Abstract
Trehalose supports the growth of Thermus thermophilus strain HB27, but the absence of obvious genes for the hydrolysis of this disaccharide in the genome led us to search for enzymes for such a purpose. We expressed a putative α-glucosidase gene (TTC0107), characterized the recombinant enzyme, and found that the preferred substrate was α,α-1,1-trehalose, a new feature among α-glucosidases. The enzyme could also hydrolyze the disaccharides kojibiose and sucrose (α-1,2 linkage), nigerose and turanose (α-1,3), leucrose (α-1,5), isomaltose and palatinose (α-1,6), and maltose (α-1,4) to a lesser extent. Trehalose was not, however, a substrate for the highly homologous α-glucosidase from T. thermophilus strain GK24. The reciprocal replacement of a peptide containing eight amino acids in the α-glucosidases from strains HB27 (LGEHNLPP) and GK24 (EPTAYHTL) reduced the ability of the former to hydrolyze trehalose and provided trehalose-hydrolytic activity to the latter, showing that LGEHNLPP is necessary for trehalose recognition. Furthermore, disruption of the α-glucosidase gene significantly affected the growth of T. thermophilus HB27 in minimal medium supplemented with trehalose, isomaltose, sucrose, or palatinose, to a lesser extent with maltose, but not with cellobiose (not a substrate for the α-glucosidase), indicating that the α-glucosidase is important for the assimilation of those four disaccharides but that it is also implicated in maltose catabolism.
α-Glucosidases (EC 3.2.1.20) are a widespread group of enzymes that catalyze the hydrolysis of the α-glucosidic bond from the nonreducing end of a chain as well as the α-glucosidic bond of free disaccharides (21, 23). Many known α-glucosidases seem to prefer the α-1,4 bonds of maltose or maltooligosaccharides (21). However, the α-glucosidases from Thermus thermophilus strain GK24 and from Bacillus sp. strain SAM1606 preferentially hydrolyze the α-1,6 bond of isomaltose over other α linkages (20, 21, 23). Trehalose is a natural disaccharide that is widespread in nature and that can serve multiple roles, namely, as a general stress protectant, a source of carbon and energy, a sensing and regulatory compound, and a structural element of the bacterial cell wall (11). The assimilation of trehalose as a carbon and energy source requires the activity of enzymes that hydrolyze the α-1,1 bond of this disaccharide, including trehalase (EC 3.2.1.28), trehalose phosphorylase (EC 2.4.1.64 and EC 2.4.1.231), and other α-glucosidases (EC 3.2.1.20) with broad specificity (14, 16). These enzymes may also play important roles in the regulation of the level of trehalose in a cell. Trehalose can also be converted into maltose by trehalose synthase (TreS; EC 5.4.99.16) and then metabolized via amylomaltase, maltose/maltodextrin phosphorylase, or other α-glucosidases with α-1,4-linkage hydrolytic activity (7, 14, 18). Although T. thermophilus HB27 lacks the genetic machinery for trehalose biosynthesis, this disaccharide is taken up from the medium (2, 24) but does not serve as a compatible solute for strain HB27, unlike the majority of the strains of this species. In this organism, mannosylglycerate is the sole osmolyte under salt stress (2), indicating that the disaccharide is used only as a carbon source.
To understand the mechanisms involved in trehalose metabolism in T. thermophilus HB27, we attempted to identify the enzyme responsible for its hydrolysis. Unlike the HB8 and GK24 strains, the HB27 strain lacks a TreS gene and should not be able to convert trehalose into maltose. Additionally, no obvious trehalase or trehalose phosphorylase genes could be found in the HB27 genome (12). Hence, we hypothesized that gene TTC0107 could encode an enzyme required for trehalose utilization, although the highly homologous enzyme from strain GK24 was shown to lack trehalose-hydrolyzing activity (21). In fact, trehalose is the preferred substrate for the HB27 enzyme, which is, to our knowledge, the first report of an α-glucosidase with this property. To examine the importance of the affinity of a distinctive peptide containing eight amino acids from the HB27 α-glucosidase that is not found in the homologous GK24 enzyme for trehalose, we created site-specific mutant α-glucosidases from both strains. We also constructed an α-glucosidase disruption mutant to investigate the specific role of this enzyme in strain HB27 and studied the organism's adaptation to specific nutritional conditions.
MATERIALS AND METHODS
Strains and plasmids.
Thermus thermophilus strains HB27 (DSM 7039) and HB8 (DSM 579) were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany, and T. thermophilus strain GK24 (27) was a gift from Richard Sharp of PHLS, Salisbury, United Kingdom. Escherichia coli DH5α and expression vector pTRC99A were used for the cloning and expression of α-glucosidase genes from T. thermophilus strains HB27 (aglHHB27), GK24 (aglHGK24), and HB8 (aglHHB8). Plasmid pGEM-T Easy was used to accommodate the disrupted HB27 aglH gene and for the disruption of this gene in the strain HB27 chromosome as described below. Plasmid pMK18 was used as the source for the kanamycin (Kan) resistance cassette (9).
Amplification and cloning of the α-glucosidase genes.
All PCRs were performed with the GC-Rich PCR system kit (Roche) according to the manufacturer's instructions. DNA from strains HB27, GK24, and HB8 was isolated and used as a template for the amplification of α-glucosidase genes (22). The primer sequences are listed in Table S1 in the supplemental material. Primers AG1 and AG2 were designed with additional EcoRI and HindIII sites, respectively, and used for the amplification of the HB27 aglH gene (TTC0107). Primer AG3 was designed based on the 5′ end of the α-glucosidase gene from strain GK24 (aglHGK24) (GenBank accession number AF096282) with an EcoRI site and used with primer AG2 for the amplification of the aglHGK24 and aglHHB8 genes. The products were cloned into pTrc99A and sequenced (AGOWA GmbH, Berlin, Germany).
Expression and purification of recombinant enzymes.
The constructs were transformed into E. coli DH5α, and the recombinant bacteria were grown and induced as previously described (26). Cells were harvested and suspended in 25 mM Bis-Tris propane buffer (BTP), pH 7.5, containing a protease inhibitor cocktail (Roche), DNase I (1 μg ml−1), and 10 mM MgCl2, followed by disruption on a French press. Cell debris was removed, and cell extracts were heat denatured to remove the majority of the host proteins.
Recombinant α-glucosidases were purified with two sequential Q-Sepharose columns (Hi-Load 16/10 Q FF) equilibrated with 25 mM BTP, pH 7.5, at a constant flow rate of 3 ml/min. Elution was carried out with linear NaCl gradients (0.0 to 1.0 M), and activity was determined as described below. Active fractions were concentrated and loaded onto a Superdex 200 column equilibrated with 50 mM BTP and 0.2 M NaCl, pH 7.5, at a constant flow rate of 1 ml/min. The purest fractions were pooled, concentrated, equilibrated with 25 mM BTP (pH 7.5), and stored at −20°C. The molecular mass of the HB27 recombinant α-glucosidase was estimated by gel filtration as previously described (8).
Enzyme assays and substrate specificity.
The activities of the recombinant α-glucosidases were determined by measuring the release of glucose using the glucose oxidase assay kit (Sigma), according to the manufacturer's instructions. Standard reaction conditions were 25 mM of BTP at pH 7 with 20 mM CaCl2 and incubation at 70°C. Mixtures containing 10 mM of substrate were incubated with 0.5 μg of pure recombinant α-glucosidase for 20 min, unless stated otherwise. Alternative reaction conditions used for the determination of activities with maltose and maltotriose were 25 mM of BTP at pH 7 with 20 mM CaCl2, 100 mM of each substrate, and 4 μg of pure enzyme incubated for 1 h at 70°C. Several di-, tri-, and polysaccharides—namely, starch, amylopectin, glycogen, and pullulan—were tested as possible substrates (Table 1). Trehalose-6-phosphate was also tested as a possible substrate for the HB27 α-glucosidase (AglHHB27). The reverse reaction was tested with different combinations of 5 mM (each) ADP-glucose, GDP-glucose, UDP-glucose, glucose, glucose-6-phosphate, or glucose-1,6-bisphosphate. To test transglucosylation, standard reaction mixtures containing trehalose, isomaltose, maltose, or glucose (5 to 500 mM) were incubated with 2 μg of pure recombinant AglHHB27 at 90°C for 24 h and cooled on ice. Products were visualized by thin-layer chromatography as previously described (5).
TABLE 1.
Substrate specificity of recombinant α-glucosidases (AglH) from Thermus thermophilus strains HB27, HB8, and GK24
| Substrate | Glycosidic linkage | Glucose released (μmol/min·mg protein)a
|
||
|---|---|---|---|---|
| AglHHB27 | AglHHB8 | AglHGK24 | ||
| Disaccharides | ||||
| α,α-Trehalose | O-α-d-Glucosyl-(1→1)-d-glucose | 126 ± 5.3 | 0.0 | 0.0 |
| α,β-Trehalose | O-α-d-Glucosyl-(1→1)-β-d-glucose | 49 ± 3.6 | 0.0 | 0.0 |
| β,β-Trehalose | O-β-d-Glucosyl-(1→1)-β-d-glucose | 0.0 | 0.0 | 0.0 |
| Kojibiose | O-α-d-Glucosyl-(1→2)-d-glucose | 26 ± 3.1 | 31 ± 3.3 | 38 ± 4.7 |
| Sucrose | O-α-d-Glucosyl-(1→2)-β-d-fructose | 10 ± 1.1 | 9 ± 4.1 | 67 ± 3.4 |
| Nigerose | O-α-d-Glucosyl-(1→3)-d-glucose | 20 ± 1.1 | 36 ± 8.1 | 56 ± 2.2 |
| Turanose | O-α-d-Glucosyl-(1→3)-d-fructose | 18 ± 3.6 | 28 ± 3.5 | 3 ± 3.4 |
| Maltose | O-α-d-Glucosyl-(1→4)-d-glucose | 12 ± 0.2b | 3.1 ± 0.2b | 1.8 ± 0.1b |
| Cellobiose | O-β-d-Glucosyl-(1→4)-d-glucose | 0.0 | 0.0 | 0.0 |
| Leucrose | O-α-d-Glucosyl-(1→5)-d-fructose | 9.3 ± 0.4 | 14 ± 2.1 | 14 ± 0.8 |
| Isomaltose | O-α-d-Glucosyl-(1→6)-d-glucose | 80 ± 3.6 | 108 ± 5.9 | 113 ± 7.7 |
| Palatinose | O-α-d-Glucosyl-(1→6)-d-fructose | 41 ± 1.6 | 45 ± 6.7 | 40 ± 1.4 |
| Gentiobiose | O-β-d-Glucosyl-(1→6)-d-glucose | 0.0 | 0.0 | 0.0 |
| Melibiose | O-α-d-Galactosyl-(1→6)-d-glucose | 0.0 | 0.0 | 0.0 |
| Trisaccharides | ||||
| Melizitose | O-α-d-Glucosyl-(1→3)-β-d-fructosyl-(2→1)-d-glucose | 0.0 | 0.0 | 0.0 |
| Maltotriose | O-α-d-Glucosyl-(1→4)-α-d-glucosyl-(1→4)-d-glucose | 18 ± 3.7b | 8.3 ± 1.2b | 5.8 ± 0.8b |
| Raffinose | O-α-d-Galactosyl-(1→6)-α-d-glucosyl-(1→2)-β-d-fructose | 0.0 | 0.0 | 0.0 |
| Panose | O-α-d-Glucosyl-(1→6)-α-d-glucosyl-(1→4)-d-glucose | 45 ± 0.2 | 45 ± 5.8 | 50 ± 2.7 |
| Isomaltotriose | O-α-d-Glucosyl(1→6)-α-d-glucosyl-(1→6)-d-glucose | 38 ± 4.5 | 49 ± 8.2 | 5 ± 4.5 |
Activity was determined with 0.5 μg of pure enzymes and 10 mM of each substrate at 70°C for 20 min, except where indicated otherwise.
Activity was determined with 4 μg of pure enzymes and 100 mM of each substrate at 70°C for 1 h.
Characterization of the HB27 recombinant α-glucosidase.
The effect of pH on enzyme activity was tested with 25 mM of each buffer, i.e., acetate (pH 4.0 to 5.5), MES (morpholineethanesulfonic acid; pH 5.5 to 6.5), BTP (pH 6.0 to 9.5), Tris-HCl (7.0 to 9.0), and TAPS [N-Tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid; pH 8.0 to 9.0], at 70°C and calculated as previously described (8). The standard mixtures containing trehalose were preheated for 2 min; the reaction was initiated by the addition of enzyme and stopped at different times by cooling on ethanol-ice. Prior to glucose quantification, the enzyme was inactivated by the addition of 0.5 M HCl (required for irreversible inhibition), incubated at room temperature for 20 min, and neutralized with 0.5 M NaOH. The temperature profile was determined by the incubation of reaction mixtures with trehalose at different temperatures (30 to 100°C). The thermal inactivation was determined by incubating α-glucosidase aliquots (30 μl of a solution of 0.5 mg ml−1 in 25 mM BTP buffer at pH 7.0) at 70, 80, and 90°C. Aliquots were withdrawn at appropriate times (up to 48 h) and examined for residual activity under standard conditions. The effect of the cations Ca2+, Fe2+, Zn2+, Sr2+, Ni2+, Mn2+, Mg2+, Cu2+, Co2+, and Ba2+ was tested using 25 mM BTP buffer, pH 7, containing cations at concentrations of 2 or 20 mM or no cations. The effect of Ca2+ was also determined at 10, 30, 40, or 80 mM. The effects of NaCl (20, 50, 100, and 500 mM) and KCl (50, 100, 500, and 1,000 mM) were also tested. To investigate the possibility of a phosphorylase reaction, the α-glucosidase activity was tested in the presence of 1 to 10 mM of ATP, ADP, and AMP or 10 mM phosphate buffer.
Kinetic parameters.
The kinetic parameters for the recombinant α-glucosidases from strains HB27 and GK24 and for the mutant enzymes created as described below were determined under standard reaction conditions, with trehalose or isomaltose (0.25 to 50 mM) as the substrate. Reactions were initiated by the addition of 0.5 μg of the enzyme and stopped at different times (up to 6 min) by cooling on ethanol-ice. The kinetic parameters for the recombinant α-glucosidases with maltose as the substrate were calculated with maltose concentrations ranging from 5 to 250 mM, 4 μg of enzyme, and incubation times up to 1.5 h. The protein content of each sample was determined (6). All experiments were performed in triplicate.
Site-directed mutagenesis and expression of mutant α-glucosidases.
The replacement of the AglHHB27 peptide (LGEHNLPP) with the AglHGK24 peptide (EPTAYHTL) was carried out by two rounds of PCR. First, we separately amplified a 1,176-bp fragment with primers AG1 and SV24Rv and a 435-bp fragment with primers SV24Fw and AG2 (see Table S1 in the supplemental material), using the DNA from strain HB27 as a template. Primer SV24Fw was designed to introduce the sequence coding for EPTAYHTL (Fig. 1), and primer SV24Rv was designed to introduce the complementary sequence for EPTAYHTL. The PCR products (1,176 bp and 435 bp), which had 24 overlapping bp (corresponding to the target EPTAYHTL sequence), were purified and used as templates in a second PCR with additional AG1 and AG2 primers. The identity of the final 1,587-bp product coding for a mutant HB27 α-glucosidase, where the LGEHNLPP peptide was replaced with the EPTAYHTL peptide, was confirmed by sequencing (AGOWA GmbH, Berlin, Germany). The substitution of the AglHGK24 peptide EPTAYHTL with the peptide LGEHNLPP of AglHHB27 was achieved by a similar strategy. Mutant α-glucosidase genes were expressed in E. coli DH5α, and the recombinant enzymes were purified as described above for the unmodified enzymes.
FIG. 1.
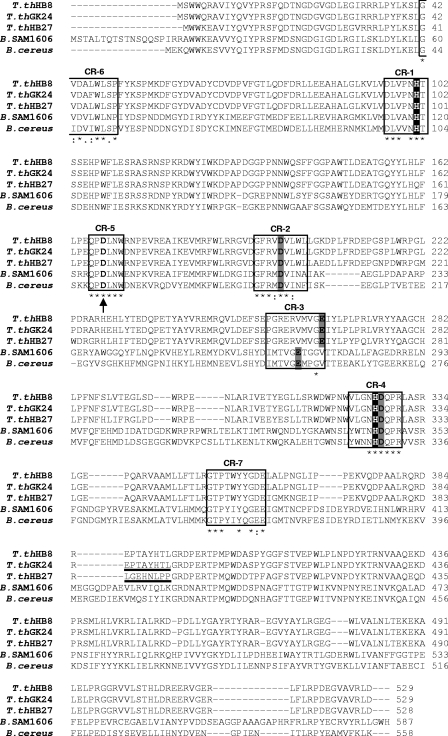
ClustalX alignment (29) of α-glucosidases from T. thermophilus strains HB27, HB8, and GK24 with homologous proteins from Bacillus species. The boxes represent the seven CRs of the α-amylase family. The boldface amino acids indicate putative catalytic and substrate-binding residues, and the underlined portions indicate an eight-amino-acid sequence, which is the target sequence for mutagenesis, that is identical in the α-glucosidases of strains GK24 and HB8 but different in the HB27 α-glucosidase. Gray shading, putative catalytic aspartates and glutamate; black shading, putative functional histidines (substrate-binding sites); arrow, Ca2+-binding aspartate; star, conserved identical amino acids; colon, conservative substitutions; period, semiconservative substitutions; T. thHB27, T. thermophilus HB27 (NCBI accession number YP_004082); T. thHB8, T. thermophilus HB8 (NCBI accession number YP_143747); T. thGK24, T. thermophilus GK24 (EMBL accession number AAD50603); B. SAM1606, Bacillus sp. strain SAM1606 (EMBL accession number CAA54266); B. cereus, Bacillus cereus ATCC 7064 (EMBL accession number CAA37583).
Construction of T. thermophilus HB27 α-glucosidase disruption mutant.
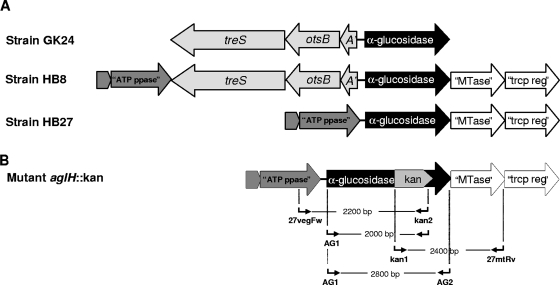
aglHHB27 was cloned into pGEM-T Easy to obtain pGEM-aglH. A 1,217-bp fragment containing the kanamycin resistance cassette was removed from pMK18 by digestion with BamHI and subcloned into the BglII site of pGEM-aglH to disrupt the α-glucosidase gene (pGEM-aglH-Kan). The orientation of the Kan insert was confirmed by restriction analysis as previously described (1). Escherichia coli DH5α carrying pGEM-aglH-Kan was grown overnight at 37°C in medium containing 30 μg ml−1 of kanamycin, and plasmids were isolated and sequenced (AGOWA GmbH, Berlin, Germany). Pure pGEM-aglH-Kan was used to transform strain HB27 and to disrupt the α-glucosidase gene as previously described (1, 9). The mutant's DNA was isolated, and successful disruption was confirmed by PCR with the following primer combinations: AG1 plus AG2; kan1 plus kan2, both designed based on the Kan cassette of pMK18; 27vegFw plus kan2; and kan1 plus 27mtRv (see Table S1 in the supplemental material and Fig. 2). Primer 27vegFw was designed based on the putative ATP pyrophosphatase gene adjacent to the α-glucosidase gene on the HB27 genome (12), and primer 27mtRv was designed based on the 3′ end of the putative methyltransferase gene located immediately downstream of the α-glucosidase gene (Fig. 2).
FIG. 2.
(A) Schematic representation of the α-glucosidase gene environment in T. thermophilus strains HB7 and HB8 (http://www.genome.ad.jp/kegg/catalog/org_list.html) and in GK24 (2); (B) strain HB27 α-glucosidase gene disruption with the kanamycin resistance cassette and representation of primers 27vegFw/kan2, AG1/kan2, AG1/AG2, and kan1/27mtRv used to confirm the expected mutation. treS, trehalose synthase gene; otsB, trehalose-6-phosphate phosphatase gene; A′, partial trehalose-phosphate synthase; “ATP ppase,” putative ATP pyrophosphatase gene; “MTase,” putative methyltransferase gene; “trcp reg,” putative transcriptional regulator gene.
Growth of α-glucosidase mutant with single carbon sources.
The wild-type HB27 strain and the aglH::Kan α-glucosidase disruption mutant were grown in complex Thermus medium or in a minimal medium (MM) containing 1.0 g liter−1 NH4Cl, 0.01 g liter−1 of yeast extract, and a vitamin solution (25). Filter-sterilized glutamate, proline, glucose, cellobiose, trehalose, isomaltose, palatinose, sucrose, or maltose (1.0 g liter−1) was independently added to the medium. The aglH::Kan mutant was grown with and without kanamycin (30 μg ml−1). Cultures were grown at 65°C as previously described (25).
RESULTS
Sequence analysis, genome environment, and functional expression of Thermus thermophilus α-glucosidases in E. coli.
The 1,587-bp gene (TTC0107) from strain HB27, coding for a polypeptide with 528 amino acids with a calculated molecular mass of 61.8 kDa, was annotated as a putative α-glucosidase/glycoside hydrolase (12). This protein exhibits high sequence homology (90% amino acid identity) with the putative oligo-1,6-glucosidase from T. thermophilus HB8 (GenPept accession number YP143747), the α-glucosidase from T. thermophilus GK24 (GenBank accession number AF096282), and the oligo-1,6-glucosidase from “Bacillus flavocaldarius” (EMBL accession number BAB18518). Lower sequence identity (43%) was detected with an α-glucosidase from Bacillus sp. strain SAM1606 (EMBL accession number CAA54266). Amplification of the α-glucosidase genes (aglH) from strains HB27, GK24, and HB8 yielded products with the expected gene sizes and sequences, and their expression in E. coli resulted in high yields of protein, which were purified to homogeneity (data not shown).
We identified in the T. thermophilus α-glucosidases the seven conserved regions (CRs) and five invariable residues common to family 13 glycoside hydrolases (GH13) (http://www.cazy.org/fam/GH13.html) that belong to the α-amylase superfamily (19). Based on a comparison with the sequences from homologous enzymes, the three catalytic residues in the α-glucosidase of strain HB27 (AglHHB27) are Asp197, Glu264, and Asp326, and the substrate-binding histidines are His100 and His325 (Fig. 1) (17, 19, 28).
In the region immediately upstream from the α-glucosidase genes of strains GK24 and HB8, we detected an incomplete operon-like structure containing genes implicated in the synthesis of trehalose, namely, a partial otsA gene and complete otsB and treS genes (2). This structure is absent from the HB27 genome, and no trehalose biosynthetic genes were found (2, 12). All genes surrounding the α-glucosidase gene, with the exception of the trehalose cluster that is absent from the HB27 chromosome, are similar in strains HB27 and HB8 (Fig. 2A). The regions downstream of treS and downstream of α-glucosidase in GK24 have not been identified.
Substrate specificity of the α-glucosidases from strains HB27, HB8, and GK24.
AglHHB27 hydrolyzed the glucosidic bonds, with declining relative activity, of the disaccharides α,α-trehalose (100%), isomaltose (63.5%), α,β-trehalose (39.3%), palatinose (32.8%), kojibiose (20.7%), nigerose (16.3%), turanose (14.3%), sucrose (8.3%), and leucrose (7.4%) and of the trisaccharides panose (35.6%) and isomaltotriose (30.1%). Maltose and maltotriose were poor substrates for this enzyme (Table 1). Trehalose-6-phosphate was not a substrate. AglHHB27 could also catalyze transglucosylation reactions only in the presence of higher levels of trehalose or isomaltose and with incubation times up to 24 h. Glucose and oligosaccharides with different molecular weights (not identified in this study) were detected by thin-layer chromatography (data not shown). Our results indicate that the recombinant enzyme exists as a dimeric protein with a molecular mass of 132 ± 5.3 kDa.
The preferred substrate of the α-glucosidases from strains GK24 and HB8 was isomaltose. Both enzymes could, however, also hydrolyze sucrose, nigerose, palatinose, kojibiose, turanose, leucrose, and the trisaccharides panose and isomaltotriose. The disaccharides α,α-trehalose or α,β-trehalose were not hydrolyzed by these enzymes under any conditions tested (Table 1). The α-glucosidases from the GK24 and HB8 strains could also hydrolyze maltose and maltotriose but only at higher substrate and enzyme concentrations as described above (Table 1). None of the enzymes were able to hydrolyze the disaccharides β,β-trehalose, cellobiose, gentiobiose, and melibiose or the trisaccharides raffinose and melizitose. None of the polysaccharides tested, namely, starch, amylopectin, glycogen, and pullulan, served as substrates for the enzymes.
Properties of the recombinant α-glucosidase from strain HB27.
The α-glucosidase from strain HB27 had maximal activity at 90°C and about 50% of maximal activity at 70°C but was inactive below 30°C. The enzyme also retained 44% of maximal activity at 100°C. The optimum pH was 6.2, and 80% of maximal activity was retained between pH 5.2 and 6.8. The enzyme had a half-life of 44 ± 9.8 h (mean ± standard deviation) at 70°C (inactivation constant [Kd] = 0.0237 ± 0.0059 min−1) and 23 ± 3.4 h at 80°C (Kd = 0.0431 ± 0.0073 min−1). At 90°C, the residual activity progressively decreased during the initial 2 h, but after this period, it remained constant, with about 35% of maximum activity for 48 h. The enzyme was completely inactivated after incubation at 100°C for 30 min. Cations were not required for enzyme activity, but 20 mM Ca2+ (153%) followed by 20 mM Sr2+ (123%) and Mn2+ (114%) stimulated activity. Mg2+ did not affect the activity of the enzyme, but Fe2+, Zn2+, Ni2+, Cu2+, Co2+, and Ba2+ were inhibitory. Increasing NaCl and KCl concentrations up to 0.5 M and 1 M, respectively, gradually inhibited the enzyme activity to about 50% (data not shown). To examine trehalose phosphorylase activity, we also determined the activity of the enzyme using trehalose and ATP, ADP, AMP, or phosphate as the substrates, with negative results (data not shown).
Kinetic studies of wild-type and mutant α-glucosidases.
Lineweaver-Burk plots allowed the calculation of apparent Km values for the substrates trehalose, isomaltose, and maltose as well as the rate constants (kcat) and catalytic efficiencies (kcat/Km) of each of the wild-type and mutant α-glucosidases with each substrate (Table 2). Our data confirm previous results reporting that trehalose was not a substrate for AglHGK24 and that isomaltose was the preferred substrate (21). However, we now show that AglHGK24 is also able to hydrolyze maltose at a low rate (Tables 1 and 2).
TABLE 2.
Effect of the reciprocal replacement of specific peptides containing eight amino acids of the α-glucosidases from T. thermophilus HB27 and GK24 on kinetic parameters determined with trehalose, isomaltose, and maltose as substratesa
| α-Glucosidase source | Trehalose
|
Isomaltose
|
Maltose
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vmax | Km | kcat | kcat/Km | Vmax | Km | kcat | kcat/Km | Vmax | Km | kcat | kcat/Km | |
| HB27 | ||||||||||||
| Wild type | 333 ± 24 | 4.9 ± 1.4 | 343 ± 39 | 71 ± 11 | 250 ± 15 | 8.7 ± 1.8 | 257 ± 20 | 31 ± 5.9 | 21 ± 0.9 | 50 ± 3.3 | 22 ± 1.8 | 0.44 ± 0.02 |
| Mutantb | 46 ± 2.5 | 17 ± 0.8 | 47 ± 4.5 | 2.8 ± 0.4 | 286 ± 19 | 8.5 ± 0.7 | 295 ± 20 | 35 ± 2.5 | 1.9 ± 0.4 | 55 ± 6.3 | 2.0 ± 0.6 | 0.04 ± 0.02 |
| GK24 | ||||||||||||
| Wild type | 0 | 0 | 0 | 0 | 357 ± 42 | 5.2 ± 0.4 | 361 ± 44 | 68 ± 16 | 11 ± 0.6 | 69 ± 7.7 | 11 ± 0.9 | 0.17 ± 0.04 |
| Mutantc | 20 ± 0.1 | 6.9 ± 0.1 | 20 ± 1.3 | 2.9 ± 0.1 | 162 ± 8.0 | 4.1 ± 0.2 | 164 ± 12 | 39 ± 0.9 | 3.4 ± 0.1 | 72 ± 6.2 | 3.5 ± 0.1 | 0.05 ± 0.01 |
Vmax values expressed as μmol/min·mg protein, Km values expressed as mM, kcat values expressed as s−1, and kcat/Km values expressed as mM−1s−1. All values are means ± standard deviations.
HB27 α-glucosidase with the EPTAYHTL peptide of the GK24 enzyme (see Fig. 1).
GK24 α-glucosidase with the LGEHNLPP peptide of the HB27 enzyme (see Fig. 1).
Amino acid substitution comprised residues 385 to 392 (LGEHNLPP) in AglHHB27 and residues 386 to 393 (EPTAYHTL) in AglHGK24 (Fig. 1). Upon the replacement of the HB27 α-glucosidase peptide by the GK24 peptide, the affinity of the HB27 enzyme for trehalose was significantly affected (the Km increased from 4.9 mM to 17 mM and the kcat decreased 7-fold), resulting in a 25-fold decrease in the catalytic efficiency (Table 2). The Km values for isomaltose and maltose between the native enzyme and the mutant AglHHB27 were not significantly altered, although isomaltose replaced trehalose as the preferred substrate of this mutant enzyme (Table 2).
The replacement of the GK24 α-glucosidase peptide (EPTAYHTL) by the peptide LGEHNLPP from the HB27 enzyme had a marked effect on substrate utilization, since the mutant AglHGK24 acquired the ability to hydrolyze trehalose (Km = 6.9 mM and kcat = 20 s−1). The Km values of isomaltose and maltose were not significantly affected for the mutant AglHGK24; however, the rate decreased more than twofold in comparison with that of the GK24 native enzyme (Table 2). Nevertheless, isomaltose was still the preferred substrate for the mutant AglHGK24.
Phenotypic analysis of the wild type and the HB27 α-glucosidase disruption mutant.
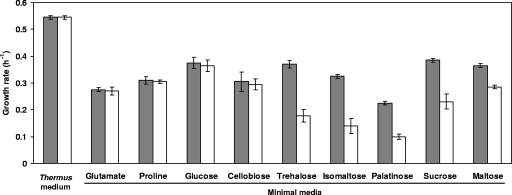
Growth rates and final cell yields of wild-type HB27 and the aglH::Kan α-glucosidase disruption mutant were similar in complex Thermus medium and in MM with glutamate, proline, glucose, or cellobiose as sources of carbon and energy. However, the growth rate of this mutant in MM containing trehalose, isomaltose, sucrose, and palatinose was about 50% lower than the growth rate of the wild type, while the growth rate of the mutant in MM with maltose was about 20% lower than that of the wild-type organism (Fig. 3). Despite the lower growth rates with these substrates, the mutant was always able to reach the final cell yields of the wild type.
FIG. 3.
Growth rates of the wild-type T. thermophilus strain HB27 (gray bars) and the aglH::Kan α-glucosidase mutant (white bars) in Thermus medium (complex) and in MM with glutamate, proline, glucose, cellobiose, trehalose, isomaltose, palatinose, sucrose, or maltose as the sole carbon source. Data are the mean values from three independent experiments.
DISCUSSION
The α-glucosidase (AglH) from T. thermophilus strain HB27 is unique among α-glucosidases (EC 3.2.1.20), since it preferentially hydrolyzes the α-1,1-glucosidic linkage of trehalose, with lower activity for disaccharides containing α-1,6, α-1,2, α-1,3, and α-1,5 linkages, such as isomaltose, palatinose, kojibiose, sucrose, nigerose, turanose, and leucrose. Moreover, this enzyme possesses much lower activity for the α-1,4 bond of maltose. This broad substrate specificity is common among α-glucosidases, but trehalose has never been shown to be the preferred substrate and is not used at all by several of these enzymes (15, 19). Surprisingly, the HB27 α-glucosidase (AglHHB27) shares 90% identical amino acids with the homologous enzymes from T. thermophilus strains HB8 and GK24 that are able to hydrolyze the α-1,2, α-1,3, α-1,5, and α-1,6 linkages of several disaccharides but cannot hydrolyze α-1,1-trehalose (21); the α-1,4 linkage of maltose was also hydrolyzed but at a lower rate. It has been suggested that enzymes with high amino acid identity but different substrate specificity might have “evolved,” through a limited number of amino acid substitutions, to give rise to enzymes with extended substrate specificity (23). Several amino acids within the CRs of the Bacillus sp. strain SAM1606 α-glucosidase, which is able to hydrolyze trehalose, are different from those of the homologous enzyme from Bacillus cereus, which is not able to hydrolyze this disaccharide. Site-specific mutagenesis demonstrated the involvement of these residues in the affinity for trehalose and confirmed that slight differences within the CRs dictate significant differences in α-glucosidase substrate specificity (13, 21). However, the α-glucosidases from T. thermophilus strains HB27, GK24, and HB8 have identical CRs, implying that other residues outside the CRs must be involved in substrate affinity. Our results clearly show that a unique peptide with eight amino acids (LGEHNLPP) in AglHHB27, located outside the CRs, was crucial for the specificity of the enzyme, since its substitution affected the preference for trehalose. The implication of this sequence in trehalose utilization was strongly supported by the acquisition of trehalose-hydrolyzing activity by the GK24 enzyme, when the specific EPTAYHTL sequence was replaced by the HB27 sequence (LGEHNLPP). Indeed, the peptide substitution significantly decreased the affinity of AglHHB27 for trehalose and the kcat value. The kinetic values of isomaltose and maltose hydrolysis were not significantly altered, indicating that the interaction with these substrates has not been affected by the substitution. The GK24 mutant enzyme acquired the ability to hydrolyze trehalose, and the Km for this substrate was similar to that of AglHHB27; however, the kcat of the GK24 mutant enzyme was much lower, which resulted in a 25-fold decrease in the kcat/Km, indicating that trehalose hydrolysis was not as efficient with the GK24 mutant enzyme as with AglHHB27 and that additional residues in the sequence were important for an enhanced catalytic rate.
The results obtained with the HB27 α-glucosidase mutant indicate that the α-glucosidase has an important role in the assimilation of trehalose, isomaltose, sucrose, and palatinose, since the growth rates of the α-glucosidase mutant were considerably affected when these disaccharides were the sole carbon sources. Surprisingly, the growth rate of the α-glucosidase mutant on maltose was also slightly affected, suggesting that the enzyme has a role in maltose degradation, despite the low affinity for this substrate in vitro. Different mechanisms exist for maltose degradation, and the one operating in T. thermophilus HB27 is unclear. In E. coli, for example, maltose is taken up and metabolized by the combined action of 4-α-glucanotransferase (MalQ), maltodextrin phosphorylase (MalP), and maltodextrin glucosidase (MalZ), leading to glucose and glucose-1-phosphate that can be utilized in glycolysis (4). Many of the mal genes from E. coli, including the global regulator malT and those encoding MalQ, MalP, and MalZ, have homologues in the genome of strain HB27 (4, 12). In strain HB8, the 4-α-glucanotransferase (MalQ) (TTC0897 in HB27) had very low activity with maltose, the maltodextrin glucosidase (MalZ) (TTC1283 in HB27) has been found to hydrolyze sucrose and maltose, and the maltodextrin phosphorylase (MalP) (TTC0808 in HB27) hydrolyzed maltooligosaccharides and glycogen but not maltose (3, 17, 30). In Bacillus species, maltose can be taken up and hydrolyzed to glucose and glucose-1-phosphate by a maltose phosphorylase, which is absent from strain HB27 (14). The difference between the growth rates of the wild-type HB27 and the α-glucosidase mutant with maltose allowed us to conclude that AglH, as deduced from the enzyme assays, is not the primary hydrolase for maltose and oligosaccharides with α-1,4 bonds but that a minor role in their hydrolysis cannot be excluded. The hypothesis that trehalose, isomaltose, palatinose, and sucrose are, to a large extent, hydrolyzed by AglH in vivo is consistent with the decrease in the growth rate of the α-glucosidase mutant with these disaccharides. Nevertheless, the organism has other enzymes to degrade the disaccharides with enough efficiency to support a lower but steady growth rate for the mutant. We searched the genome of strain HB27 for the corresponding enzymes that could possibly hydrolyze disaccharides and found a hypothetical protein (TTC0614), with a conserved domain common to trehalases, which we experimentally confirmed to have very low activity toward trehalose and isomaltose but which was not further characterized. The HB27 genome also contains other genes coding for putative disaccharide-hydrolyzing enzymes, namely, a plasmid-borne α-glucosidase gene (TTP0221) and a putative pullulanase gene (TTC1198). However, the function of the corresponding enzymes in strain HB27 can be unequivocally established only after biochemical characterization. Cellobiose with a β-1,4 linkage is not a substrate for the α-glucosidase in vitro: the lack of involvement of the α-glucosidase in cellobiose assimilation in vivo was also confirmed by the similar growth behaviors of the wild type and the α-glucosidase mutant. Other studies indicate that this disaccharide is probably hydrolyzed by a plasmid-encoded β-glycosidase (TTP0042) (10).
Previous results have shown that among the T. thermophilus strains studied, HB27 was the only strain that did not accumulate trehalose under salt stress (2, 24). The present data suggest that the trehalose taken up cannot be accumulated in HB27 to serve as a compatible solute due to the trehalose-hydrolyzing ability of the α-glucosidase from this strain. On the other hand, strains HB8 and GK24 can grow with trehalose as a carbon and energy source and concomitantly accumulate this disaccharide under salt stress, possibly because the homologous α-glucosidases are unable to hydrolyze this disaccharide.
While the mechanisms involved in the regulation of the assimilation/accumulation of trehalose in T. thermophilus strains remain elusive, our data from comparative site-specific and disruption mutagenesis revealed unique determinants for the substrate specificity and the physiological role of the HB27 α-glucosidase, allowing us to propose a central role for this enzyme in the assimilation of trehalose and other disaccharides by this particular strain of T. thermophilus.
Supplementary Material
Acknowledgments
This work was supported by Fundação para a Ciência e a Tecnologia (FCT), Portugal, and FEDER, project POCI/BIA-MIC/56511/2004. S. Alarico and N. Empadinhas acknowledge scholarships from FCT (SFRH/BD/14140/2003 and SFRH/BPD/14828/2003).
Footnotes
Published ahead of print on 25 January 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alarico, S., N. Empadinhas, A. Mingote, C. Simões, M. S. Santos, and M. S. da Costa. 2007. Mannosylglycerate is essential for osmotic adjustment in Thermus thermophilus strains HB27 and RQ-1. Extremophiles 11833-840. [DOI] [PubMed] [Google Scholar]
- 2.Alarico, S., N. Empadinhas, C. Simões, Z. Silva, A. Henne, A. Mingote, H. Santos, and M. S. da Costa. 2005. Distribution of genes for synthesis of trehalose and mannosylglycerate in Thermus spp. and direct correlation of these genes with halotolerance. Appl. Environ. Microbiol. 712460-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeck, B., and R. Schinzel. 1996. Purification and characterization of an alpha-glucan phosphorylase from the thermophilic bacterium Thermus thermophilus. Eur. J. Biochem. 239150-155. [DOI] [PubMed] [Google Scholar]
- 4.Boos, W., and H. Shuman. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brabban, A. D., E. N. Orcutt, and S. H. Zinder. 1999. Interactions between nitrogen fixation and osmoregulation in the methanogenic archaeon Methanosarcina barkeri 227. Appl. Environ. Microbiol. 651222-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 7.Cardoso, F. S., R. F. Castro, N. Borges, and H. Santos. 2007. Biochemical and genetic characterization of the pathways for trehalose metabolism in Propionibacterium freudenreichii, and their role in stress response. Microbiology 153270-280. [DOI] [PubMed] [Google Scholar]
- 8.Costa, J., N. Empadinhas, and M. S. da Costa. 2007. Glucosylglycerate biosynthesis in the deepest lineage of the Bacteria: characterization of the thermophilic proteins GpgS and GpgP from Persephonella marina. J. Bacteriol. 1891648-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Grado, M., P. Castán, and J. Berenguer. 1999. A high-transformation-efficiency cloning vector for Thermus thermophilus. Plasmid 42241-245. [DOI] [PubMed] [Google Scholar]
- 10.Dion, M., L. Fourage, J.-N. Hallet, and B. Colas. 1999. Cloning and expression of a β-glycosidase gene from Thermus thermophilus. Sequence and biochemical characterization of the encoded enzyme. Glycoconj. J. 1627-37. [DOI] [PubMed] [Google Scholar]
- 11.Elbein, A. D., Y. T. Pan, I. Pastuszak, and D. Carroll. 2003. New insights on trehalose: a multifunctional molecule. Glycobiology 1317-27. [DOI] [PubMed] [Google Scholar]
- 12.Henne, A., H. Brüggemann, C. Raasch, A. Wiezer, T. Hartsch, H. Liesegang, A. Johann, T. Lienard, O. Gohl, R. Martinez-Arias, C. Jacobi, V. Starkuviene, S. Schlenczeck, S. Dencker, R. Huber, H. P. Klenk, W. Kramer, R. Merkl, G. Gottschalk, and H. J. Fritz. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 22547-553. [DOI] [PubMed] [Google Scholar]
- 13.Inohara-Ochiai, M., T. Nakayama, R. Goto, M. Nakao, T. Ueda, and Y. Shibano. 1997. Altering substrate specificity of Bacillus sp. SAM1606 α-glucosidase by comparative site-specific mutagenesis. J. Biol. Chem. 2721601-1607. [DOI] [PubMed] [Google Scholar]
- 14.Inoue, Y., N. Yasutake, Y. Oshima, Y. Yamamoto, T. Tomita, S. Miyoshi, and T. Yatake. 2002. Cloning of the maltose phosphorylase gene from Bacillus sp. strain RK-1 and efficient production of the cloned gene and the trehalose phosphorylase gene from Bacillus stearothermophilus SK-1 in Bacillus subtilis. Biosci. Biotechnol. Biochem. 662594-2599. [DOI] [PubMed] [Google Scholar]
- 15.Janecek, S. 2002. How many conserved sequence regions are there in the α-amylase family? Biol. Bratislava 5729-41. [Google Scholar]
- 16.Jorge, C. D., M. M. Sampaio, G. Ó. Hreggvidsson, J. K. Kristjánson, and H. Santos. 2007. A highly thermostable trehalase from the thermophilic bacterium Rhodothermus marinus. Extremophiles 11115-122. [DOI] [PubMed] [Google Scholar]
- 17.Kaper, T., H. Leemhuis, J. C. Uitdehaag, B. A. van der Veen, B. W. Dijkstra, M. J. van der Maarel, and L. Dijkhuizen. 2007. Identification of acceptor substrate binding subsites +2 and +3 in the amylomaltase from Thermus thermophilus HB8. Biochemistry 465261-5269. [DOI] [PubMed] [Google Scholar]
- 18.Koh, S., J. Kim, H.-J. Shin, D. Lee, J. Bae, D. Kim, and D.-S. Lee. 2003. Mechanistic study of the intramolecular conversion of maltose to trehalose by Thermus caldophilus GK24 trehalose synthase. Carbohydr. Res. 3381339-1343. [DOI] [PubMed] [Google Scholar]
- 19.MacGregor, E. A., S. Janecek, and B. Svensson. 2001. Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim. Biophys. Acta 15461-20. [DOI] [PubMed] [Google Scholar]
- 20.Nakao, M., T. Nakayama, A. Kakudo, M. Inohara, M. Harada, F. Omura, and Y. Shibano. 1994. Structure and expression of a gene coding for thermostable alpha-glucosidase with a broad substrate specificity from Bacillus sp. SAM1606. Eur. J. Biochem. 220293-300. [DOI] [PubMed] [Google Scholar]
- 21.Nashiru, O., S. Koh, S.-Y. Lee, and D.-S. Lee. 2001. Novel α-glucosidase from extreme thermophile Thermus caldophilus GK24. J. Biochem. Mol. Biol. 34347-354. [Google Scholar]
- 22.Nielsen, P., D. Fritze, and F. G. Priest. 1995. Phenetic diversity of alkaliphilic Bacillus strains: proposal for nine new species. Microbiology 1411745-1761. [Google Scholar]
- 23.Noguchi, A., M. Yano, Y. Ohshima, H. Hemmi, M. Inohara-Ochiai, M. Okada, K.-S. Min, T. Nakayama, and T. Nishino. 2003. Deciphering the molecular basis of the broad substrate specificity of alpha-glucosidase from Bacillus sp. SAM1606. J. Biochem. 134543-550. [DOI] [PubMed] [Google Scholar]
- 24.Silva, Z., M.-M. Sampaio, A. Henne, A. Böhm, R. Gutzat, W. Boos, M. S. da Costa, and H. Santos. 2005. The high-affinity maltose/trehalose ABC transporter in the extremely thermophilic bacterium Thermus thermophilus HB27 also recognizes sucrose and palatinose. J. Bacteriol. 1871210-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva, Z., S. Alarico, A. Nobre, R. Horlacher, J. Marugg, W. Boos, A. I. Mingote, and M. S. da Costa. 2003. Osmotic adaptation of Thermus thermophilus RQ-1: lesson from a mutant deficient in synthesis of trehalose. J. Bacteriol. 1855943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva, Z., S. Alarico, and M. S. da Costa. 2005. Trehalose biosynthesis in Thermus thermophilus RQ-1: biochemical properties of the trehalose-6-phosphate synthase and trehalose-6-phoshate phosphatase. Extremophiles 929-36. [DOI] [PubMed] [Google Scholar]
- 27.Taguchi, H., M. Hamaoki, H. Matsuzawa, and T. Ohta. 1983. Heat-stable extracellular proteolytic enzyme produced by Thermus caldophilus strain GK24, an extremely thermophilic bacterium. J. Biochem. 937-13. [DOI] [PubMed] [Google Scholar]
- 28.Takata, H., T. Kuriki, S. Okada, Y. Takesada, M. Iizuca, N. Minamiura, and T. Imanaka. 1992. Action of neopullulanase. Neopullulanase catalyzes both hydrolysis and transglycosylation at alpha-(1,4)- and alpha-(1,6)-glucosidic linkages. J. Biol. Chem. 26718447-18452. [PubMed] [Google Scholar]
- 29.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 244876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, S., and S. Zhang. 1992. Purification and characterization of alpha-glucosidase from an extreme thermophile, Thermus thermophilus HB8. Wei Sheng Wu Xue Bao 3223-29. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.