Abstract
We previously published a genetic map of Gibberella zeae (Fusarium graminearum sensu lato) based on a cross between Kansas strain Z-3639 (lineage 7) and Japanese strain R-5470 (lineage 6). In this study, that genetic map was aligned with the third assembly of the genomic sequence of G. zeae strain PH-1 (lineage 7) using seven structural genes and 108 sequenced amplified fragment length polymorphism markers. Several linkage groups were combined based on the alignments, the nine original linkage groups were reduced to six groups, and the total size of the genetic map was reduced from 1,286 to 1,140 centimorgans. Nine supercontigs, comprising 99.2% of the genomic sequence assembly, were anchored to the genetic map. Eight markers (four markers from each parent) were not found in the genome assembly, and four of these markers were closely linked, suggesting that >150 kb of DNA sequence is missing from the PH-1 genome assembly. The alignments of the linkage groups and supercontigs yielded four independent sets, which is consistent with the four chromosomes reported for this fungus. Two proposed heterozygous inversions were confirmed by the alignments; otherwise, the colinearity of the genetic and physical maps was high. Two of four regions with segregation distortion were explained by the two selectable markers employed in making the cross. The average recombination rates for each chromosome were similar to those previously reported for G. zeae. Despite an inferred history of genetic isolation of lineage 6 and lineage 7, the chromosomes of these lineages remain homologous and are capable of recombination along their entire lengths, even within the inversions. This genetic map can now be used in conjunction with the physical sequence to study phenotypes (e.g., fertility and fitness) and genetic features (e.g., centromeres and recombination frequency) that do not have a known molecular signature in the genome.
Gibberella zeae (Schwein.) Petch (anamorph, Fusarium graminearum Schwabe sensu lato) is the most important causal agent of Fusarium head blight of cereals (27). Fusarium head blight has recently reemerged worldwide as a devastating disease of wheat and barley (15, 32). In addition to direct yield losses, Fusarium head blight reduces grain quality (42), and infected seed often is contaminated with mycotoxins, such as nivalenol, deoxynivalenol, and zearalenone (10, 21). In the United States, the direct and secondary monetary losses due to this disease from 1998 to 2000 were estimated to be $2.7 billion (34).
G. zeae is a homothallic (i.e., selfing) ascomycete fungus (12), but it also can outcross. For genetic studies, special methods are required to distinguish selfed progeny from outcrossed progeny (6). Bowden and Leslie (5) developed a laboratory outcrossing method using nitrate-non utilizing (nit) mutants of different G. zeae strains. Lee et al. (23) overcame the problem of homothallism in crosses by partially disabling the mating type (MAT) genes. In addition to outcrossing under laboratory conditions, high levels of genotypic heterogeneity in field populations of this fungus suggest that outcrossing occurs in nature (4, 47).
O'Donnell et al. (35) constructed a phylogeny for G. zeae based on the sequences of six genes and established seven phylogenetic lineages. Subsequent studies with more strains and more sequenced loci extended the number of lineages to 11, and the lineages were elevated to phylogenetic species status (36, 44, 46). The phylogenetic species are morphologically cryptic and were distinguished based on DNA sequence polymorphisms. Members of some lineages are cross-fertile with strains of other lineages, which suggests that the members of all of the lineages may belong to a single biological species (5).
The genome of G. zeae lineage 7 strain PH-1 was sequenced at the Broad Institute (Cambridge, MA), and an initial analysis has been published (9). The current (third) genome assembly has a total length of 36.4 Mb and contains >13,000 annotated genes, 433 >2-kb contigs, and 31 supercontigs (SCs), with nearly 11× genome coverage (http://www.broad.mit.edu). In addition, G. zeae lineage 7 strain Z-3639 has been partially sequenced with 24,072 shotgun reads representing 0.4× genome coverage (http://www.broad.mit.edu).
A genetic map of G. zeae (19) was constructed from an outcross between nit mutants of strain Z-3639 and G. zeae lineage 6 strain R-5470. This map, referred to below as the Jurgenson map, was saturated with 1,048 polymorphic markers that mapped to 468 unique loci on nine linkage groups (LGs). The total map length was 1,286 centimorgans (cM), and the average interval between loci was 2.8 map units. The map was used to locate the gene controlling toxin type (deoxynivalenol or nivalenol) near the trichodiene synthetase gene (TRI5) on LG 1. These genetic conclusions were confirmed by Lee et al. (25), who showed that TRI13 determines which toxin type is produced and that this locus is located in the trichothecene cluster near TRI5. Alexander et al. (1) used the map to locate Tri15 on LG 2, and Desjardins et al. (11) used the map to locate a diagnostic SCAR marker locus on LG 5. The genetic map also was used to map loci for pathogenicity and aggressiveness of G. zeae toward wheat (8). A single quantitative trait locus for aggressiveness of G. zeae was detected near the TRI5 locus, and this quantitative trait locus was hypothesized to reflect the differential toxicity of deoxynivalenol and nivalenol. A pathogenicity locus (PATH1) segregated qualitatively and was tightly linked to PER1 (a locus for perithecial production), PIG1 (a locus for red pigmentation), and TOX1 (a locus for the amount of toxin produced).
Several questions were raised by the Jurgenson genetic map. (i) Why do five of the nine LGs show segregation distortion? (ii) Is recombination generally suppressed in this cross? (iii) Are unusual haplotype regions in two of the LGs large heterozygous inversions? (iv) Are PATH1, PER1, PIG1, and TOX1 the same locus? (v) Can the proposed locations of the nit1 and nit3 selectable markers be confirmed?
A second map of G. zeae (14), referred to below as the Gale map, also has been published and was based on a cross between two closely related G. zeae strains, PH-1 and 00-676. This map has 235 loci with nine LGs, and the total length is 1,234 cM. One hundred sixty-eight sequence-tagged markers, many based on the genomic sequence, were used to align the Gale map with the first assembly of the genomic sequence of G. zeae. The nine LGs aligned with 22 SCs and were combined into four sets that were postulated to represent four chromosomes and to account for 99.8% of the first genome sequence assembly (14). Most of the linkages and alignments were well supported, although some depended on one or two markers. Linkage maps are inherently subject to scoring errors, statistical ambiguities, and differences in distance and/or order depending on the markers included (14). The assembly of sequence scaffolds also is subject to various errors and assumptions. Since the Gale map and the genome assembly were developed jointly (14), independent verification of their sequence is needed. The Jurgenson map was developed independently of the genome sequence and can be used to evaluate the alignment of the physical genome with the Gale map. This evaluation can be made even though the Jurgenson and Gale maps cannot be compared directly because they share no genetic markers. However, the two maps can be compared indirectly by using the genomic sequence assembly as an intermediary.
Here we report alignment of the Jurgenson map with the third genomic DNA sequence assembly of strain PH-1. Our objectives in this study were (i) to validate the genetic map of G. zeae Z-3639 × R-5470 (19) and resolve questions arising from this map, (ii) to verify the proposed sequence assembly and the alignments of LGs and SCs into chromosomes, and (iii) to compare the Jurgenson map with the Gale map.
MATERIALS AND METHODS
Genetic map and fungal strains.
We used the genetic map of G. zeae based on the cross of Kansas lineage 7 strain Z-3639 (= NRRL 29169) and Japanese lineage 6 strain R-5470 (19). The parents and 99 progeny (FGSC 8632 to FGSC 8732) are available from the Fungal Genetic Stock Center (School of Biological Sciences, University of Missouri-Kansas City, Kansas City, MO; http://www.fgsc.net). Strain PH-1 (= FGSC 9075) was obtained from Frances Trail, Michigan State University.
DNA protocols.
DNA was extracted by using a cetyltrimethylammonium bromide procedure (28). The DNA concentration was determined with a ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and adjusted to 20 ng/μl for PCRs. Standard procedures were used for restriction enzyme digestion, agarose gel electrophoresis, and Southern hybridization (41).
Isolation and reamplification of AFLP markers.
The protocol of Shan et al. (43) was modified slightly to amplify amplified fragment length polymorphism (AFLP) markers already placed on the genetic map. For each primer pair, AFLP gels were run following PCR amplification of sequences from the genomic DNA of G. zeae strains Z-3639, R-5470, and PH-1. Polymorphic bands were correlated with markers from the map. A dried AFLP gel and the autoradiograph obtained from it were aligned, and needles were used to mark the polymorphic fragments. The portion of the gel containing the targeted DNA was excised with a razor blade. Each gel slice was placed in a PCR tube with 20 μl of sterile water and incubated overnight at 4°C. Two microliters of each sample was used for reamplification with an MJ PTC-200 thermocycler (MJ Research, Watertown, MA). The reaction was performed using a 30-μl (total volume) mixture as follows: 1 min at 94°C, followed by 30 cycles of 30 s at 94°C, 1 min at 56°C, and 1 min at 72°C and then a final extension at 72°C for 10 min. The corresponding unlabeled selective AFLP primers were used to prime the reaction. A 10-μl aliquot of each PCR product was resolved on a 1.5% agarose gel to estimate the band size and number.
Sequencing of AFLP fragments and selection of clones.
Thirty-one reamplified PCR products were sequenced directly with an ABI 3700 DNA analyzer at the DNA sequencing facility at Kansas State University after purification with a PCR clean-up system (Promega, Madison, WI). Ninety-eight fragments were cloned into a pGEM-T Easy vector (Promega). Three clones were selected and sequenced for each of these 98 fragments.
For selected markers, we designed extended AFLP primers with three additional bases on each end to confirm the presence of polymorphism in the progeny. The PCRs were performed under the conditions described above with an annealing temperature of 59 to 64°C depending on the primer pair.
The program AFLPinSilico (40) was used with the G. zeae PH-1 genomic database to generate virtual AFLP marker bands. Sequenced clones were compared with bands from the virtual AFLP analysis to confirm their presence and size. Cloning of AFLP fragments revealed a systematic bias in the previous marker size estimates (19). The corrected size as a function of the previous estimate was determined by using the equation y = 0.9107x + 28.843 (R2 = 0.9831).
Mapping of nuclear genes.
The following seven structural genes were mapped: the genes encoding an elongation factor (EF-1α), phosphate permease (PHO), trichodiene synthetase (TRI5), trichothecene 3-O-acetyltransferase (TRI101), a regulator of trichothecene biosynthesis (TRI15), a putative reductase (RED), and mitogen-activated protein kinase (MGV1). TRI5 was mapped previously by using restriction fragment length polymorphisms (19). Six genes were mapped by using cleaved amplified polymorphic sequences. Each gene was amplified from both parental strains with specific primers. Except for the MGV1 primer sets, all of the primers used were primers described by O'Donnell et al. (35). The primers for MGV1 were designed for this study (forward primer, 5′-CACAAATACACCATGGGCGACCTA-3′; reverse primer, 5′-GACACCCTGGCCCTGAAGACCTC-3′) and synthesized by IDT (Coralville, IA). Amplified DNA fragments were sequenced after purification with a PCR clean-up system (Promega). Sequences from both parental strains were compared to identify polymorphic restriction sites. Cleaved amplified polymorphic sequence markers for each gene were scored for all 99 progeny of the mapping population used to construct the genetic map (19). The genes were placed on the genetic map using Map Manager QTX11 (http://mapmgr.Roswellpark.org/mm PTX) and a Macintosh G4 Power PC computer (31).
Location of nit1 and nit3.
To identify nit1 and nit3 of G. zeae in the genomic database, nit-3 (structural gene for nitrate reductase) and nit-4 (gene for nitrate assimilation transcription factor) from Neurospora crassa, the homologs of nit1 and nit3 in Fusarium (22), respectively, were used to search the Fusarium group database (www.broad.mit.edu). Candidate genes were deleted by split marker recombination (7), and deletion mutants were characterized on phenotyping media for nit mutants (28).
Alignment of maps.
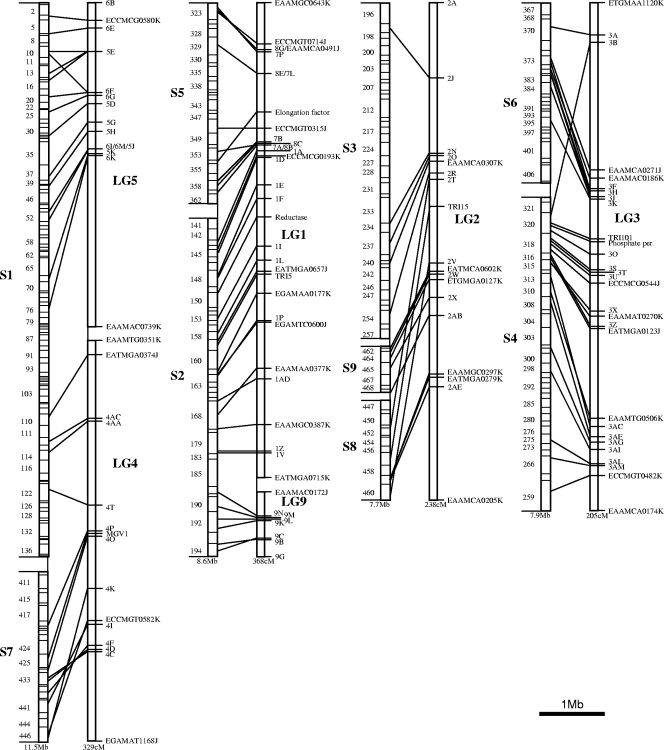
The sequenced genetic markers were found in the third genome assembly of G. zeae with BLASTN (2), and the genetic map and the genome assembly were aligned manually. Average recombination rates were obtained by dividing the total linkage distance (in cM) by the total physical length (in Mb) for each chromosome. The estimates were not adjusted for gaps between LGs or SCs or for differences in marker density. Chromosome regions with high or low recombination rates were identified by comparing physical and genetic distances (Fig. 1 and Table 1).
FIG. 1.
Alignment between physical (left) and genetic (right) maps of G. zeae. SCs from the third genome assembly are designated S1 to S9. The LGs on the genetic map are designated LG1 to LG9. The original LG 6 was combined with LG 5; LG 7 and LG 8 were combined with LG 1. LGs were drawn using a different scale for each chromosome to facilitate views of alignment. Markers at the ends of LGs are shown for reference. Marker information is shown in Table 2. phosphate per, phosphate permease.
TABLE 1.
Markers sequenced in this study and their locations in the third F. graminearum genome assembly
| Markera | Location on genetic map (cM) | Origin
|
Size (bp) | SC | Position | ||
|---|---|---|---|---|---|---|---|
| R-5470 | Z-3639 | PH-1 | |||||
| Chromosome 1 | |||||||
| EATMCA0383J | 6A (0.0) | + | − | − | 291 | No hits | |
| ECCMCG0580K | 6 (10.1) | − | + | +b | 422 | 1 | 237025-237445 |
| EAAMTG0323J | 6E (14.1) | + | − | − | 266 | No hits | |
| EATMCG0731K | 6E (14.1) | − | +b | + | 489 | 1 | 535968-536430 |
| EAAMAA0090J | 5E (28.0) | + | − | − | 65 | 1 | 840115-840179 |
| EAAMGC0435J | 5E (28.0) | + | − | − | 392 | 1 | 1139718-1140109 |
| EAAMCA0404J | 5E (28.0) | + | − | − | 373 | 1 | 1250068-1250438 |
| EAAMAC0295K | 6F (35.0) | − | +b | + | 239 | 1 | 840115-840353 |
| EAAMCA0557Jc | 6F (35.0) | + | − | − | 466 | 1 | 1600947-1601333 |
| EAAMAA0717J | 6G (36.0) | + | − | − | 610 | 1 | 1885126-1885671 |
| EGAMAT0721J | 5D (38.0) | + | − | − | 614 | 1 | 2144020-2144633 |
| EAAMCA0246J | 5G (47.0) | − | +b | + | 201 | 1 | 2618374-2618574 |
| EAAMAG0388K | 5H (51.7) | − | +b | + | 349 | 1 | 3024847-3025195 |
| EAAMAT0097J | 5J (59.7) | + | − | − | 62 | No hits | Exd |
| EAAMGC0111K | 5J (59.7) | − | +b | + | 81 | 1 | 3383282-3383362 |
| EATMCA0352J | 6M (59.7) | + | − | − | 223 | 1 | 3513107-3513327 |
| ECCMGT0362K | 6I (59.7) | − | + | +b | 243 | 1 | 3544782-3545025 |
| EATMCG0165Kc | 5K (61.7) | − | + | +b | 84 | 1 | 4490384-4490412 |
| EAAMCC0116J | 6K (62.7) | + | − | − | 82 | 1 | 4905538-4905617 |
| EATMGA0374J | 4 (6.0) | + | − | − | 327 | 1 | 6908379-6908705 |
| ECCMGT0614K | 4AC (36.6) | − | + | − | 505 | 1 | 7073041-7073543 |
| EATMGA0676K | 4AA (38.6) | − | + | +b | 503 | 1 | 7239867-7240369 |
| EAAMAC0217J | 4T (77.0) | + | − | − | 183 | 1 | 7935713-7935891 |
| EAAMAT0192K | 4P (92.0) | − | +b | + | 160 | 7 | 924207-924366 |
| MGV1 | MGV1 (94.0) | 7 | 1486778-1488707 | ||||
| EATMCA0305J | 4O (95.0) | + | − | − | 195 | 7 | 1566369-1566546 |
| EGAMTA0339Jc | 4K (112.1) | + | − | − | 280 | 7 | 2526164-2526276 |
| ECCMGT0582K | 4 (136.2) | − | + | − | 478 | 7 | 2536241-2536718 |
| EATMCA0199J | 4I (138.2) | + | − | − | 94 | 7 | 2259055-2259146 |
| EGAMAT0378Kc | 4F (148.3) | − | + | − | 361 | 7 | 1959874-1959920 |
| EGAMAA0344J | 4D (150.3) | + | − | − | 289 | 7 | 1767528-1767815 |
| EAAMGC0119K | 4C (151.3) | − | +b | + | 87 | 7 | 1706519-1706605 |
| Chromosome 2 | |||||||
| ECCMGT0714J | 8 (28.2) | + | − | − | 461 | 5 | 3023758-3024213 |
| EAAMAC0350K | 8G (32.2) | − | + | − | 294 | 5 | 3000989-3001282 |
| EAAMCA0491Jc | 7 (32.2) | + | − | − | 422 | 5 | 3044032-3044370 |
| EAAMAG0158K | 7P (33.2) | − | +b | + | 121 | 5 | 3073233-3064348 |
| EAAMGC0320J | 7L (47.2) | + | − | − | 263 | 5 | 2476839-2477096 |
| Elongation factor gene | Elongation factor gene (72.9) | 5 | 531518-532161 | ||||
| ECCMGT0315J | 7 (84.2) | + | − | − | 257 | 5 | 1360303-1360558 |
| EAAMAG0123K | 7B (93.5) | − | + | − | 90 | No hits | |
| EAAMAA0675Kc | 7B (93.5) | − | + | − | 559 | 5 | 356549-356977 |
| EGAMTC0740K | 8C (94.5) | − | + | +b | 646 | 5 | 284175-284812 |
| EGAMAT0181J | 8B (95.5) | + | − | − | 144 | 5 | 790865-791004 |
| EAAMCC0237J | 7A (95.5) | − | +b | + | 196 | 5 | 195828-196020 |
| EAAMTG0298J | 1A (99.5) | + | − | − | 240 | 2 | 5047963-5048198 |
| ECCMCG0193K | 1 (102.5) | − | + | +b | 163 | 2 | 4897838-4898000 |
| EAAMAT0263K | 1D (103.5) | − | +b | + | 214 | 2 | 4882860-4883073 |
| EATMCG0329J | 1E (122.5) | + | − | − | 271 | 2 | 4539360-4539704 |
| ECCMGT0168J | 1F (131.6) | + | − | − | 74 | 2 | 4534991-4535047 |
| Reductase gene | Reductase gene (144.6) | 2 | 4221356-4222230 | ||||
| EAAMAC0392K | 1I (164.2) | − | +b | + | 355 | 2 | 3837149-3837503 |
| EATMCA0327K | 1I (164.2) | − | + | +b | 266 | 2 | 3827713-3827978 |
| EAAMAA0347J | 1L (174.4) | + | − | − | 290 | 2 | 3607835-3608127 |
| EATMGA0657J | 1 (182.6) | + | − | − | 480 | 2 | 3402701-3403180 |
| TRI5 | TRI5 (183.6) | 2 | 3376567-3377751 | ||||
| EGAMAA0177K | 1 (197.4) | − | + | − | 139 | 2 | 2929953-2930089 |
| EAAMCA0294K | 1P (216.3) | − | +b | + | 237 | 2 | 2833524-2833760 |
| EGAMTC0600J | 1 (217.3) | + | − | − | 496 | 2 | 2889043-2889532 |
| EAAMAA0377K | 1 (249.5) | − | +b | + | 329 | 2 | 2354068-2354396 |
| EAAMTG0185K | 1AD (256.6) | − | + | − | 151 | 2 | 2505292-2505438 |
| EAAMGC0387K | 1 (289.1) | − | +b | + | 354 | 2 | 2001562-2001915 |
| EAAMAG0727K | 1Z (307.5) | − | +b | + | 624 | 2 | 1817487-1818110 |
| EAAMCA0283J | 1Z (307.5) | + | − | − | 230 | 2 | 1817882-1818110 |
| EATMCA0194K | 1V (308.5) | − | + | +b | 163 | 2 | 1676958-1677120 |
| ECCMGT0347J | 9N (17.0) | + | − | − | 229 | No hits | Ex |
| EGAMAT0360J | 9N (17.0) | + | − | − | 307 | 2 | 1096103-1096409 |
| ECCMAT0288J | 9M (18.0) | + | − | − | 180 | 2 | 764688-764863 |
| EGAMTC0432K | 9L (19.0) | − | + | − | 384 | 2 | 646379-646759 |
| ECCMGT0446J | 9K (20.0) | + | − | − | 332 | 2 | 465211-465522 |
| EAAMCA0680J | 9D (30.0) | + | − | − | 578 | No hits | |
| EAAMAT0100K | 9C (32.0) | − | +b | + | 72 | 2 | 90866-90937 |
| EATMCA0149K | 9B (33.0) | − | + | +b | 107 | 2 | 200829-200935 |
| Chromosome 3 | |||||||
| ETGMGA0314K | 2H (33.0) | − | + | − | 259 | No hits | |
| EAAMGC0142J | 2H (33.0) | + | − | − | 107 | No hits | |
| EATMGA0624K | 2I (35.0) | − | + | − | 508 | No hits | |
| EAAMAT0513K | 2J (36.0) | − | + | − | 430 | No hits | |
| EATMCG0537K | 2J (36.0) | − | + | +b | 445 | 3 | 360587-361031 |
| EATMCA0309K | 2N (72.0) | − | +b | + | 250 | 3 | 3634867-3635114 |
| EAAMAA0185J | 2O (73.0) | + | − | − | 154 | 3 | 3877869-3878022 |
| EAAMCA0307K | 2 (76.0) | − | +b | + | 249 | 3 | 4147664-4147851 |
| EATMGA0797K | 2R (82.0) | − | + | +b | 660 | 3 | 5050557-5051215 |
| EGAMAT0800K | 2T (85.0) | − | + | +b | 751 | 8 | 1160984-1161728 |
| TRI15 | TRI15 (97.0) | 8 | 589117-591752 | ||||
| ECCMCG0652K | 2V (125.0) | − | + | − | 536 | 8 | 70801-71330 |
| EATMCA0602K | 2 (129.0) | − | + | − | 492 | 9 | 668627-669118 |
| EAAMTG0174J | 2W (130.0) | + | − | − | 133 | 9 | 679601-679733 |
| ETGMGA0127K | 2 (132.0) | − | + | − | 93 | 9 | 588753-588845 |
| EAAMAG0314K | 2X (141.0) | − | + | +b | 253 | 9 | 406495-406747 |
| ECCMAT0417J | 2AB (150.0) | + | − | − | 322 | 9 | 58940-59256 |
| EGAMTC0386K | 2AB (150.0) | − | + | − | 289 | 9 | 54392-54679 |
| EAAMGC0297K | 2 (178.0) | − | +b | + | 241 | 8 | 276275-276515 |
| EATMGA0279K | 2 (179.0) | − | + | +b | 164 | 8 | 275985-276148 |
| EAAMAT0323J | 2AE (184.0) | + | − | − | 264 | 8 | 376598-376867 |
| Chromosome 4 | |||||||
| ECCMAT0358J | 3A (15.0) | + | − | − | 244 | 6 | 389556-389742 |
| EATMCA0377Jc | 3B (18.0) | + | − | − | 279 | 4 | 4567231-4567377 |
| EAAMCA0271J | 3 (70.0) | + | − | − | 221 | 6 | 1003343-1003560 |
| EAAMAC0186K | 3 (73.0) | − | +b | + | 153 | 6 | 1091170-1091323 |
| EAAMAA0119K | 3F (77.0) | − | +b | + | 96 | 6 | 1027866-1027961 |
| ECCMCG0418J | 3H (78.0) | + | − | − | 330 | 6 | 1014868-1015075 |
| EATMGA0455J | 3H (78.0) | + | − | − | 397 | 6 | 1206658-1207048 |
| ECCMAT0528J | 3J (80.0) | + | − | − | 441 | 6 | 1232353-1232789 |
| ETGMGA0145J | 3J (80.0) | + | − | − | 107 | 6 | 1425042-1425148 |
| EAAMCC0239J | 3K (81.0) | + | − | − | 196 | 6 | 2006853-2007044 |
| ECCMAT0278K | 3K (81.0) | − | + | +b | 150 | 6 | 2194012-2194161 |
| ECCMGT0373K | 3K (81.0) | − | + | +b | 270 | 6 | 2529420-2529689 |
| TRI101 | TRI101 (97.0) | 4 | 4787612-4788885 | ||||
| Phosphate permease gene | Phosphate permease gene (98.0) | 4 | 4784489-4785396 | ||||
| ECCMAT0117J | 3O (103.0) | + | − | − | 69 | 4 | 4743042-4743110 |
| ETGMGA0677K | 3S (109.0) | − | + | − | 570 | 4 | 4382673-4383728 |
| ECCMCG0627K | 3S (109.0) | − | + | +b | 520 | 4 | 4371100-4371619 |
| ECCMGT0251K | 3T (110.0) | − | + | +b | 149 | 4 | 4264059-4264205 |
| EAAMAC0786K | 3U (111.0) | − | +b | + | 734 | 4 | 4316734-4317466 |
| ECCMCG0544J | 3 (114.0) | + | − | − | 448 | 4 | 4191107-4191553 |
| EAAMAT0188J | 3X (125.0) | + | − | − | 155 | 4 | 3896166-3896295 |
| EAAMAT0270K | 3 (127.0) | − | + | − | 217 | 4 | 3917150-3917362 |
| EATMCA0212J | 3Z (131.0) | + | − | − | 102 | 4 | 4039381-4039473 |
| EATMGA0123Jc | 3 (132.0) | + | − | − | 68 | 4 | 4039234-4039279 |
| EAAMTG0506K | 3 (168.0) | − | + | − | 431 | 4 | 3644296-3644726 |
| EAAMAG0216K | 3AC (171.0) | − | +b | + | 183 | 4 | 3670012-3670194 |
| EATMGA0170K | 3AF (175.0) | − | + | − | 133 | 4 | 3293782-3293914 |
| EAAMCA0327J | 3AG (177.0) | + | − | − | 270 | 4 | 2537372-2537635 |
| EAAMCC0186K | 3AI (180.0) | − | +b | + | 151 | 4 | 2244914-2245064 |
| EATMCG0704J | 3AL (186.0) | + | − | − | 547 | 4 | 1201133-1201677 |
| EATMCA0196J | 3AM (187.0) | + | − | − | 167 | 4 | 847099-847265 |
| ECCMGT0482K | 3 (191.0) | − | +b | + | 413 | 4 | 388151-388562 |
The markers were designated in previous work (19) using the nomenclature EXXMYY0000Z, where EXX indicates the EcoRI primer (E) with two additional selective nucleotides (XX), MYY indicates the MseI primer (M) with two additional selective nucleotides (YY), the four-digit number is an estimate of the size of the band (in base pairs) (see the text for the size correction equation), and Z is either J or K and indicates the parent that was the source of the “band present” allele or was the source of the larger band of a size polymorphism.
Origin of sequenced marker band.
Sequences partially present in the PH-1 genome database.
Ex, hits on the excluded reads of the PH-1 genome database.
RESULTS
Sequencing and alignment.
One hundred twenty-nine AFLP bands were isolated from strains Z-3639, R-5470, and PH-1 and sequenced (see the supplemental material). Of the 98 markers cloned from AFLP bands, 70 were represented by three matching subclones and 28 were represented by two or three different subclones. Virtual AFLPs were used to choose the correct clone from among each of these 28 sets of fragments by comparing the sequence of the clone with bands predicted by the virtual AFLP analysis. For example, the marker band designated EAAMAA0315J from R-5470 was not observed in either Z-3639 or PH-1. This marker band yielded two subclones, one of which was 282 bp long and one of which was 283 bp long. The virtual AFLP based on the PH-1 genome did not contain the 283-bp fragment but did contain the 282-bp fragment. Thus, the 283-bp clone was associated with EAAMAA0315J.
One hundred twenty-nine AFLP sequences were blasted on the third genome assembly. One hundred nineteen AFLP sequences had hits on the genome assembly. Two AFLP sequences from R-5470 were in the excluded reads in the G. zeae sequencing project, which included highly repetitive sequences and rRNA genes. None of the AFLPs in this study had CCCTAA sequence repeats that are characteristic of fungal telomeres (29). PCR amplification with three additional bases showed that 11 of the 119 AFLP bands were monomorphic in the parents and that the wrong AFLP band had been cloned. These bands were discarded. For the 108 remaining AFLP sequences, 100 markers were present in their entirety in the genome assembly with 83 to 100% identity (Table 1). Eight markers (one from PH-1, two from Z-3639, and five from R-5470) were present as an incomplete sequence in the genome assembly (Table 1). In all cases, single copies of marker sequences were present in the genomic sequence.
The 108 AFLP markers and seven structural genes were used to align the genetic map LGs with the physical map SCs. LG 1, LG 7, LG 8, and LG 9 aligned with SC 3.2 and SC 3.5; LG 2 aligned with SC 3.3, SC 3.8, and SC 3.9; LG 3 aligned with SC 3.4 and SC 3.6; and LG 4, LG 5, and LG 6 aligned with SC 3.1 and SC 3.7 (Fig. 1). These alignments suggested that several LGs could be combined. The nine LGs identified in the Jurgenson map (19) were reanalyzed and reduced to six LGs. LGs 7 and 8 were merged into LG 1, and LG 6 was merged into LG 5 in the new genetic map (Fig. 1 and Table 2). The combination of LGs also resulted in some changes in marker order.
TABLE 2.
Chromosomes of G. zeae
Markers without hits in the genome database.
Eight AFLP markers (four from Z-3639 and four from R-5470) were not found in either the genome assembly or the excluded reads in the genomic database and did not have significant hits in either a BLASTN or BLASTX search against sequences of other organisms in the NCBI database (http://www.ncbi.nlm.nih.gov/). Two of these markers (EATMCA0383J and EAAMTG0323J) were located near the end of LG 5, which aligned with the distal end of SC 3.1 (Fig. 1). EAAMAG0123K and EAAMCA0680J were located on LG 1 and aligned with the proximal end of SC 3.5 and the distal end of SC 3.2, respectively. The other four markers (ETGMGA0314K, EAAMGC0142J, EATMGA0624K, and EAAMAT0513K) were closely linked near the end of LG 2, which aligned to the distal end of SC 3.3. All four of these markers hybridized in Southern hybridizations with genomic DNA of R-5470, Z-3639, and PH-1.
Localization of nit1 and nit3.
The best BLASTP hit in the G. zeae genome for the nitrate reductase gene of N. crassa was FGSG_01947.3 (57% identity; E value, 0.0) on SC 3.1. FGSG_01947.3 previously was annotated as the nitrate reductase gene in the genomic database. FGSG_01947.3 was deleted by split marker recombination, and deletion mutants had the nit1 phenotype, confirming that FGSG_01947.3 is nit1.
The best BLASTP hit for the nitrate assimilation transcription factor gene of N. crassa was FGSG_02799.3 (29% identity; E value, 3e-72) on SC 3.2. However, FGSG_02799.3 deletion mutants did not have the nit3 phenotype, indicating that FGSG_02799.3 is not the nit3 gene in G. zeae. Homologs of the nitrate assimilation transcription factor gene in N. crassa were annotated in the recently released genomic assemblies of Gibberella moniliformis (anamorph, Fusarium verticillioides) and Fusarium oxysporum as sequences FVEG_04246.3 and FOXG_06396.2, respectively. BLASTN analysis with FVEG_04246.3 identified homologous sequences at positions 1894169 to 1894902 (734 bp; 83% identity; E value, e-133) and 1896619 to 1896967 (349 bp; 82% identity; E value, 3e-51) on SC 3.3 of G. zeae. A 486-bp piece of the larger sequence fragment was annotated previously as the predicted protein FGSG_12715.3 gene and is homologous to the 3′ end of FVEG_04246.3. The 349-bp sequence is homologous to the 5′ end of FVEG_04246.3. The middle portion of the gene was not found in the genomic sequence. The 734- and 349-bp sequences are on the ends of contigs 3.176 and 3.177, respectively. The automated annotation of the genome using the GeneTransfers feature postulated a 1.5-kb gap between the contigs, which corresponds to the middle of the putative gene. G. moniliformis, F. oxysporum, and G. zeae all had genes similar to xlnR, which encodes a xylanase regulatory protein, immediately downstream of the gene encoding the putative nitrate assimilation transcription factor. All three species also had genes similar to CTF18 (chromosome transmission fidelity) and NAM9 (mitochondrial ribosomal component) immediately upstream of the gene. Therefore, the sequence identity and microsynteny in the region are consistent with the identification of FGSG_12715.3 as a partial sequence of the nit3 gene of G. zeae.
Recombination rates.
The average recombination rates were 28.1, 42.3, 30.9, and 25.6 cM/Mb for chromosomes 1, 2, 3, and 4, respectively. Regions with high recombination rates were noted on chromosome 1 from approximately locus 6K to locus 4T, on chromosome 2 from the reductase gene to EAAMGC0387K, on chromosome 3 from locus 2A to locus 2J, and on chromosome 4 from locus 3A to EAAMCA0271J and from TRI101 to EAAMTG0506K. Regions with low recombination rates were noted on chromosome 1 from approximately locus 5E to locus 6K, on chromosome 2 from locus 7P to the reductase gene and from locus 9N to locus 9B, on chromosome 3 from locus 2J to locus 2T, and on chromosome 4 from EAAMCA0271J to TRI101 and from EAAMTG0506K to ECCMGT0482K.
DISCUSSION
We used 108 sequenced AFLP markers and seven structural genes to align the Jurgenson genetic map (19) with the third assembly of the genomic sequence of G. zeae strain PH-1. Based on the alignments, three LGs could be combined with other LGs, which reduced the total number of LGs from nine to six and the map length from 1,286 to 1,140 cM (Table 2). The revised genetic map anchored 36.1 Mb (99.2%) of the genomic sequence assembly and arranged the LGs and SCs into the same four sets reported by Gale et al. (14). Thus, our results provide independent verification of the composition of the four chromosome haplotypes of G. zeae.
Based on patterns in progeny haplotype diagrams and an excess of even-numbered crossovers, Jurgenson et al. (19) postulated that there were heterozygous inversions from locus 2F to locus 2AJ on LG 2 (chromosome 3) and from locus 4C to locus 4P on LG 4 (chromosome 1). The alignments with the physical map showed apparent inversions for both LG 2 (locus 2T to locus 2AE) and LG 4 (locus 4C to locus 4K) that are within the regions predicted by Jurgenson et al. (Fig. 1), confirming the existence of two large heterozygous inversions in this cross. It was not possible to determine which parent has an inversion relative to the PH-1 physical sequence from the map alignment. However, inversions were not detected in the alignment of the Gale map involving parent strains PH-1 and 00-676. Z-3639, PH-1, and 00-676 all belong to lineage 7, so the inversions relative to the physical sequence probably are in R-5470, which belongs to lineage 6.
Two additional small regions of nonsynteny involving more than one marker were found on LG 1 (chromosome 2) between the ECCMGT0714J and 7P loci and on LG 3 (chromosome 4) between the 3X and EATMGA0123J loci. These regions could result from small chromosome rearrangements in either parent strain compared to strain PH-1 or from errors underlying either the linkage or the physical map. The regions are too small for the alternative explanations to be resolved by comparison to the Gale map alignment or to the progeny haplotypes.
Cumagun et al. (8) analyzed the mapping population used to generate the Jurgenson map and described a pathogenicity locus, PATH1, closely linked to loci for perithecium production (PER1), red pigmentation (PIG1), and the amount of toxin produced (TOX1). These loci map near locus 4A on LG 4 adjacent to the inversion or, with a slightly lower LOD score, near locus 4P at the other end of the inversion (8). The knockout phenotype of the mitogen-activated kinase gene, MGV1, includes pleiotropic effects on pathogenicity, perithecium formation, pigmentation, and trichothecene toxin accumulation (17). Cumagun et al. (8) postulated that MGV1 might be the same locus as PATH1, PER1, PIG1, and TOX1, with the apparent linkage distances between the loci resulting from phenotyping errors. In the present study, we mapped MGV1 near locus 4P (Table 1 and Fig. 1), which strongly supports the hypothesis of Cumagun et al. (8). Chromosome rearrangements are frequently associated with readily detected mutant phenotypes (37, 38). All four putative mutant phenotypes (low pathogenicity, lack of perithecia, lack of red pigment, and low toxin production) originated in the R-5470 parent, so the same genetic event(s) may have resulted in both the inversion and the mutant phenotypes. In such a case, the inversion on chromosome 1 would not be typical of lineage 6 strains.
With the exception of the inversions, the colinearity of the genetic map and the physical map was very high (Fig. 1). The physical locations of TRI5, TRI15, and the SCAR marker were consistent with the previous linkage analyses (1, 11, 19). The structural genes MGV1, PHO, RED, and TRI101 also were located in expected positions on the physical map. Several markers, including EF-1α, were located in nonsyntenous positions, which could be explained by linkage mapping errors, errors in cloning AFLP bands, chromosome rearrangements, or gene transposition. The proportion of nonsyntenous single markers was similar to that seen in map alignments for G. moniliformis and N. crassa, which is higher than that seen for the Gale map but which was much less than that for Magnaporthe grisea (http://www.broad.mit.edu). Different researchers may use different criteria for excluding anomalous markers, which could alter the interpretation of these comparisons. In the present study, no markers were excluded due to anomalous alignment results.
Eight AFLP markers from the cross (four from each parent) were not found in the genome assembly (Table 1). Four of these markers (three from Z-3639 and one from R-5470) occur in a region at the end of LG 2 (Table 1). Based on Southern hybridizations, Z-3639, R-5470, and PH-1 all contain these four fragments, which suggests that at least 150 kb of SC 3.3, corresponding to 5 cM of LG 2, could be missing from the genome assembly of PH-1. The other four markers (the 6A, 6E, 7B, and 9D loci) not found in the PH-1 genomic sequence were located in regions that correspond to the ends of SCs, suggesting that the genome database may be missing terminal or subterminal portions of SC 3.1, SC 3.5, and SC 3.2.
Eight AFLP markers were present as partial sequences in the PH-1 genomic sequence (Table 1). Two of these markers were cloned from the Z-3639 parent, one was cloned from PH-1, and five were cloned from R-5470. The partial sequence hit of the marker cloned from PH-1 (EATMCG0165K [= locus 5K]) suggests that the genome assembly may be incomplete near this fragment. Another partial sequence marker (EATMCA0377J [= locus 3B]) was aligned in a nonsyntenous position on chromosome 4 (Fig. 1). This result could be explained by transposition of a sequence fragment in one of the strains. Two fragmented markers (EGAMAT0378K [= locus 4F] and EGAMTA0339J [= locus 4K]), one from each parent, were located in the LG 4 inversion on chromosome 1. The partial sequence marker EAAMAA0675K (= locus 7B) was completely linked on LG 1 to marker EAAMAG0123K (= locus 7B), which was missing in the PH-1 genomic sequence. It is possible that PH-1 has a deletion involving both markers. Locus 7B aligns with contig 3.269, which also contains the MAT locus.
Segregation distortion on LG 4 and LG 5 resulted from the nit mutant selection method used to obtain the progeny (19). Progeny were selected for wild-type alleles at both nit loci, and segregation distortion of adjacent markers was expected. nit1 is located on SC 3.1 and aligns near locus 4AC on LG 4, as predicted by Jurgenson et al. (19). The map alignments combined LG 4 and LG 5 into chromosome 1, and thus selection for nit1 can explain the previously reported segregation distortion toward the Z-3639 wild-type allele at the ends of LG 4 and LG 5 near the middle of chromosome 1.
Segregation distortion on LG 2 also can be explained by the selection for nit mutants. Jurgenson et al. (19) predicted that nit3 was located at the end of LG 2 because the region from approximately locus 2A to locus 2M is highly skewed toward the R-5470 parent, which has the wild-type nit3 allele. Based on sequence identity and microsynteny, we identified the putative homolog of nit3 on SC 3.3 in a physical position that aligns with the region between the 2J and 2N loci on LG 2. Segregation distortion toward the R-5470 allele also occurred at the opposite end of LG 2. The large inversion on LG 2 had breakpoints near nit3. The crossover suppression phenotype associated with the inversion could increase the size of the distorted region, as there would be fewer surviving progeny associated with crossovers in this region than would normally be expected. This carryover effect would be more pronounced at the ends of the inverted region than in the middle. Thus, selection for a wild-type nit3 allele can account for the skewing of segregation at both ends of LG 2.
Unexplained segregation distortion remains in two regions. There is an excess of R-5470 alleles at the distal end of chromosome 1 (LG 5) near locus 5E. Similarly, moderate distortion toward the R-5470 allele on chromosome 4 (LG 3) from about locus EATMGA0123J to locus EAAMCA0174K is unexplained. This region corresponds to the distal two-thirds of SC 3.4. Gale et al. (14) found two clusters of significantly skewed markers on chromosome 1, two clusters on chromosome 3, and one cluster on chromosome 4. One of their clusters on chromosome 1 coincides with the nit1 locus. However, none of the other distorted regions on the Gale map coincides with the regions on the Jurgenson map.
Jurgenson et al. (19) reported recombination suppression of four LGs based on an excess of nonrecombinant haplotypes in the progeny (former LG 6, 62%; LG 7, 61%; LG 8, 56%; and LG 9, 75%). However, alignment of genetic and physical maps into chromosomes greatly reduced the frequency of nonrecombinant haplotypes (Table 2). The revised estimates range from 4 to 33% and are similar to estimates of nonrecombinant chromosome frequencies in the mapping cross for G. moniliformis (20). Inspection of haplotype diagrams suggested that nonrecombinant chromosome haplotypes were rare in the Gale mapping population (14).
Several regions with relatively high or low recombination rates were discernible when the genetic linkage distances were compared to the physical sequence distances (Fig. 1). The subterminal region of chromosome 1, which corresponds to the middle of LG 5, exhibited relatively little recombination in our map and the Gale map. We found a region with a high recombination rate near the middle of chromosome 1 that coincided with a region identified by Gale et al. (14). This region also is a region with high single nucleotide polymorphism (SNP) density for PH-1 and Z-3639 (9). The unadjusted average recombination rate in our map for chromosome 1 is 28.1 cM/Mb, while the corresponding estimate for the Gale map is 31.9 cM/Mb.
Chromosome 2 of our map and chromosome 2 of the Gale map have very similar recombination patterns. Both have low recombination rates in the region corresponding to SC 3.5, higher-than-average recombination rates in the middle of the chromosome, and a small region with a low rate of recombination corresponding to the subterminal portion of SC 3.2. SC 3.5 contains the MAT locus, and this might explain the low recombination rate in that region. Recombination suppression has been reported for chromosome 1 of the pseudohomothallic species Neurospora tetrasperma (16), where mating type is included in a large genomic region that does not pair properly during meiosis (13, 16, 33). Mating type locus-related recombination suppression also has been reported in the basidiomycetes Ustilago hordei (24) and Cryptococcus neoformans (26). If G. zeae has chromosomal rearrangements around the mating type locus (e.g., the rearrangement possibly associated with locus 7B), then recombination suppression such as that observed in N. tetrasperma is to be expected, especially if the rearrangement is small enough that multiple crossover events in the rearranged region are rare. The average recombination rate in our map for chromosome 2 is 42.3 cM/Mb, while the corresponding estimate for the Gale map is 45.3 cM/Mb. This recombination rate is the highest rate for any of the four chromosomes.
Chromosome 2 was the most polymorphic chromosome in our study, and the numbers of polymorphic AFLP markers per Mb of physical sequence were 25, 38, 26, and 27 for chromosomes 1, 2, 3, and 4, respectively. Cuomo et al. (9) correlated a large region with a high rate of recombination in the middle of chromosome 2 with high SNP density and suggested that interstitial regions with high SNP density could represent telomere locations of ancestral chromosome fusions. The polymorphic trichothecene cluster (25, 46), which is important for virulence to plants (39), and a quantitative trait locus for pathogen aggressiveness (8) map to the middle of chromosome 2. This region is polymorphic in both the Gale and Jurgenson maps, but toxin type (deoxynivalenol or nivalenol) and pathogen aggressiveness are known to segregate only in the Jurgenson mapping population (8, 19).
The recombination rate was high in the terminal region of chromosome 3 corresponding to the distal end of SC 3.3 and low for the middle of the chromosome comprising the rest of SC 3.3. Gale et al. (14) reported the same recombination pattern and suggested that a 2-Mb region with a very low recombination rate in the middle of chromosome 3 might contain an inversion in their cross. They found high recombination rates in the terminal region corresponding to SC 9. The large heterozygous inversion on chromosome 3 in our cross prevents accurate estimates of recombination rates for the regions corresponding to SC 8 and SC 9. Chromosome 3 had the greatest frequency of nonrecombinant chromosomes in our mapping population (33%). All of the nonrecombinant chromosomes had the R-5470 haplotype, presumably due to the nit3 selectable marker on this chromosome. The inversion on chromosome 3 apparently amplified the influence of the nit marker selection across a large region. This effect is similar to a model described by Livingstone and Rieseberg (30) in which the effects of genes responsible for reproductive isolation during the early stages of speciation are extended across entire chromosome rearrangements. The average recombination rate in our map for chromosome 3 is 30.9 cM/Mb, while the corresponding estimate for the Gale map is 30.3 cM/Mb.
Chromosome 4 had low recombination rates in the region corresponding to the middle of SC 3.6, high recombination rates near the middle of the chromosome on SC 3.4, and low recombination rates in the distal portion of SC 3.4. Our depiction of chromosome 4 (Fig. 1) is inverted compared to that presented by Gale et al., but the results are very similar. Gale et al. (14) located the rRNA gene cluster at the distal end of SC 3.6. Cuomo et al. (9) correlated the high-recombination region in the middle with high SNP diversity. The average recombination rate in our map for chromosome 4 is 25.6 cM/Mb, while the corresponding estimate for the Gale map is 29.2 cM/Mb.
O'Donnell and coworkers (14, 36, 44) used several properties of the Jurgenson map to support their conclusion that the parents of the cross differed enough to belong to two different species, F. graminearum (Z-3639) and Fusarium asiaticum (R-5470). The properties that they cited as indicative of an interspecific cross included (i) the presence of nine LGs rather than the four cytologically determined chromosomes of F. graminearum (18, 45), (ii) high levels of segregation distortion in five of nine LGs, (iii) two large chromosome inversions, (iv) differences in gene order compared to the physical sequence, (v) recombination suppression in four LGs, (vi) a significant number of marker orthologs that are not shared by parental strains of the cross, and (vii) reduced cross fertility.
The alignment of the genetic and physical maps into chromosomes and the comparison of the revised Jurgenson map to the Gale map enabled reassessment of these issues. (i) In both genetic maps, there were nine LGs that aligned the same SCs into four chromosomes, so the basic chromosome complement is the same in the two maps. In our present map the number of LGs is reduced from the nine LGs in the Jurgenson map to six LGs. (ii) Selection for nit markers could explain the observed segregation distortion except for two regions at the ends of chromosomes 1 and 4 in our map. Segregation distortion is not unusual in intraspecific crosses and also occurred in the Gale map at four marker clusters. (iii) The Jurgenson map has two confirmed inversions. (iv) Except for the two inversions, the marker colinearity of our map with the physical sequence was very good (Fig. 1), even within the inverted regions. (v) The Jurgenson map alignment has large regions with low recombination rates that correspond to similar regions of recombination suppression in the Gale map. The average recombination rates were similar in the two maps for each chromosome. Our mapping population contained more chromosomes with nonrecombinant haplotypes than the population used to construct the Gale map, but it was similar to the population seen in the G. moniliformis intraspecific mapping cross (20). (vi) O'Donnell et al. (36) suggested that a significant number of AFLP marker orthologs are not shared by parental strains of the Jurgenson mapping cross because they were not found in the PH-1 sequence. However, four of eight AFLP markers not found in the PH-1 sequence database were from Z-3639 and four were from R-5470, so AFLP bands from both parents had equal chances of “no hits” on the PH-1 database (Table 1). (vii) The fertility of our lineage 6 × lineage 7 cross was relatively low, as indicated by the low percentage (<1%) of heterozygous perithecia produced on crossing plates (19). Cross-fertility is difficult to measure in a selfing organism unless efforts are made to prevent selfing from occurring. We took no such precautions in these crosses as we were relying on the presence of prototrophic recombinant progeny to generate our mapping population. Bowden and Leslie (5) did observe intermediate to high levels of fertility, however, in crosses of strain R-5469 (= NRRL 13818; lineage 6) and strain Z-3636 (lineage 7) or Z-3639, which indicates that low fertility is not a necessary outcome of lineage 6 × lineage 7 crosses.
The only potential evidence for species-level differentiation of the Jurgenson mapping parents in the analysis described above is the two large inversions, whereas the Gale map alignment has no confirmed rearrangements. However, interpretation of the two inversions as evidence of species differentiation is problematic for several reasons. First, karyotype variation and chromosome rearrangements are not uncommon in filamentous fungi (3, 38). Asexual and homothallic sexual reproduction should be unaffected by balanced chromosome rearrangements, so G. zeae may be relatively tolerant of rearrangements compared to obligately outcrossing heterothallic fungi. Second, it is not clear that the Gale mapping population lacked chromosome rearrangements. Gale et al. (14) suggested that there might be an inversion on chromosome 3 based on the extensive recombination suppression in the middle of the chromosome, but this hypothesis has not been tested. Third, the parents of the Gale cross appeared to be very closely related and may not have adequately represented population variation for chromosome rearrangements in strains belonging to lineage 7. Fourth, the inversion on chromosome 1 of the Jurgenson map appeared to be associated with mutant phenotypes in R-5470. Thus, the inversion may not have been typical of lineage 6 strains as most field strains do not carry these mutations. Finally, there is ambiguity about which parent in the Jurgenson mapping cross harbored the inversion relative to the PH-1 sequence. More information about chromosome rearrangements at the population level is needed to determine which, if either, of these inversions is fixed in the various lineages.
Detecting and confirming chromosome rearrangements in these fungi are currently difficult. We used patterns in progeny haplotype diagrams and alignments of genetic and physical maps to identify and/or confirm rearrangements. Gale et al. (14) suggested that a large region of suppressed recombination could indicate an inversion. Unfortunately, these methods of detection are laborious and cannot detect smaller rearrangements that might be more common. Faster and easier methods are needed to survey for chromosome rearrangements in this group. Gale et al. (14) suggested that inversions in this fungus could be detected cytologically by using pachytene analysis of chromosome pairing, but cytological studies of fungal chromosomes are difficult even in well-developed systems, such as Neurospora. In Neurospora, chromosome rearrangements are routinely detected by patterns of dark-colored viable ascospores and light-colored nonviable ascospores produced in heterozygous crosses (37, 38) observed initially with a dissecting microscope. A similar strategy could be developed for G. zeae by using the relative frequencies of asci with four, six, and eight mature ascospores produced in heterozygous crosses. However, even if this strategy is successful, the rearrangements identified are likely to be relatively large. Thus, a complete inventory of chromosome rearrangements probably will not be available until there is additional whole-genome sequencing for this group.
In summary, the availability of the genomic sequence of G. zeae enabled us to compare simultaneously the Jurgenson genetic map, the PH-1 physical map, and the Gale genetic map. The Jurgenson map and the physical sequence validate each other, while they also reveal some problems in each data set. Alignments to the physical map were used to reduce the number of LGs to six and to combine the LGs into four chromosomes, while largely resolving the outstanding questions about segregation distortion and recombination suppression associated with the Jurgenson map. On the other hand, several potential gaps in the genomic sequence were identified based on missing markers from the linkage map. The physical sequence also allowed indirect comparison of two genetic maps that share few markers and that cannot be compared directly. The only two significant differences detected between the maps were the heterozygous inversions on chromosomes 1 and 3 in the Jurgenson map. The average recombination rates for each chromosome were comparable in the two studies, and regions with high and low recombination rates were located in analogous regions. Overall, the similarity between the linkage maps was remarkable considering the large differences in the degree of polymorphism, marker types, and methods of map construction. Despite an inferred history of genetic isolation between lineages 6 and 7 (35, 36), the chromosomes of these lineages are homologous and are capable of recombination along their entire lengths. The availability of two detailed genetic maps and a physical sequence for G. zeae means that this system can be used to study both basic questions (e.g., centromere structure and the regulation of recombination) and questions of applied importance (e.g., mechanisms underlying fertility, fitness, and toxigenicity).
Supplementary Material
Acknowledgments
We thank Celest McGowan for technical assistance in this study.
The results are based on work supported by the U.S. Department of Agriculture. This was a cooperative project with the U.S. Wheat & Barley Scab Initiative.
Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 8 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
Contribution number 08-38-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Alexander, N. J., S. P. McCormick, T. M. Larson, and J. E. Jurgenson. 2004. Expression of Tri15 in Fusarium sporotrichioides. Curr. Genet. 45:157-162. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Boehm, E. W. A., R. C. Ploetz, and H. C. Kistler. 1994. Statistical analysis of electrophoretic karyotype variation among vegetative compatibility groups of Fusarium oxysporum f. sp. cubense. Mol. Plant-Microbe Interact. 7:196-207. [Google Scholar]
- 4.Bowden, R. L., and J. F. Leslie. 1992. Nitrate-nonutilizing mutants of Gibberella zeae (Fusarium graminearum) and their use in determining vegetative compatibility. Exp. Mycol. 16:308-315. [Google Scholar]
- 5.Bowden, R. L., and J. F. Leslie. 1999. Sexual recombination in Gibberella zeae. Phytopathology 89:182-188. [DOI] [PubMed] [Google Scholar]
- 6.Bowden, R. L., J. F. Leslie, J. E. Jurgenson, and J. Lee. 2004. Genetic mapping in Gibberella zeae, p. 555-556. In S. M. Canty, T. Boring, J. Wardwell, and R. W. Ward (ed.), Proceedings of the 2nd International Symposium on Fusarium Head Blight, incorporating the 8th European Fusarium Seminar, 11-15 December 2004, Orlando, FL. Michigan State University, East Lansing.
- 7.Catlett, N. L., B.-N. Lee, O. C. Yoder, and B. G. Turgeon. 2003. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Newsl. 50:9-11. [Google Scholar]
- 8.Cumagun, J. R., R. L. Bowden, J. E. Jurgenson, J. F. Leslie, and T. Miedaner. 2004. Genetic mapping of pathogenicity and aggressiveness of Gibberella zeae (Fusarium graminearum) toward wheat. Phytopathology 94:520-526. [DOI] [PubMed] [Google Scholar]
- 9.Cuomo, C. A., U. Güldener, J.-R. Xu, F. Trail, B. G. Turgeon, A. Di Pietro, J. D. Walton, L.-J. Ma, S. E. Baker, M. Rep., G. Adam, J. Antoniw, T. Baldwin, S. Calvo, Y.-L. Chang, D. DeCaprio, L. R. Gale, S. Gnerre, R. S. Goswami, K. Hammond-Kosack, L. J. Harris, K. Hilburn, J. C. Kennell, S. Kroken, J. K. Magnuson, G. Mannhaupt, E. Mauceli, H.-W. Mewes, R. Mitterbauer, G. Muehlbauer, M. Münsterkötter, D. Nelson, K. O'Donnell, T. Ouellet, W. Qi, H. Quesneville, M. I. G. Roncero, K.-Y. Seong, I. V. Tetko, M. Urban, C. Waalwijk, T. J. Ward, J. Yao, B. W. Birren, and H. C. Kistler. 2007. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317:1400-1402. [DOI] [PubMed] [Google Scholar]
- 10.Desjardins, A. E. 2006. Fusarium mycotoxins: chemistry, genetics, and biology. APS Press, St. Paul, MN.
- 11.Desjardins, A. E., A. M. Jarosz, R. D. Plattner, N. J. Alexander, D. W. Brown, and J. E. Jurgenson. 2004. Patterns of trichothecene production, genetic variability, and virulence to wheat of Fusarium graminearum from smallholder farms in Nepal. J. Agric. Food Chem. 52:6341-6346. [DOI] [PubMed] [Google Scholar]
- 12.Eide, C. J. 1935. The pathogenicity and genetics of Gibberella saubinetii. Technical bulletin 105. University of Minnesota Agricultural Experiment Station, St. Paul.
- 13.Fraser, J. A., and J. Heitman. 2004. Evolution of fungal sex chromosomes. Mol. Microbiol. 51:299-306. [DOI] [PubMed] [Google Scholar]
- 14.Gale, L. R., J. D. Bryant, S. Calvo, H. Giese, T. Katan, K. O'Donnell, H. Suga, M. Taga, T. R. Usgaard, T. J. Ward, and H. C. Kistler. 2005. Chromosome complement of the fungal plant pathogen Fusarium graminearum based on genetic and physical mapping and cytological observations. Genetics 171:985-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gale, L. R., L.-F. Chen, C. A. Hernick, K. Takamura, and H. C. Kistler. 2002. Population analysis of Fusarium graminearum from wheat fields in eastern China. Phytopathology 92:1215-1322. [DOI] [PubMed] [Google Scholar]
- 16.Gallegos, A., D. J. Jacobson, N. B. Raju, M. P. Skupski, and D. O. Nativig. 2000. Suppressed recombination and a pairing anomaly on the mating-type chromosome of Neurospora tetrasperma. Genetics 154:623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou, Z., C. Xue, Y. Peng, T. Katan, H. C. Kistler, and J.-R. Xu. 2002. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant-Microbe Interact. 15:1119-1127. [DOI] [PubMed] [Google Scholar]
- 18.Howson, W. T., R. C. McGinnis, and W. L. Gordon. 1963. Cytological studies on the perfect stages of some species of Fusarium. Can. J. Genet. Cytol. 5:60-64. [Google Scholar]
- 19.Jurgenson, J. E., R. L. Bowden, K. A. Zeller, J. F. Leslie, N. J. Alexander, and R. D. Plattner. 2002. A genetic map of Gibberella zeae (Fusarium graminearum). Genetics 160:1451-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurgenson, J. E., K. A. Zeller, and J. F. Leslie. 2002. Expanded map of Gibberella moniliformis (Fusarium verticillioides). Appl. Environ. Microbiol. 68:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura, M., H. Anzai, and I. Yamaguchi. 2001. Microbial toxins in plant-pathogen interactions: biosynthesis, resistance mechanisms, and significance. J. Gen. Appl. Microbiol. 47:149-160. [DOI] [PubMed] [Google Scholar]
- 22.Klittich, C. J. R., and J. F. Leslie. 1988. Nitrate reduction mutants of Fusarium moniliforme (Gibberella fujikuroi). Genetics 118:417-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, J., T. Lee, Y.-W. Lee, S.-H. Yun, and B. G. Turgeon. 2003. Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae. Mol. Microbiol. 50:145-152. [DOI] [PubMed] [Google Scholar]
- 24.Lee, N., G. Bakkeren, K. Wong, J. E. Sherwood, and J. W. Kronstad. 1999. The mating-type and pathogenicity locus of the fungus Ustilago hordei spans a 500-kb region. Proc. Natl. Acad. Sci. USA 96:15026-15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, T., Y.-K. Han, K.-H. Kim, S.-H. Yun, and Y.-W. Lee. 2002. TRI13 and TRI7 determine deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae. Appl. Environ. Microbiol. 68:1248-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lengeler, K. B., D. S. Fox, J. A. Fraser, A. Allen, K. Forrester, F. S. Dietrich, and J. Heitman. 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1:704-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leonard, K. J., and W. R. Bushnell. 2003. Fusarium head blight of wheat and barley. APS Press, St. Paul, MN.
- 28.Leslie, J. F., and B. A. Summerell. 2006. The Fusarium laboratory manual. Blackwell Professional, Ames, IA.
- 29.Li, W., C. J. Rehmeyer, C. Staben, and M. L. Farman. 2005. TERMINUS—telomeric end-read mining in unassembled sequences. Bioinformatics 21:1695-1698. [DOI] [PubMed] [Google Scholar]
- 30.Livingstone, K., and L. Rieseberg. 2004. Chromosomal evolution and speciation: a recombination-based approach. New Phytol. 161:107-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manly, K. F., and J. M. Olson. 1999. Overview of QTL mapping software and introduction to Map Manager QT. Mamm. Genome 10:327-334. [DOI] [PubMed] [Google Scholar]
- 32.McMullen, M., R. Jones, and D. Gallenberg. 1997. Scab of wheat and barley: a re-emerging disease of devastating impact. Plant Dis. 81:1340-1348. [DOI] [PubMed] [Google Scholar]
- 33.Merino, S. T., M. A. Nelson, D. J. Jacobson, and D. O. Natvig. 1996. Pseudohomothallism and evolution of the mating-type chromosome in Neurospora tetrasperma. Genetics 143:789-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nganje, W. E., D. A. Bangsund, F. L. Leistritz, W. W. Wilson, and N. M. Tiapo. 2002. Estimating the economic impact of a crop disease: the case of Fusarium head blight in U.S. wheat and barley, p. 275-281. In 2002 National Fusarium Head Blight Forum Proceedings. Michigan State University, East Lansing.
- 35.O'Donnell, K., H. C. Kistler, B. K. Tacke, and H. H. Casper. 2000. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. USA 97:7905-7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Donnell, K., T. J. Ward, D. M. Geiser, H. C. Kistler, and T. Aoki. 2004. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 41:600-623. [DOI] [PubMed] [Google Scholar]
- 37.Perkins, D. D. 1974. The manifestation of chromosome rearrangements in unordered asci of Neurospora. Genetics 77:459-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perkins, D. D. 1997. Chromosome rearrangements in Neurospora and other filamentous fungi. Adv. Genet. 36:239-398. [DOI] [PubMed] [Google Scholar]
- 39.Proctor, R. H., T. M. Hohn, and S. P. McCormick. 1995. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant-Microbe Interact. 8:593-601. [DOI] [PubMed] [Google Scholar]
- 40.Rombauts, S., Y. V. Peer, and P. Rouze. 2003. AFLPinSilico, simulating AFLP fingerprints. Bioinformatics 19:776-777. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Seitz, L. M., W. D. Eustace, H. E. Mohr, M. D. Shogren, and W. T. Yamazaki. 1986. Cleaning, milling, and baking tests with hard red winter wheat containing deoxynivalenol. Cereal Chem. 63:146-150. [Google Scholar]
- 43.Shan, X., T. K. Blake, and L. E. Talbert. 1999. Conversion of AFLP markers to sequence-specific PCR markers in barley and wheat. Theor. Appl. Genet. 98:1072-1078. [Google Scholar]
- 44.Starkey, D. E., T. J. Ward, T. Aoki, L. R. Gale, H. C. Kistler, D. M. Geiser, H. Suga, B. Tóth, J. Varga, and K. O'Donnell. 2007. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol. 44:1191-1204. [DOI] [PubMed] [Google Scholar]
- 45.Taga, M., C. Waalwijk, W. G. Flier, and G. H. J. Kema. 2003. Cytological karyotyping of somatic chromosomes from Phytophthora infestans, Mycosphaerella graminicola, and Fusarium spp. Fungal Genet. Newsl. 50(Suppl.):149. [Google Scholar]
- 46.Ward, T. J., J. P. Bielawski, H. C. Kistler, E. Sullivan, and K. O'Donnell. 2002. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc. Natl. Acad. Sci. 99:9278-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeller, K. A., R. L. Bowden, and J. F. Leslie. 2004. Population differentiation and recombination in wheat scab populations of Gibberella zeae in the United States. Mol. Ecol. 13:563-571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.