Abstract
In this study we analyzed the membrane lipid composition of “Candidatus Nitrosopumilus maritimus,” the only cultivated representative of the cosmopolitan group I crenarchaeota and the only mesophilic isolate of the phylum Crenarchaeota. The core lipids of “Ca. Nitrosopumilus maritimus” consisted of glycerol dialkyl glycerol tetraethers (GDGTs) with zero to four cyclopentyl moieties. Crenarchaeol, a unique GDGT containing a cyclohexyl moiety in addition to four cyclopentyl moieties, was the most abundant GDGT. This confirms unambiguously that crenarchaeol is synthesized by species belonging to the group I.1a crenarchaeota. Intact polar lipid analysis revealed that the GDGTs have hexose, dihexose, and/or phosphohexose head groups. Similar polar lipids were previously found in deeply buried sediments from the Peru margin, suggesting that they were in part synthesized by group I crenarchaeota.
Crenarchaeota, one of the phyla in the domain Archaea, has long been thought to consist mostly of (hyper)thermophiles, as cultivated representatives had optimal growth temperatures of >40°C and isolates were obtained exclusively from extreme environments, such as sulfidic hot springs (reference 28 and references therein). Culture-independent DNA surveys, however, have shown that crenarchaeota occur ubiquitously in oceans (6, 9, 15), lakes (16, 21), and soils (12, 19) and thrive under moderate or even psychrophilic conditions. In fact, marine crenarchaeota are proposed to account for ca. 20% of all prokaryotic cells in the global ocean (15), suggesting that they play an important role in the biogeochemical cycles.
The metabolism of these mesophilic crenarchaeota has been a subject of ongoing debate. Several studies have shown that marine crenarchaeota can utilize dissolved inorganic carbon as a carbon source (11, 34) but are able to take up amino acids (11, 22). Recently, a diverse set of putative archaeal ammonia monooxygenase (amoA) genes were obtained from enrichment or uniarchaeal cultures (10, 36), marine waters and sediments (8, 31, 36), and soils (19). These genes were shown to be more abundant than bacterial amoA genes (19, 36), suggesting that crenarchaeota may play an important role in marine and terrestrial nitrogen cycles, possibly as ammonia oxidizers. Confirmation of the hypothesis that crenarchaeota possess the capacity for nitrification came from the first isolation of a representative of the mesophilic crenarchaeota, “Candidatus Nitrosopumilus maritimus” (18). Like bacterial ammonia oxidizers, this organism grows autotrophically and obtains its energy by oxidizing ammonium aerobically to nitrite. The chemolithotrophic physiology of “Ca. Nitrosopumilus maritimus” as a nitrifier was supported by the identification of putative amoA, amoB, and amoC genes (18).
The core membrane lipids of nonthermophilic crenarchaeota are thought to consist of glycerol dibiphytanyl glycerol tetraethers (GDGTs) based on analyses of marine waters and sediments (13, 25), lakes (23), soils (19, 33), peats (32), the uniarchaeal “culture” Cenarcheum symbiosum (27), and enrichment cultures of a marine crenarchaeote from the Wadden Sea (35, 36). The GDGTs which have been attributed to nonthermophilic crenarchaeota include GDGTs 0 to 3 (Fig. 1), which are also synthesized by other (hyper)thermophilic crenarchaeota (references 7 and 17 and references therein). In addition, a unique GDGT containing a cyclohexyl moiety (crenarchaeol) has been attributed to nonthermophilic crenarchaeota and was thought to be a unique biomarker lipid for this group (27). Recently, however, a thermophilic crenarchaeote, “Candidatus Nitrosocaldus yellowstonii,” was enriched and shown to synthesize crenarchaeol (5) in addition to large amounts of glycerol trialkyl glycerol tetraethers (GTGTs). Finally, small quantities of a regio-isomer of crenarchaeol, thought to have a parallel glycerol configuration rather than an antiparallel glycerol configuration, are often detected (25, 27, 35). So far, most research has been focused on identifying the core membrane lipids (i.e., the GDGTs), while a few studies have investigated the polar head groups which may be attached. Biddle et al. (1) analyzed deeply buried sediments in the Peru margin and found evidence of GDGTs 0 to 3 with either a hexose or a “nonitol” (a cyclohexanetetraol with a glycosidic hexose moiety [30]) head group.
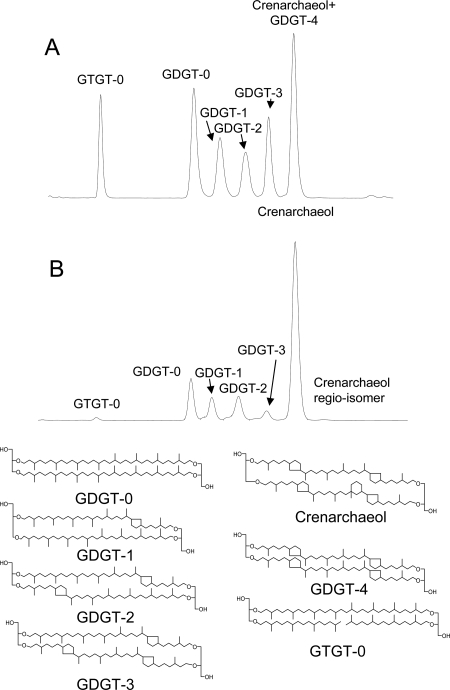
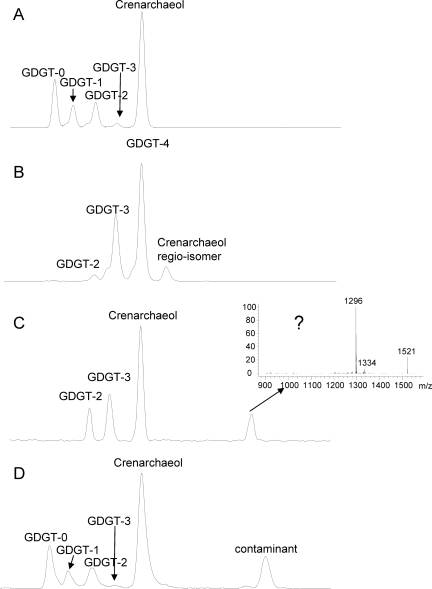
FIG. 1.
Base peak chromatograms from HPLC-APCI-MS analysis of GDGT core lipids in (A) extract released after saponification of the cell material and (B) extract released after acid hydrolysis.
Despite all these studies, there is no unambiguous evidence for biosynthesis of GDGTs 0 to 3 and crenarchaeol by nonthermophilic crenarchaeota and their putative polar head groups due to the lack of a cultivated representative. In this study we analyzed the lipid composition of the recently isolated mesophilic crenarchaeote “Ca. Nitrosopumilus maritimus” and examined both the core GDGT lipid composition and the intact polar lipid composition.
MATERIALS AND METHODS
Culture material.
“Ca. Nitrosopumilus maritimus” was cultured in chemically defined saltwater medium amended with 0.5 mM ammonium chloride as described previously (18). Cultures A and B were inoculated (10%, vol/vol) with the same prefiltered (0.45-μm syringe filter; Sartorius AG, Goettingen, Germany) culture of “Ca. Nitrosopumilus maritimus.” Both cultures were incubated at 28°C, and growth was monitored by measuring nitrite formation. Purity was tested by epifluorescence microscopy using SYBR green I-stained cells (20). Four liters of each culture was harvested by filtration using a 0.1-μm filter (PESU membrane filter; Sartorius AG, Goettingen, Germany), followed by a single washing step with sterile medium and subsequent centrifugation (30 min, 4°C, 3,360 × g). Cell pellets were transferred into precleaned glass vials and freeze-dried (Alpha2-4; Christ GmbH, Osterode, Germany). Freeze-dried cell material was stored frozen prior to analysis. Culture A was used for analysis of core lipids, while culture B was used for analysis of intact polar lipids, as described below.
Analysis of core lipids.
Cell material from culture A of “Ca. Nitrosopumilus maritimus” was base hydrolyzed by refluxing 1 M KOH in methanol for 1 h. Distilled water was added, and the pH of the hydrolyzed extract was adjusted to pH 3 using 2 M HCl-methanol (MeOH) (1/1, vol/vol). Dichloromethane (DCM) was added, the mixture was ultrasonicated and centrifuged, and the DCM layer was collected. This procedure was repeated four times. The DCM layer was dried using a sodium sulfate column to obtain the base-hydrolyzed extract. The MeOH-water layer and residue were then acid hydrolyzed by refluxing with 1 M HCl for 5 h. The MeOH-water layer was extracted with DCM three times. The DCM layer was dried using a combined sodium sulfate and sodium carbonate column to obtain the acid hydrolysis extract. Solvents were removed from the fractions released by base and acid hydrolysis by rotary evaporation. One subsample of the base hydrolysis extract was methylated using diazomethane, filtered using a column filled with silica gel, and eluted with 3 column volumes of ethyl acetate. The methylated extract was then silylated using bis-trimethyl silyl trifluoroacetamide (containing 1% trimethylchlorosilane) and pyridine for 20 min at 60°C. The derivatized extract was analyzed by gas chromatography (GC) and GC-mass spectrometry (MS) using conditions described previously (13).
For high-performance liquid chromatography (HPLC)-MS analysis the extracts were further separated into two fractions using a column filled with activated Al2O3 (2 h at 150°C) as the stationary phase and eluted with 4 column volumes of hexane-DCM (9/1, vol/vol) to collect the apolar fraction and with 3 column volumes of DCM-MeOH (1/1, vol/vol) to collect the polar fraction. Each polar fraction was dissolved in n-hexane-isopropanol (99:1, vol/vol) and filtered using a polytetafluoroethylene 0.4-μm filter prior to analysis by HPLC-MS. HPLC-MS analyses were performed as described by Hopmans et al. (14), with modifications described by Schouten et al. (26). Analyses were performed using an Agilent (Palo Alto, CA) 1100 series LC-MSD SL equipped with an autoinjector and Chemstation chromatography manager software. Separation was achieved with a Prevail Cyano column (2.1 by 150 mm; 3 μm; Alltech, Deerfield, IL) maintained at 30°C. The injection volume was 10 μl. GDGTs were eluted isocratically with 99% hexane-1% isopropanol for 5 min, followed by a linear gradient to 1.8% isopropanol in 45 min. The flow rate was 0.2 ml/min. After each analysis the column was cleaned by back-flushing it with n-hexane-isopropanol (90:10, vol/vol) at a flow rate of 0.2 ml/min for 10 min. Compounds in the eluent were detected using atmospheric pressure positive-ion chemical ionization (APCI)-MS. The conditions used for the HP 1100 APCI-MSD SL were as follows: nebulizer pressure, 60 lb/in2; vaporizer temperature, 400°C; drying gas (N2) flow rate, 6 liters/min; drying gas temperature, 200°C; capillary voltage, −3 kV; and corona, 5 μA (∼3.2 kV). GDGTs were detected by mass scanning from m/z 950 to 1450 and quantified by integration of the peak areas of their [M+H]+ and [M+H+1]+ ions. Crenarchaeol (molecular weight of protonated molecule, 1,292) coelutes with GDGT 4 (molecular weight of protonated molecule, 1,294), and thus its isotope peaks ([M+H+2]+ and [M+H+3]+) interfere with those of the [M+H]+ and [M+H+1]+ ions of GDGT 4, leading to an overestimate of GDGT 4 (32). To correct for this, the average percentage of the [M+H+2]+ isotope peak in the mass spectrum of a crenarchaeol standard was calculated and was estimated to be about 33%. The GDGT 4 peak area in our samples was corrected by this percentage to obtain an estimate of the relative abundance of GDGT 4.
Analysis of intact polar lipids.
For analysis of the intact polar lipids, cell material in culture B of “Ca. Nitrosopumilus maritimus” was extracted using a modified Bligh-Dyer procedure (2). A solvent mixture containing phosphate buffer, MeOH, and DCM (0.8/2/1, vol/vol/vol) was added to the sample. The sample was sonicated for 10 min, after which DCM and phosphate buffer were added at a ratio of 0.9/1/1 (vol/vol/vol). After centrifugation (2,500 rpm, 5 min) the DCM layer was collected. The residue was reextracted twice using the same procedure. The combined DCM layers were concentrated by evaporating solvent under an N2 stream and dried with Na2SO4, and the solvent was removed under an N2 stream. The residue was dissolved in a mixture of hexane and isopropanol (79:21, vol/vol), ultrasonicated, and filtered using a regenerated cellulose 0.45-μm filter (Alltech, Deerfield, IL) prior to analysis by HPLC-electrospray ionization (ESI)-MS.
Intact polar lipids were analyzed as described by Sturt et al. (29), with some modifications (3). An Agilent 1100 series LC (Agilent, San Jose, CA) was used; this instrument was equipped with a thermostat-controlled autoinjector and column oven and was coupled to a Thermo TSQ Quantum Ultra EM triple quadrupole mass spectrometer equipped with an Ion Max source with an ESI probe (Thermo Electron Corporation, Waltham, MA). Separation was achieved with an Inertsil diol column (250 by 2.1 mm; 5-μm particles; Alltech Associates Inc., Deerfield, IL) maintained at 30°C. The following linear gradient was used with a flow rate of 0.2 ml·min−1: 100% eluent A to 35% eluent A-65% eluent B over 45 min, which was maintained for 20 min, and then back to 100% eluent A for 20 min to reequilibrate the column, where eluent A is hexane-isopropanol-formic acid-14.8 M aqueous NH3 (79:20:0.12:0.04, vol/vol/vol/vol) and eluent B is isopropanol-water-formic acid-aqueous 14.8 M NH3 (88:10:0.12:0.04, vol/vol/vol/vol). For MS detection, source parameters were optimized using loop injection of standard phospholipids (1,2-dipalmitoyl-sn-glycero-3-phosphocholine, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine, 1,2-dipalmitoyl-sn-glycero-3-phospho-l-serine, 1,2-dipalmitoyl-sn-glycero-3-phospho-rac-glycerol, 1,2-dipalmitoyl-sn-glycero-3-phosphate, and soya l-α-phosphatidylinositol; Avanti Polar Lipids, Alabaster, AL) into a stream of eluent A at a flow rate of 0.2 ml/min. The ESI settings were as follows: capillary temperature, 250°C; sheath gas (N2) pressure, 49 arbitrary units; auxiliary gas (N2) pressure, 21 arbitrary units; spray voltage, 4.2 kV; and source collision-induced dissociation, −14 V. Mass detection was performed in single quadrupole mode by scanning from m/z 1000 to 2000 with the first quadrupole mass spectrometer.
Polar lipids were isolated by semipreparative HPLC. An HP 1100 series LC (Hewlett Packard, San Jose, CA) equipped with an autoinjector and column oven was used. Separation was achieved with an Inertsil diol column (250 by 10 mm; 5-μm particles; Alltech Associates Inc., Deerfield, IL) maintained at 30°C. The solvents and gradient used were the same as those described above for intact polar lipid analysis, but the flow rate was 3 ml/min and the reequilibration time between runs was 10 min. Fractions were collected and analyzed by flow injection analysis using an HP 1100 series HPLC coupled to a single quadrupole MS with positive-ion ESI. Aliquots (5 μl) of each collected fraction were injected into a stream (flow rate, 0.2 ml/min) containing 60% (vol/vol) eluent A and 40% (vol/vol) eluent B. The ESI settings were as follows: nebulizer pressure (N2), 20 lb/in2; drying gas (N2) flow rate, 10 liters/min; drying gas temperature, 200°C; and spray voltage, 3.5 kV. Mass detection was performed by scanning from m/z 1300 to 1800.
RESULTS
Core lipids.
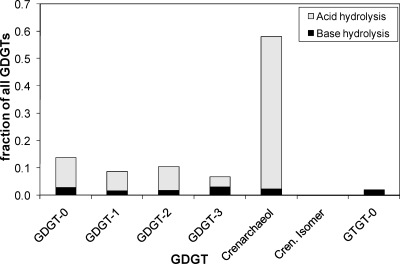
GC-MS analysis of the total extract released after base hydrolysis revealed only small amounts of diether lipids, such as archaeol, suggesting that “Ca. Nitrosopumilus maritimus” does not biosynthesize substantial amounts of diether lipids as final products. HPLC-APCI-MS analysis of the extract released upon base hydrolysis of cell material from culture A of “Ca. Nitrosopumilus maritimus” revealed the presence of a range of GDGTs (GDGTs 0 to 4) and crenarchaeol. In addition, a GTGT (GTGT 0) with one biphytane and two phytane hydrocarbon chains was detected (Fig. 1A). The regio-isomer of crenarchaeol, which is frequently detected with crenarchaeol in environmental samples, was not detected. GDGT 0 was slightly more abundant than GDGTs 1 to 3 (Fig. 2). Relatively substantial amounts of crenarchaeol and GDGT 4 (which coelute but can be quantified based on their molecular ions [see Materials and Methods]) were also present, although the levels were lower than those of the other GDGTs (Fig. 2). Acid hydrolysis of the residue released the same GDGTs, but their distribution was different; the residue was dominated by crenarchaeol, and the amounts of GDGTs 0 to 3 and GTGT 0 were relatively small (Fig. 1B). Small amounts of the regio-isomer of crenarchaeol were detected, but the presence of GDGT 4 could not be established. Quantification of the relative amounts of GDGTs released by both hydrolysis steps showed that the amounts of GDGTs released by acid hydrolysis were substantially larger than the amounts released by base hydrolysis (Fig. 2).
FIG. 2.
Relative contributions of GDGTs to cell material of “Ca. Nitrosopumilus maritimus” released by base hydrolysis and acid hydrolysis.
Intact polar lipids.
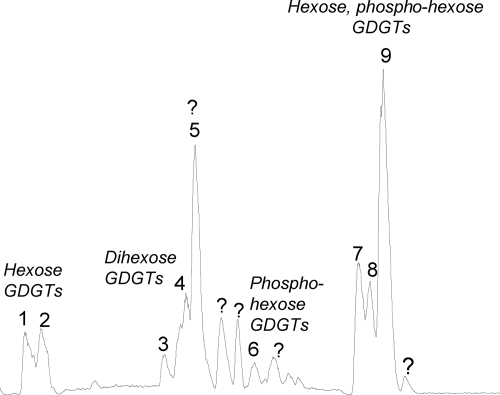
HPLC-ESI-MS analysis of the Bligh-Dyer extract of cell material from culture B of “Ca. Nitrosopumilus maritimus” revealed three major clusters of peaks and a number of minor peaks (Fig. 3). The polar lipids were all tentatively identified by mass spectral interpretation (see below). Supporting evidence for the structural identification was obtained from the isolation of four clusters of peaks (peak 1 plus peak 2, peak 3 plus peak 4, peak 5, and peaks 7 to 9 [Fig. 3]) using preparative HPLC. The amounts isolated were not sufficient for nuclear magnetic resonance (NMR) analysis, but the core GDGTs could be identified by HPLC-MS analysis of GDGT core lipids after removal of the polar head groups by acid hydrolysis.
FIG. 3.
Base peak chromatogram for intact polar lipids of “Ca. Nitrosopumilus maritimus” analyzed by HPLC-ESI-MS.
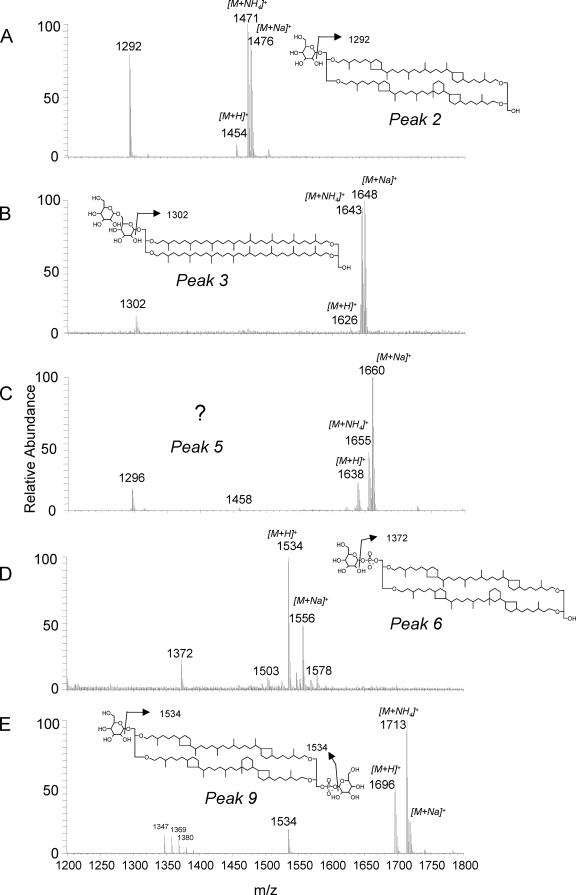
The compounds in the first eluting cluster of polar lipids (peaks 1 and 2 [Fig. 3]) were tentatively identified as GDGTs with a glycosidically bound hexose head group. For example, the mass spectrum for peak 2 was characterized by a dominant ion at m/z 1292, the [M+H]+ ion of crenarchaeol, suggesting that crenarchaeol was the core GDGT of this polar lipid (Fig. 4A and Table 1). In addition, there was a cluster of ions at m/z 1454, 1471, and 1476 (Fig. 4A). This distribution of ions suggests that m/z 1454 represents the [M+H]+ ion of the intact polar lipid, while the ions at m/z 1471 and 1476 represent the ammonia and sodium adducts, respectively, a result typically obtained for polar lipids with the HPLC-ESI conditions used (29). The protonated molecule ion at m/z 1454 is in agreement with the presence of a glycosidic head group (i.e., a hexose moiety glycosidically bound to crenarchaeol) (29). The mass spectrum for peak 1 was dominated by molecular ions in agreement with GDGTs 0 and 2 and GTGT 0 with a hexose head group (Table 1). Interestingly, GDGTs having a glycosidically bound hexose moiety and having similar mass spectrahave been found in deep subsurface sediments from the Peru margin (1). Compounds eluting in peaks 1 and 2 were isolated by preparative HPLC and subjected to acid hydrolysis to remove the glycosidically bound hexose moiety. GDGT analysis showed that the core lipids of these components consisted of GDGTs 0 to 3 and crenarchaeol, with crenarchaeol predominating (Fig. 5A), in full agreement with the identification described above. Unfortunately, the mass spectra of the polar lipids did not allow the structure of the hexose moiety to be inferred, and sufficient quantities that would allow GC analysis were not released in the acid-hydrolyzed fractions.
FIG. 4.
ESI mass spectra (corrected for background) of (A) tentatively identified crenarchaeol with a hexose head group, (B) tentatively identified GDGT 0 with two hexose moieties as the head group, (C) an unknown polar lipid, (D) tentatively identified crenarchaeol with a phosphoglycerol head group, and (E) tentatively identified crenarchaeol with hexose and phosphoglycerol head groups in a polar lipid extract of “Ca. Nitrosopumilus maritimus.” Note that the positions and structures of the hexose moieties are tentative as the position and structure cannot be established from mass spectral data.
TABLE 1.
Composition of intact polar lipids in “Ca. Nitrosopumilus maritimus”
| Peaka | Main ions (m/z) | Head group(s) | Core lipid(s) |
|---|---|---|---|
| 1 | 1298-1304, 1477-1488 | Hexose | GDGTs 0 to 2, GTGT 0 |
| 2 | 1292, 1471, 1476 | Hexose | Crenarchaeol |
| 3 | 1302, 1643, 1648 | Dihexose | GDGT 0 |
| 4 | 1294-1300, 1635-1646 | Dihexose | GDGTs 1 to 4 |
| 5 | 1296-1298, 1638-1640, 1655-1657, 1660-1662 | Hexose and unknown | GDGTs 2 and 3 and unknown |
| 6 | 1372, 1534, 1556 | Phosphohexose | Crenarchaeol |
| 7 | 1544, 1706, 1723, 1728 | Hexose, phosphohexose | GDGT 0 |
| 8 | 1540-1542, 1702-1704, 1719-1726 | Hexose, phosphohexose | GDGTs 1 and 2 |
| 9 | 1534, 1696, 1713, 1718 | Hexose, phosphohexose | Crenarchaeol |
The peak numbers are the peak numbers shown in Fig. 3.
FIG. 5.
HPLC-APCI-MS base peak chromatograms of GDGTs released by acid hydrolysis of intact polar lipids eluting in (A) peaks 1 and 2, (B) peaks 3 and 4, (C) peak 5, and (D) peaks 7 to 9 in the HPLC-ESI-MS base peak chromatogram shown in Fig. 4.
The second cluster of peaks in the HPLC-MS chromatogram (peaks 3 to 5 [Fig. 3]) was formed by two types of polar lipids. Mass spectra for peaks 3 and 4 (Fig. 4B and Table 1) indicated that these peaks were formed by GDGTs with two hexose moieties attached. Based on the fragment ions in the mass spectra of these diglycosides, their core lipids consisted of GDGTs 2 to 4 (Table 1). Indeed, acid-hydrolyzed preparations of the compounds eluting in peak 4 were dominated by GDGTs 2 to 4, with GDGT 4 predominating (Fig. 5B). The levels of ions indicative of crenarchaeol with two hexose moieties were relatively low, and these ions coeluted with peak 5. It was difficult to establish whether the two hexose moieties were attached as a disaccharide to one glycerol unit, as shown in Fig. 4B, or whether one hexose moiety was glycosidically bound to each glycerol unit. However, in the latter case a higher level of a [M+H-hexose]+ fragment (e.g., m/z 1464 for the mass spectrum in Fig. 4B) would have been expected, and thus we tentatively suggest that the head group is diglycosidic. The mass spectrum for peak 5 was different from that for peaks 3 and 4 (Fig. 4C and Table 1). Based on the dominant presence of the m/z 1296 ion in the mass spectrum, the core GDGT was likely predominantly GDGT 3, while the presence of the m/z 1458 ion suggests that a hexose moiety was present. The protonated molecule at m/z 1638, established by the presence of ammonia and sodium adduct ions, suggests that an additional unknown head group with a molecular mass of 180 Da was present (Fig. 4C). Interestingly, unknown GDGTs with identical mass spectra have been reported by Sturt et al. (29) and Biddle et al. (1) for deep subsurface sediments from the Peru margin. HPLC-MS analysis of the GDGT core lipids released by acid hydrolysis of these intact polar lipids showed that they contained, as expected, relatively high levels of GDGT 2 and GDGT 3 and also crenarchaeol, likely derived from coelution of crenarchaeol with two hexose moieties attached (Fig. 5C). Intriguingly, an unknown compound eluted at much later retention times and had a mass spectrum characterized by a base peak at m/z 1296, suggestive of GDGT 3, and also by an ion at m/z 1521. This unknown GDGT may have represented the core GDGT of the unknown intact polar lipid eluting in peak 5. Further research involving isolation and NMR analysis of these components is required to identify their full structures.
The third cluster of peaks was formed in part by unknown compounds (indicated by question marks in Fig. 3), and there were no ions diagnostic for the presence of GDGT core lipids in their mass spectra. The mass spectrum for peak 6, however, did contain fragment ions suggestive of polar lipids with a GDGT core lipid structure (Fig. 4D), and the findings suggested that this polar lipid was crenarchaeol with a phosphohexose head group.
The compounds in the last eluting cluster, peaks 7 to 9, had mass spectra suggestive of GDGTs with a glycosidically bound hexose head group and a phosphohexose head group (Fig. 4E and Table 1). For example, the molecular ion at m/z 1696 and the fragment ion at m/z 1534 of peak 9 suggested that it was composed of crenarchaeol with a glycosidically bound hexose moiety and a phosphohexose head group (Fig. 4E). The mass spectra for peaks 7 and 8 (Table 1) also suggested that they represented GDGTs 0 to 2 with a glycosidically bound hexose head group and a phosphohexose head group. Acid hydrolysis of the prepped cluster of peaks followed by HPLC-MS analysis confirmed that the core GDGTs were GDGTs 0 to 2 and crenarchaeol, with crenarchaeol predominating (Fig. 5D).
To summarize, our HPLC analysis of the intact polar lipids of “Ca. Nitrosopumilus maritimus,” in combination with the hydrolysis of isolated compounds, showed that these lipids are predominantly GDGTs with glycosidic and phosphohexose head groups. It should be emphasized that the position of the head groups on the GDGTs could not be established from our data, nor could the exact structure of the hexose moieties. Large-scale cultivation of “Ca. Nitrosopumilus maritimus” followed by preparative HPLC and NMR analysis is needed to fully identify the tentative structures proposed here.
DISCUSSION
Our results unambiguously show that “Ca. Nitrosopumilus maritimus” synthesizes GDGTs and, importantly, crenarchaeol as the major membrane lipids. Thus, the group I crenarchaeota, of which “Ca. Nitrosopumilus maritimus” is an isolated representative, are likely one of the dominant sources of GDGTs in oceans, lakes, and soils. A detailed comparison of the GDGT composition of “Ca. Nitrosopumilus maritimus” with that of C. symbiosum (27) and that of an enrichment culture from the North Sea (35) showed that the compositions of the dominant GDGTs are relatively similar to that of “Ca. Nitrosopumilus maritimus” (i.e., GDGTs 0 to 3 and crenarchaeol and its regio-isomer). In contrast to the findings for “Ca. Nitrosopumilus maritimus,” GTGT 0 and GDGT 4 were not reported to be present in the enrichment cultures. This likely has to do with the fact that in these enrichment cultures the biomass was either directly extracted (35) or directly acid hydrolyzed (27) and not, as done here, subjected to a base hydrolysis step. In these cases the relative levels of GTGT 0 and GDGT 4 may have been low, thereby escaping detection. Alternatively, the differences are real and GDGT 0 and GDGT 4 are not synthesized by C. symbiosum and the enrichment culture from the North Sea. The GDGT composition of an enrichment culture of the thermophilic crenarchaeote “Ca. Nitrosocaldus yellowstonii” was quite different from that of “Ca. Nitrosopumilus maritimus”; the former organism synthesizes larger amounts of GTGT 0 and GTGTs with one to three cyclopentane rings and relatively smaller amounts of GDGT 0 and crenarchaeol (5).
Regardless of the minor differences between the GDGT compositions of “Ca. Nitrosopumilus maritimus,” C. symbiosum, and the uniarchaeal enrichment culture from the North Sea, all these organisms synthesize crenarchaeol as the dominant GDGT. All these archaea belong to the group I.1a crenarchaeota (18, 24, 36), suggesting that at least some species in this phylogenetic cluster possess the capacity to synthesize this specific GDGT. However, Coolen et al. (4) suggested that, based on crenarchaeol levels and 16S rRNA sequencing data for the Black Sea, even within the group I.1a crenarchaeota there may be differences in the level of crenarchaeol synthesis. It has also not been unambiguously established if species falling in other phylogenetic clusters in the group I crenarchaeota, such as members of groups I.1b and I.1c (24), possess the capacity to synthesize crenarchaeol. Analyses of the core lipids of isolated representatives of these groups of Archaea are required to establish a phylogenetic pattern for the synthesis of crenarchaeol within the group I crenarchaeota.
The head groups of the intact polar lipids of “Ca. Nitrosopumilus maritimus” mainly include hexose, dihexose, and phosphohexose moieties. Such polar lipid head groups are relatively common in cultivated (hyper)thermophilic crenarchaeota (17) and thus are not unique for nonthermophilic crenarchaeota. It should be noted that all identifications of the polar lipids are tentative and that large-scale cultivation of “Ca. Nitrosopumilus maritimus” and isolation of these lipids are needed to rigorously establish their structure. Nevertheless, the tentative identifications should help workers identify these lipids in environmental samples using HPLC-MS. For example, our results support the inferred crenarchaeotal origin of some of the GDGTs with glycosidically bound hexose moieties in deep subsurface sediments in the Peru margin (1). However, Biddle et al. (1) reported the presence of only GDGTs (including GDGTs with a crenarchaeol core lipid structure) with two hexose moieties and not GDGTs with a phosphohexose moiety. This possibly indicates that different nonthermophilic crenarchaeota produce GDGTs with different head group compositions or that there is a partial contribution from fossil remains. Interestingly, 16S rRNA gene sequences obtained from the Peru margin sediments did not include sequences of group I.1a crenarchaeota but were mainly sequences of members of marine benthic group B and the miscellaneous crenarchaeotal group (1). Furthermore, Biddle et al. (1) and Sturt et al. (29) also reported the presence of minor amounts of GDGTs with a “nonitol” head group, a head group that typically occurs in cultivated hyperthermophilic species belonging to the order Sulfolobales (7, 17). However, we did not find peaks with retention times and mass spectra indicative of intact polar lipids with a “nonitol” head group (29) in “Ca. Nitrosopumilus maritimus.” This suggests that these lipids originate from Archaea different than the group I crenarchaeota in the Peru margin sediments.
Acknowledgments
The HPLC-ESI-MS instrument was obtained with an equipment grant from The Netherlands Organization for Scientific Research (NWO) to E.C.H. S.S. was financially supported by the Deutsche Forschungsgemeinschaft (DFG). Support for D.A.S. was provided by NSF Microbial Interactions Program grant MCB-0604448 and NSF Biological Oceanography grant OCE-0623174.
Footnotes
Published ahead of print on 22 February 2008.
REFERENCES
- 1.Biddle, J. F., J. S. Lipp, M. A. Lever, K. G. Lloyd, K. B. Sorensen, R. Anderson, H. F. Fredricks, M. Elvert, T. J. Kelly, D. P. Schrag, M. L. Sogin, J. E. Brenchley, A. Teske, C. H. House, and K. U. Hinrichs. 2006. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. USA 103:3846-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 3.Boumann, H. A., E. C. Hopmans, H. Op Den Camp, M. Strous, M. S. M. Jetten, J. S. Sinninghe Damsté, and S. Schouten. 2006. Ladderane phospolipids in anammox bacteria comprise phosphocholine and phosphoethanolamine headgroups. FEMS Microbiol. Lett. 258:297-304. [DOI] [PubMed] [Google Scholar]
- 4.Coolen, M. J. L., B. Abbas, J. van Bleiswijk, E. C. Hopmans, M. M. M. Kuypers, S. G. Wakeham, and J. S. Sinninghe Damsté. 2007. Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ. Microbiol. 9:1001-1016. [DOI] [PubMed] [Google Scholar]
- 5.de la Torre, J. R., C. B. Walker, A. E. Ingalls, M. Könneke, and D. A. Stahl. 2008. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10:810-818. [DOI] [PubMed] [Google Scholar]
- 6.DeLong, E. F., K. Y. Wu, B. B. Prézelin, and R. V. M. Jovine. 1994. High abundance of Archaea in Antarctic marine picoplankton. Nature 371:695-697. [DOI] [PubMed] [Google Scholar]
- 7.De Rosa, M., and A. Gambacorta. 1988. The lipids of archaeabacteria. Prog. Lipid Res. 27:153-175. [DOI] [PubMed] [Google Scholar]
- 8.Francis, C. A., K. J. Roberts, J. M. Beman, A. E. Santoro, and B. B. Oakley. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 102:14683-14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1992. Novel major archaebacterial group from marine plankton. Nature 356:148-149. [DOI] [PubMed] [Google Scholar]
- 10.Hallam, S. J., T. J. Mincer, C. Schleper, C. M. Preston, K. Roberts, and P. M. Richardson. 2006. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine crenarchaeota. PLoS Biol. 4:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herndl, G. J., T. Reinthaler, E. Teira, E. H. van Aken, C. Veth, A. Pernthaler, and J. Pernthaler. 2005. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 71:2303-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hershberger, K. L., S. M. Barns, A. L. Reysenbach, S. C. Dawson, and N. R. Pace. 1996. Wide diversity of Crenarchaeota. Nature 384:420. [DOI] [PubMed] [Google Scholar]
- 13.Hoefs, M. J. L., S. Schouten, L. King, S. G. Wakeham, J. W. de Leeuw, and J. S. Sinninghe Damsté. 1997. Ether lipids of planktonic archaea in the marine water column. Appl. Environ. Microbiol. 63:3090-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopmans, E. C., S. Schouten, R. D. Pancost, M. T. J. van der Meer, and J. S. Sinninghe Damsté. 2000. Analysis of intact tetraether lipids in archaeal cell material and sediments by high performance liquid chromatography/atmospheric pressure chemical ionization mass spectrometry. Rapid Commun. Mass Spectrom. 14:585-589. [DOI] [PubMed] [Google Scholar]
- 15.Karner, M., E. F. DeLong, and D. M. Karl. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507-510. [DOI] [PubMed] [Google Scholar]
- 16.Keough, B. P., T. M. Schmidt, and R. E. Hicks. 2003. Archaeal nucleic acids in picoplankton from great lakes on three continents. Microb. Ecol. 46:238-248. [DOI] [PubMed] [Google Scholar]
- 17.Koga, Y., and H. Morii. 2005. Recent advances in structural research on ether lipids from archaea including comparative and physiological aspects. Biosci. Biotechnol. Biochem. 69:2019-2034. [DOI] [PubMed] [Google Scholar]
- 18.Könneke, M., A. E. Bernhard, J. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 19.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, J. I. Prosser, S. C. Schuster, and C. Schleper. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 20.Lunau, M., A. Lemke, K. Walther, W. Martens-Habbena, and M. Simon. 2005. An improved method for counting bacteria from sediments and turbid environments by epifluorescence microscopy. Environ. Microbiol. 7:961-968. [DOI] [PubMed] [Google Scholar]
- 21.MacGregor, B. J., D. P. Moser, D. W. Alm, K. H. Nealson, and D. A. Stahl. 1997. Crenarchaeota in Lake Michigan sediment. Appl. Environ. Microbiol. 63:1178-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouverney, C. C., and J. A. Fuhrman. 2000. Marine planktonic archaea take up amino acids. Appl. Environ Microbiol. 66:4829-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powers, L. A., J. P. Werne, T. C. Johnson, E. C. Hopmans, J. S. Sinninghe Damsté, and S. Schouten. 2004. Crenarchaeotal lipids in lake sediments: a new paleotemperature proxy for continental paleoclimate reconstruction? Geology 32:613-616. [Google Scholar]
- 24.Schleper, C., G. Jurgens, and M. Jonuscheit. 2005. Genomic studies of uncultivated archaea. Nat. Rev. Microbiol. 3:479-488. [DOI] [PubMed] [Google Scholar]
- 25.Schouten, S., E. C. Hopmans, R. D. Pancost, and J. S. Sinninghe Damsté. 2000. Widespread occurrence of structurally diverse tetraether membrane lipids: evidence for the ubiquitous presence of low-temperature relatives of hyperthermophiles. Proc. Natl. Acad. Sci. USA 97:14421-14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schouten, S., C. Huguet, E. C. Hopmans, M. V. M. Kienhuis, and J. S. Sinninghe Damsté. 2007. Improved analytical methodology for TEX86 paleothermometry by high performance liquid chromatography/atmospheric pressure chemical ionization-mass spectrometry. Anal. Chem. 79:2940-2944. [DOI] [PubMed] [Google Scholar]
- 27.Sinninghe Damsté, J. S., E. C. Hopmans, S. Schouten, A. C. T. van Duin, and J. A. J. Geenevasen. 2002. Crenarchaeol: the characteristic core glycerol dibiphytanyl glycerol tetraether membrane lipid of cosmopolitan pelagic crenarchaeota. J. Lipid Res. 43:1641-1651. [DOI] [PubMed] [Google Scholar]
- 28.Stetter, K. O. 1998. Hyperthermophiles: isolation, classification and properties, p. 1-17. In K. Horikoshi and W. D. Grant (ed.), Extremophiles: microbial life in extreme environments. Wiley Series in Ecological and Applied Microbiology. Wiley-Liss, New York, NY.
- 29.Sturt, H. F., R. E. Summons, K. Smith, M. Elvert, and K. U. Hinrichs. 2004. Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry—new biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrom. 18:617-628. [DOI] [PubMed] [Google Scholar]
- 30.Sugai, A., R. Sakuma, I. Fukuda, Y. H. Itoh, K. Kon, S. Ando, and T. Itoh. 1995. The structure of the core polyol of the ether lipids from Sulfolobus acidocaldarius. Lipids 30:339-344. [DOI] [PubMed] [Google Scholar]
- 31.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Y. Wu, L. Paulsen, K. E. Nelson, et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 32.Weijers, J. W. H., S. Schouten, M. van der Linden, B. van Geel, and J. S. Sinninghe Damsté. 2004. Water table related variations in the abundance of intact archaeal membrane lipids in a Swedish peat bog. FEMS Microbiol. Lett. 239:51-56. [DOI] [PubMed] [Google Scholar]
- 33.Weijers, J. W. H., S. Schouten, O. C. Spaargaren, and J. S. Sinninghe Damsté. 2006. Occurrence and distribution of tetraether membrane in soils: implications for the use of the BIT index and the TEX86 SST proxy. Org. Geochem. 37:1680-1693. [Google Scholar]
- 34.Wuchter C., S. Schouten, H. T. S. Boschker, and J. S. Sinninghe Damsté. 2003. Bicarbonate uptake by marine Crenarchaeota. FEMS Microbiol. Lett. 219:203-208. [DOI] [PubMed] [Google Scholar]
- 35.Wuchter, C., S. Schouten, M. J. L. Coolen, and J. S. Sinninghe Damsté. 2004. Temperature-dependent variation in the distribution of tetraether membrane lipids of marine Crenarchaeota: implications for TEX86 paleothermometry. Paleoceanography 19:PA4028. [Google Scholar]
- 36.Wuchter, C., B. Abbas, M. J. L. Coolen, L. Herfort, P. Timmers, M. Strous, J. van Bleijswijk, E. Teira, G. J. Herndl, J. J. Middelburg, S. Schouten, and J. S. Sinninghe Damsté. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. USA 103:12317-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]