouts, N. J. Tourasse, S. V. Angiuoli, J. Kolonay, W. C. Nelson, A. B. Kolsto, C. M. Fraser, and T. D. Read. 2004. The genome sequence of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to Bacillus anthracis pXO1. Nucleic Acids Res. 32:977-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasko, D. A., M. J. Rosovitz, O. A. Okstad, D. E. Fouts, L. Jiang, R. Z. Cer, A. B. Kolsto, S. R. Gill, and J. Ravel. 2007. Complete sequence analysis of novel plasmids from emetic and periodontal Bacillus cereus isolates reveals a common evolutionary history among the B. cereus-group plasmids, including Bacillus anthracis pXO1. J. Bacteriol. 189:52-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenquist, H., L. Smidt, S. R. Andersen, G. B. Jensen, and A. Wilcks. 2005. Occurrence and significance of Bacillus cereus and Bacillus thuringiensis in ready-to-eat food. FEMS Microbiol. Lett. 250:129-136. [DOI] [PubMed] [Google Scholar]
- 37.Sissons, C. H., H. E. Perinpanayagam, E. M. Hancock, and T. W. Cutress. 1990. pH regulation of urease levels in Streptococcus salivarius. J. Dent. Res. 69:1131-1137. [DOI] [PubMed] [Google Scholar]
- 38.Soriano, A., G. J. Colpas, and R. P. Hausinger. 2000. UreE stimulation of GTP-dependent urease activation in the UreD-UreF-UreG-urease apoprotein complex. Biochemistry 39:12435-12440. [DOI] [PubMed] [Google Scholar]
- 39.Spira, W. M., and J. M. Goepfert. 1975. Biological characteristics of an enterotoxin produced by Bacillus cereus. Can. J. Microbiol. 21:1236-1246. [DOI] [PubMed] [Google Scholar]
- 40.Stenfors, L. P., and P. E. Granum. 2001. Psychrotolerant species from the Bacillus cereus group are not necessarily Bacillus weihenstephanensis. FEMS Microbiol. Lett. 197:223-228. [DOI] [PubMed] [Google Scholar]
- 41.Sumner, J. B. 1926. The isolation and crystallization of the enzyme urease. J. Biol. Chem. 69:435-441. [Google Scholar]
- 42.Tsuda, M., M. Karita, M. G. Morshed, K. Okita, and T. Nakazawa. 1994. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 62:3586-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turnbull, P. C. 1981. Bacillus cereus toxins. Pharmacol. Ther. 13:453-505. [DOI] [PubMed] [Google Scholar]
- 44.van Schaik, W., M. H. Tempelaars, J. A. Wouters, W. M. de Vos, and T. Abee. 2004. The alternative sigma factor σB of Bacillus cereus: response to stress and role in heat adaptation. J. Bacteriol. 186:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Schaik, W., M. H. Tempelaars, M. H. Zwietering, W. M. de Vos, and T. Abee. 2005. Analysis of the role of RsbV, RsbW, and RsbY in regulating σB activity in Bacillus cereus. J. Bacteriol. 187:5846-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vilain, S., Y. Luo, M. B. Hildreth, and V. S. Brozel. 2006. Analysis of the life cycle of the soil saprophyte Bacillus cereus in liquid soil extract and in soil. Appl. Environ. Microbiol. 72:4970-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wijnands, L. M., J. B. Dufrenne, M. H. Zwietering, and F. M. van Leusden. 2006. Spores from mesophilic Bacillus cereus strains germinate better and grow faster in simulated gastro-intestinal conditions than spores from psychrotrophic strains. Int. J. Food Microbiol. 112:120-128. [DOI] [PubMed] [Google Scholar]
- 48.Williams, C. L., T. Preston, M. Hossack, C. Slater, and K. E. McColl. 1996. Helicobacter pylori utilises urea for amino acid synthesis. FEMS Immunol. Med. Microbiol. 13:87-94. [DOI] [PubMed] [Google Scholar]
- 49.Wray, L. V., Jr., A. E. Ferson, and S. H. Fisher. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 179:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young, G. M., D. Amid, and V. L. Miller. 1996. A bifunctional urease enhances survival of pathogenic Yersinia enterocolitica and Morganella morganii at low pH. J. Bacteriol. 178:6487-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]