Abstract
Two chemical signaling systems, quorum sensing (QS) and 3′,5′-cyclic diguanylic acid (c-di-GMP), reciprocally control biofilm formation in Vibrio cholerae. QS is the process by which bacteria communicate via the secretion and detection of autoinducers, and in V. cholerae, QS represses biofilm formation. c-di-GMP is an intracellular second messenger that contains information regarding local environmental conditions, and in V. cholerae, c-di-GMP activates biofilm formation. Here we show that HapR, a major regulator of QS, represses biofilm formation in V. cholerae through two distinct mechanisms. HapR controls the transcription of 14 genes encoding a group of proteins that synthesize and degrade c-di-GMP. The net effect of this transcriptional program is a reduction in cellular c-di-GMP levels at high cell density and, consequently, a decrease in biofilm formation. Increasing the c-di-GMP concentration at high cell density to the level present in the low-cell-density QS state restores biofilm formation, showing that c-di-GMP is epistatic to QS in the control of biofilm formation in V. cholerae. In addition, HapR binds to and directly represses the expression of the biofilm transcriptional activator, vpsT. Together, our results suggest that V. cholerae integrates information about the vicinal bacterial community contained in extracellular QS autoinducers with the intracellular environmental information encoded in c-di-GMP to control biofilm formation.
Vibrio cholerae, the causative agent of the diarrheal disease cholera, exists primarily in marine environments, and infection of humans usually occurs through ingestion of contaminated water (45). After traversing the stomach, V. cholerae expresses an array of virulence factors that enable colonization of the host intestinal epithelium. The major colonization factor, toxin coregulated pilus, promotes adherence to the intestinal lining, and the subsequent secretion of cholera toxin leads to severe diarrhea and release of the bacterium into the environment (14). Critical to its infection cycle is V. cholerae's ability to alternate between expression of virulence traits essential for survival inside the host and expression of traits such as biofilm formation that are necessary for survival in its ex vivo marine environment (13, 22, 23, 51, 59). Two chemical signaling systems, quorum sensing (QS) via autoinducer (AI) molecules (see below) and 3′,5′-cyclic diguanylic acid (c-di-GMP) signaling, control the transition between these two lifestyles.
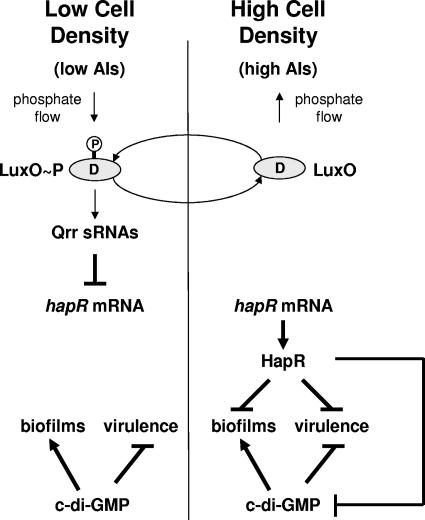
QS is a cell-cell communication process involving the production, secretion, and detection of chemical signal molecules known as AIs that allows bacteria to synchronize the behavior of the population (54). V. cholerae produces two AIs and responds to them by using parallel phosphorelay signaling systems (36). The two AI molecules are termed CAI-1, (S)-3-hydroxytridecan-4-one (19), and AI-2, (2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran borate (5, 36). In the low-cell density state (i.e., when AI levels are low), the AI receptors function as kinases, and funnel phosphate to the response regulator, LuxO (Fig. 1). LuxO∼P activates the expression of four genes encoding the Qrr small regulatory RNAs (sRNAs) (31, 52). The Qrr sRNAs destabilize the mRNA encoding a major regulator of QS, HapR (25, 31). This relay culminates in the expression of low-cell density specific genes, including genes required for biofilm formation and virulence factor production (17, 36, 59, 60) (Fig. 1, left side). When the cell density increases, the AIs accumulate, bind their cognate receptors, and switch the receptors to phosphatases. Phosphatase activity leads to dephosphorylation of LuxO and termination of qrr expression. The mRNA encoding HapR is stabilized, and HapR protein is produced (31, 36). HapR is a DNA-binding transcription factor that initiates a program of gene expression that switches the cells from the individual, low-cell-density state to the high-cell-density state (Fig. 1, right side). When HapR is produced, biofilm and virulence genes are repressed, which is proposed to promote dispersal of V. cholerae (17, 36, 59). The molecular mechanism connecting QS to virulence factor expression is well defined (27, 28). However, the regulatory mechanism linking QS to repression of genes controlling biofilm formation has not been established.
FIG. 1.
Simplified model of the interaction between QS and c-di-GMP in the regulation of gene expression in V. cholerae. When concentrations of AIs are low (left side), the response regulator LuxO is phosphorylated, resulting in expression of multiple genes encoding the Qrr sRNAs that repress translation of the master transcriptional regulator, HapR. When concentrations of AIs are high (right side), LuxO is dephosphorylated which leads to termination of qrr expression. In the absence of the Qrr sRNAs, HapR is produced. HapR represses both biofilm formation and virulence factor expression. Like HapR, c-di-GMP also represses virulence factor expression, but unlike HapR, c-di-GMP activates biofilm formation. Here, HapR is shown to repress biofilm formation both directly (via control of vpsT) and indirectly by reducing the levels of c-di-GMP.
The intracellular second messenger molecule c-di-GMP regulates the transition from a motile lifestyle to a sessile state in numerous bacteria (reviewed in references 7, 24, 39, and 42). Proteins containing domains with GGDEF motifs synthesize c-di-GMP by the cyclization of two GTP molecules and the loss of two pyrophosphate moieties. Degradation of c-di-GMP is carried out by proteins containing domains with EAL or HD-GYP motifs. V. cholerae possesses 62 genes that encode proteins with domains predicted to be involved in governing c-di-GMP levels (16). Consistent with what is known in other bacteria, in V. cholerae, high levels of c-di-GMP enhance biofilm formation and repress virulence factor expression and motility, while low levels of c-di-GMP repress biofilm formation and induce virulence factor expression and motility (2, 50, 51) (Fig. 1). Therefore, c-di-GMP is proposed to mediate the transition of V. cholerae from the in vivo (low c-di-GMP levels) to the ex vivo (high c-di-GMP levels) lifestyle (50, 51).
Here, we show that HapR production at high cell density in V. cholerae represses biofilm formation through two distinct mechanisms. First, HapR alters the expression of 14 genes encoding proteins with GGDEF and/or EAL domains. QS control of these genes results in increased intracellular levels of c-di-GMP at low cell density and decreased c-di-GMP levels at high cell density. The major consequence of increased c-di-GMP in the low-cell-density state is induction of the biofilm transcriptional activator vpsT (4). Second, we show that HapR directly binds to and represses the expression of vpsT, contributing to the reduction in biofilm formation that occurs at high cell density. Artificial production of c-di-GMP in high-cell-density strains to levels that naturally occur in the low-cell-density state induces biofilm production, while artificial reduction of c-di-GMP in the low-cell-density state represses biofilm formation, indicating that c-di-GMP is epistatic to QS in the control of biofilm formation in V. cholerae. We propose that the integration of the c-di-GMP and QS relays allows V. cholerae to combine information about the surrounding bacterial population contained in extracellular chemical QS signals with information regarding local environmental conditions contained in the intracellular chemical signal c-di-GMP to control biofilm formation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All V. cholerae strains used in the present study were derived from the wild-type (WT) El Tor biotype strain C6706str2 (49). The strains carrying mutations in the QS circuit are luxO(D47E) (SLS340) and ΔluxO (SLS349). ΔvpsL mutants were constructed as previously described (48) and are designated CW2034 (ΔvpsL), CW2035 [luxO(D47E) ΔvpsL], CW2036 (ΔhapR ΔvpsL), and CW2037(ΔluxO ΔvpsL). V. cholerae was grown in Luria-Bertani medium (LB) at 37°C with shaking in all cases except in bioluminescence experiments when 30°C growth with shaking was used. Antibiotics were obtained from Sigma and used at the following concentrations (in μg/ml): ampicillin, 100; kanamycin, 100; and chloramphenicol, 10 (unless stated otherwise). qrgB, qrgB* (GGDEF→AAEEF), VC1086, and VC1086* (EAL→AAL) were induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Sigma). The E. coli strains S17-λpir (8) was used for general cloning procedures, while BL21 (35) was used to determine QrgB toxicity in E. coli (data not shown).
DNA manipulations.
DNA manipulation was performed by using standard procedures (44). T4 DNA ligase and restriction enzymes were purchased from New England Biolabs (NEB). PCRs were carried out with iProof DNA polymerase (NEB). qrgB was PCR amplified from V. harveyi WT strain BB120, and VC1086 was amplified from WT V. cholerae. Both were cloned into the EcoRI and BamHI sites of pEVS143 (12). gfp transcriptional fusions were constructed in the plasmid pCMW1 (55) by insertion into the SphI and SalI restriction sites. The qrgB* (AAEEF) and VC1086* (AAL) alleles were constructed by using a QuikChange II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The sequences and designations of all plasmids and primers used are available on request.
gfp and bioluminescence measurements.
V. cholerae strains CW2035 and CW2037 carrying the GGDEF- and/or EAL-gfp transcriptional fusions were grown overnight and diluted 1/100 in triplicate in LB broth with kanamycin. gfp expression was determined by using a BD FACSAria (Becton Dickinson) flow cytometer at 6 h (prestationary phase, optical density = 1.0) and 20 h (stationary phase, optical density = 2.5) by diluting 40 μl of the culture in 1 ml of phosphate-buffered saline. The flow cytometer settings were as follows: forward scatter (log) voltage 400, side scatter (log) voltage 505, and fluorescein isothiocyanate (log) voltage 626. Only genes that showed reproducible twofold changes in regulation between the low- and high-cell-density locked mutants were considered QS regulated. The expression analyses of vpsT, vpsR, and vpsL were performed in Costar 96-well black-bottom plates (catalog no. 3904). The vector control was the plasmid pEVS141 (12). Overnight cultures were diluted 1/100 in triplicate in LB broth with kanamycin (10 μg/ml), chloramphenicol (1 μg/ml), and IPTG, and bioluminescence was determined on an EnVision Multilabel 2103 plate reader (Perkin-Elmer) after 7 h of growth.
Purification of HapR and gel mobility shift assays.
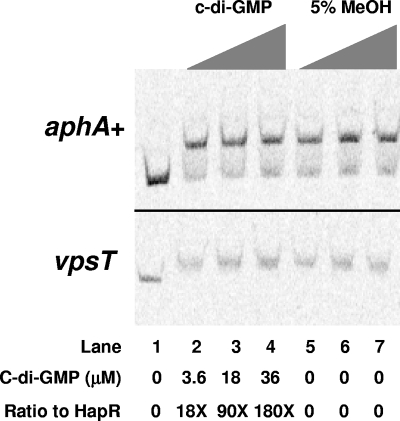
HapR was purified with the IMPACT protein purification system (NEB) using the expression plasmid pTYB11 and the protocol described in the manufacturer's instructions. Purified HapR was stored in 20 mM Tris (pH 7.5), 1 mM EDTA, 10 mM NaCl, and 0.1 mM dithiothreitol with 20% glycerol as previously described (33). DNA probes for gel mobility shift analyses were generated by using 5′-tagged fluorescent primers in a standard PCR and purified by using the Zymoclean gel DNA recovery kit (Zymo Research). Each probe (10 nM) was incubated with the indicated amount of HapR (42 to 1,000 nM) and 1 μl of 1 μg of poly(dI-dC)/ml in a final volume of 20 μl at 30°C for 15 min. Gel mobility shifts were performed on a 5% TAE-polyacrylamide gel and visualized by using a Storm 860 imaging system (Molecular Dynamics). For the experiment shown in Fig. 8, purified c-di-GMP, or an equivalent amount of 5% methanol was preincubated with 200 nM HapR at the indicated concentrations for 15 min at 30°C prior to the addition of the DNA probe.
FIG. 8.
C-di-GMP does not alter HapR DNA binding. HapR binding to the aphA (top panel) and vpsT promoter (bottom panel) was determined in the presence of increasing amounts of c-di-GMP (lanes 2 to 4) or a 5% methanol buffer control (lanes 5 to 7). The concentration of c-di-GMP and the molar ratio of c-di-GMP to HapR are indicated below the panels. Lane 1 contains no HapR, while lanes 2 to 7 have 200 nM HapR.
Determination of intracellular c-di-GMP levels.
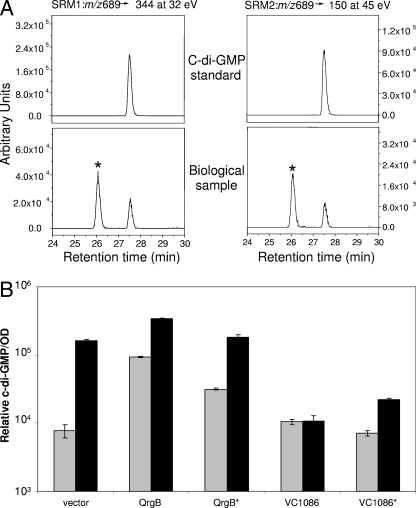
c-di-GMP levels were determined in V. cholerae WT and luxO(D47E) strains containing the vector pEVS141 (12) or overexpressing qrgB, qrgB*, VC1086, and VC1086*. Each strain was inoculated from a frozen stock in 3 ml of LB with kanamycin in an 18X150 mM glass tube and grown overnight with shaking at 37°C. Inoculation of overnight cultures with a previous overnight culture showed lower fold differences in c-di-GMP than direct inoculation from the frozen stock culture (∼3-fold versus 20-fold, respectively). This difference between inoculating techniques may indicate that the c-di-GMP levels decrease over time in stationary phase. C-di-GMP was also measured at early times in the growth curve (optical density at 600 nm of 0.1 to 0.5), but only low level c-di-GMP was detected, and no significant differences were observed between strains. Analysis at low cell densities requires a long centrifugation step that may result in a reduction in c-di-GMP signal. Frozen stock cultures were prepared by diluting 750 μl of an overnight culture with 250 μl of 80% glycerol. After 16 h of growth, 1.5 ml of each culture was pelleted in a benchtop microcentrifuge at 16,100 relative centrifugal force for 30 s. The resulting supernatant was removed, 300 μl of lysis buffer (40% acetonitrile, 40% methanol, 0.1% formic acid) (38) was added, and the pellet was resuspended with a pipette and mixed for 30 s by vortexing, followed by 15 min of incubation on ice. Insoluble material was removed by benchtop centrifugation at 16,100 relative centrifugal force for 5 min at 4°C. The resulting supernatant was collected and analyzed by using liquid chromatography-tandem mass spectrometry on a Finnigan TSQ Quantum DiscoveryMax triple quadrupole mass spectrometer (Thermo Electron Corp., San Jose, CA), coupled with an LC-20AD HPLC system (Shimadzu, Columbia, MD). c-di-GMP was detected by using selected reaction monitoring analyses (SRM) in negative ionization mode. Two SRM were monitored simultaneously, m/z 689→344 at 32 eV and m/z 689→150 at 45 eV, which gave a signal ratio of 1:0.4 (Fig. 4A). The mass spectrometry parameters were as follows: spray voltage, 3,000 V; nitrogen as sheath gas at 30 lb/in2 and auxiliary gas at 10 lb/in2; argon as the collision gas at 1.5 mtorr, and capillary temperature of 325°C. The scan time for each SRM was 0.1 s with a scan width of 1 m/z.
FIG. 4.
c-di-GMP levels in V. cholerae strains. (A) In vivo c-di-GMP levels were determined by measuring ionization fragments with SRM1 (m/z 689→344 at 32 eV [left]) and SRM2 (m/z 689→150 at 45 eV [right]). The top panels show the purified standard at 2 μg/ml in 50:50 methanol-H2O, and the bottom panels show the biological sample. c-di-GMP has a retention time of 27.5 min. The peak labeled with an asterisk in the biological sample is an unknown interfering compound that is neither a conformer nor an isomer of c-di-GMP. (B) Relative levels of c-di-GMP in V. cholerae WT (░⃞) and the luxO(D47E) mutant (▪) containing a vector control or overexpressing QrgB, QrgB* (GGEEF→AAEEF), VC1086, or VC1086* (EAL→AAL) were determined by tandem high-pressure liquid chromatography-mass spectrophotometric analysis. Each strain was analyzed in triplicate. Error bars indicate the standard errors of the mean.
Chromatography conditions for c-di-GMP analyses were reverse phase with the ion pairing agent tributylamine (34). Liquid chromatography separation was achieved on a Synergi Hydro-RP column (4-μm particle size, 150 by 2 mm; Phenomenex, Torrance, CA) at an acidic pH. Solvent A was 10 mM tributylamine plus 15 mM acetic acid in 97:3 water-methanol. Solvent B was methanol. The gradient was as follows: t = 0, 0% solvent B; t = 5 min, 0% solvent B; t = 10 min, 20% solvent B; t = 15 min, 20% solvent B; t = 30 min, 65% solvent B; t = 33 min, 95% solvent B; t = 37 min, 95% solvent B; t = 38 min, 0% solvent B; and t = 45 min, 0% solvent B. Each sample had a running time of 45 min. Under this chromatography condition, c-di-GMP had a retention time of 27.5 min. Other liquid chromatography parameters were as follows: autosampler temperature, 4°C; column temperature, 25°C; injection volume, 10 μl; and solvent flow rate, 200 μl/min. The signal for c-di-GMP in biological samples was defined as the observed peak area on the corresponding chromatography trace. All of the samples had 1 μg of isotopically labeled ATP/μl (>98% 13C, >98% 15N [Medical Isotopes, Pelham, NH]; SRM m/z 521→423 at 21 eV) as the internal standard; no systematic variation in the internal standard was observed. Chemically synthesized c-di-GMP (GLSynthesis, Inc., Worcester, MA, using a modification of methods described previously [26, 40]) was used to determine the retention time and fragmentation pattern of genuine c-di-GMP.
This method had a limit of detection of 5 ng/ml. Running the c-di-GMP standard dissolved in 50:50 methanol-H2O in a series of concentrations (5 ng/ml, 10 ng/ml, 100 ng/ml, 1 μg/ml, and 10 μg/ml) showed that the assay is linear from 5 ng/ml to 10 μg/ml with an R2 of 0.999. In Fig. 4A, representative chromatography traces for the c-di-GMP purified standard and for a typical biological sample are shown. In the biological sample, in addition to c-di-GMP (retention time, 27.6 min), there is also a peak at 26.1 min corresponding to an unknown interfering compound. Accurate mass measurement of the biological sample in positive ion mode was made on an Orbitrap mass spectrometer (Thermo Electron Corp.) and revealed masses for two peaks of 691.1356 and 691.1011 with associated mass errors relative to protonated c-di-GMP (C20H25N10O10P2+; m/z 691.1021) of 48 and 1.5 ppm (the typical mass error on the Orbitrap is <2 ppm) (21). These mass data confirm that the latter peak corresponds to c-di-GMP, while the first peak does not match the chemical formula of c-di-GMP.
Biofilm formation.
Static cultures were grown for 24 h from a 1/100 dilution of an overnight culture that was previously grown shaking at 37°C. Glass tubes (18 by 150 mm) containing 5 ml of LB with kanamycin and IPTG were used, and biofilm formation was monitored by the generation of pellicles at the air-liquid interface.
RESULTS
QS regulates the expression of 14 genes encoding proteins with GGDEF and EAL domains.
Two genes encoding proteins with GGDEF domains, qrgA and qrgB (quorum-regulated GGDEF, denoted promoters 275 and 342 [55]; gene peptide identifications ABU70180.1 and ABU69224.1, respectively), were identified as repressed at high cell density in the model QS bacterium Vibrio harveyi. V. harveyi only poorly produces biofilms, so although the initial link between QS and c-di-GMP regulators was made in V. harveyi, examining their inputs into biofilm formation was difficult to pursue. For this reason, we chose to explore the link between QS, c-di-GMP, and biofilm formation in the related bacterium, V. cholerae, which produces robust biofilms (17, 53, 58, 59).
We inferred, based on the V. harveyi finding, that QS might influence the transcription of genes involved in the production of c-di-GMP in V. cholerae. V. cholerae possesses 31 genes encoding proteins containing GGDEF domains, 10 genes encoding proteins containing GGDEF and EAL domains, and 12 genes encoding proteins with only EAL domains. In addition, V. cholerae has nine genes encoding proteins with HD-GYP domains, a family of proteins that have also been shown to degrade c-di-GMP (11, 16, 41). To determine the extent of QS regulation of genes involved in c-di-GMP signaling in V. cholerae, gfp transcriptional fusions were constructed to 52 of the 53 genes encoding GGDEF- or EAL-containing proteins. Multiple attempts to PCR amplify the promoter of VC0515 were unsuccessful, which suggests it is either not present or is altered in the genome of the V. cholerae strain used here (El Tor biotype strain C6706str2) compared to the sequenced strain (El Tor N16961). For genes encoded in operons, the promoter preceding the first gene was cloned and analyzed. Analysis of the regulation of HD-GYP proteins by QS will be reported elsewhere (B. Hammer and B. Bassler, unpublished data).
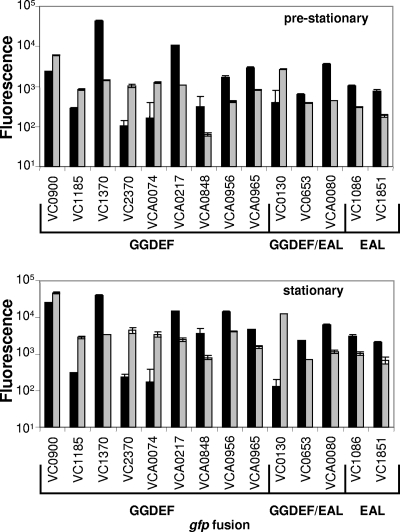
Flow cytometry was used to measure expression of each gfp promoter fusion in a V. cholerae luxO(D47E) mutant that constitutively mimics the low-cell-density state versus the expression in a V. cholerae ΔluxO mutant that constitutively mimics the high-cell-density state. The V. cholerae luxO(D47E) mutant (15) locks the cells at low cell density because it harbors a gain of function allele of luxO that constitutively mimics LuxO∼P, leading to constitutive Qrr sRNA expression and repression of hapR (31) (see Fig. 1, left side). In contrast, the V. cholerae ΔluxO mutant is locked in high cell density because the Qrr sRNAs are not transcribed and, consequently, the hapR mRNA is not destabilized (31) (Fig. 1, right side). The gfp expression of each promoter fusion was determined at two time points, and 14 of the 52 promoters examined exhibited QS regulation (Fig. 2). QS regulation of each of these promoters depended on HapR (data not shown), suggesting that all of these genes lie downstream of the known QS machinery. VCA0074 was previously identified in a microarray analysis aimed at identifying QS regulated genes (59). VCA0074 was also identified in experiments examining the V. cholerae smooth-rugose colony transition and was named cgdA (32).
FIG. 2.
QS regulates 14 genes involved in the control of c-di-GMP. The expression of gfp transcriptional fusions to the promoter of each predicted GGDEF and EAL encoding gene was measured in the high-cell-density ΔluxO strain (▪) and the low-cell-density luxO(D47E) strain (░⃞) at two time points (prestationary, top; and stationary, bottom). Fourteen genes showed twofold or greater regulation. The QS-controlled genes containing only a GGDEF motif (GGDEF), a GGDEF and EAL motif (GGDEF/EAL) and only an EAL motif (EAL) are indicated on the x axis. The error bars show the standard deviation of three independent cultures.
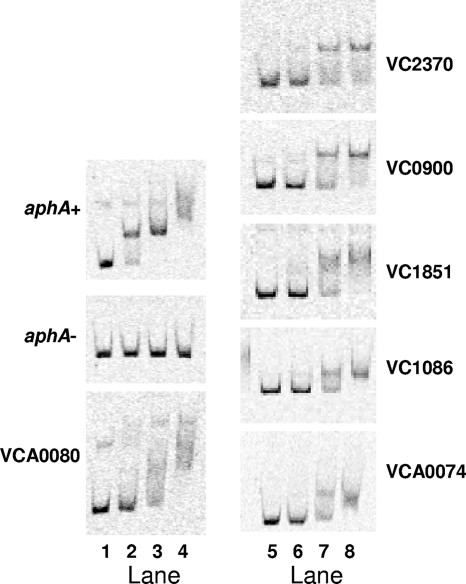
HapR both directly and indirectly controls gene expression (17, 27, 28, 33, 36, 59). For this reason, gel mobility shift assays were used to determine which of the QS-regulated promoters of genes encoding GGDEF and EAL proteins are directly controlled by HapR. Regions of the aphA promoter containing a known HapR binding site (aphA+) and lacking the binding site (aphA−) were used as controls for HapR binding specificity (28). As expected, purified HapR bound and shifted the aphA upstream DNA region containing the HapR binding site but not the DNA lacking the site (Fig. 3). Six of the fourteen QS-regulated promoters driving genes encoding proteins containing GGDEF and/or EAL domains interacted with HapR. Five promoters bound HapR at levels similar to those required for binding to the aphA promoter (right panels, VC2370, VC0900, VC1851, VC1086, and VCA0074), while one promoter required higher levels of HapR and also showed larger, less-specific mobility shifts (left panel, VCA0080). Both QS-activated and QS-repressed promoters were directly bound by HapR. There was no correlation between the fold regulation of transcription and direct interaction with HapR. For example, VC1370 is strongly induced by QS (30.1- and 11.9-fold at the late-exponential phase and stationary phase, respectively) but shows no HapR binding.
FIG. 3.
HapR binds six promoters preceding genes encoding GGDEF and EAL proteins. Gel mobility shifts are shown for the promoters of genes encoding GGDEF and EAL containing proteins that interacted with HapR. HapR concentrations are as follows: lanes; 1 and 5, 0 nM; 2, 80 nM; 3, 420 nM; 4, 1,000 nM; 6, 40 nM; 7, 200 nM; and 8, 500 nM. aphA+, contains the HapR binding site; aphA−, lacks the HapR binding site.
These results indicate that QS, either directly or indirectly through HapR, significantly influences the expression of multiple genes encoding proteins with GGDEF and/or EAL domains. Based on previous studies showing that QS and c-di-GMP reciprocally control biofilm formation in V. cholerae (2, 17, 50, 59) (Fig. 1), we would predict that GGDEF-containing proteins that synthesize c-di-GMP would be repressed by QS and therefore expressed at low cell density (to promote biofilm formation), whereas EAL-containing proteins that degrade c-di-GMP would be activated by QS and dominate at a high cell density (to repress biofilm formation). However, an overall pattern of regulation is not obvious. Of the nine genes encoding proteins with exclusively GGDEF domains, four are repressed by QS, while five are activated. Likewise, one gene encoding a protein with both GGDEF and EAL domains was repressed, while two were activated. Consistent with our prediction, two genes encoding proteins containing only EAL domains were activated by QS. A V. cholerae gene encoding a GGDEF protein (VCA0939) was recently shown to be directly activated posttranscriptionally by the Qrr sRNAs acting independently of HapR (18). QS regulation of the VCA0939-gfp fusion was not observed in the present study, highlighting that the gfp fusions used here report transcriptional, not posttranscriptional, control.
QS controls the intracellular level of c-di-GMP in V. cholerae.
Although HapR-mediated regulation of the transcription of genes encoding GGDEF- and EAL-type proteins did not exhibit a clear pattern, the net result of the transcriptional alterations we observed could translate into modulation of the intracellular pool of c-di-GMP, which in turn could control biofilm formation. To examine this possibility, we developed a high-pressure liquid chromatography-tandem mass spectrometry method to measure the relative concentration of c-di-GMP in bacterial lysates. We measured c-di-GMP in the V. cholerae WT strain to determine the level present at high cell density and in the luxO(D47E) locked low-cell density mutant to determine the level present at low cell density. The retention time and fragmentation pattern of authentic c-di-GMP was determined by using a chemically synthesized standard (a generous gift from George O'Toole). A peak with identical properties to the c-di-GMP standard was observed in each strain (Fig. 4A). Importantly, the luxO(D47E) locked low-cell-density mutant contained 20-fold more c-di-GMP than the WT grown to high cell density (Fig. 4B, vector control). Thus, QS control of transcription of genes encoding GGDEF and EAL proteins indeed alters the intracellular level of c-di-GMP. This result agrees with a previous finding that shows a ΔhapR rugose colony morphotype of V. cholerae (i.e., that also mimics low cell density) has higher levels of c-di-GMP than the smooth morphotype (32). High levels of c-di-GMP at the low-cell-density QS state are consistent with biofilm formation, while decreased c-di-GMP concentrations at the high-cell-density state correspond to reduced biofilm formation.
c-di-GMP is epistatic to QS in control of biofilm formation in V. cholerae.
Increased c-di-GMP at the low-cell-density state relative to the high-cell-density state suggests that QS could control biofilm formation in V. cholerae through modulation of c-di-GMP levels. If so, we reasoned that increasing c-di-GMP levels in V. cholerae at a high cell density by overexpression of an active GGDEF type protein could reestablish biofilm formation under this condition. To test this, we used the GGDEF protein, QrgB, which we had previously studied in V. harveyi (identified as AI-regulated clone 342, ABU69224.1) (55). QrgB was chosen for two reasons. First, overexpression of QrgB was toxic to the E. coli strain BL21, indicating that QrgB functions as a diguanylate cyclase under laboratory conditions (43). Mutation of the QrgB active site (QrgB*, GGEEF→AAEEF) alleviated toxicity, showing that, indeed, toxicity is due to c-di-GMP production (data not shown). Second, V. cholerae strain C6706str2 does not possess a homolog for qrgB. Thus, we reasoned that phenotypes associated with overexpression of qrgB would most likely result from c-di-GMP production rather than another property of the protein, such as interaction with other cellular components.
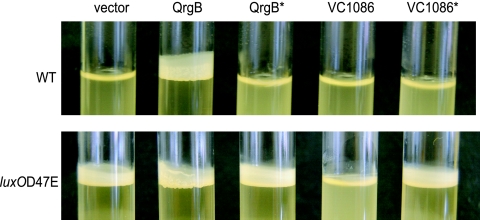
To examine what effect increasing c-di-GMP has on V. cholerae biofilm formation at high cell density, we assayed pellicle formation in the WT strain grown to high cell density and in the locked low-cell-density luxO(D47E) mutant with or without qrgB overexpression (Fig. 5). V. cholerae WT does not normally produce pellicles (17); however, overexpression of qrgB induced biofilm formation in this strain (Fig. 5). This activity was dependent on c-di-GMP production by QrgB because overexpression of the QrgB* active site mutant did not induce biofilm formation (Fig. 5). As expected, the luxO(D47E) strain formed biofilms under each of these conditions.
FIG. 5.
c-di-GMP is epistatic to QS in the control of biofilms. Biofilm formation in overnight static cultures of V. cholerae WT and luxO(D47E) strains containing either the vector alone (denoted vector) or overexpressing QrgB, QrgB* (GGEEF→AAEEF), VC1086, or VC1086* (EAL→AAL).
Using the same logic, we hypothesized that reducing c-di-GMP levels in the luxO(D47E) mutant strain through overexpression of an EAL-type protein should inhibit biofilm formation. For these experiments, the EAL encoded by the locus VC1086 was overexpressed. VC1086 was chosen for two reasons. First, expression of VC1086 is activated directly by HapR (Fig. 2 and 3), suggesting it may be involved in the natural reduction of c-di-GMP levels that occurs at high cell density. Second, VC1086 encodes the key amino acid residues that are predicted to be critical in enzymatically active EAL domains (46). As predicted, overexpression of VC1086 resulted in poor biofilm formation in the luxO(D47E) mutant (Fig. 5). The biofilm reduction caused by VC1086 depended on its c-di-GMP degradation activity because overexpression of an active site mutant of VC1086 (VC1086*, EAL→AAL) did not eliminate biofilm formation (Fig. 5). No biofilms were formed under any condition in a ΔvpsL mutant strain, suggesting the biofilms observed require the known vps extracellular polysaccharide biosynthetic operon (data not shown). Because the production of c-di-GMP at high cell density or removal of c-di-GMP at low cell density overrides QS control of biofilm formation, we conclude that c-di-GMP is epistatic to QS in the control of biofilm formation in V. cholerae.
We measured the corresponding alterations in c-di-GMP that occur after the overexpression of QrgB and VC1086 in the WT strain and the luxO(D47E) mutant (Fig. 4B). The amount of c-di-GMP produced after QrgB overexpression in the WT strain is roughly equivalent to that produced endogenously in locked low-cell-density strains (see second gray bar in Fig. 4B). Thus, we infer that the effect of increasing c-di-GMP on biofilm formation through overexpression of QrgB that we observe at high cell density (Fig. 5) is physiologically relevant and not due to unnaturally high levels of c-di-GMP. A modest but reproducible increase in c-di-GMP levels occurred when the QrgB* active site mutant protein was overexpressed in the WT strain, but this change was significantly less than that after overexpression of WT QrgB. Although we do not understand this result, we note that QrgB encodes a RXXD product inhibition feedback motif (6), which would likely remain functional in the QrgB* allele. One possibility is that QrgB* could bind c-di-GMP at the RXXD site reducing the free intracellular c-di-GMP pool. This reduction in free c-di-GMP may alleviate product inhibition in other GGDEF proteins in the cell, slightly increasing their diguanylate cyclase activities. Overexpression of VC1086 in the luxO(D47E) mutant reduced the c-di-GMP concentration consistent with the ability of VC1086 to repress biofilm formation in this strain. Overexpression of the active site allele of VC1086 (VC1086*) in the luxO(D47E) low-cell-density mutant only partially restored normal c-di-GMP levels. Apparently, this level of c-di-GMP is sufficient to promote biofilm formation in the luxO(D47E) mutant (Fig. 5). Importantly, Fig. 4B shows that similar levels of c-di-GMP are present when the QrgB* allele is overexpressed in the WT strain, and the VC1086* allele is overexpressed in the luxO(D47E) strain. However, the biofilm phenotypes of these two strains are dramatically different (Fig. 5). One notable difference between these two strains is that the WT produces HapR, whereas the luxO(D47E) mutant does not (Fig. 1). This result suggests that, in addition to modulating c-di-GMP levels through the activation and/or repression of transcription of genes encoding GGDEF and EAL proteins, HapR could control biofilm formation through additional mechanisms.
HapR and c-di-GMP reciprocally control vpsT expression.
Biofilm formation in V. cholerae is primarily controlled by two transcriptional activators, VpsR and VpsT, which are required for the expression of the Vibrio polysaccharide biosynthesis operons, vpsA-K and vpsL-Q (4, 56). We previously showed that QS regulates biofilm formation specifically through repression of vpsT (30). QS repression of vpsR has also been reported (57), but we have not observed this (17). Increased c-di-GMP is reported to activate both vpsT and vpsR (2, 50). Because HapR reduces c-di-GMP levels via altering overall GGDEF and EAL activity and because c-di-GMP is epistatic to HapR in the control of biofilm formation (Fig. 5), we predict that HapR and c-di-GMP must function in the same regulatory pathway. Furthermore, because c-di-GMP and HapR have opposite effects on biofilm formation (Fig. 1), we predict that HapR and c-di-GMP must act reciprocally. Given previous studies, the obvious candidate(s) for the point at which QS and c-di-GMP integrate information into biofilm formation is vpsT and/or vpsR.
To examine these hypotheses, we analyzed expression of vpsR, vpsT, and vpsL transcriptional fusions to luciferase in both WT (Fig. 6, gray bars) and the luxO(D47E) mutant (Fig. 6, black bars) V. cholerae strains. We also measured their transcription in the presence of high levels of c-di-GMP (via overexpression of QrgB) and low c-di-GMP levels (via overexpression of VC1086). Overexpression of the QrgB* and VC1086* active-site mutant alleles had no effect on the expression of the vps genes compared to the vector control (data not shown). We note that increasing c-di-GMP levels in the luxO(D47E) mutant causes V. cholerae to aggregate, making quantification of gene expression difficult. For this reason, each of the strains used in this set of experiments carried an in-frame deletion of vpsL to eliminate polysaccharide production and subsequent aggregation.
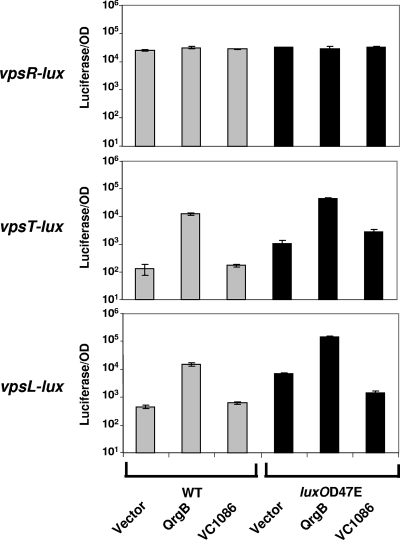
FIG. 6.
QS and c-di-GMP reciprocally regulate vpsT and vpsL. Expression of vpsR-lux (top), vpsT-lux (middle), and vpsL-lux (bottom) was determined in WT (gray bars) and luxO(D47E) (black bars) V. cholerae strains containing a vector control or overexpressing QrgB or VC1086. Error bars indicate the standard deviations of three independent cultures.
Consistent with our previous work showing that QS represses biofilm formation, vpsT and vpsL expression are repressed in the WT strain grown to high cell density relative to the luxO(D47E) locked low-cell-density mutant, whereas no QS regulation of vpsR occurred (Fig. 6, vector controls). In contrast, increasing c-di-GMP activated expression of vpsT and vpsL in both the WT and the luxO(D47E) genetic backgrounds. In the reciprocal experiment, overexpression of VC1086 reduced expression of vpsL-lux (fivefold) in the luxO(D47E) mutant strain, a finding consistent with a reduction in biofilm formation under this condition. Interestingly, overexpression of VC1086 had no effect on vpsT-lux expression. No difference in vpsL or vpsT expression occurred upon overproduction of VC1086 in the WT. This final result is not unexpected since the WT strain has naturally low levels of c-di-GMP (Fig. 4B). Unlike previously published results (2), we observed no differences in vpsR-lux expression upon changes in c-di-GMP levels. From these results, we conclude that the major influence of QS and c-di-GMP on the regulation of biofilm formation occurs through transcriptional changes in vpsT and vpsL. Since VpsT activates the expression of vpsL, the QS and c-di-GMP effects on vpsL transcription are likely due, in a large part, to downstream consequences of regulation of vpsT. However, because overexpression of VC1086 reduced vpsL but not vpsT expression, c-di-GMP levels may also control expression of vpsL through a vpsT-independent mechanism.
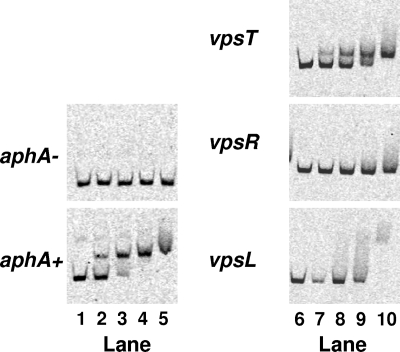
We considered two possible mechanisms for how HapR and c-di-GMP regulate vpsT. First, the effect of HapR on vpsT expression could be exclusively through control of c-di-GMP levels. In this scenario, HapR-controlled reductions in c-di-GMP levels at high cell density are translated (by an unknown mechanism) into transcriptional repression of vpsT expression. However, our results demonstrate that artificially reducing c-di-GMP in the absence of HapR is not sufficient to fully repress vpsT (Fig. 4B and 6). A second possibility is that HapR controls c-di-GMP levels which impinge on vpsT expression, and HapR also regulates the vpsT promoter. To investigate this second possibility, we determined whether HapR could bind to the vpsT (and vpsR and vpsL) promoter by using gel mobility shift assays. Indeed, HapR directly binds the vpsT promoter (Fig. 7). At high concentrations of HapR, we also observed a shift of the vpsL promoter, but we do not know whether this interaction is physiologically relevant. HapR showed no interaction with the vpsR promoter at any concentration examined. From these data, we conclude that, in addition to modulating the levels of c-di-GMP, HapR directly represses vpsT expression. We note that these results support the findings in Fig. 6 that QS regulates biofilm formation exclusively through the repression of vpsT (and not via the repression of vpsR).
FIG. 7.
HapR binds the vpsT promoter. Gel mobility shift analyses of the vpsT, vpsR, and vpsL promoters with purified HapR. HapR amounts are as follows: lanes 1 and 6, 0 nM; 2 and 7, 40 nM; 3 and 8, 120 nM; 4 and 9, 250 nM; and 5 and 10, 500 nM. aphA−, no HapR binding site; aphA+, contains the HapR binding site.
HapR is a member of the TetR family of DNA repressors. This family of proteins typically possesses an N-terminal DNA-binding domain and a C-terminal sensory domain that interacts with a small molecule to modulate its DNA-binding activity (10, 20, 37, 47). Consistent with these findings, the recently reported crystal structure of HapR shows it contains a water-soluble channel in its C-terminal sensory domain that could bind a small molecule ligand (9). A mechanism for reciprocal regulation of vpsT by HapR and c-di-GMP that incorporates these new structural results and our above findings is that HapR binds to c-di-GMP, and this prevents HapR from binding the vpsT promoter. To test this possibility, binding of HapR to the aphA and vpsT promoters was examined in the presence or absence of chemically synthesized c-di-GMP. No difference in DNA binding to either promoter occurred at any c-di-GMP concentration examined (Fig. 8). Therefore, we conclude that HapR represses vpsT expression both by reducing the cellular concentration of c-di-GMP via altering transcription of genes encoding GGDEF and EAL proteins and by directly binding to the vpsT promoter.
DISCUSSION
Bacteria use both intra- and extracellular chemical signals to couple information about their physiology, the surrounding environment, and the vicinal bacterial community into changes in gene expression (3). Detection of this sensory information is vital to direct proper responses to fluctuating environmental conditions. For V. cholerae, this response can be simplified to a transition between in vivo (virulence) or ex vivo (biofilm) developmental pathways. Although virulence factors such as the toxin-coregulated pilus and cholera toxin promote adhesion and nutrient acquisition inside the host and subsequently, dissemination into the environment at high numbers, biofilm formation enables V. cholerae to survive in nutrient-poor conditions outside of the host (22, 23, 59). Biofilm formation also increases the infectivity of V. cholerae, but, importantly, dispersal of biofilms is thought to occur once the bacteria colonize the host (13, 59). Two chemical signaling pathways converge to control the transition between these two critical developmental fates, QS and c-di-GMP (17, 36, 50, 51).
Based on our previous discovery of a link between QS and transcriptional regulation of GGDEF proteins in V. harveyi (55), we hypothesized that QS might regulate biofilm formation in V. cholerae through modulation of intracellular c-di-GMP levels. A number of lines of evidence suggest that this model is correct. First, HapR regulates the transcription of 14 genes encoding proteins harboring GGDEF and/or EAL domains. Second, the intracellular concentration of c-di-GMP is higher in locked low-cell-density strains than in locked high-cell-density strains. Third, production of c-di-GMP at high cell density at levels analogous to those present in the low-cell-density state induces biofilm formation, showing that c-di-GMP is epistatic to HapR in the control of biofilms. Finally, HapR and c-di-GMP converge to control the identical regulatory component in the biofilm transcriptional cascade, vpsT. In addition to regulating vpsT through the modulation of c-di-GMP levels, HapR directly represses vpsT expression.
The influence of HapR on biofilm formation through the transcriptional regulation of GGDEF- and EAL-type proteins likely depends heavily on environmental stimuli that control GGDEF and EAL activities. In contrast, direct repression of vpsT by HapR is likely only responsive to changes in the surrounding bacterial population and thus less sensitive to prevailing environmental conditions. Repression of vpsT requires the simultaneous presence of HapR and low c-di-GMP levels because either the absence of HapR or high c-di-GMP levels induces expression of vpsT (Fig. 6). As mentioned above, c-di-GMP signaling is epistatic to QS signaling because high c-di-GMP levels, which presumably reflect ex vivo conditions, override HapR-directed repression of vpsT (Fig. 6), and ultimately biofilm formation (Fig. 5). This final finding suggests that, under environmental conditions favoring high c-di-GMP levels, V. cholerae could produce biofilms irrespective of cell density.
How changes in c-di-GMP concentrations are coupled to changes in the transcription of target genes in bacteria is not known. We examined whether HapR could be a component of this unknown regulatory cascade, but our results showed no alteration in HapR DNA binding in the presence of c-di-GMP. It remains possible that HapR does bind c-di-GMP and that binding modulates the activity of HapR without disrupting gross DNA binding, which would not be detected in our assay. The bet and mer systems in E. coli both contain transcriptional repressors whose activities are modulated by interaction with cognate ligands while DNA binding is not affected (1, 29).
Although QS regulates the transcription of 14 GGDEF- and/or EAL-encoding proteins, we predict that this is but one layer of regulation modulating intracellular c-di-GMP concentrations. Most of these proteins have N-terminal sensory domains that are hypothesized to recognize specific environmental cues (7, 42). Recognition of these cues elicits changes in the enzymatic activities of the corresponding C-terminal GGDEF or EAL domains. Therefore, environmental conditions greatly influence c-di-GMP levels. Our results demonstrate that global HapR transcriptional control of genes encoding GGDEF- and EAL-type proteins indeed alters c-di-GMP levels, suggesting that, at least in some cases, changes in transcription lead directly to changes in net c-di-GMP biosynthesis or degradation. This finding implies that at least some of the cues recognized by the 14 GGDEF/EAL proteins controlled by QS are present in our experimental setup. However, our experiments do not sample changes in the local environment that would normally be experienced by V. cholerae transitioning between aquatic reservoirs in the environment and in vivo niches inside of a host. These environmental changes, coupled with QS-controlled transcriptional regulation of genes encoding GGDEF and EAL proteins, could uniquely set the optimal intracellular c-di-GMP concentrations required for survival in various environments, as well as during the transitions between them. Because we are not currently able to simulate these environments, many of the cues detected by proteins controlling c-di-GMP levels are likely not present under laboratory conditions. Thus, we suspect that the results we show here, and specifically the changes in c-di-GMP we observe in cells, likely underestimate the full range of c-di-GMP concentrations, and thus the full range of phenotypic changes, the cells naturally experience.
QS and c-di-GMP are two important chemical signaling relays in V. cholerae. Each of these relays alternates between two distinct states. Low- and high-cell-density QS states reflect the density and complexity of the surrounding bacteria in the local environment, while low and high intracellular levels of c-di-GMP potentially reflect in vivo versus ex vivo environments (51). These two signaling relays converge to regulate biofilms and virulence factor expression, processes fundamental for V. cholerae survival ex vivo and in vivo, respectively. Interestingly, although each signaling relay transmits information into both processes, QS and c-di-GMP function antagonistically to control biofilm formation, while they act synergistically to repress virulence. Likewise, although high c-di-GMP levels override QS control of biofilm formation, it is not known which process dominates during virulence factor expression. Apparently, V. cholerae parses the unique information carried by these two chemical signaling processes appropriately to regulate particular behaviors depending on the relative importance of cell density and environmental inputs. The plasticity with which V. cholerae can convert these two chemical inputs into unique gene expression patterns could provide V. cholerae tremendous flexibility in adapting to a fluctuating environment.
Acknowledgments
We thank George O'Toole for providing chemically synthesized c-di-GMP and B. Hammer, W. Ng, S. Svenningsen, and K. Tu for strain construction and critical reading of the manuscript.
This study was supported by HHMI, NIH grant 5R01GM065859, and NSF grant MCB-0343821 to B.L.B. and by NIH postdoctoral fellowship F32-AI0586492 to C.M.W.
Footnotes
Published ahead of print on 25 January 2008.
REFERENCES
- 1.Ansari, A. Z., J. E. Bradner, and T. V. O'Halloran. 1995. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature 374:371-375. [DOI] [PubMed] [Google Scholar]
- 2.Beyhan, S., A. D. Tischler, A. Camilli, and F. H. Yildiz. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 188:3600-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camilli, A., and B. L. Bassler. 2006. Bacterial small-molecule signaling pathways. Science 311:1113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186:1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 6.Christen, B., M. Christen, R. Paul, F. Schmid, M. Folcher, P. Jenoe, M. Meuwly, and U. Jenal. 2006. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 281:32015-32024. [DOI] [PubMed] [Google Scholar]
- 7.Cotter, P. A., and S. Stibitz. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 10:17-23. [DOI] [PubMed] [Google Scholar]
- 8.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 9.De Silva, R. S., G. Kovacikova, W. Lin, R. K. Taylor, K. Skorupski, and F. J. Kull. 2007. Crystal structure of the Vibrio cholerae quorum sensing regulatory protein HapR. J. Bacteriol. 189:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dover, L. G., P. E. Corsino, I. R. Daniels, S. L. Cocklin, V. Tatituri, G. S. Besra, and K. Futterer. 2004. Crystal structure of the TetR/CamR family repressor Mycobacterium tuberculosis EthR implicated in ethionamide resistance. J. Mol. Biol. 340:1095-1105. [DOI] [PubMed] [Google Scholar]
- 11.Dow, J. M., Y. Fouhy, J. F. Lucey, and R. P. Ryan. 2006. The HD-GYP domain, cyclic di-GMP signaling, and bacterial virulence to plants. Mol. Plant-Microbe Interact. 19:1378-1384. [DOI] [PubMed] [Google Scholar]
- 12.Dunn, A. K., D. S. Millikan, D. M. Adin, J. L. Bose, and E. V. Stabb. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faruque, S. M., K. Biswas, S. M. Udden, Q. S. Ahmad, D. A. Sack, G. B. Nair, and J. J. Mekalanos. 2006. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. USA 103:6350-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faruque, S. M., and G. B. Nair. 2002. Molecular ecology of toxigenic Vibrio cholerae. Microbiol. Immunol. 46:59-66. [DOI] [PubMed] [Google Scholar]
- 15.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31:665-677. [DOI] [PubMed] [Google Scholar]
- 16.Galperin, M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6:552-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 18.Hammer, B. K., and B. L. Bassler. 2007. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. USA. 104:11145-11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgins, D. A., M. E. Pomianek, C. M. Kraml, R. K. Taylor, M. F. Semmelhack, and B. L. Bassler. 2007. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450:883-886. [DOI] [PubMed] [Google Scholar]
- 20.Hinrichs, W., C. Kisker, M. Duvel, A. Muller, K. Tovar, W. Hillen, and W. Saenger. 1994. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science 264:418-420. [DOI] [PubMed] [Google Scholar]
- 21.Hu, Q., R. J. Noll, H. Li, A. Makarov, M. Hardman, and R. G. Cooks. 2005. The Orbitrap: a new mass spectrometer. J. Mass Spectrom. 40:430-443. [DOI] [PubMed] [Google Scholar]
- 22.Huq, A., E. B. Small, P. A. West, M. I. Huq, R. Rahman, and R. R. Colwell. 1983. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 45:275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huq, A., P. A. West, E. B. Small, M. I. Huq, and R. R. Colwell. 1984. Influence of water temperature, salinity, and pH on survival and growth of toxigenic Vibrio cholerae serovar 01 associated with live copepods in laboratory microcosms. Appl. Environ. Microbiol. 48:420-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenal, U., and J. Malone. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385-407. [DOI] [PubMed] [Google Scholar]
- 25.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023-1034. [DOI] [PubMed] [Google Scholar]
- 26.Kawai, R., R. Nagata, A. Hirata, and Y. Hayakawa. 2003. A new synthetic approach to cyclic bis(3′→5′)diguanylic acid. Nucleic Acids Res. Suppl. 3:103-104. [DOI] [PubMed] [Google Scholar]
- 27.Kovacikova, G., W. Lin, and K. Skorupski. 2003. The virulence activator AphA links quorum sensing to pathogenesis and physiology in Vibrio cholerae by repressing the expression of a penicillin amidase gene on the small chromosome. J. Bacteriol. 185:4825-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovacikova, G., and K. Skorupski. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 46:1135-1147. [DOI] [PubMed] [Google Scholar]
- 29.Lamark, T., T. P. Rokenes, J. McDougall, and A. R. Strom. 1996. The complex bet promoters of Escherichia coli: regulation by oxygen (ArcA), choline (BetI), and osmotic stress. J. Bacteriol. 178:1655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenz, D. H., M. B. Miller, J. Zhu, R. V. Kulkarni, and B. L. Bassler. 2005. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 58:1186-1202. [DOI] [PubMed] [Google Scholar]
- 31.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 32.Lim, B., S. Beyhan, J. Meir, and F. H. Yildiz. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60:331-348. [DOI] [PubMed] [Google Scholar]
- 33.Lin, W., G. Kovacikova, and K. Skorupski. 2005. Requirements for Vibrio cholerae HapR binding and transcriptional repression at the hapR promoter are distinct from those at the aphA promoter. J. Bacteriol. 187:3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo, B., K. Groenke, R. Takors, C. Wandrey, and M. Oldiges. 2007. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway, and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J. Chromatogr. A 1147:153-164. [DOI] [PubMed] [Google Scholar]
- 35.McClain, M. S., I. C. Blomfield, K. J. Eberhardt, and B. I. Eisenstein. 1993. Inversion-independent phase variation of type 1 fimbriae in Escherichia coli. J. Bacteriol. 175:4335-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 37.Natsume, R., R. Takeshita, M. Sugiyama, Y. Ohnishi, T. Senda, and S. Horinouchi. 2003. Crystallization of CprB, an autoregulator-receptor protein from Streptomyces coelicolor A3(2). Acta Crystallogr. D Biol. Crystallogr. 59:2313-2315. [DOI] [PubMed] [Google Scholar]
- 38.Rabinowitz, J. D., and E. Kimball. 2007. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal. Chem. 79:6167-6173. [DOI] [PubMed] [Google Scholar]
- 39.Romling, U., M. Gomelsky, and M. Y. Galperin. 2005. C-di-GMP: the dawning of a novel bacterial signaling system. Mol. Microbiol. 57:629-639. [DOI] [PubMed] [Google Scholar]
- 40.Ross, P., R. Mayer, H. Weinhouse, D. Amikam, Y. Huggirat, M. Benziman, E. de Vroom, A. Fidder, P. de Paus, L. A. Sliedregt, et al. 1990. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinu: chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J. Biol. Chem. 265:18933-18943. [PubMed] [Google Scholar]
- 41.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 103:6712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Ryan, R. P., Y. Fouhy, J. F. Lucey, and J. M. Dow. 2006. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J. Bacteriol. 188:8327-8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Sanchez, J., and J. Holmgren. 2005. Virulence factors, pathogenesis and vaccine protection in cholera and ETEC diarrhea. Curr. Opin. Immunol. 17:388-398. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt, A. J., D. A. Ryjenkov, and M. Gomelsky. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2002. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 21:1210-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 49.Thelin, K. H., and R. K. Taylor. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 64:2853-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tischler, A. D., and A. Camilli. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73:5873-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tu, K. C., and B. L. Bassler. 2007. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 21:221-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 55.Waters, C. M., and B. L. Bassler. 2006. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 20:2754-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497-515. [DOI] [PubMed] [Google Scholar]
- 58.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647-656. [DOI] [PubMed] [Google Scholar]
- 60.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]