Abstract
Sequence-specific degradation of mRNA by short interfering RNA (siRNA) allows the selective inhibition of viral proteins that are critical for human immunodeficiency virus type 1 (HIV-1) replication. The aim of this study was to characterize the potency and durability of virus-specific RNA interference (RNAi) in cell lines that stably express short hairpin RNA (shRNA) targeting the HIV-1 transactivator protein gene tat. We found that the antiviral activity of tat shRNA was abolished due to the emergence of viral quasispecies harboring a point mutation in the shRNA target region. Our results suggest that, in order for RNAi to durably suppress HIV-1 replication, it may be necessary to target highly conserved regions of the viral genome. Alternatively, similar to present antiviral drug therapy paradigms, DNA constructs expressing multiple siRNAs need to be developed that target different regions of the viral genome, thereby reducing the probability of generating escape mutants.
Silencing of gene expression by RNA interference (RNAi) offers a new tool with potential therapeutic applications for the treatment of chronic viral infections such as those with human immunodeficiency virus type 1 (HIV-1) (3, 4, 6-8, 10, 11). Sequence-specific degradation of mRNA by short interfering RNA (siRNA) allows the selective inhibition of viral proteins that are critical for HIV-1 replication. Thus far, virus-specific RNAi has been achieved through transient transfection of either synthetic siRNA molecules or vectors containing siRNA expression cassettes. Development of efficient vector delivery systems capable of mediating stable siRNA expression in CD4+ T lymphocytes or progenitor stem cells will be required for RNAi to be used as a therapeutic modality against HIV-1. Furthermore, siRNA must be capable of sustained inhibition of the rapid replication kinetics of HIV-1 infection (9). The aim of this study was to characterize the potency and durability of HIV-1-specific RNAi.
MATERIALS AND METHODS
Synthetic siRNA transfection.
293T cells were trypsinized and plated at 4 × 105 cells per well in six-well plates 24 h prior to siRNA transfection in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS). A 200-pmol amount of the indicated siRNA was cotransfected with 1 μg of HIV-1NL4.3 with 6 μl of Lipofectamine 2000 (Invitrogen, Carlsbad, Calif.) per reaction. Cell-free supernatant was collected on day 4, and viral p24 antigen level was quantified by enzyme-linked immunosorbent assay (ELISA; Beckman-Coulter, Fullerton, Calif.) according to the manufacturer's instructions. siRNAs with the indicated sense and antisense sequences were used: tat (sense), 5′-CUG CUU GUA CCA AUU GCU AdTdT-3′; tat (antisense), 5′-UAG CAA UUG GUA CAA GCA GdTdT-3′; non-HIV (sense), 5′-UGG CCC AGC GUA AGA AUG CdTdT-3′; non-HIV (antisense), 5′-GCA UUC UUA CGC UGG GCC AdTdT-3′. All siRNA molecules were purchased from Dharmacon Research (Lafayette, Colo.) and were deprotected and annealed according to the manufacturer's instructions.
Construction of H1-driven tat short hairpin RNA (shRNA) expression cassette.
The H1 promoter was PCR amplified from human genomic DNA isolated from HeLa cells with primers 5′-CCA TGG AAT TCG AAC GCT GA-3′ and 5′-GGG AAA GAG TGG TCT CAT AC-3′. The resulting PCR product was subjected to a second PCR with primers 5′-GTG ACA GCG GCC GCC CAT GGA ATT CGA ACG CT-3′ and 5′CAA CAG GCG GCC GCA TCG ATG TTA GGA GAT CTA AAA ACT CGA GAT TTC ATC TAG AGG GAA AGA GTG GTC TCA-3′. The gel-purified product was digested with NotI and cloned into the NotI site of shuttle vector pCMV-MCS (Stratagene, Valencia, Calif.), resulting in pCMV-H1-MCS. A neomycin resistance cassette including the simian virus 40 (SV40) early promoter and the SV40 early polyadenylation signal was cloned into BglII and ClaI sites of pCMV-H1-MCS. The resulting CMV-H1-Neo vector was subsequently digested with NotI, and the released Not-H1-Neo DNA cassette was transferred into pAAV-MCS. Oligonucleotides containing 21-nucleotide tat sense and antisense sequences separated by a 6-nucleotide spacer were annealed and cloned into XbaI and XhoI sites of pAAV-H1-Neo (Fig. 1). pAAV-tat was generated by using oligonucleotides 5′-CTA GAA CTG CTT GTA CCA ATT GCT ATT AGG ATC AAT AGC AAT TGG TAC AAG CAG TTT TTC-3′ and 5′-TCG AGA AAA ACT GCT TGT ACC AAT TGC TAT TGA TCC TAA TAG CAA TTG GTA CAA GCA GTT-3′. pAAV-tatmut containing a point mutation at position 9 (indicated in boldface) of the tat target sequence was created with oligonucleotides 5′-CTA GAA CTG CTT GTT CCA ATT GCT ATT AGG ATC AAT AGC AAT TGG AAC AAG CAG TTT TTT C-3′ and 5′-TCG AGA AAA AAC TGC TTG TTC CAA TTG CTA TTG ATC CTA ATA GCA ATT GGA ACA AGC AGT T-3′. A control plasmid (pAAV-Δtat) was constructed by deleting tat-specific sequences in the shRNA expression cassette. All constructs were sequence verified.
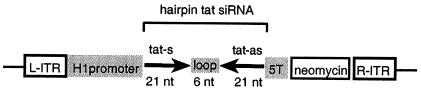
FIG. 1.
Schematic representation of a short hairpin siRNA expression cassette introduced within the ITRs of a recombinant AAV-2 DNA vector. The H1 promoter was used to express tat siRNA in the form of a hairpin transcript consisting of a 21-nucleotide (nt) sense and 21-nucleotide antisense sequence separated by a hexaloop (5′-CTG CTT GTA CCA ATT GCT ATT AGG ATC AAT AGC AAT TGG TAC AAG CAG TTT TT-3′). A five-thymidine transcription termination signal was placed downstream of the hairpin sequence. To allow the generation of shRNA-expressing cell lines, an SV40 early promoter-driven neomycin resistance gene was introduced downstream of the hairpin expression cassette.
Transfection with shRNA expression vectors.
293T cells were trypsinized and plated at 4 × 105 cells per well in six-well plates 24 h prior to transfection in DMEM containing 10% FBS. One microgram of DNA of pAAV-tat, pAAV-tatmut, and pAAV-Δtat (control mock vector) was cotransfected with 1 μg of HIV-1NL4.3 or HIV-1NL4.3mut with 6 μl of Lipofectamine 2000 per reaction. On day 2 supernatant was collected for p24 ELISA.
Generation of shRNA-expressing cell lines.
We used the adeno-associated virus (AAV) Helper-Free system (Stratagene) to produce recombinant AAV particles containing shRNA expression cassettes. The AAV transduction system consists of pAAV expression, pAAV-RC packaging, and pAAV helper plasmids. The original expression plasmid pAAV-MCS contains the inverted terminal repeats (ITRs) necessary for chromosomal integration of AAV type 2 (AAV-2) and a cytomegalovirus promoter-driven cassette for the expression of the desired gene product. The AAV packaging plasmid encodes rep and cap gene products in trans. The AAV helper plasmid contains the adenovirus genes E2A and E4, which are critical for virus particle production. Upon cotransfection into 293T cells, which stably express the adenovirus E1A and E1B proteins, replication-deficient, infective viral particles are produced. The AAV expression vector was modified by removing the cytomegalovirus promoter-controlled expression cassette flanked by the ITRs. This region was replaced with an H1-driven tat shRNA expression cassette followed by the neomycin selection gene (Fig. 1). 293T cells were plated at 5 × 105 cells per well in 2 ml of DMEM containing 10% FBS in six-well plates 24 h prior to transfection. A 1.5-μg amount of each plasmid (recombinant pAAV expression plasmid, pAAV-RC, and pAAV-Helper) was combined, precipitated, and eluted in 20 μl of elution buffer and used to transfect 293T cells. Cells were harvested 72 h posttransfection and subjected to four rounds of freeze-thaw in dry ice-ethanol and a 37°C water bath, respectively. Cell debris was removed by centrifugation at 5,000 × g for 10 min, and viral particles from the supernatant were used for the transduction of target cells.
H9 cells were collected and resuspended in 1 ml of RPMI medium supplemented with 10% FBS and chemically induced with 100 mM hydroxyurea and 3 mM sodium butyrate for 6 h at 37°C. Cells were washed and resuspended in 500 μl of RPMI medium containing 5 μg of Polybrene/ml and transduced with recombinant AAV at a multiplicity of infection of 0.3 at 37°C for 2 h. Two days later, cells were washed and resuspended in RPMI medium containing 600 μg of neomycin/ml and maintained for 4 weeks. Subsequently, cells were challenged with 100 50% tissue culture infective doses (TCID50) of HIV-1NL4.3, and p24 antigen levels were assessed over a 2-month period.
Northern blot analysis.
Northern blot analysis was performed with 10 to 50 μg of total RNA isolated from H9 cells with Trizol reagent (Invitrogen). RNA was denatured in glyoxal sample loading dye (Ambion, Austin, Tex.) according to the manufacturer's protocol and loaded onto a 1.2% agarose gel. RNA was blotted on a Zeta-Probe GT membrane (Bio-Rad Laboratories, Hercules, Calif.) using the Turboblotter system (Schleicher & Schuell, Dassel, Germany) and immobilized by UV fixation. The hybridization was carried out at 42°C with Ultrahyb-Oligo hybridization buffer (Ambion) and a 32P-labeled tat-specific antisense oligonucleotide probe, 5′-AAT AGC AAT TGG TAC AAG CAG T-3′. The membranes were washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate and 0.2× SSC-0.1% sodium dodecyl sulfate at 37°C. HIV tat RNA in vitro transcripts of various sizes were used as markers.
Genotypic analysis of the tat siRNA target region of HIV-1NL4.3.
Viral RNA was isolated from cell-free culture supernatant with the QIAamp viral RNA kit (Qiagen) according to the manufacturer's instructions. Five microliters of RNA was used in a reverse transcriptase reaction including Powerscript reverse transcriptase (Clontech, Palo Alto, Calif.), 1 mM (each) deoxynucleoside triphosphates, 1× First-Strand buffer (Clontech), 200 ng of random hexamers (Promega, Madison, Wis.), and 10 U of RNasin (Promega). Reverse transcription was performed at 42°C for 1 h followed by heat inactivation of the reverse transcriptase enzyme at 70°C for 15 min. Two microliters of cDNA was added to 48 μl of PCR mix including 1× Qiagen TaqPCR buffer including 1.5 mM MgCl2, 20 pmol of sense primer Tat-A (5′-ATG GAG CCA GTA GAT CCT A-3′) and antisense primer Tat-B (5′-TGC TTT GAT AGA GAA ACT TGA TG-3′), 1 mM deoxynucleoside triphosphates, and 2.5 U of Taq polymerase (Qiagen). PCR was carried out in a Mastercycler gradient PCR cycler (Eppendorf, Hamburg, Germany) with a thermal program of 95°C for 1 min; 35 cycles of 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s; and 72°C for 5 min. The PCR product representing the 215-bp tat exon 1 was analyzed on a 1% agarose gel and column purified with the QIAquick PCR kit (Qiagen). Cycle sequencing was performed using dye-labeled terminator chemistry.
Generation of escape mutant HIV-1NL4.3mut and rechallenge of wild-type tat shRNA-expressing cells.
The introduction of a point mutation (A→T) at position 9 of the tat target sequence in the infectious molecular clone pNL4.3 was achieved by recombinational overlap extension PCR. Two separate PCRs were performed on pNL4.3 DNA involving primer pair 5′-GGC AGG AGT GGA AGC CAT A-3′ and 5′-GCA ATT GGA ACA AGC AGT TTT AGG C-3′ and primer pair 5′-ACT GCT TGT TCC AAT TGC TAT TGT AAA AAG TG-3′ and 5′-CCC TCC TGA GGA TTG CTT AAA GA-3′. The DNA of the gel-purified PCR products was amplified with primers 5′-GGC AGG AGT GGA AGC CAT A-3′ and 5′-CCC TCC TGA GGA TTG CTT AAA GA-3′. The column-purified PCR product was digested with EcoRI and SalI and introduced in the equally digested pNL4.3. The mutant molecular clone was sequence verified.
Stable cell lines H9-tat and H9-mock (Δtat) were challenged with either wild-type virus HIV-1NL4.3 or mutant virus HIV-1NL4.3mut. Two million cells were infected with 20 ng of p24 of each virus for 3 h at 37°C. Infected cells were washed once with phosphate-buffered saline and resuspended in RPMI medium. The course of the infection was monitored over a 10-day period by p24 ELISA measurements.
RESULTS AND DISCUSSION
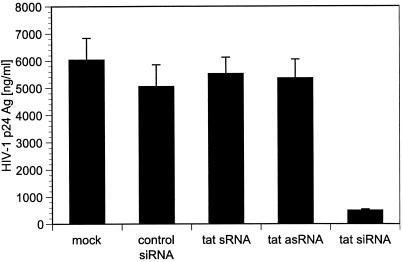
Our initial investigations used a synthetic siRNA molecule that specifically targets exon 1 of the HIV-1 tat gene. tat encodes a transactivator protein that is essential for viral replication. The antiviral potency of tat-specific siRNA was assessed by transient cotransfection of 293T cells with tat siRNA and an infectious molecular clone of HIV-1 (HIV-1NL4.3). The levels of HIV-1 p24 antigen were measured in the culture supernatant by ELISA 4 days after transfection, and tat siRNA suppressed HIV-1 replication by 91% compared to relevant controls (Fig. 2).
FIG. 2.
Cotransfection of various synthetic RNA species including tat-specific sense (s), antisense (as), and short interfering (si) molecules with HIV-1NL4.3 in 293T cells. An siRNA with no sequence homology to HIV-1 was included as a control. Cell-free supernatant was assayed for HIV-1 p24 antigen (Ag) levels on day 4 posttransfection and demonstrated 91% reduction of p24 in cells transfected with tat siRNA compared to controls. Values represent averages of three independent experiments, with the ranges indicated.
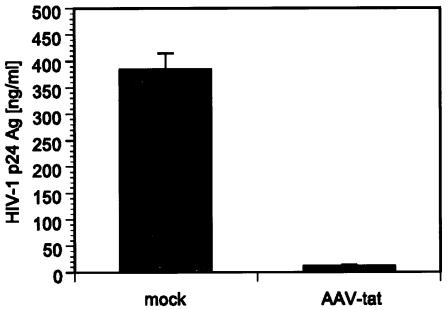
To achieve cellular expression of tat siRNA, a recombinant AAV DNA vector (pAAV-tat) was created that contained an H1 promoter-driven shRNA expression cassette (Fig. 1). The H1 promoter was chosen based on recent evidence that it can effectively drive shRNA expression with retroviral vectors (2). A second vector, devoid of tat-specific sequences in the shRNA expression cassette (pAAV-Δtat), served as a control. Each vector was cotransfected with HIV-1NL4.3 in 293T cells, and HIV-1 p24 antigen levels were quantified in cell-free supernatant 48 h after transfection. HIV-1 p24 antigen level was reduced by 97% in cells expressing tat shRNA compared to control cells transfected with a mock construct, pAAV-Δtat (Fig. 3). Analysis of total intracellular HIV-1 RNA levels by Northern blotting and quantitative real-time PCR confirmed a similar reduction in virus replication (data not shown).
FIG. 3.
To verify the antiviral activity of H1 promoter-driven tat siRNA, 293T cells were cotransfected with HIV-1NL4.3 and pAAV DNA constructs. Quantification of viral p24 antigen levels 48 h posttransfection in cell-free supernatant revealed a 97% decrease in cells expressing tat shRNA (pAAV-tat) compared to cells that were transfected with HIV-1NL4.3 and control mock vector lacking tat-specific sequences (pAAV-Δtat). Values represent averages of three independent experiments, with the ranges indicated.
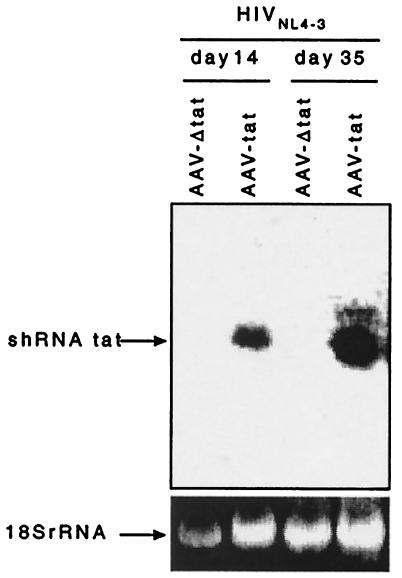
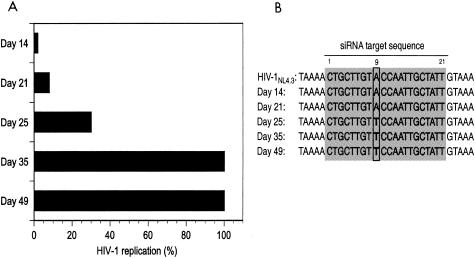
To create a cell line that stably expresses shRNA, AAV-tat and AAV-Δtat vectors were introduced into an AAV-2 transduction system. Recombinant viral particles were generated and used to infect H9 cells, which are derived from a human cutaneous T-cell lymphoma. H9 cells are permissive for HIV-1 replication, thereby facilitating determinations of the magnitude and durability of antiviral RNAi. Stable expression of tat shRNA in H9 cells was confirmed by Northern blot analysis (Fig. 4). Genomic integration of AAV-tat was verified by Southern blot analysis (data not shown). The cells were then infected with 100 TCID50 of the infectious molecular clone HIV-1NL4.3, and p24 antigen levels were quantified in cell-free supernatant at weekly intervals over a 2-month period. During the first 3 weeks, HIV-1 replication was reduced by 95% in cultures of cells expressing tat shRNA compared to control cells (Fig. 5A). By day 25, however, p24 levels had increased, indicating loss of tat shRNA-mediated antiviral activity. Northern blot analysis revealed continued intracellular production of shRNA (Fig. 4). To determine whether HIV-1 escape from RNAi was due to the emergence of mutations within the tat gene, viral RNA was extracted from sequential samples of culture supernatant and the tat siRNA target region was PCR amplified and sequenced. During the first 3 weeks, the sequence of the tat target region was identical to the expressed shRNA. On day 25, a viral species emerged with a nonsynonymous mutation at nucleotide position 9 of the targeted sequence leading to an amino acid change from threonine (ACC) to serine (TCC) (Fig. 5B).
FIG. 4.
Northern blot analysis of tat shRNA expression in HIV-1NL4.3-infected H9 cells transduced with AAV-tat or AAV-Δtat. Continuous expression of tat shRNA was observed on days 14 and 35 after HIV-1NL4.3 infection. rRNA expression was used as a loading control.
FIG. 5.
(A) Inhibition of HIV-1 replication in H9 cells stably expressing tat shRNA. Cells were challenged with 100 TCID50 of HIV-1NL4.3, and p24 antigen levels were assessed over a 2-month period. Viral replication was suppressed by 95% until day 21 compared to control cells. Viral escape started to emerge on day 25, and viral replication was no longer suppressed by day 35, with observed p24 antigen levels being similar to those in control cells. (B) Sequence analysis of HIV-1NL4.3 in culture supernatant at indicated time points. A mutation in position 9 of the tat target sequence emerged at day 25, coincident with the loss of tat shRNA antiviral activity.
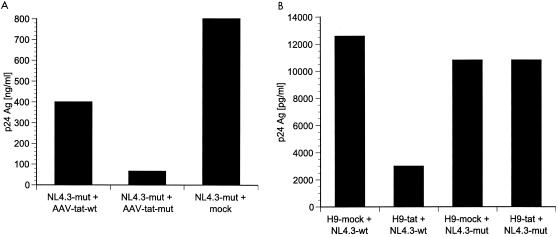
To determine whether the viral escape mutant was indeed resistant to tat shRNA, we created virus HIV-1NL4.3mut and shRNA pAAV-tatmut with a point mutation corresponding to position 9 of the tat target sequence. Cotransfection of 293T cells with HIV-1NL4.3mut and pAAV-tatwt or pAAV-tatmut, respectively, resulted in well-suppressed viral p24 production with pAAV-tatmut (92%) opposed to only moderately reduced virus production with pAAV-tatwt (50%) (Fig. 6A). Next, we infected tat shRNA-expressing (H9-tat) and control (H9-mock) cells with HIV-1NL4.3mut and HIV-1NL4.3wt. Viral challenge of the tat shRNA-expressing cell line with mutant virus HIV-1NL4.3mut did not result in any suppression of viral replication whereas challenge of the same cell line (H9-tat) with wild-type virus HIV-1NL4.3wt showed inhibition of HIV-1 replication by day 10 (Fig. 6B).
FIG. 6.
(A) Cotransfection of HIV-1NL4.3mut with pAAV-tatwt, pAAV-tatmut, and mock vector (pAAV-Δtat). When the targeted tat sequence and shRNA were homologous (HIV-1NL4.3-mut/tat-mut), virus production was reduced by 92%. When the targeted tat sequence and shRNA were not homologous (HIV-1NL4.3-mut/tat-wt), virus production was suppressed by only 50%. (B) H9 cells stably expressing tat shRNA (H9-tat) or Δtat (H9-mock) were infected with 20 ng of p24 of HIV-1NL4.3 or HIV-1NL4.3mut. On day 10 viral replication was reduced with wild-type virus (H9-tat/NL4.3-wt) but not with mutant virus (H9-tat/NL4.3-mut), demonstrating the importance of complete sequence homology between the viral target sequence and the expressed shRNA.
Our results demonstrate a critical obstacle to using RNAi as an antiretroviral agent. The rapid replication kinetics of HIV-1 and the error-prone nature of viral reverse transcriptase led to the emergence and selection of viral quasispecies that were genotypically distinct in the tat shRNA region, thereby eliminating the antiviral effect. The point mutation observed in this study occurred in the middle of the tat target sequence. Previous work has demonstrated that transfection with siRNA containing mismatches to the target sequence in the middle of the siRNA molecule reduces the efficiency of gene silencing (1, 2, 5). Our findings suggest that, in order for RNAi to durably suppress HIV-1 replication, more potent shRNAs need to be designed which target highly conserved regions of the viral genome (e.g., gag and pol) essential for the viral life cycle. Alternatively, similar to present antiviral drug therapy paradigms, RNAi constructs coexpressing multiple shRNAs need to be developed that simultaneously target different regions of the viral genome, thereby reducing the probability of generating shRNA escape mutants.
Acknowledgments
D. Boden and O. Pusch contributed equally to the preparation of the manuscript.
This work was supported by a Clinical Scientist Development Award from the Doris Duke Charitable Foundation, a Daland Fellowship in Clinical Investigation from the American Philosophical Society, and a Career Development Grant from NIAID/NIH (B.R.).
We thank the Brown/Tufts/Lifespan CFAR Retrovirology Core Laboratory for assay support. We acknowledge the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, for supplying H9 cells (contributed by Robert Gallo) and HIV-1NL4.3 (contributed by Malcolm Martin).
REFERENCES
- 1.Amarzguioui, M., T. Holen, E. Babaie, and H. Prydz. 2003. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 31:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2:243-247. [DOI] [PubMed] [Google Scholar]
- 3.Capodici, J., K. Kariko, and D. Weissman. 2002. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J. Immunol. 169:5196-5201. [DOI] [PubMed] [Google Scholar]
- 4.Coburn, G. A., and B. R. Cullen. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76:9225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gitlin, L., S. Karelsky, and R. Andino. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430-434. [DOI] [PubMed] [Google Scholar]
- 6.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20:500-505. [DOI] [PubMed] [Google Scholar]
- 8.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 9.Ramratnam, B., S. Bonhoeffer, J. Binley, A. Hurley, L. Zhang, J. E. Mittler, M. Markowitz, J. P. Moore, A. S. Perelson, and D. D. Ho. 1999. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet 354:1782-1785. [DOI] [PubMed] [Google Scholar]
- 10.Surabhi, R. M., and R. B. Gaynor. 2002. RNA interference directed against viral and cellular targets inhibits human immunodeficiency virus type 1 replication. J. Virol. 76:12963-12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto, T., S. Omoto, M. Mizuguchi, H. Mizukami, H. Okuyama, N. Okada, N. K. Saksena, E. A. Brisibe, K. Otake, and Y. R. Fuji. 2002. Double-stranded nef RNA interferes with human immunodeficiency virus type 1 replication. Microbiol. Immunol. 46:809-817. [DOI] [PubMed] [Google Scholar]