Abstract
When Escherichia coli O157:H7 bacteria are added to alfalfa sprouts growing in water, the bacteria bind tightly to the sprouts. In contrast, laboratory K-12 strains of E. coli do not bind to sprouts under similar conditions. The roles of E. coli O157:H7 lipopolysaccharide (LPS), capsular polysaccharide, and exopolysaccharides in binding to sprouts were examined. An LPS mutant had no effect on the binding of the pathogenic strain. Cellulose synthase mutants showed a significant reduction in binding; colanic acid mutants were more severely reduced, and binding by poly-β-1,6-N-acetylglucosamine (PGA) mutants was barely detectable. The addition of a plasmid carrying a cellulose synthase gene to K-12 strains allowed them to bind to sprouts. A plasmid carrying the Bps biosynthesis genes had only a marginal effect on the binding of K-12 bacteria. However, the introduction of the same plasmid allowed Sinorhizobium meliloti and a nonbinding mutant of Agrobacterium tumefaciens to bind to tomato root segments. These results suggest that although multiple redundant protein adhesins are involved in the binding of E. coli O157:H7 to sprouts, the polysaccharides required for binding are not redundant and each polysaccharide may play a distinct role. PGA, colanic acid, and cellulose were also required for biofilm formation by a K-12 strain on plastic, but not for the binding of E. coli O157:H7 to mammalian cells.
Infections with Escherichia coli O157:H7 result in bloody diarrhea and can progress to hemolytic-uremic syndrome. The disease was first characterized in people who had acquired the bacteria by eating undercooked hamburger meat (5). In recent years, there have been several outbreaks of foodborne illness caused by E. coli O157:H7 and Salmonella enteritidis carried on plant surfaces, as well as on meat products. The bacteria have been associated particularly with alfalfa and radish sprouts and lettuce and spinach leaves (4, 18). The bacteria bind tightly to the plant surface and cannot be removed by water washing (1, 14). Thus, they pose a risk to consumers of those vegetables which are not generally cooked prior to consumption.
S. enteritidis binds rapidly to the surface of sprouts and grows about 3 to 4 logs in their presence (1, 6). E. coli O157:H7 binds very slowly to sprouts and grows less well (2 to 3 logs) in their presence (1, 6, 14). Both species of bacteria appear to prefer roots of sprouts to shoots as a binding site (1, 14). E. coli has not been observed in the internal tissues of water-grown sprouts in studies using bacteria carrying the green fluorescent protein gene (14). Although all pathogenic strains of E. coli tested bind to sprouts, laboratory K-12 strains fail to bind (14). The roles of some of the protein adhesins known to be made by pathogenic strains in binding to sprouts have been examined. Mutations in genes coding for the synthesis of curli or the calcium-binding adhesins Cah-1 and Cah-2 do not affect the ability of O157:H7 strains to bind to sprouts (29). Bacteria with triple mutations in genes encoding these surface-associated adhesins also show no reduction in binding. However, the introduction of plasmids carrying clones of these genes into K-12 strains allows the bacteria to bind to sprouts (29). These results suggest that O157 strains possess several redundant protein adhesins, each of which is sufficient to mediate bacterial binding to sprouts by K-12 strains.
The roles of these protein adhesins in binding to animal cells and plastic were also examined. The binding of the O157:H7 strain 86-24 to Caco-2 human cells was unchanged in Cah-1 and Cah-2 double mutants and was increased in mutants unable to make curli (29). Biofilm formation on polyvinylchloride (PVC) by both the Cah and the curli mutants was reduced. However, the introduction of the genes encoding curli biosynthesis or Cah-1 into K-12 strains ER2267 and DH5α did not increase biofilm formation on PVC (29). Thus, these protein adhesins play different roles in bacterial binding to plant, animal, and abiotic surfaces.
Previous studies have not examined the role of polysaccharides in E. coli binding to sprouts or to mammalian cells. Polysaccharides are known to be involved in the binding of several species of bacteria to plants, including cellulose in the binding of Agrobacterium tumefaciens to dicot hosts (20), succinoglycan in the interaction of Sinorhizobium meliloti with alfalfa roots (7), and alginate in the binding of Pseudomonas fluorescens to carrot roots (11). Polysaccharides are also generally important in the formation of bacterial biofilms on many surfaces, including plants and plastic (26, 27, 33). Diarrheagenic E. coli strains have the potential to make several polysaccharides which are exposed on the surface and so could be involved in bacterial binding; these include the O antigen of lipopolysaccharide (LPS), colanic acid, cellulose, and poly-β-1,6-N-acetyl-d-glucosamine (PGA). These surface polysaccharides are known to be made by some strains under the conditions in which alfalfa sprouts are grown: namely, 25°C and low-osmotic-strength, nutrient-limited medium (12, 13, 34). In this study, we examined the roles of these polysaccharides in bacterial binding to sprouts, mammalian cells, and plastic surfaces.
MATERIALS AND METHODS
Bacterial strains and media.
The strains and plasmids used in this study are listed in Table 1. Bacteria were routinely grown in Luria-Bertani (LB) broth or on LB agar at 37°C. Antibiotics were added to the medium at the following concentrations: kanamycin at 20 μg/ml, chloramphenicol at 30 μg/ml, streptomycin at 100 μg/ml, tetracycline at 12.5 μg/ml, carbenicillin at 100 μg/ml, and neomycin at 20 μg/ml in liquid and 60 μg/ml in solid medium.
TABLE 1.
Properties of bacterial strains and plasmids used in this research
| Bacterial strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| E. coli strains | ||
| 86-24 | E. coli O157:H7 Smr Nalr | A. G. Torres |
| P6C6E25 | 86-24 waaI::Tnp Kmr (truncated LPS) | 32 |
| 8624N | yhjN Carbr (cellulose deficient) | This study |
| 8624C | wcaD Carbr (colonic acid deficient) | This study |
| 8624P | pgaC Carbr (PGA deficient) | This study |
| DEC4A | E. coli O157:H7 | DEC collectiona |
| DEC4AN | yhjN Carbr (cellulose deficient) | This study |
| DEC4AC | wcaD Carbr (colonic acid deficient) | This study |
| DEC4AP | pgaC Carbr (PGA deficient) | This study |
| ER2267 | K-12 F′ proA+B+lacIqΔ(lacZ)M15 zzf::mini-Tn10 (Kanr)/Δ(argF-lacZ)U169 glnV44 e14−(McrA−) rfbD1? recA1 relA1? endA1 spoT1? thi-1 Δ(mcrC-mrr)114::IS10 Kmr | New England Biolabs |
| ER2267O | yhjO Carbr (cellulose deficient) | This study |
| ER2267W | wcaD Carbr (colonic acid deficient) | This study |
| ER2267P | pgaC Carbr (PGA deficient) | This study |
| DH5α | K-12 endA1 hsdR17(rK− mK−) supE44 thi1 recA1 gyrA relA1Δ(lacZYA-argF)U169 deoR[f80dlac Δ(lacZ)M15] | Lab collection |
| Agrobacterium tumefaciens A1045 | C58 wild-type derivative, nonbinding mutant, chvB mutant; Neor | 10 |
| Sinorhizobium meliloti 1021 | Wild type (cellulose deficient) | Lab collection |
| Plasmids | ||
| pBBR1mcs | Cloning vector; Cmr | 16 |
| pBBR1mcs-5 | Cloning vector; Gentr | 15 |
| pBBRcelA | pBBR1mcs-5 Gentr contains the celA gene of A. tumefaciens C58 cloned behind the lac promoter | 21 |
| pMM11 | pBBR1mcs Cmr contains the bps operon of Bordetella bronchiseptica cloned behind the lac promoter | 24 |
DEC strains were obtained from the DEC collection, Thomas Whittam, Michigan State University. Strain descriptions are listed at www.shigatox.net/cgi-bin/deca.
Recombinant DNA techniques.
Plasmid DNA was isolated from 3 ml of overnight bacterial culture by using a Qiagen QIAprep plasmid preparation kit (Qiagen, Valencia, CA) following the manufacturers’ suggested procedure. Plasmid DNA was introduced by transformation into CaCl2-treated competent E. coli strains (28). Standard molecular methods were used for restriction endonuclease analyses and the construction of recombinant plasmids. Mutations in pgaC, yhjO, yhjN, and wcaD were constructed as described by Matthysse and Jeter (14). Briefly, a fragment of the gene obtained by PCR was cloned into pUTgfpΔXho which has a pir-dependent origin of replication. The resulting plasmid was then introduced into the target strain, and a mutant resulting from homologous recombination between the cloned fragment and the bacterial gene was isolated by selecting for carbenicillin resistance carried on the plasmid. The PCR primer pairs used were as follows: pgaC (5′-GCAGGTACCGCGATGCGTTATTAGACCGC-3′ and 5′-CGATCTAGAGCCCTGAGCCCAGCGCAGGCGC-3′), yjhN (5′-GTAACATTCTGTTCCGTGGTGAAAGCG-3′ and 5′-CGCATCGTACGGCTTACGCGGTAGCAGCGG-3′), yhjO (5′-GCCTGCAGTAGATCTGCGAAAGCAGGCAACATCAACAATGCG-3′ and 5′-GCGAGCTCTCCCGGGCCTTACGGGCTGTTCTTCACCTTGTAC-3′), and wcaD (5′-GATGATTGCCTTAGGGATTGGCG-3′ and 5′-CGTCTAGACGCGATTACCACACCAACCAGG-3′).
Assays of binding to alfalfa sprouts and seed coat.
Measurements of bacterial binding to alfalfa sprouts were carried out as previously described (14). Alfalfa sprouts were obtained by germinating seeds. In order to ensure that the alfalfa sprouts were axenic, the seeds were surface sterilized by soaking them briefly in 80% ethanol followed by 20 min in 2.6% sodium hypochlorite solution containing Tween 80 as a wetting agent. The seeds were then washed three times with sterile deionized water and germinated in the dark at 25°C. About 1 in 30 seeds in our present seed lot has bacteria and fungi on the inside of the seed coat (this number is typical of most alfalfa seed lots) (25); these organisms cannot be killed without harming the plant. For most experiments, seeds were germinated in small batches (less than 10 seeds) and those batches which were contaminated were discarded. Contamination was determined by visual and microscopic examination and by plating a sample of the water surrounding the germinating seeds on LB agar.
After 1 day of germination, sprouts (between 0.3 and 0.8 cm long) were placed in plastic dishes in groups of four with 5 ml of sterile deionized water. Bacteria were diluted and added to the germinated seeds to a final concentration of approximately 5 × 103 bacteria per ml. In some cases, 0.2 μg/ml isopropyl-β-d-thiogalactopyranoside (IPTG) was added to induce the expression of genes cloned behind the lac promoter. The inoculated sprouts were incubated at 25°C for 3 days. This time was chosen because previous research has shown that the number of E. coli which can be recovered from sprouts or seed coats reaches a plateau after 2 to 3 days of incubation (14). Less than 10 bacteria per sprout were bound after 1 day (14). The number of free bacteria increased to between 104 and 105 per ml after 1 day and to 105 to 106 after 2 days. At 3 days, the number of free bacteria was between 106 and 107 per ml for the wild type and its mutant derivatives. The number of wild-type bacteria bound was between 104 and 105 per sprout and between 105and 106 per open seed coat. There was 1.2 ml of water present for each sprout. Sprouts and open seed coats were analyzed separately. Each was washed twice by vigorous inversion in 5 ml sterile deionized water. The sprout or seed coat was then homogenized in 5 ml of washing buffer (19) in a Teflon glass homogenizer and plated on McConkey agar containing antibiotics as appropriate to determine viable cell counts. The numbers given are the mean log10 ± the standard deviation of the number of viable bacteria recovered from the homogenate of washed sprouts or seed coats (numbers calculated per sprout or seed coat). All numbers are the results of a minimum of three separate experiments. The average sprout weighed about 50 ± 5 mg (mean ± standard deviation), and the average opened seed coat about 5 ± 3 mg wet weight.
The numbers of bacteria bound would include any internalized bacteria since the sprouts and seed coats were ground before the viable-bacterial-cell count was made. However, in previous experiments we observed no internal E. coli bacteria using strains marked with a plasmid carrying the gfp gene (14). In addition, when the numbers of bacteria released by grinding were compared with the numbers released from sprouts by sonication in a bath sonicator, no significant difference was observed (data not shown). Sonication leaves the sprouts intact and thus would not be expected to release internal bacteria. This is in contrast to the results obtained using older sprouts which were grown on agar, in which Salmonella enteritidis and E. coli bacteria were internalized into the sprouts, apparently largely through the sites of emergence of lateral roots (9). None of our sprouts had any visible lateral roots. In all cases (except for the pgaC mutant, for which few bound bacteria were recovered) the number of bacteria recovered from the wash water (second wash) as measured by viable cell counts was less than 1% of the number recovered from the sprout or seed coat.
Binding to tomato root segments.
Tomato seeds (cv. Rutgers) were surface sterilized and germinated in sterile water as previously described (22). Bacterial binding to root segments was measured as previously described (22). Briefly, when the roots were 5 to 10 cm long, segments approximately 1 cm long were cut from them, placed in 5 ml of sterile water, inoculated with approximately 5 × 103 bacteria per ml (Agrobacterium tumefaciens) or 106 bacteria per ml (Sinorhizobium meliloti), and incubated for 2 or 18 h, respectively, at 25°C. Bound bacteria were released from washed roots by sonication in a bath sonicator and enumerated by viable cell counts on LB agar containing appropriate antibiotics as previously described (22). This method does not reduce the viable-bacterial-cell count but does remove more than 99% of the bacteria from the root surface (22).
Bacterial adhesion assays.
The ability of the E. coli O157:H7 strains to adhere to Caco-2 (human adenocarcinoma intestinal epithelial cells) cells was assessed as previously described (32). The Caco-2 cells were grown to confluence in 24-well plates at 37°C in 5% CO2. Before use, the cells were washed with sterile phosphate-buffered saline (PBS) (pH 7.4) and replenished with Dulbecco's modified Eagle's medium. The cells were then infected with the individual E. coli strains (multiplicity of infection of approximately 100:1) for 3 h at 37°C. To quantify the adherence of the E. coli strains, the infected cells were washed twice with PBS and lysed with 200 μl of 0.1% Triton X-100 in PBS buffer. The adherent bacteria were recovered and plated on LB agar plates. The plates were incubated at 37°C overnight, the colonies counted, and the results expressed as the mean ± standard error of the mean of the results of three independent experiments. Statistical differences were considered significant if the P value was <0.05 as determined by analysis of variance followed by Student's t test.
Biofilm formation on PVC.
The growth of biofilms on plastic was analyzed by the method optimized by Torres et al. (30). Briefly, bacterial strains were grown in polyvinyl chloride (PVC) 96-well plates without shaking in LB or M63 minimal glucose broth at 30°C for a period of 2 to 3 days. The biofilms formed in LB or M63 medium were rinsed and stained with 1% (wt/vol) crystal violet. To quantify the formation of biofilms, the crystal violet-stained cells were solubilized in dimethyl sulfoxide or ethanol, and the optical density (570 nm) of each crystal violet-stained sample was determined by using a microtiter plate reader or a spectrophotometer. The values for at least six independently grown biofilms were used to calculate the average results for each sample. P values were calculated by a paired t test.
RESULTS
We used two strategies to investigate the role of surface carbohydrates in the binding of E. coli O157:H7 to alfalfa sprouts. The first was the examination of the effect of mutations in genes required for the production of various carbohydrates on binding, and the second strategy was the introduction of genes encoding the enzymes required for the synthesis of carbohydrates cloned behind the inducible lac promoter into nonbinding K-12 strains and other nonbinding plant-associated bacteria. In order to obtain a broader picture of the roles of these polysaccharides in the adhesion of the bacteria, the abilities of these strains to bind to mammalian cells and to PVC plastic were also examined.
Effects of mutations in genes required for the synthesis of surface carbohydrates on binding to sprouts.
The O antigen of LPS is one of the prominent carbohydrates exposed on the surface of gram-negative bacteria. However, truncation of the O antigen in the waaI mutant of E. coli O157 strain 86-24 had no effect on binding, suggesting that this polysaccharide is not required for binding (Table 2). Cellulose, colanic acid, and PGA are all reported to be made by various strains of E. coli under conditions similar to those used in studying bacterial binding to sprouts: room temperature (23 to 25°C), low-ionic-strength medium, and nutrient limitation. A mutation in the cellulose synthesis operon (yhjN, also called bscB) reduced the binding of strain 86-24 by approximately 100-fold. A lack of cellulose synthesis had a smaller effect on the binding of DEC4A (another E. coli O157:H7 strain); the binding of the mutant was reduced 10-fold (Table 2). A mutation which prevented the synthesis of colanic acid (wcaD) also reduced the binding of both strain 86-24 (1,000-fold) and strain DEC4A (100-fold). A mutation in pgaC which prevents the synthesis of PGA blocked binding to sprouts. In both the 86-24 and the DEC4A strain, bacteria with the pgaC mutation showed no detectable binding to sprouts and only marginal levels of binding to open seed coats (Table 2). Thus, although PGA appears to be essential for binding to sprouts, both cellulose and colanic acid are required in addition to PGA for maximum binding of E. coli O157. The growth of the parent strain and that of its mutant derivatives in the water surrounding the sprouts were the same, resulting in populations of about 107 bacteria/ml at the end of the incubation. The parent strain shows diffuse binding to sprouts, typically binding end on as individual bacteria or as loose clusters of 5 to 15 bacteria (see Fig. 1 and 2 in reference 14). No bacterial colonies were observed on the surface of sprouts incubated with the parent or mutant strains in these experiments (data not shown).
TABLE 2.
Effects of mutations on binding of E. coli O157:H7 to sprouts and open seed coats
| Bacterial strain | Mutation or genotype (relevant phenotype) | Bacterial binding toa:
|
|
|---|---|---|---|
| Alfalfa sproutsb | Open seed coats | ||
| 86-24 | None (wild type) | 4.7 ± 0.6 | 5.6 ± 0.2 |
| P6C6E25 | waaI (truncated LPS) | 5.7 ± 0.1 | 6.3 ± 0.4 |
| 8624N | yhjN (cellulose deficient) | 2.9 ± 0.7c | 3.5 ± 0.6c |
| 8624C | wcaD (colonic acid deficient) | 1.8 ± 0.7c | 2.4 ± 0.5c |
| 8624P | pgaC (PGA deficient) | <1.0c | 1.0 ± 1.0c |
| DEC4A | None (wild type) | 5.6 ± 0.2 | 6.1 ± 0.3 |
| DEC4AN | yhjN (cellulose deficient) | 4.8 ± 0.8d | 4.1 ± 0.8d |
| DEC4AC | wcaD (colonic acid deficient) | 3.9 ± 0.5c | 4.8 ± 0.8d |
| DEC4AP | pgaC (PGA deficient) | <1.0c | 1.2 ± 0.7c |
Mean ± standard deviation of a minimum of three measurements of the number (log10) of cells bound after 3 days.
Sprouts were washed before measurement.
Significantly different from the wild type: P < 0.01.
Significantly different from the wild type: P < 0.05.
FIG. 1.
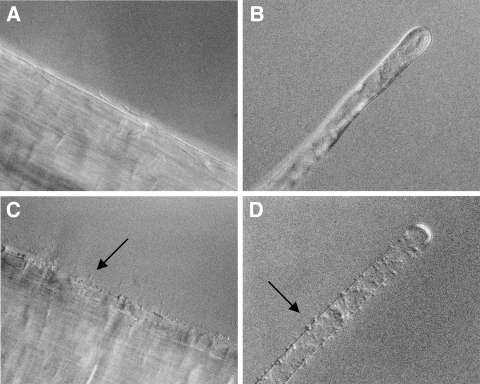
A. tumefaciens A1045 (A and B) and A1045 (pMM11) (C and D) binding to tomato root epidermis (A and C) and individual root hairs (B and D). The arrows point to bound bacteria.
FIG. 2.
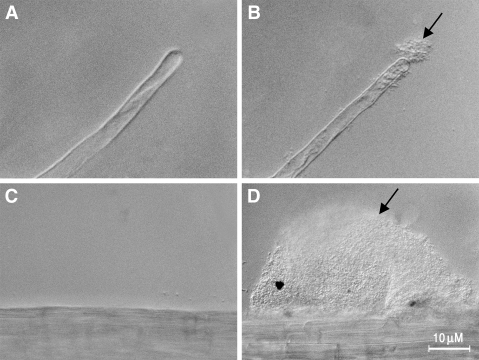
Binding of S. meliloti 1021 (A and C) and S. meliloti 1021(pMM11) (B and D) to tomato root hairs and epidermis. Panels A and B show a single tomato root hair. Panels C and D show the surface of an epidermal cell. The arrows point to bound bacteria.
Effects of introduction of plasmids carrying genes encoding enzymes involved in carbohydrate biosynthesis on binding to sprouts.
Although K-12 strains of E. coli carry genes for the biosynthesis of the polysaccharides found to aid in the binding of O157:H7 strains to sprouts (colanic acid, cellulose, and PGA) the bacteria may not make PGA or cellulose under the conditions used for binding to sprouts. Binding to sprouts was carried out at room temperature in low-ionic-strength medium. Colanic acid is known to be made under these conditions (12). However, the regulation of the synthesis of cellulose and PGA is not well understood. It is possible that the inability of K-12 bacteria to make one or both of these polysaccharides under these conditions is responsible for their failure to bind. When a plasmid carrying the cellulose synthase gene from Agrobacterium tumefaciens (celA; Atu3309) (21) cloned behind the lacZ promoter (plasmid pBBRcelA) was introduced into E. coli K-12 ER2267 or DH5α, the resulting bacteria showed more than a 100-fold increase in binding to sprouts after gene induction with IPTG (Table 3). The introduction of a plasmid carrying the Bps polysaccharide biosynthesis operon (plasmid pMM11) from Bordetella bronchiseptica (24) resulted in a smaller but significant increase in binding to sprouts. The bps locus from B. bronchiseptica encodes a polysaccharide that is biochemically and antigenically similar to the PGA polysaccharides from other bacteria, including the PGA of E. coli (24). As previously described (24), we utilized immunoblotting to confirm that the E. coli strains harboring pMM11 produced the Bps polysaccharide (data not shown). The failure of the production of PGA to allow the binding of K-12 strains to sprouts suggests that PGA is not sufficient by itself to mediate such binding.
TABLE 3.
Effects of plasmids carrying genes for the synthesis of cellulose or PGA on the binding of E. coli K-12 to sprouts
| Bacterial strain | Plasmid (relevant phenotype) | Bacterial binding to alfalfa sproutsa |
|---|---|---|
| ER2267 | None | 0.2 ± 0.2 |
| ER2267 | pBBR1mcs (vector) | 0.5 ± 0.4 |
| ER2267 | pBBRmcs-5 (vector) | 0.3 ± 0.2 |
| ER2267 | pBBRcelA (cellulose synthesis) | 3.4 ± 0.5b |
| ER2267 | pMM11 (PGA synthesis) | 1.5 ± 0.4c |
| DH5α | None | <0.7 |
| DH5α | pBBRcelA (cellulose synthesis) | 3.6 ± 0.2b |
Mean ± standard deviation of a minimum of three measurements of the number (log10) of cells bound after 3 days. Sprouts were washed before measurement.
Significantly different from the wild type: P < 0.01.
Significantly different from the wild type: P < 0.05.
Ability of PGA biosynthesis genes to promote binding of other bacteria to plants.
Since the introduction of a plasmid encoding the PGA biosynthesis operon resulted in only a marginal increase in the binding of K-12 strains, we were unable to use this method to assess the role of PGA in binding to root surfaces. PGA could be directly involved in binding to the root surface, or its role could be indirect; for example, it could be involved in the presentation of other surface molecules in an orientation suitable for their participation in binding. For this reason, we turned to other plant-associated bacteria which fail to bind to roots to examine the possible role of PGA in binding. The chvB mutant of A. tumefaciens fails to bind to a variety of plant surfaces, including tomato root segments, at temperatures above 22°C (3). The ability of this mutant to bind can be restored by the overproduction of cellulose caused by mutations in genes which regulate cellulose biosynthesis (21). Thus, it seemed possible that the synthesis of another exopolysaccharide, PGA, might also be able to restore binding ability to this mutant. Sinorhizobium meliloti 1021 binds to alfalfa sprouts and causes the formation of root nodules. However, it fails to bind to distantly related nonlegumes, such as tomato. Both A. tumefaciens and S. meliloti lack genes for PGA synthesis. In addition, S. meliloti strain 1021 lacks cellulose biosynthesis genes. The plasmid pMM11 carrying the PGA biosynthesis operon was introduced into these two bacteria, and their abilities to bind to tomato root segments were measured. Both bacteria showed significant binding to tomato roots only when they carried pMM11 (Table 4). Microscopic observation showed that strain A1045 bound as single cells to the epidermis and root hairs (Fig. 1). S. meliloti 1021 appeared to be bound as large aggregates on the root hairs and epidermis (Fig. 2). These results suggest that PGA may be directly involved in binding to plant surfaces.
TABLE 4.
Effect of a plasmid carrying genes for the synthesis of PGA on the binding of A. tumefaciens A1045 and S. meliloti 1021 to tomato root segments
| Bacterial strain | Plasmid (relevant phenotype) | Binding to tomato root segmentsa
|
|
|---|---|---|---|
| % of inoculum bound after 2 h | No. (log10) of bacteria bound after 18 h | ||
| A. tumefaciens A1045; chvB mutant | None | 5 ± 5 | |
| A. tumefaciens A1045; chvB mutant | pBBR1mcs (vector) | 5 ± 5 | |
| A. tumefaciens A1045; chvB mutant | pMM11 (PGA synthesis) | 19 ± 6b | |
| S. meliloti 1021 | None | None detected | |
| S. meliloti 1021 | pBBR1mcs (vector) | None detected | |
| S. meliloti 1021 | pMM11 (PGA synthesis) | 5.6 ± 0.5b | |
Mean ± standard deviation of a minimum of 3 measurements.
Significantly different from the parent: P < 0.01.
Effects of mutations in genes required for carbohydrate synthesis on binding to animal cells.
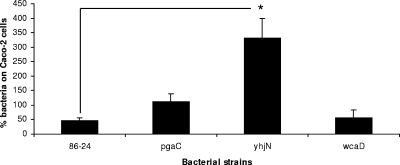
Because E. coli O157:H7 bacteria colonize the colonic intestinal epithelia, we determined whether mutations in genes encoding proteins required for carbohydrate synthesis affect adherence to Caco-2 cells. Our results indicate that mutations in the pgaC and yhjN genes produced increases in adherence compared with the adherence of the wild-type strain (2.4-fold in the case of pgaC [P > 0.05] and 7.2-fold for yhjN [P < 0.05]), which suggests that the absence of PGA and, particularly, the absence of cellulose synthesis have direct effects on the ability of E. coli O157:H7 to bind colonic cells (Fig. 3). In contrast, no change in adherence was observed when the wcaD gene was disrupted.
FIG. 3.
Proportions of E. coli O157 strains 86-24, 8624P (pgaD), 8624N (yhjN), and 8624C (wcaD) adherent to cultured Caco-2 cells after 3 h of incubation. The proportions were calculated in relation to the concentration of the initial inoculum. The error bars indicate the standard deviations of the means. *, P < 0.05.
Effects of mutations in genes required for carbohydrate synthesis on biofilm formation on PVC.
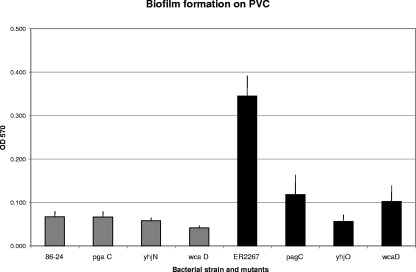
E. coli O157 strain 86-24 grown in LB broth made only a very small biofilm on PVC. The amount of biofilm formed was not reduced in the polysaccharide mutants (Fig. 4). Another O157 strain, DEC4A, failed to form a detectable biofilm on PVC (the optical density at 540 nm of the crystal violet-stained biofilm cells was less than 0.01). The introduction of pMM11 into strain 86-24 failed to significantly increase the biofilm formed, as did using minimal medium instead of LB broth (data not shown). The K-12 strain ER2267 grown in LB broth did form a significant biofilm on PVC. The biofilms of the polysaccharide mutants tested, pgaC, wcaD, and yhjO, were reduced to approximately one-third the amount formed by the parent strain (Fig. 4). Thus, PGA, cellulose, and colanic acid each play a role in biofilm formation by K-12 strains on PVC plastic. All of the strains tested showed approximately equal growth in the planktonic phase.
FIG. 4.
Measurements of crystal violet staining of biofilms formed on PVC plastic by O157:H7 strain 86-24 and its mutant derivatives (gray bars) and by K-12 strain ER2267 and its mutant derivatives (black bars). Numbers shown are the means of the results of at least three separate experiments. Error bars indicate standard errors of the means. The reduction in biofilm formation in the pgaC, yhjO, and wcaD mutants of ER2267 is significant at a P value of <0.05. OD 570, optical density at 570 nm.
DISCUSSION
In contrast to mutants altered in their ability to make surface proteins, bacteria with mutations in genes required for the production of three of the four polysaccharides examined had reductions in their ability to bind to sprouts. It was not surprising that the mutation which results in a truncated O antigen had no effect, since DEC strains with a variety of O and H antigens are able to bind to sprouts (14). However, the other three polysaccharides examined all appeared to play a role in bacterial binding to sprouts and tomato root segments. These roles must be distinct to some extent since mutants with single mutations in each of the polysaccharides were reduced in binding. Although all three polysaccharides were required for maximum binding to plants, mutants unable to produce them were either unaffected (colonic acid) or showed increases in binding to mammalian cells (cellulose). The lack of effect of a mutation in colanic acid biosynthesis was expected since this polysaccharide is typically made by E. coli grown at 25°C and not at 37°C (12). None of these polysaccharides appeared to be required for the small biofilm formed by strain 86-24 on PVC plastic. However, all of them were required for the more-extensive biofilm formed by the nonpathogenic E. coli strain ER2267. PGA has been shown previously to be involved in the binding of E. coli K-12 to plastic (33).
Alfalfa sprouts incubated in water provide only very limited nutrients to bacteria incubated with them. Sugars, dicarboxylic acids, and amino acids secreted by the sprouts are estimated to be present at approximately 1 mM when dense populations of sprouts are germinated (13). More than 90% of the nutrients which were released by the germinating sprouts were released during the first 3 days after germination. After this time, very few nutrients were detected in the water surrounding the sprouts (13). In our experiments, sprouts were present at 1 sprout per 1.25 ml of water, and so the concentration of nutrients would be expected to be less than 1 mM. Under these conditions, the pathogenic strain and its mutants grew from an initial inoculum of approximately 104/ml to about 107/ml. As previously observed, the K-12 strain ER2267 failed to grow or bind (14). Earlier observations suggest that growth is a result of binding rather than vice versa. In mixed populations of ER2267 carrying plasmids which do and do not allow binding to sprouts, only those bacteria which bind grow (14).
When glucose and salts were added to the water, ER2267 bacteria grew but still failed to bind to the sprouts or seed coats (14). ER2267 bacteria carrying plasmids which allowed binding to sprouts grew to 106 to 107 bacteria/ml (14). The addition of pBBRcelA to strain ER2267 caused the bacteria to bind to sprouts and to grow to 104/ml, which represents approximately a 10-fold increase in the number of bacteria present.
The roles of the O antigen, colanic acid, and cellulose in the binding of E. coli bacteria contrast with those reported for Salmonella enterica in which mutations in genes required for the biosynthesis of the O antigen or cellulose disrupt binding to alfalfa sprouts (2). A mutation in colanic acid biosynthesis had no effect on the binding of S. enterica bacteria. These bacteria lack the genes for the biosynthesis of PGA. Thus, S. enterica uses the O antigen and cellulose for binding, while E. coli uses colanic acid, cellulose, and PGA. Interestingly, plants are capable of interacting with E. coli O157:H7 LPS, which has been shown recently to play an important role in plant innate immunity against bacterial infection (23). Treatment of Arabidopsis thaliana leaf discs with E. coli O157:H7 LPS caused the closure of the stomata. However, in contrast to the situation with many plant pathogenic bacteria, which are able to cause the plant to reopen its stomata, the stomata remained closed after contact with E. coli. This response may lead to restricted access of E. coli to the stomata.
In S. enterica, mutations in colanic acid and cellulose biosynthesis genes disrupt biofilm formation on HEp-2 cells but have no effect on biofilm formation on plastic (17). In E. coli, the genes for colanic acid, cellulose, and PGA biosynthesis were all required for biofilm formation on plastic, but none of them were required for binding to Caco-2 cells; the reasons for this difference are unknown. It is possible that the presence of genes for the synthesis of PGA in E. coli may influence the production and/or roles of cellulose and colanic acid in this organism. Data generated by other research groups and ours support the role of fimbrial and afimbrial adhesins as the main mediators of binding to intestinal epithelial cells (31). The role of polysaccharides in the initial binding to human cells seems to be accessory, and it is overshadowed by distinct adhesins expressed upon contact with the cells. Once the initial attachment has been established, it is possible that colonic acid, cellulose, and PGA could play a role during the formation of a biofilm or in persistence in the intestine. We have just recently begun to investigate this possibility.
One of the major differences between plant and animal cells is the hydrophilic polysaccharide plant cell wall which covers the more hydrophobic plasma membrane. In the shoot, the cell wall is covered by the hydrophobic cuticle, but in the root, the hydrophilic cell wall is exposed. The surfaces of animal cells and of PVC plastic are hydrophobic. All three of the polysaccharides involved in binding to plants are hydrophilic and could potentially bind to the hydrophilic surface of the plant root. On animal cells, the presence of the hydrophilic polysaccharide cellulose or PGA appears to inhibit bacterial binding to the hydrophobic cell surface. These data confirm our previous conclusion that E. coli O157:H7 binding to plants and human cells appears to be mediated by overlapping but distinct sets of surface-exposed factors encoded in the genome of this organism (29). The ability of PGA to mediate the binding of other plant-associated bacteria (A. tumefaciens and S. meliloti) to the root surface suggests that it may be able to bind directly to this surface. Microscopic examination of the location of the bound bacteria suggested that binding was not associated with a particular site on the root. The bacteria bound to the epidermis and to root hairs. No preference for epidermal cells in any particular location was observed. Similar observations were made with the nonbinding A. tumefaciens chvB mutant into which the plasmid pBBRcelA was introduced (21). Thus, it appears that both cellulose and PGA mediate binding to the surface of the root in general without any site specificity.
In addition to mediating binding to surfaces, these polysaccharides may be involved in the binding of bacteria to each other to form a biofilm. The role of cellulose in forming aggregates of A. tumefaciens on root surfaces is well documented (20). Danese et al. (8) showed that the role of colanic acid in biofilm formation on plastic is largely to aid in the formation of a deep three-dimensional biofilm. PGA has been shown to participate in binding E. coli K-12 cells to each other as well as in the adherence of the bacteria to plastic (33). Thus, it appears that a likely role for these hydrophilic carbohydrates in biofilm formation on the hydrophobic surface of PVC is in the development of a three-dimensional biofilm.
Acknowledgments
We thank Elizabeth Moulton for assistance with some of the early experiments with colanic acid mutants. We are grateful to Gerald B. Pier for the generous gift of the d-PNAG antibody.
This research was supported by USDA grant 2003-35201-13693 to A.G.M., a grant from Wake Forest University Health Sciences, USDA grant 35604-16874, NIH grant R21 AI071054, an American Heart Association Mid-Atlantic Affiliate grant to R.D., and a grant from the UTMB John Sealy Memorial Endowment Fund for Biomedical Research to A.G.T.
Footnotes
Published ahead of print on 29 February 2008.
REFERENCES
- 1.Barak, J. D., L. C. Whitehand, and A. O. Charkowski. 2002. Differences in attachment of Salmonella enterica serovars and Escherichia coli O157:H7 to alfalfa sprouts. Appl. Environ. Microbiol. 68:4758-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barak, J. D., C. Jahn, D. L. Gibson, and A. O. Charkowski. 2007. The role of cellulose and O-antigen capsule in the colonization of plants by Salmonella enterica. Mol. Plant-Microbe Interact. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 3.Bash, R., and A. G. Matthysse. 2002. Attachment to roots and virulence of a chvB mutant of Agrobacterium tumefaciens are temperature sensitive. Mol. Plant-Microbe Interact. 15:160-163. [DOI] [PubMed] [Google Scholar]
- 4.Breuer, T., D. H. Benkel, R. L. Shapiro, W. N. Hall, M. M. Winnett, M. J. Linn, J. Neimann, T. J. Barrett, S. Dietrich, F. P. Downes, D. M. Toney, J. L. Pearson, H. Rolka, L. Slutsker, and P. M. Griffin. 2001. A multistate outbreak of Escherichia coli O157:H7 infections linked to alfalfa sprouts grown from contaminated seeds. Emerg. Infect. Dis. 7:977-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1997. Outbreaks of Escherichia coli O157:H7 infection associated with eating alfalfa sprouts—Michigan and Virginia, June-July 1997. JAMA 278:809-810. [PubMed] [Google Scholar]
- 6.Charkowski, A. O., J. D. Barak, C. Z. Sarreal, and R. E. Mandrell. 2002. Differences in growth of Salmonella enterica and Escherichia coli O157:H7 on alfalfa sprouts. Appl. Environ. Microbiol. 68:3114-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, H., and G. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180:5183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danese, P., L. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, Y., A. L. Iniguez, B. M. Ahmer, and E. W. Triplett. 2003. Kinetics and strain specificity of rhizosphere and endophytic colonization by enteric bacteria on seedlings of Medicago sativa and Medicago truncatula. Appl. Environ. Microbiol. 69:1783-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas, C. J., W. Halperin, and E. W. Nester. 1982. Agrobacterium tumefaciens mutants affected in attachment to plant cells. J. Bacteriol. 152:1265-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675-690. [DOI] [PubMed] [Google Scholar]
- 12.Gottesman, S., and V. Stout. 1991. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12. Mol. Microbiol. 5:1599-1606. [DOI] [PubMed] [Google Scholar]
- 13.Howard, M. B., and S. W. Hutcheson. 2003. Growth dynamics of Salmonella enterica strains on alfalfa sprouts and in waste seed irrigation water. Appl. Environ. Microbiol. 69:548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeter, C., and A. G. Matthysse. 2005. Characterization of the binding of diarrheagenic strains of E. coli to plant surfaces and the role of curli in the interaction of the bacteria with alfalfa sprouts. Mol. Plant-Microbe Interact. 18:1235-1242. [DOI] [PubMed] [Google Scholar]
- 15.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 16.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 17.Ledeboer, N. A., and B. D. Jones. 2005. Exopolysaccharide sugars contribute to biofilm formation by Salmonella enterica serovar Typhimurium on HEp-2 cells and chicken intestinal epithelium. J. Bacteriol. 187:3214-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lodato, R. J. 2002. Sprout-associated outbreaks. Ann. Intern. Med. 137:372-373. [DOI] [PubMed] [Google Scholar]
- 19.Loper, J., T. Suslow, and M. Schroth. 1984. Lognormal distribution of bacterial populations in the rhizosphere. Phytopathology 74:1454-1460. [Google Scholar]
- 20.Matthysse, A. G. 1983. Role of bacterial cellulose fibrils in Agrobacterium tumefaciens infection. J. Bacteriol. 154:906-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthysse, A. G., M. Marry, L. Krall, M. Kaye, B. E. Ramey, C. Fuqua, and A. R. White. 2005. The effect of cellulose overproduction on binding and biofilm formation on roots by Agrobacterium tumefaciens. Mol. Plant-Microbe Interact. 18:1002-1010. [DOI] [PubMed] [Google Scholar]
- 22.Matthysse, A. G., H. A. Yarnall, and N. Young. 1996. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J. Bacteriol. 178:5302-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melotto, M., W. Underwood, J. Koczan, K. Nomura, and S. He. 2006. Plant stomata function in innate immunity against bacterial invasion. Cell 126:969-980. [DOI] [PubMed] [Google Scholar]
- 24.Parise, G., M. Mishra, Y. Itoh, T. Romeo, and R. Deora. 2007. Role of a putative polysaccharide locus in Bordetella biofilm development. J. Bacteriol. 189:750-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prokopowich, D., and G. Blank. 1991. Microbiological evaluation of vegetable sprouts and seeds. J. Food Prot. 54:560-562. [DOI] [PubMed] [Google Scholar]
- 26.Ramey, B., M. Koutsoudis, S. von Bodman, and C. Fuqua. 2004. Biofilm formation in plant-microbe associations. Curr. Opin. Microbiol. 7:602-609. [DOI] [PubMed] [Google Scholar]
- 27.Ryu, J., H. Kim, and L. R. Beuchat. 2004. Attachment and biofilm formation by Escherichia coli O157:H7 on stainless steel as influenced by exopolysaccharide production, nutrient availability, and temperature. J. Food Prot. 67:2123-2131. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Torres, A. G., C. Jeter, W. Langley, and A. G. Matthysse. 2005. Differential binding of Escherichia coli O157:H7 to alfalfa, human epithelial cells, and plastic is mediated by a variety of surface structures. Appl. Environ. Microbiol. 71:8008-8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres, A. G., N. T. Perna, V. Burland, A. Ruknudin, F. R. Blattner, and J. B. Kaper. 2002. Characterization of Cah, a calcium-binding and heat-extractable autotransporter protein of enterohaemorrhagic Escherichia coli. Mol. Microbiol. 45:951-966. [DOI] [PubMed] [Google Scholar]
- 31.Torres, A. G., X. Zhou, and J. B. Kaper. 2005. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 73:18-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torres, A. G., and J. B. Kaper. 2003. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect. Immun. 71:4985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, X., J. F. Preston III, and T. Romeo. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]