Abstract
The denitrifying betaproteobacterium “Aromatoleum aromaticum” strain EbN1 degrades several aromatic compounds, including ethylbenzene, toluene, p-cresol, and phenol, under anoxic conditions. The hydrophobicity of these aromatic solvents determines their toxic properties. Here, we investigated the response of strain EbN1 to aromatic substrates at semi-inhibitory (about 50% growth inhibition) concentrations under two different conditions: first, during anaerobic growth with ethylbenzene (0.32 mM) or toluene (0.74 mM); and second, when anaerobic succinate-utilizing cultures were shocked with ethylbenzene (0.5 mM), toluene (1.2 mM), p-cresol (3.0 mM), and phenol (6.5 mM) as single stressors or as a mixture (total solvent concentration, 2.7 mM). Under all tested conditions impaired growth was paralleled by decelerated nitrate-nitrite consumption. Additionally, alkylbenzene-utilizing cultures accumulated poly(3-hydroxybutyrate) (PHB) up to 10% of the cell dry weight. These physiological responses were also reflected on the proteomic level (as determined by two-dimensional difference gel electrophoresis), e.g., up-regulation of PHB granule-associated phasins, cytochrome cd1 nitrite reductase of denitrification, and several proteins involved in oxidative (e.g., SodB) and general (e.g., ClpB) stress responses.
BTEX (benzene, toluene, ethylbenzene, and xylenes) and phenolic compounds are of environmental concern. They represent major constituents of crude oil (in the case of BTEX) (27) and are widely used as solvents and starting compounds in chemical synthesis. Due to accidental spillage during production, storage, and transport, these compounds are increasingly released into the natural environment (13). Contamination with BTEX or phenolic compounds in the vicinity of groundwater aquifers is of particular concern, since these compounds also exhibit considerable water solubility. For example, 1:20 mixtures of gasoline and water contain 3.2 mg liter−1 ethylbenzene, 70 mg liter−1 toluene, and altogether 130 mg liter−1 BTEX (10). The frequent occurrence of oxygen-limited or even anoxic conditions in such environments emphasizes the significance of anaerobic biodegradation. In past years many novel bacterial strains that are able to degrade aromatic compounds under anoxic conditions employing many novel and intriguing biochemical reactions have been isolated (18, 51).
The denitrifying bacterium strain EbN1 is able to degrade toluene and ethylbenzene under anoxic conditions and also directly from crude oil (36). This organism belongs to a novel cluster of anaerobic degraders within the Betaproteobacteria, the proposed new genus “Aromatoleum,” which is distinct from the plant-associated Azoarcus sensu stricto species (R. Rabus, L. Wöhlbrand, D. Lange, B. Reinhold-Hurek, F. Widdel, and P. Kämpfer, unpublished). In addition, strain EbN1 also utilizes p-cresol, phenol, and some other polar aromatic compounds under anoxic conditions. Recently, the complete genome sequence of strain EbN1 was determined, allowing detailed reconstruction of its metabolic network and in-depth proteomic analysis of substrate-dependent regulation of individual degradation pathways (28, 35, 54). Strain EbN1 represents a promising model organism for in situ processes, since toluene-amended microcosms from oil-contaminated and pristine sites displayed rapid consumption of nitrate (31) and contained high numbers of bacteria related to the phylogenetic cluster harboring strain EbN1 (20, 31). Correspondingly, strain EbN1 removes toluene with high efficiency in laboratory two-dimensional aquifer microcosms which mimic in situ concentration gradients (2). Moreover, the detection of bssA expression (BssA is the catalytic subunit of toluene-activating benzylsuccinate synthase) in contaminated aquifers (3, 53) further emphasized the significance of anaerobic degradation for in situ bioremediation.
The toxicity of aromatic compounds generally correlates with their hydrophobicity, which is described by the logarithm of their partition coefficients in a mixture of n-octanol and water (log PO/W). Compounds with a log PO/W between 1 and 4 are cytotoxic, since they preferentially dissolve in biological membranes (40). Consequently, membrane fluidity increases, which leads to a loss of ions, ATP, and other cellular metabolites. Furthermore, dissipation of the proton motive force and denaturation of membrane proteins (e.g., respiratory complexes or nutrient transporters) result in severe energetic problems in solvent-exposed cells (for an overview, see reference 41).
Despite the recent insights into the organisms, biochemistry, and genetics of anaerobic aromatic compound degradation, little is known about the strategies that the novel isolates use to cope with the toxicity of their growth substrates. Until now, bacterial solvent tolerance was studied mainly with aerobic, biotechnologically relevant microorganisms. The solvent stress response of these organisms involved mainly solvent efflux pumps, heat shock proteins, and modifications of the cytoplasmic membrane (19, 22). Here, we investigated for the first time the physiological and proteomic responses of the denitrifying bacterium “Aromatoleum aromaticum” strain EbN1 to semi-inhibitory concentrations of its aromatic growth substrates.
MATERIALS AND METHODS
Medium and cultivation.
The denitrifying bacterium strain EbN1 was cultivated under nitrate-reducing conditions as previously described (36). Cultivation was carried out in 500-ml flat glass bottles containing 400 ml medium that were anoxically sealed with butyl rubber stoppers under an N2-CO2 (90/10%, vol/vol) atmosphere. The alkylbenzene substrates for anaerobic growth were provided as dilutions in 20 ml of deaerated 2,2,4,4,6,8,8-heptamethylnonan (HMN) as an inert carrier phase. Succinate was directly added to the medium from sterile stock solutions. The chemicals used were analytical grade.
Physiological experiments.
The present study was based on two different lines of experiments comprising a total of seven different solvent stress conditions. (i) Strain EbN1 was adapted to “standard” substrate concentrations of 5% (vol/vol) ethylbenzene in HMN and 2% (vol/vol) toluene in HMN for at least five passages. These cultures served as inocula for subsequent cultivation at low, standard, and semi-inhibitory (about 50% growth inhibition) substrate concentrations (see Table S1 in the supplemental material). In the case of ethylbenzene these concentrations were 2, 5, and 8% (corresponding to determined equilibrium concentrations of 0.08, 0.21, and 0.32 mM, respectively, in the culture medium), and in the case of toluene they were 0.5, 2, and 5.5% (corresponding to 0.07, 0.24, and 0.74 mM, respectively). In this first line of experiments the alkylbenzenes served as growth substrates. (ii) Cultures of strain EbN1 adapted for at least five passages to anaerobic growth with 10 mM succinate were suddenly exposed (shocked) to ethylbenzene, toluene, p-cresol, or phenol as a single stressor and to a mixture of all four compounds during early linear growth with succinate. Application of the solvent mixture should have mimicked an in situ multiple-stress situation. In the second line of experiments succinate served as the growth substrate. The aromatic stressors were added directly to the aqueous medium, and the culture bottles were shaken vigorously to dissolve the hydrophobic compounds. The determined semi-inhibitory concentrations of the single solvents in the culture medium were 0.5 mM ethylbenzene, 1.2 mM toluene, 3.0 mM p-cresol, and 6.5 mM phenol (see Table S1 in the supplemental material). The solvent mixture had a semi-inhibitory effect at a total solvent concentration of 2.7 mM, corresponding to about one-quarter of the individual solvent concentrations mentioned above.
The same sampling procedure was used for all seven experiments. Samples were taken from the aqueous phase of the cultures using N2-flushed, sterile syringes to determine the concentration(s) of the aromatic compound(s), the nitrate-nitrite concentrations, and the optical density. Carrier phase-containing culture bottles were inverted to avoid retrieving HMN together with the sample. Contact between the inert carrier phase and rubber stoppers was generally avoided. Each sample (2.5 ml) was divided into two subsamples (1 and 1.5 ml), which were processed as follows. The 1-ml subsamples containing alkylbenzenes were filtered (Spartan 13/0.2 RC; Schleicher & Schuell, Dassel, Germany) into 1 ml methanol (high-performance liquid chromatography [HPLC] grade) and stored in Teflon-sealed vials at 4°C. The 1-ml subsamples containing phenol or p-cresol were not diluted with methanol, but otherwise were treated in the same way. The second subsample (1.5 ml) was first used to record growth by measuring the optical density at 660 nm (UV-mini 1240; Shimadzu, Duisburg, Germany). Subsequently, cells were removed by filtration (Spartan 13/0.2 RC), and the filtrates were stored at 4°C for HPLC analysis of nitrate and nitrite. Parallel cultures (≥3) yielded very similar time courses of growth and concentrations of aromatic compounds, nitrate, and nitrite (data not shown). The concentrations of aromatic compounds in the aqueous medium remained essentially constant during the incubation time (data not shown). Controls lacking either the aromatic compounds or inoculum were treated in the same way.
Theoretical membrane concentrations of the aromatic solvents were calculated based on their log PO/W values (obtained from www.inchem.org) as described previously (40).
Mass cultivation.
Mass cultivation was performed to obtain sufficient cell material for proteomic and polyhydroxyalkanoate (PHA) analysis. Cultures grown with alkylbenzenes and cultures grown with succinate were treated differently, as follows. To obtain compact cell pellets from alkylbenzene-utilizing cultures, a phosphate-buffered mineral medium supplemented with NaCl (1 g liter−1) was used (46). Twelve parallel cultures for each of the three different concentrations of ethylbenzene and toluene were inoculated with correspondingly adapted precultures. All cultures were harvested at an optical density at 660 nm of around 0.2, corresponding to the half-maximal optical density during growth with 0.32 mM ethylbenzene. The latter culture yielded the lowest maximal optical density among the cultures containing the six alkylbenzene concentrations used. Six parallel succinate-utilizing cultures were harvested for each aromatic stressor and time point (45, 250, and 600 min after stressor addition). Unexposed succinate-utilizing cultures were harvested at the same time points. Harvesting and storage of cells were performed as described previously (8).
Chemical analysis.
Concentrations of the alkylbenzenes, p-cresol, and phenol were determined with an HPLC system (Sykam, Fürstenfeldbruck, Germany) using a reversed-phase column as described previously (36). The eluent was composed of either 80% (vol/vol) acetonitrile (for analysis of alkylbenzenes) or 45% (vol/vol) acetonitrile (for analysis of p-cresol and phenol) and 0.75 mM phosphoric acid. In both cases the flow rate was 1 ml min−1. The aromatic compounds were detected at 215 nm (alkylbenzenes), 254 nm (phenol), and 270 nm (p-cresol) and had retention times of 4.4 min (phenol), 4.5 min (toluene), 5.1 min (ethylbenzene), and 5.4 min (p-cresol).
Nitrate and nitrite samples were analyzed with an HPLC system (Sykam) as previously described (36).
2D DIGE.
Cells were disrupted with a PlusOne sample grinding kit (GE Healthcare, Munich, Germany), and protein extracts were prepared as recently reported (14). The protein concentration was determined as described by Bradford (5). Isoelectric focusing was performed using an IPGphor (GE Healthcare) and commercial 24-cm IPG strips with a nonlinear pH 3 to 10 gradient (GE Healthcare) as described previously (14). The EttanDalt II system (GE Healthcare) was used for separation according to molecular mass in 12.5% Duracryl (Genomic Solution, Ann Arbor, MI) or acrylamide gels. Two-dimensional difference gel electrophoresis (2D DIGE) was carried out essentially as described previously (14). Preelectrophoretic labeling with different fluorescent dyes allowed coseparation of three samples in a single gel, representing the reference state, the test state, and an internal standard. The compositions of these three states varied in the two different lines of experiments. (i) Protein extracts from cultures grown with 0.21 mM ethylbenzene or 0.24 mM toluene (“standard” equilibrium concentrations in the aqueous phase) served as reference states and were labeled with Cy5. Protein extracts from cultures adapted to 0.08 or 0.32 mM ethylbenzene and to 0.07 or 0.74 mM toluene represented the four test states and were each labeled with Cy3. Each test state was related to the corresponding reference state. (ii) Protein extracts from untreated succinate-utilizing cultures harvested 45 min after addition of the aromatic stressor(s) to the test cultures served as the reference state and were labeled with Cy5. This reference state was chosen since only a few proteins displayed changed abundances relative to those in untreated cells harvested after 250 min (see Table S3 in the supplemental material). In each experiment protein extracts obtained at two or three different time points (45, 250, and 600 min) after addition of the aromatic stressor(s) served as test states. For example, succinate-utilizing cultures harvested 45, 250, and 600 min after addition of 0.5 mM ethylbenzene represented three independent test states that were each related to the reference state. Each test state was labeled with Cy3. The preparations in the two lines of experiments also contained different internal standards, which consisted of equal amounts of the corresponding reference and test states and were labeled with Cy2. To achieve statistical confidence, a minimum of four parallel gels were included in the analysis for each test state.
2D DIGE gels were scanned immediately after electrophoresis with a Typhoon 9400 scanner (GE Healthcare). Cropped images were analyzed with the DeCyder software (version 5.0; GE Healthcare). The parameters used for codetection of spots were described recently (54). Spot matching was manually controlled for differentially regulated spots, which had to fulfill the following criteria: average ratio (fold change) of less than −2.5-fold or more than 2.5-fold, t test value of less than 10−4, and matched in at least 75% of the gels. Altogether, 23 and 52 2D DIGE gels (corresponding to 69 and 156 gel images) were analyzed for alkylbenzene- and succinate-utilizing cells, respectively. Differentially regulated protein spots were manually excised either from the same 2D DIGE gel (alkylbenzene-utilizing and -shocked cells and phenol-shocked cells) or from separate preparative gels (p-cresol- and solvent mixture-shocked cells), both stained with colloidal Coomassie brilliant blue using the method described by Doherty et al. (11). The high reproducibility of spot patterns between 2D DIGE and colloidal Coomassie brilliant blue-stained gels was demonstrated previously with strain EbN1 (54).
Protein identification by mass spectrometry.
Tryptic digestion of excised proteins was performed as described previously (23). Peptide masses were determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry (TopLab GmbH, Martinsried, Germany). The protein identification and genome analysis were based on the previously published list of annotated genes in the genome sequence of strain EbN1 (35). Peptide mass fingerprints were mapped to the in silico digests of the predicted proteins by using the MS-Digest program (9).
Analysis of PHA.
PHA were analyzed using whole-cell samples. For this, about 5 mg (dry weight) of cell matter was subjected to methanolysis in the presence of 15% (wt/vol) sulfuric acid. The methyl esters of 3-hydroxybutyric acid were analyzed by gas chromatography with a Hewlett Packard type GC6850 gas chromatograph (Hewlett Packard, Waldbronn, Germany) equipped with a hydrophobic capillary column (SGE GmbH, Darmstadt, Germany), an autosampler (Agilent Technologies Inc., Palo Alto, CA), and a flame ionization detector (6, 44). Retention times and peaks were analyzed using the Agilent Cerity software (version A.02.01; Agilent Technologies Inc.), and the total amount of poly(3-hydroxybutyrate) (PHB) in samples was analyzed in relation to methyl esters obtained from purchased sodium salt of 3-hydroxybutyric acid.
To analyze polyesters other than PHB, samples were also analyzed by combined gas chromatography and mass spectrometry. The solutions of 1-methylesters were injected into a Hewlett Packard series 6890 GC capillary gas chromatograph equipped with a Hewlett Packard series 5973 electron ionization mass selective detector and a capillary column (identical to the column described above). Data were collected using the program HP MSD Productivity ChemStations (revision B.01.00). Single fragments were analyzed with the program AMDIS (automated mass spectral deconvolution and identification system; version 2.1) and by the NIST mass spectral search program (version 1.6d ed.; S. Stein, A. Levitsky, O. Fateev, and G. Mallard).
RESULTS AND DISCUSSION
The ability to utilize ethylbenzene and other aromatic compounds under nitrate-reducing conditions is a distinct feature of strain EbN1. In this study we investigated the stress response of strain EbN1 exposed to experimentally determined semi-inhibitory (about 50% growth inhibition) (see Table S1 in the supplemental material) concentrations of aromatic growth substrates under two different conditions. First, during anaerobic growth with 0.32 mM ethylbenzene or 0.74 mM toluene the corresponding degradation pathways were operative. Second, when succinate-utilizing cells were suddenly exposed to 0.5 mM ethylbenzene, 1.2 mM toluene, 3.0 mM p-cresol, and 6.5 mM phenol applied as single stressors or as a mixture (total solvent concentration, 2.7 mM), the corresponding degradation pathways were not operative. Additional physiological and proteomic data for each individual experiment are provided in the supplemental material, as indicated in Table 1.
TABLE 1.
Proteomic responses of strain EbN1 to semi-inhibitory concentrations of aromatic solvents
| Proteinb | Predicted functionb | Fold changea
|
||||||
|---|---|---|---|---|---|---|---|---|
| Alkylbenzene-utilizing cellsc
|
Succinate-utilizing cells 250 min after exposure toc:
|
|||||||
| Ethylbenzene (0.32 mM)d | Toluene (0.74 mM)d | Ethylbenzene (0.5 mM)e | Toluene (1.2 mM)e | p-Cresol (3.0 mM)f | Phenol (6.5 mM)f | Solvent mixture (2.7 mM)f,g | ||
| Denitrification | ||||||||
| NirS | Cytochrome cd1 nitrite reductase | 3.0 | 3.2 | 2.3 | 1.9 | 1.7 | 1.1 | 3.0 |
| NorQ | Putative chaperone required for nitric oxide reductase (NorCB) | (−) | (−) | 1.6 | 1.3 | 1.0 | 1.0 | 2.6 |
| NosZ | Nitrous oxide reductase | (−) | (−) | 5.1 | 5.2 | 1.9 | 2.0 | 2.6 |
| Ethylbenzene degradation | ||||||||
| EbdA | Ethylbenzene dehydrogenase, alpha subunit | −1.3 | 2.7 | − | − | − | − | − |
| Apc1 | Acetophenone carboxylase, subunit 1 | −2.4 | 1.7 | − | − | − | − | 5.9 |
| Apc3 | Acetophenone carboxylase, subunit 3 | −1.9 | 1.6 | (−) | (−) | − | − | 2.6 |
| Apc4 | Acetophenone carboxylase, subunit 4 | −1.6 | 1.8 | (−) | (−) | − | − | 6.1 |
| Orf84 | Putative methyltransferase | 1.6 | 3.6 | − | − | − | − | 8.3 |
| Toluene degradation | ||||||||
| BssA | Benzylsuccinate synthase, alpha subunit | 1.6 | 3.3 | − | − | − | − | − |
| BssE | Benzylsuccinate synthase, chaperone | 2.0 | 3.1 | − | − | − | − | − |
| BbsG | (R)-Benzylsuccinyl-CoA dehydrogenase | −1.0 | 1.5 | − | − | − | − | − |
| BbsH | Phenylitaconyl-CoA hydratase | −1.3 | −1.1 | − | − | − | − | − |
| BbsA | Benzoylsuccinyl-CoA thiolase, alpha subunit | 1.0 | −1.1 | − | − | − | − | − |
| PHA synthesis | ||||||||
| PhbB | Acetoacetyl-CoA reductase | 1.3 | 3.1 | − | − | − | − | (−) |
| PhaC | Putative poly(3-hydroxyalkanoate) synthase | 1.0 | 3.5 | − | − | − | (−) | (−) |
| EbA1323 | Predicted phasin | 5.7 | 5.5 | −1.6 | −1.3 | 1.2 | 1.3 | 1.4 |
| EbA6852 | Probable phasin | 4.0 | 27.4 | (−) | (−) | (−) | (−) | (−) |
| EbA5033 | Probable phasin | 5.4 | 6.6 | −1.2 | −1.2 | − | − | − |
| Oxidative stress-related proteins | ||||||||
| SodB | Superoxide dismutase (Fe) | 1.6 | 2.8 | −1.1 | −1.1 | −1.6 | 2.1 | 2.3 |
| KatA | Catalase | (−) | (−) | 2.5 | 1.4 | 2.0 | 1.6 | 5.3 |
| Dps | DNA-binding ferritin-like protein | 1.3 | 1.4 | 2.0 | 2.0 | 2.4 | 1.3 | 2.6 |
| NorVW | Flavorubredoxin with associated reductase | (−) | (−) | 2.3 | −1.3 | (−) | 2.4 | 7.0 |
| EbA1861 | Putative high-affinity iron transporter | −9.9 | −37.3 | − | − | − | − | − |
| EbA4918 | Putative iron-binding protein of ABC iron transporter | −2.6 | −4.5 | − | − | − | − | − |
| AcnA | Aconitase A | 2.1 | 5.9 | − | − | − | − | − |
| AcnA2 | Aconitase A2 | 1.8 | 4.5 | − | − | − | − | − |
| General stress-related proteins | ||||||||
| BetB | Betaine aldehyde dehydrogenase | 2.3 | 3.5 | (−) | (−) | (−) | (−) | 2.2 |
| WbjB | Involved in O-antigen chain biosynthesis | − | − | 4.5 | 2.0 | − | − | − |
| HtpG | Heat shock protein | (−) | (−) | 2.6 | 1.9 | 1.3 | 1.5 | 1.2 |
| EbB88 | Putative heat shock protein | 1.0 | 1.2 | 3.3 | 2.1 | 1.5 | 4.8 | 3.6 |
| GrpE | Putative Hsp-70 cofactor | (−) | (−) | 1.8 | 1.4 | − | − | 4.7 |
| ClpB | Chaperone | 1.3 | 1.4 | 3.6 | 2.0 | 2.2 | 4.1 | 3.4 |
| OmpC | Outer membrane protein (porin) | − | − | 1.0 | −1.3 | 32.4 | 21.9 | 23.4 |
Fold change in protein abundance as determined by 2D DIGE (see Fig. 2).
Protein designations and predicted functions as described previously (35; http://www.micro-genomes.mpg.de/ebn1). Proteins may belong to more than one group.
Comparison of fold changes between alkylbenzene- and succinate-utilizing cells in absolute terms is not recommended, since the 2D DIGE working packages had different compositions. Visual comparison of relative spot positions of identified proteins (>|2.5|-fold change) was used to match protein spots with fold changes below the threshold in other DIGE work spaces. −, proteins that were absent on 2D electrophoresis gels; (−), uncertain matches due to ambiguous protein spot patterns. Values marked in bold represent changes above the set threshold of significance.
See Tables S1 and S2 in the supplemental material.
See Tables S1 and S3 in the supplemental material.
See Tables S1 and S4 in the supplemental material.
The total solvent concentration of a mixture of ethylbenzene, toluene, p-cresol, and phenol in the culture medium was 2.7 mM.
Growth behavior.
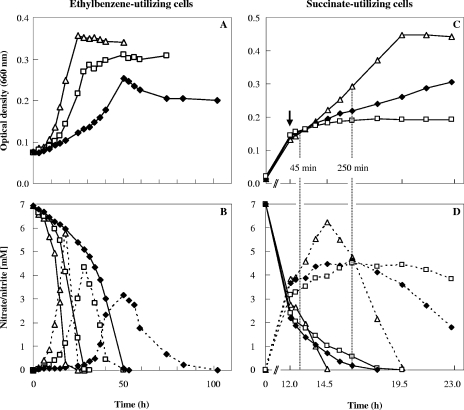
Concentration-dependent impairment of growth was observed under all stressor conditions applied (Fig. 1; see Table S1 in the supplemental material). The growth rates and maximum optical densities were significantly reduced at semi-inhibitory concentrations of aromatic solvents, as exemplified by growth with ethylbenzene (Fig. 1A) and by the growth of succinate-utilizing cultures shocked with the solvent mixture (Fig. 1C). Remarkably, in the latter case about one-quarter of the applied semi-inhibitory concentration of each single solvent sufficed to reduce growth by one-half, suggesting an additive effect of the different toxic properties of the individual solvents.
FIG. 1.
Physiological response of strain EbN1 to solvent stress during anaerobic growth with ethylbenzene (A and B) and succinate (C and D). (A) Growth of ethylbenzene-adapted cells with 0.08 mM ▵, 0.21 mM □, and 0.32 mM ⧫ ethylbenzene. These values represent the equilibrium concentrations in the aqueous medium when the ethylbenzene was supplied in an inert carrier phase. (C) Growth of succinate-utilizing cells suddenly exposed (indicated by an arrow) to a solvent mixture (containing ethylbenzene, toluene, p-cresol, and phenol) having a total solvent concentration of 0 mM ▵, 2.7 mM ⧫, or 3.0 mM □ in the culture medium. The vertical dotted lines indicate time points when mass cultures were harvested for proteomic studies (45 or 250 min after solvent addition). Concentrations of nitrate (solid lines) and nitrite (dashed lines) are indicated in panels B and D.
Growth with alkylbenzenes was completely inhibited at 0.48 mM ethylbenzene and 0.86 mM toluene (equilibrium concentrations in aqueous medium) (see Table S1 in the supplemental material). Complete growth inhibition of succinate-utilizing cultures was observed upon shock with 0.6 mM ethylbenzene, 3.0 mM toluene, 4.0 mM p-cresol, or 8.0 mM phenol singly or with 3.0 mM solvent mixture. The growth-inhibiting concentrations determined indicated that ethylbenzene was the most toxic and phenol was the least toxic of the four tested compounds, agreeing well with the theoretical membrane concentrations of the compounds (40) (e.g., 175 mM ethylbenzene and 48 mM phenol when these compounds were present in the aqueous medium at concentrations of 0.6 and 8.0 mM, respectively).
Petroleum-contaminated groundwater often contains ethylbenzene and toluene at a concentration of 1 mg liter−1 (9.4 and 10.9 μM, respectively) (17). In addition, combined concentrations of phenol and alkyl-substituted phenols up to 68 g liter−1 have been determined for industrial wastewaters (4). Considering its markedly higher solvent tolerance (e.g., growth with <0.48 mM ethylbenzene), strain EbN1 should be able to survive and proliferate in most contaminated environments. Interestingly, the semi-inhibitory alkylbenzene concentrations determined for succinate-utilizing cells of strain EbN1 were higher (at least 2.5-fold) than those reported for other anaerobic degraders (Thauera aromatica, Desulfococcus multivorans, and Geobacter sulfurreducens) (12). Indeed, the ethylbenzene tolerance of strain EbN1 is more similar to that of the aerobic bacterium Pseudomonas putida (12).
Denitrification.
During anaerobic growth with increasing concentrations of ethylbenzene, nitrate consumption and turnover of intermediary formed nitrite decelerated. With 0.21 and 0.32 mM ethylbenzene (Fig. 1B), as well as with 0.74 mM toluene (data not shown), the transition to stationary growth phase coincided with complete depletion of nitrate and maximum formation of nitrite. In contrast, succinate-utilizing, solvent-shocked cultures (Fig. 1D) continued to grow when nitrate was depleted, although denitrification was similarly impaired.
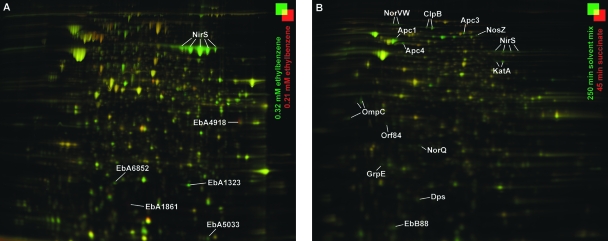
In accordance with the physiological behavior described above, the abundance of several denitrification enzymes changed in response to solvent stress (Table 1 and Fig. 2). In cells growing with 0.32 mM ethylbenzene and 0.74 mM toluene, periplasmic cytochrome cd1 nitrite reductase (NirS) was 3.0- and 3.2-fold more abundant, respectively. Notably, NirS was among the most abundant protein spots observed in 2D DIGE gels generated from alkylbenzene-utilizing cultures, while it was far less abundant in succinate-utilizing cultures. Recently, increased NirS abundance was also observed during anaerobic growth of strain EbN1 with other aromatic substrates (54). One may speculate that aromatic compounds could interfere with electron flow to or within NirS, e.g., by inhibiting NirS dimerization or the NirS-cytochrome c interaction. An impaired electron flow was reported to result in NirS inactivation by irreversible NO binding in Pseudomonas nautica (29). Hence, an increased NirS concentration might compensate for solvent-induced NirS inactivation in strain EbN1.
FIG. 2.
Proteomic response of anaerobically growing strain EbN1 to solvent stress. (A) Ethylbenzene-utilizing cells (0.21 mM ethylbenzene; reference state) were shifted to a semi-inhibitory ethylbenzene concentration (0.32 mM). Cells were harvested at an optical density at 660 nm of 0.2. (B) Succinate-utilizing cells were shocked with a solvent mixture containing ethylbenzene, toluene, p-cresol, and phenol (total concentration, 2.7 mM). The solvent-shocked cells were harvested 250 min after solvent addition. Untreated, succinate-utilizing cells were harvested 45 min (reference state) after addition of the solvent mixture to test cultures. Proteins whose abundance was increased or decreased are indicated by green or red spots, respectively. Selected, mass spectrometrically identified proteins regulated more than |2.5|-fold are indicated. Fold changes in abundance are shown in Table 1.
Unstressed, succinate-utilizing cells displayed growth phase-dependent increases in the abundance of NirS (5.7- and 5.9-fold increases in the linear [250 min] and stationary [600 min] growth phases, respectively) (see Table S3 in the supplemental material). Interestingly, upon shock with the single aromatic stressors the NirS abundance remained unchanged, whereas only the solvent mixture led to a 3.0-fold increase in NirS abundance 250 min after solvent addition. Moreover, periplasmic nitrous oxide reductase (NosZ), which performs the last reaction in denitrification, was 5.1-, 5.2-, and 2.6-fold up-regulated after shock with ethylbenzene, toluene, and the solvent mixture, respectively. The abundance of the NO reductase-related chaperone NorQ also was increased (2.6-fold) in succinate-utilizing cells shocked with the solvent mixture. Inactivation of the norQ homolog nirQ in Pseudomonas stutzeri resulted in an inactive NO reductase complex (24). Thus, up-regulation of NorQ in strain EbN1 might point to an impaired NO reductase and elevated NO levels. Accordingly, a 7.0-fold-increased abundance of the NO-detoxifying flavorubredoxin (NorVW) was observed in cells shocked with the solvent mixture. In Escherichia coli norVW expression was induced at submicromolar NO concentrations (15). Generally, fine-tuned cooperation of denitrification enzymes is thought to ensure low concentrations of toxic NO in unchallenged cells (16).
Degradation pathways.
Proteins involved in the anaerobic degradation of ethylbenzene, toluene, p-cresol, and phenol have been identified in previous proteomic studies with strain EbN1 (28, 54). Here, 11 and 12 2D electrophoresis-separated protein spots could be correlated with ethylbenzene and toluene degradation pathways, respectively (see Table S2 in the supplemental material). Interestingly, only in cells utilizing 0.74 mM toulene did a few of these protein spots reveal slightly increased abundances (e.g., 3.3- and 3.1-fold increases for the catalytic subunit [BssA] and the chaperone [BssE] of benzylsuccinate synthase, respectively). Ethylbenzene-utilizing cells did not show increased abundance of catabolic enzymes. It is noteworthy that the initial enzyme of anaerobic ethylbenzene degradation, ethylbenzene dehydrogenase, exhibits a very high affinity (Km, 0.4 μM) for ethylbenzene (43). Indeed, the lowest ethylbenzene concentration (0.08 mM) used in this study is 200 times higher than this Km. Thus, alkylbenzene degradation should not be enhanced by slightly increased levels of the catabolic enzymes and therefore should not contribute to the solvent tolerance of strain EbN1 under the conditions used. Remarkably, the toluene tolerance of P. putida strain DOT-T1E did not change when toluene degradation was disabled by deletion of the alpha subunit of toluene dioxygenase (30).
In succinate-utilizing, solvent-shocked cells enzymes involved in the degradation of the aromatic compounds were not detected during the incubation time. The only exception was observed with cells exposed to the solvent mixture, where the levels of three subunits of acetophenone carboxylase (Apc1, Apc3, and Apc4; 5.9-, 2.6-, and 6.1-fold, respectively) and of a putative methyltransferase (Orf84; 8.3-fold) were increased; both proteins are involved in ethylbenzene degradation. The physiological meaning of this regulatory behavior is unclear, since it was not observed when the single aromatic stressors were used.
PHA formation.
Many bacteria accumulate PHA as an insoluble energy and carbon storage compound when an excess of a carbon source is accompanied by a limitation in another nutrient. In Ralstonia eutropha, the initial formation of acetoacetyl-coenzyme A (acetoacetyl-CoA) from two acetyl-CoA units is catalyzed by acetyl-CoA acetyltransferase (PhbA). Subsequent reduction catalyzed by acetoacetyl-CoA reductase (PhbB) yields d-3-hydroxyacyl-CoA, which is polymerized to PHB by PHA synthase (PhaC). The product accumulates as granules which are coated with phospholipids and proteins, mostly phasins. PHA-forming cells synthesize phasins only in amounts that can be bound to the granules (52). Therefore, the occurrence of phasins can be used as a marker for PHA synthesis (55). In the present proteomic analysis, the levels of PhbB and PhaC were increased 3.0-fold during growth with 0.74 mM toluene. In addition, three phasin-like proteins (EbA1323, EbA6852, and EbA5033) were up-regulated in the same cultures (up to 27.4-fold) and also during growth with 0.32 mM ethylbenzene (up to 5.7-fold). The sequence similarities of these predicted phasins to their orthologs PhaP1 to PhaP4 in R. eutropha (33) are 42 to 66%. In succinate-utilizing, solvent-shocked cultures of strain EbN1, where the aromatic compounds were not catabolized, either PHB-related proteins were not detected or their abundances were not increased.
The proteomic findings were corroborated by chemical determination of PHA levels. Only during growth with 0.21 and 0.32 mM ethylbenzene and with 0.74 mM toluene did PHB accumulate at levels that accounted for up to 2.0, 10.3, and 5.2% of the cell dry weight, respectively. PHAs other than PHB were not observed. However, the absence of PHB in ethylbenzene- and toluene-shocked cells may have been due to an insufficient incubation time (maximum of 10 h after alkylbenzene addition) (see Table S3 in the supplemental material), since phasin-like proteins were first detected after 20 h in cultures utilizing 0.07 mM toluene.
Under all applied growth conditions, the potential electron supply provided by the organic substrate exceeded the electron-accepting capacity of nitrate (>19-fold, >4-fold, and 4-fold in ethylbenzene-, toluene-, and succinate-utilizing cells, respectively). PHB accumulation was observed only in alkylbenzene-utilizing cultures displaying reduced growth, nitrate consumption, and nitrite turnover. Importantly, in these cultures the electron-accepting nitrate and nitrite were not limiting. Thus, PHB formation presumably does not result from imbalanced nutrient supply but rather results from an impaired coupling of alkylbenzene catabolism and denitrification (see above). Thus, one may speculate that in alkylbenzene-utilizing cells of strain EbN1 alkylbenzene-derived acetyl-CoA is rerouted from oxidation via the tricarboxylic acid cycle to PHB synthesis accompanied by a decrease in the NAD(P)H pool and recycling of free CoA. In such a scenario, PHB could be considered a sink for reducing equivalents, ensuring continuous alkylbenzene degradation.
PHB could also function as a kind of hydrophobic trap or sink for aromatic compounds, since PHB granules are known to accumulate hydrophobic compounds, such as the fluorescent dye Nile red (42). Considering the absence of PHB and related proteins in succinate-utilizing, solvent-shocked cells of strain EbN1, this would be restricted to long-term solvent exposure and/or utilization, if it occurs at all.
Oxidative stress-related proteins.
Oxidative stress responses have been reported in aerobic bacteria challenged with aromatic solvents (e.g., phenol-stressed P. putida [38]), where highly reactive oxygen species can be generated from impaired oxygen respiration. Even though dioxygen and superoxide are not present in anoxic media, highly reactive NO compounds may also be generated during denitrification. In particular, the radical-containing intermediate NO may be accidentally converted to the highly reactive nitroxyl anion (NO−) via several intracellular routes (21), resulting in oxidative stress. The observed increased abundance of superoxide dismutase (SodB; 2.8-fold) during anaerobic growth with 0.74 mM toluene, as well as the increased abundance of catalase (KatA; 5.3-fold), a DNA-binding protein related to oxidative stress (Dps; 2.6-fold), and NorVW (7.0-fold) (see above), in succinate-utilizing cultures shocked with the solvent mixture may have been due to the presence of such reactive NO species. Accordingly, nitrate has been reported to increase SodB activity in Porphyromonas gingivalis (1) and to enhance sodB expression in E. coli (34). The latter result was not observed in a nitrate reductase-deficient E. coli strain (34). Indeed, NO− is a known substrate of superoxide dismutase, which reoxidizes it to NO (37). Notably, constitutive formation of SodB and KatA in anaerobically growing strain EbN1 seems likely, since the corresponding protein spots were detected on almost all 2D electrophoresis gels in this and previous studies (28, 54).
Besides up-regulation of antioxidative defense proteins, large decreases in the abundances of two predicted iron uptake proteins, EbA1861 and EbA4918, were observed in alkylbenzene-utilizing cells (up to a 37.3-fold decrease with 0.74 mM toluene). The current evidence for coordinated regulation of antioxidative defense and cellular iron homeostasis in bacteria (45) suggests that a similar regulatory circuit might be present in strain EbN1. EbA1861 is related to the small COG 3470 group, which comprises uncharacterized proteins assumed to function in high-affinity Fe2+ uptake. EbA4918 is similar to the multimember COG 1840 group of periplasmic components of ABC-type iron/thiamine transporters. In P. putida KT2440 the abundance of a similar protein (HitA; 46% similarity to EbA4918) decreased upon phenol exposure (38). Down-regulation of EbA1861 and EbA4918 in strain EbN1 might result in reduced intracellular iron availability, which would agree with the up-regulation of aconitase A and A2 (5.9- and 4.5-fold, respectively, during growth with 0.74 mM toluene). AcnA is known to replace the primary housekeeping aconitase AcnB in E. coli under oxidative stress or iron starvation conditions, since it contains a more stable [4Fe-4S] cluster (47).
General stress-related proteins.
Proteins generally associated with various types of stresses (e.g., osmotic or temperature stress) were identified mainly in succinate-utilizing, solvent-shocked cultures. Only betaine aldehyde dehydrogenase (BetB), catalyzing the last biosynthetic reaction of osmotically active betaine, displayed a 3.5-fold increase in abundance during growth with 0.74 mM toluene. Osmotically stressed cells of Oceanomonas baumannii also increased betaine synthesis when they were treated with phenol (7).
The abundance of WbjB in succinate-utilizing cultures shocked with 0.5 mM ethylbenzene increased maximally (6.5-fold) 45 min after addition of the stressor (see Table S3 in the supplemental material). WbjB orthologs in Pseudomonas aeruginosa (WbjB) and Staphylococcus aureus (Cap5E) are involved in biosynthesis of N-acetyl-l-fucosamine (26), which is a constituent of surface polysaccharide structures (O antigens of lipopolysaccharide [LPS] and capsules, respectively). Differences in solvent tolerance have been correlated with LPS structure or composition in Pseudomonas strains; e.g., o-xylene-tolerant P. putida strain Idaho had rough LPS, while o-xylene-sensitive P. putida strain MW1200 had smooth LPS (32). This and similar observations with other microorganisms led to the assumption that outer membrane LPS might confer a certain degree of solvent tolerance (50). It is noteworthy, however, that knockout of wbpL, encoding the initial enzyme in O-antigen biosynthesis in P. putida strain DOT-T1E, did not alter toluene tolerance (25).
Solvent-induced formation of protein folding catalysts has been observed in bacteria exposed to, e.g., phenol (38) or toluene (39). In this study, the abundance of several heat shock proteins (HtpG, GrpE, EbB88, and EbA2730) and the chaperone ClpB was increased in succinate-utilizing cultures upon shock with all aromatic compounds except p-cresol (Table 1; see Tables S3 and S4 in the supplemental material). In alkylbenzene-utilizing cells (at least) ClpB is constitutively formed, which was also observed during aerobic and anaerobic growth of strain EbN1 with other aromatic compounds (54).
The abundance of the outer membrane porin OmpC was increased 32.4-, 21.9-, and 23.4-fold in succinate-utilizing cells of strain EbN1 shocked with p-cresol, phenol, and the solvent mixture, respectively. Similarly, solvent-tolerant P. putida S12 up-regulated the OprH porin (12-fold) upon toluene stress, probably to stabilize the outer membrane (48). Furthermore, the occurrence of OmpC as several 2D electrophoresis-separated spots with different pI values, as well as molecular weights (Fig. 2), might indicate that there was proteolysis of OmpC in the p-cresol-, phenol-, and solvent mixture-shocked cells of strain EbN1. Notably, misfolded or unfolded outer membrane porins were reported to trigger the σE-dependent envelope stress response in E. coli (49). One may speculate that OmpC or its degradation products might be involved in a similar mechanism in solvent-stressed cells of strain EbN1. The solvent shock specificity of increased OmpC abundance is supported by the unchanged OmpC abundance and 2D electrophoresis separation as a single spot in cultures of strain EbN1 adapted to anaerobic growth with noninhibitory concentrations of p-cresol or phenol (54).
Supplementary Material
Acknowledgments
We are grateful to D. Lange for experimental assistance and to K. Affeldt, R. Meyer, J. Palfi, M. Wietz, and D. Radovan for laboratory assistance.
This work was supported by the Max-Planck-Gesellschaft and the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 8 February 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amano, A., T. Ishimoto, H. Tamagawa, and S. Shizukuishi. 1992. Role of superoxide dismutase in resistance of Porphyromonas gingivalis to killing by polymorphonuclear leukocytes. Infect. Immun. 60:712-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer, R. D., P. Maloszewski, Y. Zhang, R. U. Meckenstock, and C. Griebler. 2008. Mixing-controlled biodegradation in a toluene plume—results from two-dimensional laboratory experiments. J. Contam. Hydrol. 96:150-168. [DOI] [PubMed] [Google Scholar]
- 3.Beller, H. R. 2002. Analysis of benzylsuccinates in groundwater by liquid chromatography/tandem mass spectrometry and its use for monitoring in situ BTEX biodegradation. Environ. Sci. Technol. 36:2724-2728. [DOI] [PubMed] [Google Scholar]
- 4.Berne, F., and J. Cordonnier. 1995. Industrial water treatment: refining, petrochemicals and gas processing techniques, p. 79-87. Gulf Publishing Company, Houston, TX.
- 5.Bradford, M. M. 1976. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Brandl, H., R. A. Gross, R. W. Lenz, and R. C. Fuller. 1988. Pseudomonas oleovorans as a source of poly(β-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl. Environ. Microbiol. 54:1977-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, G. R., I. C. Sutcliffe, D. Bendell, and S. P. Cummings. 2000. The modification of the membrane of Oceanomonas baumanniiT when subjected to both osmotic and organic solvent stress. FEMS Microbiol. Lett. 189:149-154. [DOI] [PubMed] [Google Scholar]
- 8.Champion, K. M., K. Zengler, and R. Rabus. 1999. Anaerobic degradation of ethylbenzene and toluene in denitrifying strain EbN1 proceeds via independent substrate-induced pathways. J. Mol. Microbiol. Biotechnol. 1:157-164. [PubMed] [Google Scholar]
- 9.Clauser, K. R., P. Baker, and A. L. Burlingame. 1999. Role of accurate mass measurement (±10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71:2871-2882. [DOI] [PubMed] [Google Scholar]
- 10.Cline, P. V., J. J. Delfino, and P. S. C. Rao. 1991. Partitioning of aromatic constituents into water from gasoline and other complex solvent mixtures. Environ. Sci. Technol. 25:914-920. [Google Scholar]
- 11.Doherty, N., B. Littman, K. Reilly, A. Swindell, J. Buss, and N. Anderson. 1998. Analysis of changes in acute-phase plasma proteins in an acute inflammatory response and in rheumatoid arthritis using two-dimensional gel electrophoresis. Electrophoresis 19:355-363. [DOI] [PubMed] [Google Scholar]
- 12.Duldhardt, I., I. Nijenhuis, F. Schauer, and H. J. Heipieper. 2007. Anaerobically grown Thauera aromatica, Desulfococcus multivorans, Geobacter sulfurreducens are more sensitive towards organic solvents than aerobic bacteria. Appl. Microbiol. Biotechnol. 77:705-711. [DOI] [PubMed] [Google Scholar]
- 13.Environmental Protection Agency. 1986. Underground motor fuel storage tanks: a national survey. U.S. Environmental Protection Agency, Washington, DC.
- 14.Gade, D., J. Thierman, D. Markowsky, and R. Rabus. 2003. Evaluation of two-dimensional difference gel electrophoresis for protein profiling. Soluble proteins of the marine bacterium Pirellula sp. strain 1. J. Mol. Microbiol. Biotechnol. 5:240-251. [DOI] [PubMed] [Google Scholar]
- 15.Gardner, A. M., C. R. Gessner, and P. R. Gardner. 2003. Regulation of the nitric oxide reduction operon (norRVW) in Escherichia coli. J. Biol. Chem. 278:10081-10086. [DOI] [PubMed] [Google Scholar]
- 16.Goretski, J., O. Zafiriou, and T. Hollocher. 1990. Steady-state nitric oxide concentrations during denitrification. J. Biol. Chem. 265:11535-11538. [PubMed] [Google Scholar]
- 17.Gülensoy, N., and P. J. J. Alvarez. 1999. Diversity and correlation of specific aromatic hydrocarbon biodegradation capabilities. Biodegradation 10:331-340. [DOI] [PubMed] [Google Scholar]
- 18.Heider, J. 2007. Adding handles to unhandy substrates: anaerobic hydrocarbon activation mechanisms. Curr. Opin. Chem. Biol. 11:188-194. [DOI] [PubMed] [Google Scholar]
- 19.Heipieper, H. J., G. Neumann, S. Cornelissen, and F. Meinhardt. 2007. Solvent-tolerant bacteria for biotransformations in two-phase fermentation systems. Appl. Microbiol. Biotechnol. 74:961-973. [DOI] [PubMed] [Google Scholar]
- 20.Hess, A., B. Zarda, D. Hahn, A. Häner, D. Stax, P. Höhener, and J. Zeyer. 1997. In situ analysis of denitrifying toluene- and m-xylene-degrading bacteria in a diesel fuel-contaminated laboratory aquifer column. Appl. Environ. Microbiol. 63:2136-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes, M. N. 1999. Relationships between nitric oxide, nitroxyl ion, nitrosonium cation and peroxynitrite. Biochim. Biophys. Acta 1411:263-272. [DOI] [PubMed] [Google Scholar]
- 22.Isken, S., and J. A. M. de Bont. 1998. Bacteria tolerant to organic solvents. Extremophiles 2:229-238. [DOI] [PubMed] [Google Scholar]
- 23.Jenö, P., T. Mini, S. Moes, E. Hintermann, and M. Horst. 1995. Internal sequences from proteins digested in polyacrylamide gels. Anal. Biochem. 224:75-82. [DOI] [PubMed] [Google Scholar]
- 24.Jüngst, A., and W. G. Zumft. 1992. Interdependence of respiratory NO reduction and nitrite reduction revealed by mutagenesis of nirQ, a novel gene in the denitrification gene cluster of Pseudomonas stutzeri. FEBS Lett. 314:308-314. [DOI] [PubMed] [Google Scholar]
- 25.Junker, F., J. J. Rodriguez-Herva, E. Duque, M. Ramos-Gonzalez, M. Llamas, and J. L. Ramos. 2001. A WbpL mutant of Pseudomonas putida DOT-T1E strain, which lacks the O-antigenic side chain of lipopolysaccharides, is tolerant to organic solvent shocks. Extremophiles 5:93-99. [DOI] [PubMed] [Google Scholar]
- 26.Kneidinger, B., K. O'Riordan, J. Li, J.-R. Brisson, J. C. Lee, and J. S. Lam. 2003. Three highly conserved proteins catalyze the conversion of UDP-N-acetyl-d-glucosamine to precursors for the biosynthesis of O antigen in Pseudomonas aeruginosa O11 and capsule in Staphylococcus aureus type 5. J. Biol. Chem. 278:3615-3627. [DOI] [PubMed] [Google Scholar]
- 27.Koch, R., and B. O. Wagner. 1989. Umweltchemikalien: physikalisch-chemische Daten, Toxizitäten, Grenz- und Richtwerte, Umweltverhalten. VCH-Verlagsgesellschaft, Weinheim, Germany.
- 28.Kühner, S., L. Wöhlbrand, I. Fritz, W. Wruck, C. Hultschig, P. Hufnagel, M. Kube, R. Reinhardt, and R. Rabus. 2005. Substrate-dependent regulation of anaerobic degradation pathways for toluene and ethylbenzene in a denitrifying bacterium, strain EbN1. J. Bacteriol. 187:1493-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopes, H., S. Besson, I. Moura, and J. J. G. Moura. 2001. Kinetics of inter- and intramolecular electron transfer of Pseudomonas nautica cytochrome cd1 nitrite reductase: regulation of the NO-bound end product. J. Biol. Inorg. Chem. 6:55-62. [DOI] [PubMed] [Google Scholar]
- 30.Mosqueda, G., M.-I. Ramos-Gonzalez, and J. L. Ramos. 1999. Toluene metabolism by the solvent-tolerant Pseudomonas putida DOT-T1 strain, and its role in solvent impermeabilization. Gene 232:69-76. [DOI] [PubMed] [Google Scholar]
- 31.Pelz, O., A. Chatzinotas, N. Andersen, S. M. Bernasconi, C. Hesse, W.-R. Abraham, and J. Zeyer. 2001. Use of isotopic and molecular techniques to link toluene degradation in denitrifying aquifer microcosms to specific microbial populations. Arch. Microbiol. 175:270-281. [DOI] [PubMed] [Google Scholar]
- 32.Pinkart, H., J. Wolfram, R. Rogers, and D. White. 1996. Cell envelope changes in solvent-tolerant and solvent-sensitive Pseudomonas putida strains following exposure to o-xylene. Appl. Environ. Microbiol. 62:1129-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pötter, M., H. Müller, F. Reinecke, R. Wieczorek, F. Fricke, B. Bowien, B. Friedrich, and A. Steinbüchel. 2004. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology 150:2301-2311. [DOI] [PubMed] [Google Scholar]
- 34.Privalle, C. T., and I. Fridovich. 1991. Anaerobic inductions of the active forms of superoxide dismutases in Escherichia coli. Free Radic. Res. Commun. 12-13:419-428. [DOI] [PubMed] [Google Scholar]
- 35.Rabus, R., M. Kube, J. Heider, A. Beck, K. Heitmann, F. Widdel, and R. Reinhardt. 2005. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch. Microbiol. 183:27-36. [DOI] [PubMed] [Google Scholar]
- 36.Rabus, R., and F. Widdel. 1995. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch. Microbiol. 163:96-103. [DOI] [PubMed] [Google Scholar]
- 37.Reif, A., L. Zecca, P. Riederer, M. Feelisch, and H. H. H. W. Schmidt. 2001. Nitroxyl oxidizes NADPH in a superoxide dismutase inhibitable manner. Free Radic. Biol. Med. 30:803-808. [DOI] [PubMed] [Google Scholar]
- 38.Santos, P. M., D. Benndorf, and I. Sá-Correia. 2004. Insights into Pseudomonas putida KT2440 response to phenol-induced stress by quantitative proteomics. Proteomics 4:2640-2652. [DOI] [PubMed] [Google Scholar]
- 39.Segura, A., P. Godoy, P. van Dillewijn, A. Hurtado, N. Arroyo, S. Santacruz, and J.-L. Ramos. 2005. Proteomic analysis reveals the participation of energy- and stress-related proteins in the response of Pseudomonas putida DOT-T1E to toluene. J. Bacteriol. 187:5937-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sikkema, J., J. A. M. de Bont, and B. Poolman. 1994. Interactions of cyclic hydrocarbons with biological membranes. J. Biol. Chem. 269:8022-8028. [PubMed] [Google Scholar]
- 41.Sikkema, J., J. A. M. de Bont, and B. Poolman. 1995. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 59:201-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spiekermann, P., B. H. A. Rehm, R. Kalscheuer, D. Baumeister, and A. Steinbüchel. 1999. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 171:73-80. [DOI] [PubMed] [Google Scholar]
- 43.Szaleniec, M., C. Hagel, M. Menke, P. Nowak, M. Witko, and J. Heider. 2007. Kinetics and mechanism of oxygen-independent hydrocarbon hydroxylation by ethylbenzene dehydrogenase. Biochemistry 46:7637-7646. [DOI] [PubMed] [Google Scholar]
- 44.Timm, A., D. Byrom, and A. Steinbüchel. 1990. Formation of blends of various poly(3-hydroxyalkanoic) acids by a recombinant strain of Pseudomonas oleovorans. Appl. Microbiol. Biotechnol. 33:296-301. [Google Scholar]
- 45.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 46.Tschech, A., and G. Fuchs. 1987. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch. Microbiol. 148:213-217. [DOI] [PubMed] [Google Scholar]
- 47.Varghese, S., Y. Tang, and J. A. Imlay. 2003. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J. Bacteriol. 185:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volkers, R. J. M., A. L. de Jong, A. G. Hulst, B. L. M. van Baar, J. A. M. de Bont, and J. Wery. 2006. Chemostat-based proteomic analysis of toluene-affected Pseudomonas putida S12. Environ. Microbiol. 8:1674-1679. [DOI] [PubMed] [Google Scholar]
- 49.Walsh, N. P., B. M. Alba, B. Bose, C. A. Gross, and R. T. Sauer. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113:61-71. [DOI] [PubMed] [Google Scholar]
- 50.Weber, F. J., and J. A. M. de Bont. 1996. Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochim. Biophys. Acta 1286:225-245. [DOI] [PubMed] [Google Scholar]
- 51.Widdel, F., and R. Rabus. 2001. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12:259-276. [DOI] [PubMed] [Google Scholar]
- 52.Wieczorek, R., A. Pries, A. Steinbüchel, and F. Mayer. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 177:2425-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winderl, C., S. Schaefer, and T. Lueders. 2007. Detection of anaerobic toluene and hydrocarbon degraders in contaminated aquifers using benzylsuccinate synthase (bssA) genes as a functional marker. Environ. Microbiol. 9:1035-1046. [DOI] [PubMed] [Google Scholar]
- 54.Wöhlbrand, L., B. Kallerhoff, D. Lange, P. Hufnagel, J. Thiermann, R. Reinhardt, and R. Rabus. 2007. Functional proteomic view of metabolic regulation in “Aromatoleum aromaticum” strain EbN1. Proteomics 7:2222-2239. [DOI] [PubMed] [Google Scholar]
- 55.York, G. M., B. H. Junker, J. Stubbe, and A. J. Sinskey. 2001. Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J. Bacteriol. 183:4217-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.