Abstract
The identification of sites resulting in cross-contamination of poultry flocks in the abattoir and determination of the survival and persistence of campylobacters at these sites are essential for the development of intervention strategies aimed at reducing the microbial burden on poultry at retail. A novel molecule-based method, using strain- and genus-specific oligonucleotide probes, was developed to detect and enumerate specific campylobacter strains in mixed populations. Strain-specific oligonucleotide probes were designed for the short variable regions (SVR) of the flaA gene in individual Campylobacter jejuni strains. A 16S rRNA Campylobacter genus-specific probe was also used. Both types of probes were used to investigate populations of campylobacters by colony lift hybridization. The specificity and proof of principle of the method were tested using strains with closely related SVR sequences and mixtures of these strains. Colony lifts of campylobacters were hybridized sequentially with up to two labeled strain-specific probes, followed by the generic 16S rRNA probe. SVR probes were highly specific, differentiating down to 1 nucleotide in the target sequence, and were sufficiently sensitive to detect colonies of a single strain in a mixed population. The 16S rRNA probe detected all Campylobacter spp. tested but not closely related species, such as Arcobacter skirrowi and Helicobacter pullorum. Preliminary field studies demonstrated the application of this technique to target strains isolated from poultry transport crate wash tank water. This method is quantitative, sensitive, and highly specific and allows the identification and enumeration of selected strains among all of the campylobacters in environmental samples.
Campylobacter jejuni is a major reported cause of acute bacterial diarrheal disease in humans in the industrialized world. In England and Wales, the prevalence of human campylobacteriosis peaked in 2000, when 57,674 cases were reported to the Health Protection Agency; since then there has been a decline, and early provisional data for 2006 indicate that there were 46,603 notified cases of this disease (http://www.hpa.org.uk/infections). In the United States, an estimated 2.4 million people are affected each year (27). Although campylobacteriosis is generally self-limiting, the socioeconomic cost of this disease is high in terms of the burden on health resources and in terms of the cost to industry associated with time away from work (39). Thus, control and prevention of campylobacteriosis are essential to reduce this significant public health problem.
C. jejuni can asymptomatically colonize most wild and domestic birds. As highlighted by epidemiological studies, the handling and consumption of raw or undercooked poultry meat are major sources of campylobacteriosis. Once the first birds in a flock become infected, campylobacters spread rapidly. Thus, in some countries, including the United Kingdom, up to 90% of broiler flocks can be campylobacter positive at slaughter (5, 17, 22). The level of cecal colonization in broilers can be up to 109 organisms per g of cecal contents. Carcass contamination is related to the within-flock prevalence of campylobacter colonization (1). During processing, defeathering (1) and evisceration (7, 40) may increase the levels of contamination of poultry carcasses and the abattoir environment. Subsequently, carcasses from Campylobacter-negative flocks can also become contaminated either when they are placed in soiled transport crates (47) or via cross-contamination when they follow carcasses from a Campylobacter-positive flock through the abattoir (20). Determining the survival and persistence of Campylobacter at different sites is essential for the development of farm-to-fork strategies for the control and prevention of food-borne campylobacteriosis and, in particular, to inform quantitative risk assessment models. This determination needs to be performed at the strain level, preferably using a rapid and cost-effective method.
Molecular approaches have been used extensively in attempts to understand the epidemiology of campylobacteriosis. C. jejuni is both phenotypically and genotypically diverse, and a range of genotypic methods have been developed for this organism, including fla restriction fragment length polymorphism typing (2, 10), pulsed-field gel electrophoresis (PFGE) (24, 52), flaA short variable region (SVR) sequence typing (29), and multilocus sequence typing (MLST) (15). These methods and others have all revealed the presence of diverse Campylobacter genotypes in many environments, such as abattoirs (31) and poultry farms (13, 23, 38). It may be anticipated that the persistence and survival of C. jejuni in such environments vary between strains (31). Thus, for accurate quantitative risk assessment models, it is necessary to determine not only the total number of campylobacters persisting throughout an abattoir but also the survival characteristics of individual strains entering the abattoir.
Evidence from such molecular epidemiological studies indicates that the majority of conventionally reared broiler flocks in the United Kingdom are colonized with only one or two Campylobacter strains (4). This finding is supported by data from Swedish and Dutch flocks (6, 22), although it may not be true for all conventionally reared flocks (21, 36). The hypothesis developed for the present investigation was that specific strains from individual conventionally reared broiler flocks could be tracked and enumerated on chicken carcasses and at various sites in the abattoir during processing. The approach used for this study was to design and develop both genus- and strain-specific oligonucleotide probes detectable by colony hybridization. The specificity and sensitivity of these probes for enumerating both the total viable campylobacters and specific test strains were determined under control conditions using reference strains and mixed populations. Finally, the proof of principle was demonstrated using strains from washed poultry crates.
MATERIALS AND METHODS
Bacteria and culture conditions.
C. jejuni strains isolated from ceca or feces were used in this study along with the reference strains listed in Table 1. The strains were grown on blood agar (BA) (CM0271; Oxoid Ltd., Basingstoke, United Kingdom) with 5% defibrinated horse blood (SR0050; Oxoid Ltd.) or on modified charcoal cefoperazone deoxycholate agar (mCCDA) (CM0739 and SR0155; Oxoid Ltd.). Plates were incubated at either 37 or 41.5°C for 24 or 48 h, as indicated below, in a microaerobic atmosphere generated by using a CampyGen (Oxoid Ltd.) gas-generating kit. Arcobacter spp. (Table 1) were also grown on BA under aerobic conditions at their optimum growth temperature (25°C) and also in experiments at 37°C microaerobically. Strains were inoculated onto plates either using a 1-μl loop or by conventional spread plating. In addition, 50 strains from the CampyNet international strain set (http://campynet.vetinst.dk/; 19) were grown on BA with a selective supplement (BA+) (BA as described above with SR0155; Oxoid Ltd.).
TABLE 1.
Strains used in this study
| Farm isolate or reference taxon | Source or strain | fla allele identification (Oxford database) |
|---|---|---|
| D2/T/3 5a | Ceca | 117 |
| E/C/1 3b | Ceca | 191 |
| E/C/1 4b | Ceca | 16 |
| E2/5/1 28a | Feces | 92 |
| F/6/2 36a | Feces | 10 |
| G/3/6 35b | Feces | 521 |
| J2/C/5 2 | Ceca | 70 |
| L2/C/6 2a | Ceca | 52 |
| O/C/5 11b | Ceca | 21 |
| P/C/6 6b | Ceca | 18 |
| Arcobacter skirrowii BT25/06 | VLAa | |
| Arcobacter cryaerophilus subsp. skirrowii BT59/06 | VLA | |
| Arcobacter skirrowii BT170/06 | VLA | |
| Campylobacter sputorum subsp. bubulus | NCTC11367 | |
| Campylobacter hyointestinalis | NCTC11608 | |
| Campylobacter coli | NCTC12143 | |
| Campylobacter fetus subsp. fetus | NCTC10842 | |
| Campylobacter jejuni | NCTC11351 | |
| Campylobacter jejuni | RM1221 | |
| Campylobacter jejuni | NCTC11168 | |
| Helicobacter pullorum | NCTC12824 |
VLA, Veterinary Laboratories Agency, United Kingdom.
Flagellin (flaA) gene SVR as a strain-specific probe.
The Campylobacter flaA gene was selected because it is known to be highly variable (28) and has been used in several genotyping schemes (35, 48). Genomic DNA was isolated from all campylobacters using a NucleoSpin blood kit (Macherey-Nagel, Duren, Germany). The flaA SVR was amplified using primers FLA4F (5′-GGATTTCGTATTAACACAAATGGTGC-3′) and FLA625RU (5′-CAAG[AT]CCTGTTCC[AT]ACTGAAG). The forward primer is a refinement (11) of the primer described previously (28). The PCR were carried out using 25-μl reaction mixtures containing Qiagen HotStar Taq Master Mix (12.5 μl of HotStar Taq Master Mix, each deoxynucleoside triphosphate at a concentration of 200 μM, 1.5 mM MgCl2), each primer at a concentration of 0.3 μM, and 150 ng of genomic DNA. The cycling conditions were an initial activation step of 15 min at 95°C, followed by 35 cycles of denaturation for 30 s at 94°C, annealing for 1 min at 58°C, and extension for 1 min at 72°C. The 621-bp amplified fragment was sequenced with primers FLA4F and FLA625RU by the Sequencing Service at the University of Dundee. The forward and reverse chromatogram sequence results were confirmed by assembling them in ChromasPro 1.34 (Technelysium Pty Ltd., Cologne, Germany). Confirmed sequences were aligned using CLC Free Workbench 3.2 (CLC bio A/S; http://www.clcbio.com).
flaA sequences deposited at http://hercules.medawar.ox.ac.uk/flaA/ were aligned using CLC Free Workbench in order to identify a region of mismatches suitable for designing strain-specific probes. Probes were designed using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) for the complementary strand and synthesized by Sigma Genosys (Gillingham, United Kingdom).
16S rRNA as a genus-specific probe.
The genus-specific region of the genome has already been identified in the 16S rRNA, which is used routinely for PCR-based identification of campylobacters (34). The 16S rRNA probe (CAMP653; 5′-CTGCCTCTCCCTYACTCT-3′) for detection of the genus Campylobacter was selected from previously described probes (44), and its specificity was checked using BLAST at the NCBI website, the European rRNA database (51), and the ProbeMatch tool in RDP-II (12).
Probe labeling.
The oligonucleotide probes (100 pmol) were enzymatically labeled at the 3′ end with terminal transferase incorporating a single digoxigenin (DIG)-labeled ddUTP using a DIG oligonucleotide 3′ end labeling kit (Roche Applied Science, Burgess Hill, United Kingdom) according to the manufacturer's instructions. The probes were stored on ice until they were required, and for longer storage they were stored at −20°C with an equal volume of DIG Easy Hyb (Roche Applied Science). The labeling efficiency was determined by preparing specific dilutions of the labeled probe.
Colony lifts, hybridization, and chemiluminescent detection.
Plates (mCCDA or BA) of Campylobacter colonies were precooled at 4°C for at least 30 min prior to transfer of the colonies onto Hybond-N membranes (83 or 132 mm; Amersham Biosciences, Little Chalfont, United Kingdom). The colonies were lysed, and genomic material was bound to the membranes using the method described by Sambrook and Russell (42). The membranes were air dried, and the DNA was fixed by UV cross-linking (10 s at 70,000 μJ/cm2) using an HL-200 Hybri Linker (UVP Lab Products, Cambridge, United Kingdom). Membranes were used immediately or stored at −20°C until they were required.
Membranes were prehybridized in 3.5 ml (for 82-mm membranes) or 5 ml (for 132-mm membranes) of DIG Easy Hyb (Roche Applied Science) preheated to 10°C below the melting temperature (Tm) of each oligonucleotide probe (Table 2) for between 15 min and 1 h. Membranes were hybridized overnight at the same probe-specific temperature (Table 2) with 10 pmol of DIG-labeled probe in 3.5 or 5 ml of fresh DIG Easy Hyb. The DIG-labeled 16S rRNA probe was hybridized at 46°C. After hybridization, membranes were washed twice in 50 ml of 2× SSC-0.1% sodium dodecyl sulfate (SDS) at room temperature for 5 min and then twice in 50 ml of 0.5× SSC-0.1% SDS at 55°C for 15 min (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
TABLE 2.
SVR probes designed and used in this study
| Probe | Binding position | Strain(s) | Length (nucleotides) | Tm (°C)a | No. of strains with one nucleotide mismatch/total no. | No. of strains with two nucleotide mismatches/total no. | Sequence (5′-3′) |
|---|---|---|---|---|---|---|---|
| SVR4 | 560-542 | D2/T/3 5a | 19 | 48 | 2/9 | 3/9 | TGCACTTCGCCACTAGATG |
| SVR6 | 564-542 | E/C/1 3b, P/C/6 6b | 23 | 54 | 1/8 | 1/8 | AAATTGTACTTCACCACCAGATG |
| SVR10 | 559-540 | F/6/2 36a | 20 | 50 | 0/9 | 0/9 | GCACTTCACCACCAACTGAA |
| SVR12 | 553-532 | E2/5/1 28a | 22 | 54 | 2/9 | 1/9 | CACCTGAACTCACACTTTGTGA |
| SVR13 | 561-539 | G/3/6 35b | 23 | 54 | 0/9 | 0/9 | TTGCACTTCACCACTAGATGAAA |
For oligomers up to 25 nucleotides long, the Tm was calculated using the following formula: 2°C × (A+T) + 4°C × (G+C).
The DIG-labeled probes were detected immunologically with the disodium 3-{4-methoxyspiro[1,2-dioxetane-3,2-(5-chloro)tricyclo(3.3.1.13,7)decan]-4-yl} phenyl phosphate (CSPD) chemiluminescent substrate using a DIG luminescent detection kit (Roche Applied Science) and a DIG wash and block buffer set (Roche Applied Science) according to the manufacturers' instructions and using 50 and 100 ml of washing and blocking solutions for the 82- and 132-mm membranes, respectively, and 30 or 50 ml of antibody solution. The membranes were exposed to Kodak BioMax Light autoradiography film (Sigma-Aldrich, Poole, United Kingdom) for 30 min for the 16S rRNA probe and for 2.5 h for the SVR probes.
Stripping and reprobing.
The probed membranes were stripped by completely covering them with 0.1% (wt/vol) boiling SDS. The membranes were left in this solution with gentle agitation for 15 min. The solution was then replaced with fresh boiling 0.1% (wt/vol) SDS, and the preparations were incubated for a further 15 min. Then the membranes were rinsed briefly in 2× SSC, and removal of the probe was checked using the chemiluminescent detection procedure described above. Membranes were then hybridized with a second SVR probe overnight using the appropriate conditions, starting with prehybridization as described above. The 16S rRNA probe was used for the final hybridization.
Effect of SVR probe position on specificity.
The effect of the probe position in the identified 40-nucleotide variable region was investigated for one strain (G/3/6 35b) by designing five probes (probes G1 to G5) (20 nucleotides) that vary in position by 5 nucleotides and span the region (Fig. 1b). Fivefold dilutions of heat-denatured target genomic DNA (1 μg to 320 pg) were prepared and pipetted onto Hybond-N membranes, which was followed by UV cross-linking. The membranes were prehybridized and hybridized (10 pmol of labeled probe) in 3.5 ml of DIG Easy Hyb overnight at the calculated Tm for each probe and detected as described above. The results were interpreted by analyzing the secondary structure of the 40-nucleotide region using mfold (http://www.bioinfo.rpi.edu/applications/mfold; 53) with 50 mM Na+ and 1.5 mM Mg2+ at each Tm.
FIG. 1.
(a) Direct sequence alignment of 10 C. jejuni flaA SVR sequences showing the region from nucleotide 530 to nucleotide 570 which was used to design the probes. Nucleotides that differ from nucleotides in the consensus sequence are shaded, and the shaded sequences indicate the positions of the SVR probes. Nucleotide positions are based on the ATG start site of C. jejuni NCTC11168 and the C. jejuni RM1221 flaA gene sequence. (b) Sequence alignment showing the positions and sequences of probes G1 to G5 in the 40-nucleotide sequence of strain G/3/6 35b. The Tm of probes G1, G2, G4, and G5 is 44°C, and the Tm of G3 is 48°C.
Probe sensitivity and the limit of detection.
The limit of detection of the assay was determined by also preparing fivefold dilutions of target genomic DNA (1 μg to 320 pg) that were pipetted onto membranes, which was followed by UV cross-linking. The membranes were prehybridized and hybridized in 3.5 ml of DIG Easy Hyb at the Tm of the probe (Table 2). Membranes were hybridized with 10 pmol of labeled probe overnight, and the probe was detected as described above. To investigate whether there was variation in sensitivity between probes, each probe was hybridized to its complementary sequence at the following concentrations: 1, 0.2, and 0.1 pmol and 50, 25, and 10 fmol.
Specificity of the strain-specific probe.
Each SVR probe was analyzed using BLAST with the Campylobacter flaA variable region database maintained by Oxford (http://hercules.medawar.ox.ac.uk/flaA/). The number of alleles that contained the exact probe sequence was expressed as a percentage of the total number of alleles in the database at the time that the analysis was performed (January 2007). This percentage was used to calculate the expected number of CampyNet strains that might also contain each SVR probe sequence. In this way the specificity of each SVR probe could be ranked. To test these in silico results, 50 CampyNet strains that are genetically well defined and known not to be epidemiologically related were selected. Each strain was inoculated onto a BA+ plate (50 strains per plate) using a 1-μl inoculation loop in a grid and grown overnight microaerobically at 41.5°C. Five plates were inoculated, one for each probe. Colony lifts were removed and hybridized with probe SVR4, SVR6, SVR10, SVR12, or SVR13. Each probe was tested with three independent replicates.
Artificial mixtures of strains.
Five strains (D2/T/3, G/3/6 35b, L2/C/6 2a, E/C/1 3b, and F/6/2 36a) were each suspended in 200 μl of maximum recovery diluent (Oxoid Ltd.) and diluted 10-fold. Equal volumes of the strains were mixed together, and 100 μl of the suspension was plated onto mCCDA. All plates were incubated microaerobically at 41.5°C for 48 h, after which colonies were transferred onto Hybond-N membranes, lysed, denatured, and fixed as described above. The membranes were probed sequentially with the SVR10, SVR6, SVR4, and 16S rRNA probes, stripping with 0.1% boiling SDS between SVR probes but not before the final 16S rRNA probe.
Validation of the technique in a field situation.
Water samples from poultry transport crate wash tanks were stored at −80°C in 15% (vol/vol) glycerol. Using the original unstored sample, appropriate dilutions of the wash water were prepared, and aliquots (100 μl) were directly plated onto mCCDA and incubated at 41.5°C microaerobically for 48 h. Ten single colonies were picked at random and streaked onto BA+. These isolates were stored on beads at −80°C (Microbank; Prolab Diagnostics, South Wirral, United Kingdom). In addition, DNA was extracted from each of the 10 isolates using a NucleoSpin blood kit. Colony lifts (132-mm Hybond-N membranes) were removed from the mCCDA plates, and the colonies were lysed as described previously. The membranes were stored at −20°C until the probe(s) had been synthesized. For each of the isolates, PCR and sequencing of the flaA SVR were carried out using primers FLA4F and FLA625RU as described above. The sequences were aligned, and probes for the SVR were designed as described above. The membranes were removed from storage and prehybridized in 5 ml of DIG Hyb at the Tm of the probe. Membranes were hybridized sequentially with the SVR probes, stripping with 0.1% boiling SDS between probes, and finally with the 16S rRNA probe.
RESULTS
Flagellin gene (flaA) as the strain-specific probe.
An alignment of the region of the SVR sequences of the 10 test strains used in this study is shown in Fig. 1a. In conjunction with the alignment of flaA sequences from the Oxford database, this alignment identified a short segment with high variability. This region with a variable sequence (nucleotides 530 to 570) was then identified in the alignment of the sequences from the 10 C. jejuni strains with closely related SVR sequences shown in Table 1. Five probes were designed for the complementary strand of this region, where there was 34.1% identity between strains (Table 2). Preliminary investigations had shown that antisense probes were much more sensitive than sense probes, presumably because they could hybridize to both DNA and RNA (data not shown). Each probe was designed so that it was specific for one strain (except strains E/C/1 3b and P/C/6b, which had identical sequences in the 40-nucleotide region [Fig. 1a]) and had one or more mismatches with the other strains (Table 2). The conditions used for treating the colony lifts gave good hybridization signals, and probes could be effectively removed by boiling in 0.1% SDS.
Using experimentally determined stringent hybridization and washing conditions, the specificity of each strain-specific oligonucleotide probe was tested by hybridizing it to identical membranes from plate lifts of the 10 Campylobacter strains grown on BA and incubated at 37°C for 24 h. Figure 2 shows the hybridization of probe SVR4. At a lower-stringency wash temperature (46°C) some cross-reactivity was observed (Fig. 2A), but this was eliminated by increasing the temperature to 55°C (Fig. 2B). The posthybridization wash temperature was determined experimentally by using three membranes loaded with 1 μg of genomic DNA of each of the 10 Campylobacter strains, probing with SVR6, and washing one membrane at 40°C, one membrane at 50°C, and one membrane at 60°C. At 40°C, there was cross-reactivity with three strains, at 50°C there was cross-reactivity with one strain, and at 60°C no reactivity was detected (data not shown). The temperature selected based on this experiment was 55°C, and this temperature was subsequently tested with colony lifts of the strains probed with SVR4 (Fig. 2B). The other SVR probes tested (SVR10, SVR12, and SVR13) gave similar results and specifically detected the homologous strain with no cross-reactivity with other strains at a wash temperature of 55°C (data not shown).
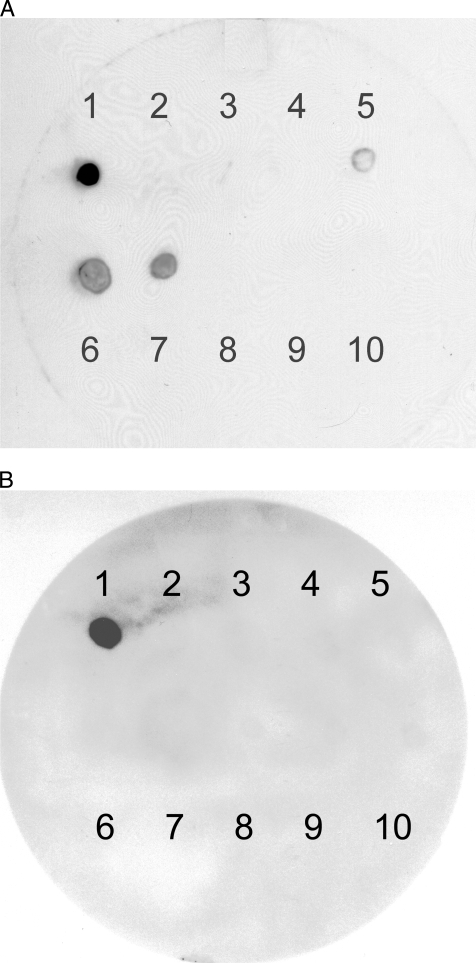
FIG. 2.
Autoradiograms demonstrating the specificity of oligonucleotide probe SVR4 with 10 C. jejuni strains using plate lifts. (A) Probe SVR4 was hybridized at 48°C and washed at 46°C. This probe was homologous to the strain D2/T/3 5a sequence (position 1), with some cross-reactivity with one nucleotide mismatch to G/3/6 35b (position 6) and J2/C/5 2 (position 7) and with four mismatches to F/6/2/36a (position 5). (B) Probe SVR4 was hybridized at 48°C and washed at 55°C. At this posthybridization wash temperature there was no cross-reactivity with other strains, and the probe bound only to strain D2/T/3 5a (position 1). The following strains were used: 1, D2/T/3 5a; 2, E/C/1/3b with two mismatches to strain D2/T/s 5a; 3, E/C/1 4b with 11 mismatches; 4, E2/5/1 28a with 12 mismatches; 5, F/6/2/36a with four mismatches; 6, G/3/6 35b with one mismatch; 7, J2/C/5 2 with one mismatch; 8, L2/C/6 2a with two mismatches; 9, O/C/5/11b with two mismatches; and 10, P/C/6 6b with three mismatches.
Effect of probe position on specificity.
The binding positions and sequences of probes G1 to G5 are shown in Fig. 1b. Hybridization of these probes to genomic DNA showed that there were differences in the sensitivities of the probes depending on the hybridization position. Probes G1, G2, and G3 gave strong hybridization with 40 ng of target and could detect amounts as small as 8 ng, whereas probes G4 and G5 showed strong hybridization with 200 ng and could detect 40 ng of target (i.e., there was a fivefold difference in sensitivity). Differences in the secondary structure of the single-stranded DNA or RNA is the most likely explanation for these specific patterns of annealing. Analysis of the secondary structures was predicted using Mfold, and the binding sites of the probes to these folded structures revealed no clear structural features that distinguished accessible sites from inaccessible sites. The best-performing probes did not, for example, correspond to linear areas in the predicted structures.
16S rRNA probe does not detect Arcobacter spp. or Helicobacter pullorum.
In contrast to a strain-specific probe which detects and tracks only one strain, the genus-specific probe was designed in order to quantify the total numbers of Campylobacter spp. in the environment. In silico BLAST analyses, using the NCBI website, the European rRNA database, and the ProbeMatch tool in RDP-II of the 16S rRNA probes described previously, showed that these probes are unique and specific for the genus Campylobacter. Mfold was used to confirm the accessibility of the selected target region.
The 16S rRNA probe (44) correctly detected all strains belonging to the genus Campylobacter investigated that were grown on BA plates at 37°C for 24 h (Fig. 3 and Table 1). Both Arcobacter spp. and H. pullorum, which are genetically related to C. jejuni, may be recovered from chicken carcasses under routine culture conditions. The 16S rRNA probe did not hybridize to Arcobacter spp. or to H. pullorum (Fig. 3).
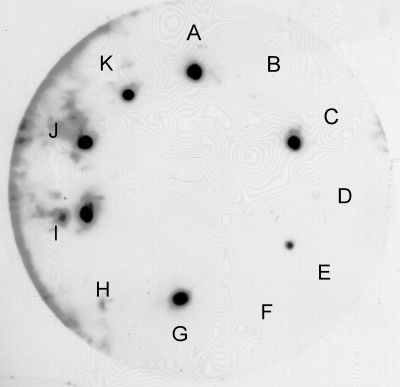
FIG. 3.
16S rRNA specifically detects Campylobacter spp. and not closely related organisms. The plate lift contained the following organisms: A, C. jejuni NCTC11168; B, Arcobacter skirrowii BT25/06; C, C. coli NCTC12143; D, H. pullorum NCTC12824; E, C. jejuni NCTC11351; F, A. skirrowii BT170/06; G, C. jejuni RM1221; H, Arcobacter cryaerophilus subsp. skirrowii BT59/06; I, Campylobacter sputorum subsp. bubulus; J, C. hyointestinalis NCTC11608; and K, C. fetus subsp fetus.
Probe sensitivities and the limit of detection.
The nonradioactive DIG-based labeling method was found to be sensitive enough for detection of nucleic acid in a single colony. The labeling reaction was always efficient, and labeled probe could be stored for long periods of time at −20°C. The 16S rRNA probe was more sensitive than the SVR probes, requiring only 20 min of exposure to X-ray film. This was mainly due to the fact that there was 100- to 1,000-fold more 16S rRNA than SVR mRNA in the genome. The optimum template DNA concentration required by the SVR probes to obtain a satisfactory signal was 8 ng, but the limit of detection of the 16S rRNA probe for genomic DNA was 1.6 ng of target (data not shown). The probes exhibited similar sensitivities when they were hybridized to the complementary sequence; e.g., SVR4, SVR6, SVR10, SVR12, and SVR13 gave good signals when they were hybridized to 50 fmol of target (data not shown).
Application of SVR probes to a mixture of strains.
Individual SVR probes could detect specific strains in a mixture of five strains grown on mCCDA (Fig. 4). The colonies were transferred to a Hybond-N membrane, which was sequentially hybridized with two different SVR probes and finally with the 16S rRNA probe. The membrane was stripped with boiling 0.1% SDS between SVR probe hybridizations. Each probe specifically detected and enumerated strain-specific target colonies in the mixture of strains, and the 16S rRNA probe revealed the total numbers of campylobacters.
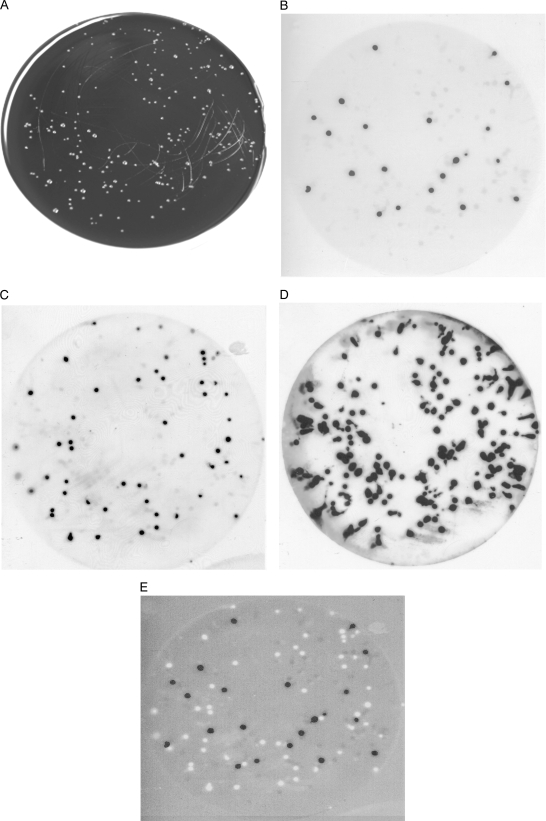
FIG. 4.
Sequential hybridization of Campylobacter with the SVR probes. A mixture of five Campylobacter strains on mCCDA (A) was probed first with SVR4 (B), which was followed by stripping with boiling 0.1% SDS, a second hybridization with SVR6 (C), and finally enumeration of all campylobacters with 16S rRNA (D). (E) Merged image of probe results for SVR4 (black dots) and SVR6 (white dots).
Specificity of the strain-specific probes.
It is anticipated that in an abattoir environment there are other Campylobacter strains in addition to the target flock strain. The ability of probes to identify the target strains depends on whether the other strains present have similar SVR sequences. By performing a BLAST analysis with the probes and available flaA sequences, some indication of the population distribution of these specific sequences in Campylobacter strains could be obtained. At present, the true diversity of flaA sequences in Campylobacter populations that could be encountered in an abattoir environment is not known; therefore, the specificity of the probes could be assessed only relative to presently determined flaA sequences. The Oxford database (http://hercules.medawar.ox.ac.uk/flaA/) represents the largest available data set and contained 907 sequences in January 2007. These sequences included all the sequences identified in the GenBank database at the time of analysis. The sequences stored in the Oxford database were used to predict the levels of positive reactions of the SVR probes (Table 3) in a population that resembled the population represented in the database. For example, SVR6 had 22 matches to strains in the database apparently sharing the SVR6 sequence. However, according to FlaA allele identification these 22 sequences are sequences from different strains based on nucleotide differences. Assuming from this finding that the 22 matches are independent strains that happen to have the same SVR6 sequence, the probe would identify them as identical strains when in fact they are genetically distinct. From this it can be calculated that SVR6 would have only a 2.4% chance (22/907 strains) of grouping unrelated strains as “identical,” giving an acceptable specificity of 97.6%. Using the Oxford database as a reference for diversity, ranking the specificity for the probes investigated resulted in the following order: SVR6 > SVR10 > SVR12 > SVR13 > SVR4. The poorest performers, SVR13 and SVR4, had specificities of 87.5 and 81.1%, respectively. A specificity below a confidence interval of 95% would be considered insufficient. However, the specificity depends on the diversity of the strains present in the sample set, and to date it is not known if the strains in the Oxford database display the same, more, or less genetic diversity than the diversity that can be expected in a poultry farm or slaughterhouse setting.
TABLE 3.
Experimental and predicted positive reactions of the SVR probes with 50 CampyNet strains
| Probe | No. of positive hits using BLAST analysis with Oxford database (%)a | No. of positive hits with 50 CampyNet strains
|
No. of CampyNet strains found to be positive (flaA allele identification) | Strain (origin) | |
|---|---|---|---|---|---|
| Expected | Experimentally confirmedb | ||||
| SVR4 | 171 (18.9) | 9 | 7 | 19 (526) | C. coli (chicken, The Netherlands) |
| 31 (ND)c | C. jejuni (human, The Netherlands) | ||||
| 45 (ND) | C. jejuni (human, Denmark) | ||||
| 56 (22) | C. jejuni (cattle, Denmark) | ||||
| 60 (ND) | C. jejuni (sheep, United Kingdom) | ||||
| 64 (ND) | C. coli (chicken, Denmark) | ||||
| 105 (37) | C. jejuni (sheep, United Kingdom) | ||||
| SVR6 | 22 (2.4) | 1 | 1 | 16 (222) | C. jejuni (chicken, The Netherlands) |
| SVR10 | 24 (2.6) | 1 | 2 | 22 (14) | C. jejuni (chicken, Denmark) |
| 32 (ND) | C. jejuni (human, The Netherlands) | ||||
| SVR12 | 25 (2.8) | 1 | 1 | 41 (372) | C. jejuni (human, Sweden) |
| SVR13 | 113 (12.5) | 6 | 2 | 14 (ND) | C. jejuni (chicken, The Netherlands) |
| 84 (67) | C. jejuni (chicken, Northern Ireland) | ||||
At the time of analysis, the Oxford database contained 907 entries.
The experiment was performed in triplicate.
ND, flaA allele identification not determined.
To validate this in silico approach, the number of matches in the Oxford database (January 2007) was used as a measure to calculate the expected number of positive results for a set of 50 strains in the CampyNet international strain set (19). These strains (8 Campylobacter coli strains and 42 C. jejuni strains) were selected because they were randomly collected from various host sources (human, sheep, and chickens), in nine European countries, and at different times. The number of strains that the SVR probes detected in this strain set was then determined experimentally. For four of five probes, the expected and confirmed numbers of positive results were comparable, whereas probe SVR13 detected less than the expected number of strains (Table 3). The deviation from expected scores could have been caused by differences in strain set bias between the Oxford database and the CampyNet strain collection. Seven of the CampyNet strains detected with the SVR probes were selected for flaA sequence typing. The resulting sequence data were subjected to a BLAST analysis with the Oxford database to obtain a flaA allele identification. All of the sequenced strains had different allele identifications (Table 3), and in all but one strain the probe sequence was conserved. The exception was CampyNet strain 19, which had a mismatch in one nucleotide of the probe (Fig. 5). Alignment of the sequence data showed that for all strains, the complete 40-nucleotide SVRs were identical (Fig. 5). The difference in the flaA allele was due to sequence variation outside this region.
FIG. 5.
Alignment of the SVR of selected CampyNet strains with the region of the probe match strain and probe identified. The data show that the regions for which the probes were designed are the same for these strains even though they are “unrelated” strains when they are classified by flaA sequence type. The sequences are from the strains indicated, detected with probes (from top to bottom) SVR4, SVR6, SVR10, SVR12, and SVR13.
Application of the probe technique to poultry transport crate wash tank water.
The transport crate wash tank water was culture positive for Campylobacter. Ten C. jejuni isolates were randomly selected from mCCDA plates containing directly plated wash water samples. The flaA genes of each strain were amplified by PCR and sequenced, and the sequences were aligned. Analysis of the sequences showed that in the 10 isolates there were two different strains (data not shown). Probe SVR6 matched one of these strains. For the other strain, a new probe was designed, SVR15 (5′-CGCCACTAGTTGAAATTCTTCC-3′; Tm, 54°C). When the probes were sequentially hybridized to colony lifts from the wash water samples, probes SVR6 and SVR15 detected different sets of colonies (Fig. 6). These results showed that each probe could detect specific strains in the naturally occurring mixture of strains present in a wash water sample. The number of colonies detected with the SVR probes was less than the total number detected by the 16S rRNA, suggesting that other strains were also present in the sample.
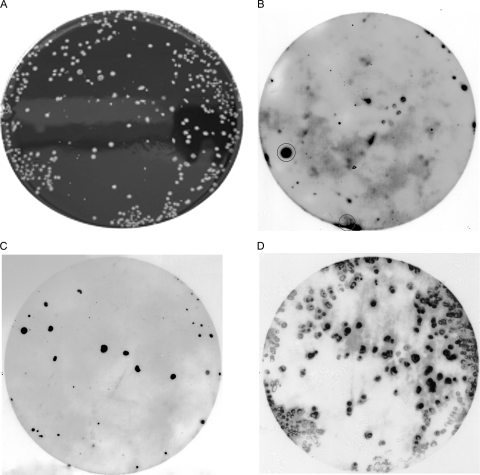
FIG. 6.
Sequential hybridization of membrane-bound colonies from poultry crate wash tank water samples. (A) Campylobacter colonies from the water sample on mCCDA. (B) Hybridization of colony lift with the first probe, SVR6. (C) Hybridization of colony lift with the second probe, SVR15, after the first probe was stripped with 0.1% boiling SDS. (D) Final hybridization with the 16S rRNA probe. Probes SVR6 and SVR15 detected different sets of colonies. The large spot and smear that are circled in panel B are background contamination.
DISCUSSION
Tracking any microorganism through the food chain is a difficult enough task. Determining the persistence and survival of a specific campylobacter strain from one flock on chicken carcasses during processing would be confounded by the multitude of other campylobacters present from previous flocks. In addition, populations are likely to shift during the production process as strains are variably resistant to various selection pressures. Methods for campylobacter strain differentiation, coupled with appropriate enumeration techniques suitable for multiple samples, are therefore essential. A number of genotyping techniques exploiting differences in the DNA sequences between strains, such as real-time PCR (14), restriction fragment length polymorphism typing (35), MLST (26), amplified fragment length polymorphism analysis (46), and PFGE (18, 24), have been developed for epidemiological investigations. The choice of the technique used depends predominately on the epidemiological questions posed, the sample investigated, and the sensitivity and specificity criteria used (49). None of these techniques allow enumeration of specific strains in a mixed population. Changes in the carriage of campylobacters during broiler and turkey processing investigated using PFGE and Fla typing have shown that some strains have a greater ability to survive in the slaughterhouse environment than other strains (3, 31, 37). To address the issue of strain persistence and survival during abattoir processing, a novel approach was developed, which combined the use of strain-specific and genus-specific hybridization probes to identify and thereby enumerate, using hybridization, colonies of one strain type recovered from environments containing mixed-strain populations.
This colony hybridization method has the advantage of being appropriate for application in real time and suitable for investigating a large number of isolates from each sample. In contrast, the previously described genotyping methods (e.g., PFGE and MLST) require individual strains to be isolated before analysis, which consumes resources and limits the number of isolates that can be investigated. The colony hybridization approach is economic, and the number of isolates investigated is limited only by the number of colonies distinguishable on a plate and the number of plates which can be handled. Moreover, this approach avoids the potential for preferential strain selection following enrichment.
The major objectives of this approach were (i) to detect and enumerate all campylobacters present in a potentially heavily microbiologically contaminated environmental sample and (ii) to identify the strain-specific campylobacters present in the sample. Achievement of these goals was dependent on the specificity of the genus-specific and strain-specific probes.
The genus-specific probe, based on the 16S rRNA, was highly specific, as demonstrated by in silico investigations and as shown in the laboratory using a number of species of the genus Campylobacter. Genetically related Arcobacter spp. and H. pullorum possibly present in the targeted environment were not detected. Our data substantiate the specificity of this probe previously reported by Schmid and colleagues (44), who developed it for florescence in situ hybridization and showed that it can be used with DIG labeling. The 16S rRNA probe also specifically enumerated Campylobacter spp. on selective plates, on which Campylobacter colonies can be indistinguishable from colonies of other microorganisms (particularly Arcobacter spp.). 16S rRNA has been shown to be a good target for identifying specific species and strains in a number of different labeling and colony hybridization systems. For example, Braun-Howland and colleagues (8) used radiolabeled 16S rRNA probes for identifying a range of common environmental organisms in water samples. A 16S rRNA probe that specifically detected Bifidobacterium adolescentis was used as an indicator of human fecal pollution (25), and radioactive isotope-labeled 16S rRNA probes have also been used for distinguishing Campylobacter fetus and Campylobacter hyointestinalis (50).
The strain-specific probes were designed for the SVR of the flaA gene. The genetic variability of flaA is well recognized and has been used previously for strain subtyping (28, 29); therefore, it is a good target for identifying and tracking a specific flock strain. The work reported here showed that after optimization of the hybridization and stringent wash conditions, individual SVR probes were highly specific, differentiating down to a single nucleotide difference in the target sequence, and sufficiently sensitive to detect colonies of a single strain in a mixture of strains. The results indicate that the selected 40-nucleotide region of the SVR is suitable for the development and selection of strain-specific probes. The theoretical sensitivity of the hybridization method, assuming that the technique detects 1 to 10 CFU per membrane in a 1-in-10 dilution of sample (i.e., 25 g of neck and breast skin in 250 ml of maximum recovery diluent), corresponds to about 1 in 1,000 CFU per g of skin. The sensitivity of the method determined experimentally by hybridizing probes to known concentrations of target DNA and the similar sensitivities of different probes when they are hybridized to the complementary sequences suggest that this level of sensitivity is appropriate for detecting specific strains in the heterogeneous environment of the abattoir. One possible issue with this method is the cross-reactivity of a probe with other strains in the abattoir that could have the same sequence in the same region of the SVR. However, the probability of this can be reduced by tracking only flock strains whose probes have high confidence intervals. Since the true diversity of Campylobacter populations likely to be encountered in the abattoir is unknown, confidence intervals can at best be assessed with reference to the extensive flaA allele sequence databases, such as that maintained at Oxford. Alternatively, a second probe could be developed for another variable region of the Campylobacter genome, such as porA; thus, the combined discriminatory power of two probes would increase the certainty that the same flock strain is being identified. As this method is flexible, in that the same membranes can be probed sequentially with different probes, detection of the flaA SVR could be followed by detection of porA.
The DIG labeling technique chosen was also shown to be suitable for short oligonucleotide probes, providing sufficient sensitivity to detect 8 ng of genomic material, which is equivalent to 106 molecules or the average number of cells in a single colony. The DIG system also has advantages over radioactive labeling, such as shorter exposure times and improved safety since nonhazardous materials are used. In addition, the probes are reusable, easily stored, and stripped from the membranes. Previous researchers have used DIG-labeled probes for the detection of Staphylococcus aureus (33) and Yersinia enterocolitica in foods (16). Similarly, randomly primed DIG labeling of a probe specific for C. jejuni subsp. jejuni and C. jejuni subsp. doylei, but not other Campylobacter or Arcobacter spp., was also used to directly enumerate organisms in food and chicken samples (32). DIG labeling of four different probes used in colony hybridization analyses for enumeration of Legionella was found to be equivalent to conventional plating and could detect Legionella colonies obscured by growth of contaminating organisms (43).
The hypothesis examined in this study was that a novel technique could be developed to track and enumerate at various sites in the abattoir during processing specific Campylobacter strains from individual conventionally reared broiler flocks. The proof of this principle was obtained using samples of the wastewater from washing poultry transport crates, which are generally contaminated by several strains (47). In this study, two different strains were identified and enumerated using two strain-specific SVR probes. Therefore, this approach appears to be feasible for studies of the abattoir environment, but it might also be appropriate in other relevant situations. For example, molecular epidemiological investigations at the farm have highlighted the genetic diversity of campylobacters in broilers, broiler houses, and the outdoor environment. Such studies generally aim to identify the sources of infection and the extent and persistence of cross-contamination (30, 41). However, to date, a specific source has not been identified, and evidence indicates that pets, insects, water, and neighboring poultry houses are involved (9, 22, 23), but previous flocks in the same house are not involved (22, 45). Using strain-specific detection approaches may provide information to improve and target intervention strategies on the farm.
In conclusion, the novel method developed, using genus- and strain-specific oligonucleotide probes in combination with colony hybridization, has been shown to be appropriate for the tracking and enumeration of a specific flock strain during abattoir processing. This method is now being used to determine the extent and persistence of individual Campylobacter strains in the abattoir. Such information is necessary to improve the accuracy of quantitative risk assessment models and to develop strategies to minimize cross-contamination during the postharvest period. Finally, the specificity and sensitivity of this approach, as demonstrated in this study, suggest that similar strategies could be effectively used in other environments and with other organisms, where the identification of single strains in mixed populations would be advantageous.
Acknowledgments
This work was financially supported by the Food Standards Agency (project code MO1037).
We thank Rick Meinersmann for helpful discussions and the University of Dundee for its sequencing service.
Footnotes
Published ahead of print on 15 February 2008.
REFERENCES
- 1.Allen, V. M., S. A. Bull., J. E. L. Corry, G. Dominque, F. Jørgensen, J. A. Frost, R. Whyte, A. Gonzalez, N. Elviss, and T. J. Humphrey. 2007. Campylobacter spp. contamination of chicken carcasses during processing in relation to flock colonisation. Int. J. Food Microbiol. 113:54-61. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., P. Guerry, and T. J. Trust. 1993. Distribution and polymorphism of the flagellin genes from isolates of Campylobacter coli and Campylobacter jejuni. J. Bacteriol. 175:3051-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alter, T., F. Gaull, A. Froeb, and K. Fehlhaber. 2005. Distribution of Campylobacter jejuni strains at different stages of a turkey slaughter line. Food Microbiol. 22:345-351. [Google Scholar]
- 4.Ayling, R. D., M. J. Woodward, S. Evans, and D. G. Newell. 1996. Restriction fragment length polymorphism of polymerase chain reaction products applied to the differentiation of poultry campylobacters for epidemiological investigations. Res. Vet. Sci. 60:168-172. [DOI] [PubMed] [Google Scholar]
- 5.Berndtson, E., M. L. Danielsson-Tham, and A. Engvall. 1996. Campylobacter incidence on a chicken farm and the spread of Campylobacter during the slaughter process. Int. J. Food Microbiol. 32:35-47. [DOI] [PubMed] [Google Scholar]
- 6.Berndtson, E., U. Emanuelson, A. Engvall, and M. L. Danielsson-Tham. 1996. A 1-year epidemiological study of campylobacters in 18 Swedish chicken farms. Prev. Vet. Med. 26:167-185. [Google Scholar]
- 7.Berrang, M. E., R. J. Buhr, J. A. Cason, and J. A. Dickens. 2001. Broiler carcass contamination with Campylobacter from feces during defeathering. J. Food Prot. 64:2063-2066. [DOI] [PubMed] [Google Scholar]
- 8.Braun-Howland, E. B., P. A. Vescio, and S. A. Nierzwicki-Bauer. 1993. Use of a simplified cell blot technique and 16S rRNA-directed probes for identification of common environmental isolates. Appl. Environ. Microbiol. 59:3219-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bull., S. A., V. M. Allen, G. Domingue, F. J.ørgensen, J. A. Frost, R. Ure, R. Whyte, D. Tinker, J. E. L. Corry, J. Gillard-King, and T. J. Humphrey. 2006. Sources of Campylobacter spp. colonizing housed broiler flocks during rearing. Appl. Environ. Microbiol. 72:645-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnens, A. P., J. Wagner, H. Lior, J. Nicolet, and J. Frey. 1995. Restriction fragment length polymorphisms among the flagellar genes of the Lior heat-labile serogroup reference strains and field strains of Campylobacter jejuni and C. coli. Epidemiol. Infect 114:423-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callicott, K., K. L. Hiett, N. J. Stern, J. Reiersen, E. Berndtson, V. Fridricksdottir, E. Gunnarsson, F. Georgsson, and R. Lowan. 2005. Abstr. 105th Gen. Meet. Am. Soc. Microbiol., abstr. D-209.
- 12.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colles, F. M., K. Jones, R. M. Harding, and M. C. Maiden. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Debretsion, A., T. Habtemariam, S. Wilson, D. Nganwa, and T. Yehualaeshet. 2007. Real-time PCR assay for rapid detection and quantification of Campylobacter jejuni on chicken rinses from poultry processing plant. Mol. Cell. Probes 21:177-181. [DOI] [PubMed] [Google Scholar]
- 15.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durisin, M. D., A. Ibrahim, and M. W. Griffiths. 1997. Detection of pathogenic Yersinia enterocolitica in milk and pork using a DIG-labelled probe targeted against the yst gene. Int. J. Food Microbiol. 37:103-112. [DOI] [PubMed] [Google Scholar]
- 17.Evans, S. J., and A. R. Sayers. 2000. A longitudinal study of campylobacter infection of broiler flocks in Great Britain. Prev. Vet. Med. 46:209-223. [DOI] [PubMed] [Google Scholar]
- 18.Ge, B., W. Girard, S. Zhao, S. Friedman, S. A. Gaines, and J. Meng. 2006. Genotyping of Campylobacter spp. from retail meats by pulsed-field gel electrophoresis and ribotyping. J. Appl. Microbiol. 100:175-184. [DOI] [PubMed] [Google Scholar]
- 19.Harrington, C. S., L. Moran, A. M. Ridley, D. G. Newell, and R. H. Madden. 2003. Inter-laboratory evaluation of three flagellin PCR/RFLP methods for typing Campylobacter jejuni and C. coli: the CAMPYNET experience. J. Appl. Microbiol. 95:1321-1333. [DOI] [PubMed] [Google Scholar]
- 20.Herman, L., M. Heyndrickx, K. Grijspeerdt, D. Vandekerchove, I. Rollier, and L. De Zutter. 2003. Routes for Campylobacter contamination of poultry meat: epidemiological study from hatchery to slaughterhouse. Epidemiol. Infect. 131:1169-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiett, K. L., N. J. Stern, P. Fedorka-Cray, N. A. Cox, M. T. Musgrove, and S. Ladely. 2002. Molecular subtype analyses of Campylobacter spp. from Arkansas and California poultry operations. Appl. Environ. Microbiol. 68:6220-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs-Reitsma, W. F., A. W. van de Giessen, N. M. Bolder, and R. W. A. W. Mulder. 1995. Epidemiology of Campylobacter spp. at two Dutch broiler farms. Epidemiol. Infect. 114:413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnsen, G., H. Kruse, and M. Hofshagen. 2006. Genetic diversity and description of transmission routes for Campylobacter on broiler farms by amplified-fragment length polymorphism. J. Appl. Microbiol. 101:1130-1139. [DOI] [PubMed] [Google Scholar]
- 24.Lienau, J.-A., L. Ellerbroek, and G. Klein. 2007. Tracing flock-related Campylobacter clones from broiler farms through slaughter to retail products by pulsed-field gel electrophoresis. J. Food Prot. 70:536-542. [DOI] [PubMed] [Google Scholar]
- 25.Lynch, P. A., B. J. Gilpin, L. W. Sinton, and M. G. Savill. 2002. The detection of Bifidobacterium adolescentis by colony hybridization as an indicator of human faecal pollution. J. Appl. Microbiol. 92:526-533. [DOI] [PubMed] [Google Scholar]
- 26.Manning, G., C. G. Dowson, M. C. Bagnall, I. H. Ahmed, M. West, and D. G. Newell. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meinersmann, R. J., L. O. Helsel, P. I. Fields, and K. L. Hiett. 1997. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J. Clin. Microbiol. 35:2810-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meinersmann, R. J., R. W. Phillips, K. L. Hiett, and P. Fedorka-Cray. 2005. Differentiation of campylobacter populations as demonstrated by flagellin short variable region sequences. Appl. Environ. Microbiol. 71:6368-6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newell, D. G., and C. Fearnley. 2003. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69:4343-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newell, D. G., J. E. Shreeve, M. Toszeghy, G. Domingue, S. Bull, T. Humphrey, and G. Mead. 2001. Changes in the carriage of Campylobacter strains by poultry carcasses during processing in abattoirs. Appl. Environ. Microbiol. 67:2636-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng, L. K., C. I. Kingombe, W. Yan, D. E. Taylor, K. Hiratsuka, N. Malik, and M. M. Garcia. 1997. Specific detection and confirmation of Campylobacter jejuni by DNA hybridization and PCR. Appl. Environ. Microbiol. 63:4558-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padmapriya, B. P., A. Ramesh, A. Chandrashekar, and M. C. Varadaraj. 2003. Staphylococcal accessory gene regulator (sar) as a signature gene to detect enterotoxigenic staphylococci. J. Appl. Microbiol. 95:974-981. [DOI] [PubMed] [Google Scholar]
- 34.Perelle, S., M. Josefsen, J. Hoorfar, F. Dilasser, J. Grout, and P. Fach. 2004. A LightCycler real-time PCR hybridization probe assay for detecting food-borne thermophilic Campylobacter. Mol. Cell. Probes 18:321-327. [DOI] [PubMed] [Google Scholar]
- 35.Petersen, L., and D. G. Newell. 2001. The ability of Fla-typing schemes to discriminate between strains of Campylobacter jejuni. J. Appl. Microbiol. 91:217-224. [DOI] [PubMed] [Google Scholar]
- 36.Petersen, L., E. M. Nielsen, and S. L. On. 2001. Serotype and genotype diversity and hatchery transmission of Campylobacter jejuni in commercial poultry flocks. Vet. Microbiol. 82:141-154. [DOI] [PubMed] [Google Scholar]
- 37.Rasschaert, G., K. Houf, J. V. Hende, and L. D. Zutter. 2006. Campylobacter contamination during poultry slaughter in Belgium. J. Food Prot. 69:27-33. [DOI] [PubMed] [Google Scholar]
- 38.Rivoal, K., C. Ragimbeau, G. Salvat, P. Colin, and G. Ermel. 2005. Genomic diversity of Campylobacter coli and Campylobacter jejuni isolates recovered from free-range broiler farms and comparison with isolates of various origins. Appl. Environ. Microbiol. 71:6216-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts, J. A., P. Cumberland, P. N. Sockett, J. Wheeler, L. C. Rodrigues, D. Sethi, and P. J. Roderick. 2003. The study of infectious intestinal disease in England: socio-economic impact. Epidemiol. Infect. 130:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenquist, H., H. M. Sommer, N. L. Nielsen, and B. B. Christensen. 2006. The effect of slaughter operations on the contamination of chicken carcasses with thermotolerant Campylobacter. Int. J. Food Microbiol. 108:226-232. [DOI] [PubMed] [Google Scholar]
- 41.Sahin, O., T. Y. Morishita, and Q. Zhang. 2002. Campylobacter colonization in poultry: sources of infection and modes of transmission. Anim. Health Res. Rev. 3:95-105. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Satoh, W., M. Nakata, H. Yamamoto, T. Ezaki, and K. Hiramatsu. 2002. Enumeration of Legionella CFU by colony hybridization using specific DNA probes. Appl. Environ. Microbiol. 68:6466-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmid, M. W., A. Lehner, R. Stephan, K.-H. Schleifer, and H. Meier. 2005. Development and application of oligonucleotide probes for in situ detection of thermotolerant Campylobacter in chicken faecal and liver samples. Int. J. Food Microbiol. 105:245-255. [DOI] [PubMed] [Google Scholar]
- 45.Shreeve, J. E., M. Toszeghy, A. Ridley, and D. G. Newell. 2002. The carry-over of Campylobacter isolates between sequential poultry flocks. Avian Dis. 46:378-385. [DOI] [PubMed] [Google Scholar]
- 46.Siemer, B. L., C. S. Harrington, E. M. Nielsen, B. Borck, N. L. Nielsen, J. Engberg, and S. L. W. On. 2004. Genetic relatedness among Campylobacter jejuni serotyped isolates of diverse origin as determined by numerical analysis of amplified fragment length polymorphism (AFLP) profiles. J. Appl. Microbiol. 96:795-802. [DOI] [PubMed] [Google Scholar]
- 47.Slader, J., G. Domingue, F. Jorgensen, K. McAlpine, R. J. Owen, F. J. Bolton, and T. J. Humphrey. 2002. Impact of transport crate reuse and of catching and processing on Campylobacter and Salmonella contamination of broiler chickens. Appl. Environ. Microbiol. 68:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi, R., F. Shahada, T. Chuma, and K. Okamoto. 2006. Analysis of Campylobacter spp. contamination in broilers from the farm to the final meat cuts by using restriction fragment length polymorphism of the polymerase chain reaction products. Int. J. Food Microbiol. 110:240-245. [DOI] [PubMed] [Google Scholar]
- 49.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wesley, I. V., R. D. Wesley, M. Cardella, F. E. Dewhirst, and B. J. Paster. 1991. Oligodeoxynucleotide probes for Campylobacter fetus and Campylobacter hyointestinalis based on 16S rRNA sequences. J. Clin. Microbiol. 29:1812-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wuyts, J., G. Perrière, and Y. V. D. Peer. 2004. The European ribosomal RNA database. Nucleic Acids Res. 32:D101-D103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan, W., N. Chang, and D. E. Taylor. 1991. Pulsed-field gel electrophoresis of Campylobacter jejuni and Campylobacter coli genomic DNA and its epidemiologic application. J. Infect. Dis. 163:1068-1072. [DOI] [PubMed] [Google Scholar]
- 53.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]