Abstract
The natural diversity of the elt operons, encoding the heat-labile toxin LT-I (LT), carried by enterotoxigenic Escherichia coli (ETEC) strains isolated from humans was investigated. For many years, LT was supposed to be represented by a rather conserved toxin, and one derivative, produced by the reference H10407 strain, was intensively studied either as a virulence factor or as a vaccine adjuvant. Amplicons encompassing the two LT-encoding genes (eltA and eltB) of 51 human-derived ETEC strains, either LT+ (25 strains) only or LT+/ST+ (26 strains), isolated from asymptomatic (24 strains) or diarrheic (27 strains) subjects, were subjected to restriction fragment length polymorphism (RFLP) analysis and DNA sequencing. Seven polymorphic RFLP types of the H10407 strain were detected with six (BsaI, DdeI, HhaI, HincII, HphI, and MspI) restriction enzymes. Additionally, the single-nucleotide polymorphic analysis revealed 50 base changes in the elt operon, including 21 polymorphic sites at eltA and 9 at eltB. Based on the deduced amino acid sequences, 16 LT types were identified, including LT1, expressed by the H10407 strain and 23 other strains belonging to seven different serotypes, and LT2, expressed by 11 strains of six different serotypes. In vitro experiments carried out with purified toxins indicated that no significant differences in GM1-binding affinity could be detected among LT1, LT2, and LT4. However, LT4, but not other toxin types, showed reduced toxic activities measured either in vitro with cultured cells (Y-1 cells) or in vivo in rabbit ligated ileal loops. Collectively, these results indicate that the natural diversity of LTs produced by wild-type ETEC strains isolated from human hosts is considerably larger than previously assumed and may impact the pathogeneses of the strains and the epidemiology of the disease.
Enterotoxigenic Escherichia coli (ETEC)-associated diarrhea represents a major cause of mortality and morbidity among children and travelers, respectively, in most developing countries in Latin America, Africa, and Asia (3, 33). ETEC secretory diarrhea involves a rather straightforward pathogenesis plan, requiring first colonization of small intestine epithelial cells by means of filamentous adhesins collectively known as colonization factors (CFs) and, at a second stage, production of at least one out of two enterotoxin types, the heat-stable toxin (ST) and/or the heat-labile toxin (LT) (28, 36). One of the most complex aspects of ETEC pathogenesis is the remarkable antigen heterogeneity. At least 150 O:H serotypes have been found among ETEC strains isolated from humans, although a more restricted number of serotype combinations is detected among strains isolated from patients requiring medical intervention, also characterized, in some cases, by a conserved set of virulence-associated factors and a common clonal origin (29, 30, 46). Moreover, the ETEC phenotypic heterogeneity is also well illustrated by the encoded virulence-associated factors, including more than 20 known CFs and production of LT, ST, or both enterotoxins (10, 33, 46).
Two types of ST, STa and STb (also known as ST-I and ST-II), have been differentiated based on biological and chemical features (7, 11). Similarly, LTs produced by ETEC strains are also a heterogeneous group of toxins. Two major LT families have been identified, LT-I and LT-II. LT-II is rarely found among human-derived ETEC strains, but two natural variants have been reported, LT-IIa and LT-IIb, based on differences in the subunit sequences (14, 16). LT-I shows a rather high similarity with cholera toxin (CT) (over 80% amino acid identity), and both have been intensively studied as virulence factors and modulators of immune responses in mammalian species, including humans (18, 28).
The known natural variability of LT-I toxins expressed by ETEC strains has been mainly restricted to the differences detected between LTs produced by human (LTh)- and pig (LTp)-derived strains. Initial evidence based on the antigenicities and electrophoretic mobilities of LTh and LTp indicated that the toxins differ in their primary amino acid sequences (19, 42). Sequencing of the elt operons encoding LTh and LTp revealed differences in the primary sequences of the toxins, which share over 95% identity along the complete amino acid sequence (45). Altogether, six amino acid replacements were detected between the A subunits (K4R, K213E, and N238D) and the B subunits (S4T, A46E, and E102K) of LTh and LTp derived from the H10407 and EWD299 strains, respectively (25, 48, 50). At a time when DNA sequencing was not available to most laboratories, application of restriction fragment length polymorphism (RFLP) typing to a larger number of strains showed that a single HhaI restriction site was not detected in the elt operon derived from pig-derived ETEC strains, thus easily discriminating LTh and LTp (5, 45). In the case of an elt operon from a chicken-derived ETEC strain, no difference from the reference human-derived H10407 strain was found (21).
LTs produced by human-derived ETEC strains apparently have reduced natural diversity, probably reflecting the limited number of fully sequenced elt operons. So far, the LT sequences produced by two human-derived ETEC strains (H74-114 and H10407) have been determined (17, 25). Based on the nucleotide sequences of the elt operons present in these two strains, five polymorphic sites have been detected, leading to four amino acid replacements: three in the A subunit (K212R, E213K, and D238N) and one in the B subunit (H13R). More recently, one LT variant with five polymorphic sites in the A subunit and one in the B subunit was reported to be encoded by a chromosomally integrated elt operon of a strain recovered from a Japanese tourist (20). Thus, a better knowledge of LT diversity among ETEC strains isolated from humans awaits a more detailed scrutiny of elt operons carried by strains belonging to different clonal groups or with different geographic origins.
In the present study, we searched for the natural genetic diversity of LTs expressed by 51 ETEC strains isolated from humans, mostly children living in three major cities in Brazil. The screened strain set included 25 strains producing LT only and 26 LT/ST-producing (LT+/ST+) strains recovered from asymptomatic (24 strains) or diarrheic (27 strains) subjects. Our results, based on RFLP and single-nucleotide polymorphism analyses, revealed that LTs produced by human-derived ETEC strains, particularly among strains producing LT only, show significant genetic diversity, and 16 LT types have been identified. The finding of rather large natural diversity among LTs produced by ETEC strains may have significant impacts on studies of both ETEC pathogenesis and the epidemiology of the disease.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The 51 tested LT-producing ETEC strains were derived from case-control studies of acute endemic childhood diarrhea in Brazil in children under the age of 5 in the cities of São Paulo, Rio de Janeiro, and Recife (12, 15, 34, 35) (Table 1). Twenty-four ETEC strains were isolated from asymptomatic children, including 20 strains producing LT only and 4 LT+/ST+ strains. Twenty-seven ETEC strains were isolated from diarrheic subjects requiring medical assistance, including 5 strains producing LT only and 22 LT+/ST+ strains. The reference LT+/ST+ ETEC H10407 strain was isolated from a patient with severe cholera-like disease in Bangladesh (9). The 258909-3 (O128:H−) strain was also isolated in Bangladesh from a symptomatic case (13), and both strains were kindly supplied by Ann Mari Svennerholm (Göteborg University, Göteborg, Sweden). Table 1 lists all tested ETEC strains and the respective clinical features, serotypes, toxigenic profiles, and geographic origins. Cultures were routinely prepared in CAYE medium (2% Casamino Acids, 0.6% yeast extract, 43 mM NaCl, 38 mM K2HPO4, and 0.1% trace salt solution consisting of 203 mM MgSO4, 25 mM MnCl2, and 18 mM FeCl3) (20) and incubated for 18 h at 37°C with vigorous agitation (200 rpm in a rotary shaker).
TABLE 1.
Serotypes and relevant phenotypic features of ETEC strains analyzed in the present study
| ETEC strain | Serotype | Toxin type | Sourcea | Originb | Reference |
|---|---|---|---|---|---|
| PE0215 | O7:H18 | LT-I | AC | Brazil (northeast) | Laboratory collection |
| 121-I | O48:H21 | LT-I | AC | Brazil (southeast) | 34 |
| 4101-1 | O114:H− | LT-I | DC | Brazil (southeast) | 12 |
| PE0534 | O106:H40 | LT-I | AC | Brazil (northeast) | Laboratory collection |
| 225-IV | O148:H28 | LT-I | AC | Brazil (southeast) | 34 |
| PE0415 | O152:H2 | LT-I | AC | Brazil (northeast) | Laboratory collection |
| 4702-1 | O67:H5 | LT-I | AC | Brazil (southeast) | 12 |
| 1372-1 | O23:H28 | LT-I | AC | Brazil (southeast) | 12 |
| 214-III | Ont:Hnt | LT-I | AC | Brazil (southeast) | 34 |
| 4692-1 | Ont:H− | LT-I | AC | Brazil (southeast) | 12 |
| 4321-1 | Ont:H− | LT-I | DC | Brazil (southeast) | 12 |
| PE0690 | Ont:H8 | LT-I | AC | Brazil (northeast) | Laboratory collection |
| PE0615 | Ont:H9 | LT-I | DC | Brazil (northeast) | Laboratory collection |
| 4092-7 | Ont:H21 | LT-I | AC | Brazil (southeast) | 12 |
| PE0260 | O8:H9 | LT-I | DC | Brazil (northeast) | Laboratory collection |
| PE0262 | O8:H9 | LT-I | DC | Brazil (northeast) | Laboratory collection |
| 136-II | O88:H25 | LT-I | AC | Brazil (southeast) | 34 |
| 136-I | O88:H25 | LT-I | AC | Brazil (southeast) | 34 |
| 136-III | O88:H25 | LT-I | AC | Brazil (southeast) | 34 |
| PE0323 | O159:H4 | LT-I | AC | Brazil (southeast) | Laboratory collection |
| 63-V | O159:H17 | LT-I | AC | Brazil (southeast) | 34 |
| 4652-2 | O159:H34 | LT-I | AC | Brazil (southeast) | 12 |
| 36-IV | O159:H21 | LT-I | AC | Brazil (southeast) | 34 |
| 187-V | O159:H21 | LT-I | AC | Brazil (southeast) | 34 |
| 36-III | O159:H21 | LT-I | AC | Brazil (southeast) | 34 |
| H10407 | O78:H11 | LT-I/ST | DC | Bangladesh | 9 |
| 258909-3 | O128:H− | LT-I/ST | DC | Bangladesh | 13 |
| 3981-3 | O23:H16 | LT-I/ST | DC | Brazil (southeast) | 12 |
| 0781-3 | O23:H16 | LT-I/ST | DC | Brazil (southeast) | 12 |
| 373F/2 | O23:H33/45 | LT-I/ST | DC | Brazil (southeast) | 15 |
| 25A-1 | O78:H12 | LT-I/ST | DC | Brazil (southeast) | 15 |
| 61A-4 | O78:H12 | LT-I/ST | DC | Brazil (southeast) | 15 |
| 2781-5 | O78:H12 | LT-I/ST | DC | Brazil (southeast) | 12 |
| PE0347 | O78:H12 | LT-I/ST | AC | Brazil (northeast) | Laboratory collection |
| PE0351 | O78:H12 | LT-I/ST | DC | Brazil (northeast) | Laboratory collection |
| PE0379 | O78:H12 | LT-I/ST | DC | Brazil (northeast) | Laboratory collection |
| 38A-4 | O6:H16 | LT-I/ST | DC | Brazil (southeast) | 15 |
| 1111-1 | O6:H16 | LT-I/ST | DC | Brazil (southeast) | 12 |
| C13/4 | O6:H16 | LT-I/ST | DC | Brazil (southeast) | 15 |
| 4372-4 | O6:H16 | LT-I/ST | AC | Brazil (southeast) | 12 |
| 4021-5 | O6:H16 | LT-I/ST | DC | Brazil (southeast) | 12 |
| TR298/1 | O6:H16 | LT-I/ST | DC | Brazil (southeast) | 35 |
| TR274 | O6:H16 | LT-I/ST | DC | Brazil (southeast) | 35 |
| TR131 | O6:H16 | LT-I/ST | DC | Brazil (southeast) | 35 |
| 1661-1 | O6:H16 | LT-I/ST | DC | Brazil (southeast) | 12 |
| 4261-2 | O6:H16 | LT-I/ST | DC | Brazil (southeast) | 12 |
| PE0016 | O6:H16 | LT-I/ST | DC | Brazil (northeast) | Laboratory collection |
| PE0064 | O6:H16 | LT-I/ST | AC | Brazil (northeast) | Laboratory collection |
| 4441-1 | O6:H16 | LT-I/ST | DC | Brazil (southeast) | 12 |
| 4291-1 | O6:H16 | LT-I/ST | DC | Brazil (southeast) | 12 |
| 4292-1 | O6:H16 | LT-I/ST | AC | Brazil (southeast) | 12 |
AC, asymptomatic child; DC, diarrheic child.
Northeast, samples isolated from Pernambuco State (northeastern region of Brazil); southeast, samples isolated from São Paulo or Rio de Janeiro State (southeastern region of Brazil).
RFLP analysis of the elt operon.
The presence of the elt operon carried by the ETEC strains was initially confirmed by colony blot assays under previously reported experimental conditions (27). Total DNA of the tested ETEC strains was isolated according the method described by Llop and colleagues (26). Amplification of the elt operon was carried out with specific primers complementary to the sequence available at GenBank (GI 408994) (50). LTI-A1 (5′-AAACAAAACAAGTGGCG) and LTI-B2 (5′-GTTGTTATATAGGTTCCTAGC) primers (annealing at positions −65 to −49 and at positions 1172 to 1192 of the elt operon, respectively) were employed for the specific amplification of a 1,257-bp amplicon containing the complete elt operon. Amplification reactions were performed in a total volume of 50 μl containing 1 mM deoxynucleoside triphosphates, 10 pmol of each primer, 2 mM MgCl2, 5 μl of 10-fold-concentrated polymerase synthesis buffer, and 1 U of Platinum Taq DNA Polymerase High Fidelity (Invitrogen). After an initial denaturation step of 4 min at 94°C, the samples were subjected to 30 cycles of denaturation (94°C; 45 s), annealing (51.7°C; 90 s), and extension (72°C; 60 s), followed by a single final extension step of 5 min at 72°C. The PCR products were analyzed on a 0.8% agarose gel and visualized by staining them with ethidium bromide.
DNA sequence analysis of the elt operon.
The complete DNA sequences of the elt operons carried by the tested ETEC strains were determined in an ABI 3100 (Perkin-Elmer Applied Biosystems) automated capillary DNA sequencer with reagents of the BigDye terminator DNA Sequencing 2 (Perkin-Elmer Applied Biosystems, Warrington, England). The sequencing reactions were performed with seven specific primers annealing at different positions in the elt amplicon: LTI-A2 (5′-CGAGGCATACGTGTATCT), LT-A2.2A (5′-CTGCCTCTTAACTTTTGATTG), LTI-A2.2B (5′-GTTCTGTAATAGACTGGGGAGC), LTI-B1.2A (5′-GGTGATACTTGTAATGAGG), and LTI-B1.2B (5′-GACTATCAGTCAGA GGTTG) and the two primers (LTI-A1 and LTI-B2) used in the amplification of the elt operon. The recovered DNA sequences were edited and assembled in a single continuous sequence using the Lasergene program (DNAStar Inc., Madison, WI). The sequencing of the elt operon amplified from each ETEC strain was repeated at least twice to ensure the accuracy of the final results. The locations of the detected polymorphic amino acid residues on the LT structural model, available at the Protein Data Bank (PDB) (code 1LTS), were generated with the Pymol program (http://www.pymol.org). The amino acid sequence relationships of the detected LT variants (AB subunits) were represented by an unrooted tree generated with the PHYLIP program based on neighbor-joining methods. The amino acid sequence alignments were carried out with the ClustalW program, and the tree was visualized using the Treeview program.
RAPD-PCR analysis.
Randomly amplified polymorphic DNA (RAPD)-PCR analyses were carried out with the arbitrary 10-mer primer 1254 (5′-CCGCAGCCAA). Template preparation and PCR amplification with primer 1254 were performed as described previously (31). The reproducibility of the banding patterns was checked at least twice using different DNA preparations. The reaction products were analyzed in 1.2% agarose gels stained with ethidium bromide. RAPD-PCR profiles were inspected visually and defined according to the presence or absence and intensities of polymorphic bands. A 100-bp DNA ladder (Invitrogen) was used as a molecular-size marker.
Purification of recombinant LTs.
A recombinant E. coli DH5α strain harboring the pBSPKS(−) vector (38) carrying the complete elt operon of the H10407 strain under the control of the native promoter was used for the expression of the LT1 type. Similarly, the genes encoding the LT2 and LT4 types from strains 25A-1 and 1372-1, respectively, were cloned into PstI and KpnI sites of the pBSPKS(−) vector under the control of their native promoters. The toxins were purified by affinity chromatography on immobilized d-galactose columns (Pierce), as previously described (24). The purified toxins were monitored in sodium dodecyl sulfate-containing 15% polyacrylamide gels, and the protein concentration was determined by the Bradford assay (Bio-Rad Laboratories) with bovine serum albumin (Sigma-Aldrich) as a standard. The proportions of the A and B subunits of different purified LT variants were evaluated in Coomassie blue-stained polyacrylamide gels in which equal toxin amounts (1.5 μg/lane), boiled or not for 5 min, were loaded.
Competitive GM1-binding assay.
The affinities of purified LT variants for GM1 were evaluated by competitive enzyme-linked immunosorbent assay (ELISA) using biotinylated cholera toxin B subunit as an inhibitor reagent (Sigma-Aldrich), according to the method of Bäckström and coworkers (1). Briefly, wells of a polystyrene 96-well microtiter plate (Nalge Nunc) were coated with phosphate-buffered saline (PBS)-diluted GM1 ganglioside (0.05 μg ml−1; Sigma-Aldrich) and incubated overnight at room temperature. The following day, the plates were washed and then blocked with 0.1% bovine serum albumin (BSA) in PBS (PBS-BSA) for 30 min at 37°C. After the plates were washed with PBS, each purified LT was diluted to 4 μg ml−1 in 200 μl of PBS-BSA in duplicate wells and twofold serially diluted. Peroxidase-labeled cholera toxin B subunit at 0.2 μg ml−1 in 50 μl was added to each well and incubated for 1 h at room temperature. Then, color reactions were developed with ο-phenylenediamine and H2O2. After 20 min at room temperature, the reactions were interrupted by the addition of 2 M H2SO4 (50 μl/well), and the A492 was measured in a microplate reader (Multiskan EX; ThermoLabsystems). CT (Calbiochem) was used as a positive control.
LT quantification by capture ELISA.
Determination of LT concentrations in whole-cell extracts was carried out by the capture ELISA method as described previously (24). Briefly, microtiter plates (Nunc Maxisorp; Nalge Nunc, Roskilde, Denmark) were coated with 100 μl/well of rabbit anti-CT serum diluted in PBS (1:1,000), followed by overnight incubation at 4°C. The plates were washed twice with PBS containing 0.05% Tween 20 (PBS-T) and blocked by incubation with 5% skim milk in PBS-T for 1 h at 37°C. After additional washings, 100 μl of crude extract and serially twofold-diluted samples were added to the wells and incubated for 2 h at room temperature. After the plates were washed, mouse anti-LT serum diluted in PBS-T (1:5,000) was added to the wells and incubated for 90 min. Detection of bound antibodies was carried out with 100 μl of PBS-T-diluted (1:3,000) horseradish peroxidase-conjugated antimouse-immunoglobulin G (Sigma-Aldrich, Poole, United Kingdom) incubated for 90 min at room temperature. After a final washing step, color reactions were developed with o-phenylenediamine and H2O2. After 20 min at room temperature, the reactions were interrupted by the addition of 2 M H2SO4 (50 μl/well), and the optical density at 492 nm was measured in a microplate reader (Multiskan EX; ThermoLabsystems). The final reaction values were obtained after deduction of background absorbance measured in control wells filled with cell extracts of an LT-negative derivative of the 4611-4 strain.
Determination of in the vitro cytotonic activity of the LT.
The adrenal Y-1 cell line was cultivated at 37°C to 5 × 104 cells/well, and 100 μl of serial twofold dilutions of the bacterial filtrates obtained by sonic disruption or purified toxins, at concentration from 75 μg ml−1, were inoculated into a microtiter plate for cell assays, as described previously (23). The microtiter plates were incubated at 37°C in a 5% CO2 atmosphere and examined daily for the characteristic LT effect. The endpoint was taken as the highest dilution of the sample that changed 50% of the Y-1 cell morphology after incubation. Each sample was tested at least in triplicate, and the results were analyzed by phase-contrast microscopy.
Rabbit ligated ileal loop assay.
Tests in rabbit ligated ileal loops were performed as previously described (24) with approximately1.5-kg male New Zealand White rabbits. The rabbits were fasted for 48 h prior to surgery, except for ingestion of water with glucose. Laparotomy was carried out aseptically to externalize the intestines of the animals, which were kept under anesthesia by intramuscular administration of telazol (20 mg kg of body weight−1) and nilperidol (0.1 mg kg−1). Isolated 5-cm duodenum loops were spaced by 2-cm interposing loops with ligatures. Whole-cell extracts, generated after sonic disruption of the bacterial cells, or purified protein samples containing the same amount of toxin (150 ng) were injected into each tied ileal loop, followed by intestine internalization and incision closure. The volumes of accumulated fluids in each loop were measured 18 h after the inoculation of the bacterial strains. A plasmid-cured LT− strain (strain 4611-4) was employed as a negative control. All strains were cultivated in CAYE broth overnight before sonic disruption.
Nucleotid sequence accession numbers.
The complete nucleotide sequences of the elt operons of 14 LT types out of the 16 studied were submitted to GenBank and were given the following accession numbers: EU113242 (LT3), EU113243 (LT4), EU113244 (LT5), EU113245 (LT6), EU113246 (LT7), EU113247 (LT8), EU113248 (LT9), EU113249 (LT10), EU113250 (LT11), EU113251 (LT12), EU113252 (LT13), EU113253 (LT14), EU113254 (LT15), and EU113255 (LT16).
RESULTS
RFLP analysis of the elt operons from human-derived ETEC strains.
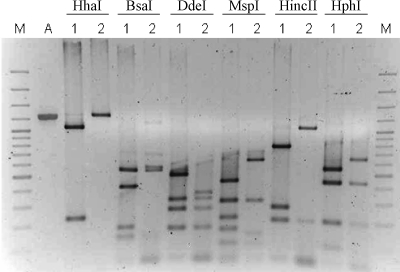
Amplification of the elt operon yielded a 1,257-bp fragment common to all 51 tested ETEC strains, including 25 strains producing LT only, 25 LT+/ST+ strains, and the reference LT+/ST+ H10407 strain. Restriction of the amplified fragment was carried out with nine restriction enzymes (BsaI, DdeI, HhaI, HincII, HinfI, HphI, TacI, MspI, and NdeI) with cleavage sites found in the reported sequence of the H10407 elt operon. Polymorphic band profiles were detected with six enzymes (BsaI, DdeI, HhaI, HincII, HphI, and MspI), resulting in seven defined RFLP types named RFLP-I to RFLP-VII. RFLP-I included the reference H10407 strain and all tested strains of serotypes O6:H16 (15 strains), O88:H25 (3 strains), and O8:H9 (2 strains), as well as 5 strains producing LT only and expressing different serotypes (Table 2). RFLP-II was found in a single O152:H2 strain (PE0415) in which one BsaI restriction site was lost concomitantly with the acquisition of a new DdeI site. RFLP-III was detected in the 4692-1 (Ont:H− [nt indicates not typeable by conventional methods]) strain and consisted of the simultaneous acquisition of a new MspI restriction site and the loss of another site for the same restriction enzyme at the eltA and eltB cistrons, respectively. RFLP-IV encompassed six LT+ ETEC strains belonging to serotypes O159:H34 (4652-2), O23:H16 (0781-3 and 3981-3), O23:H33/45 (373F/2), Ont:Hnt (214-III), and O7:H18 (PE0215) and consisted of the loss one of two HincII restriction sites in the eltA cistron. RFLP-V included all tested O159:H21 (three) and O78:H12 (six) strains, the 258909-3 strain, and three ETEC strains producing LT only belonging to different serotypes and involved the loss of the unique HphI restriction site (Table 2). RFLP-VI was characterized by the lost of the single HhaI restriction site in the eltB coding region. This RFLP type was shared by two human-derived LT+ ETEC strains belonging to different serotypes (O148:H28 [225-IV] and O23:H28 [1372-1]) (Fig. 1 and Table 2). Notably, the same RFLP type has been proposed to differentiate LTs produced by ETEC strains isolated from pigs (LTp) from those isolated from humans (45). The last RFLP group (RFLP-VII) included two LT+ ETEC strains (4092-7 and PE0690) in which the polymorphic band profiles resulted from loss of both HhaI and HphI restriction sites of the elt amplicon.
TABLE 2.
RFLP patterns of the elt operons detected among the ETEC strains studied
| PCR-RFLP typea | ETEC serotype (strain)b | Nucleotide alterationc | LT typed |
|---|---|---|---|
| I | O78:H11 (H10407) | LT1 | |
| O114:H− (4101-1) | LT1 | ||
| O8:H9 (PE0260, PE0262) | LT1 | ||
| O6:H16 (15 strains) | LT1 | ||
| O88:H25 (136-I, 136-II, 136-III) | LT3/LT5 | ||
| O48:H21 (121-I) | LT5 | ||
| O167:H5 (4702-1) | LT9 | ||
| Ont:H− (4321-1) | LT12 | ||
| Ont:H9 (PE0615) | LT13 | ||
| II (BsaI-/DdeI+) | O152:H2 (PE0415) | BsaI−, nt 661 of eltA (A to G) | LT8 |
| DdeI+, nt 842 of eltA/B (C to T) | |||
| III (MspI-/MspI+) | Ont:H− (4692-1) | Msp+, nt 93 of eltA (A to G) | LT11 |
| Msp−, nt 995 of eltB (G to T) | |||
| IV (HincII−) | O159:H34 (4652-2) | Nt 747 of eltA (C to T) | LT1 |
| O23:H16 (0781-3) | LT1 | ||
| O7:H18 (PE0215) | LT1 | ||
| O23:H16 (3981-3) | LT1 | ||
| O23:H33/45 (373F/2) | LT1 | ||
| Ont:Hnt (214-III) | LT10 | ||
| V (HphI−) | O128:H− (258909-3) | Nt 641 of eltA (G to A) | LT2 |
| O159:H4 (PE0323) | LT2 | ||
| O159:H21 (3 strains) | LT2 | ||
| O106:H40 (PE0534) | LT7 | ||
| O159:H17 (63-V) | LT14 | ||
| O78:H12 (6 strains) | LT2/LT15/LT16 | ||
| VI (HhaI−) | O23:H28 (1372-1) | Nt 973 of eltB (C to A) | LT4 |
| O148:H28 (225-IV) | LT6 | ||
| VII (HphI−/HhaI−) | Ont:H8 (PE0690) | HphI−, nt 641 of eltA (G to A) | LT2 |
| Ont:H21 (4092-7) | HhaI−, nt 971 of eltB (C to T) |
PCR-RFLP patterns detected among the 51 ETEC strains analyzed. Restriction enzymes revealing the distinct RFLP types are indicated in parentheses. RFLP-II and -III types involve the concomitant loss of one restriction site and the acquisition of another one. The RFLP-VII type involves the concomitant loss of two restriction sites.
ETEC serotypes and strains sharing the same PCR-RFLP type. Specific tested strains are indicated in parentheses.
Specific nucleotide changes at the elt operon (1,257 bp) leading to the altered PCR-RFLP patterns. Nucleotide (nt) 1 encompasses the first structural codon of the elt operon. All changes were based on the nucleotide sequence of the H10407 elt operon (PCR-RFLP type I).
LT types, defined by specific amino acid replacements, in each RFLP type.
FIG. 1.
PCR-RFLP patterns of the elt operons derived from different ETEC strains. Lane M, 100-bp molecular-size ladder (Invitrogen); lane A, elt operon amplified from the H10407 strain. PCR products digested with HhaI, BsaI, DdeI, HincII, MspI, and HphI are indicated. Lanes 1, elt operons amplified from the reference strain H10407 treated with the indicated restriction endonuclease; lanes 2, elt operons amplified from polymorphic ETEC strains treated with the indicated restriction endonuclease (HhaI, strain 1372-1; BsaI, strain PE0415; DdeI, strain PE0415; MspI, strain 4692-1; HincII, strain 4652-2; and HphI, strain 36-III).
Single-nucleotide polymorphism analysis of the elt operon.
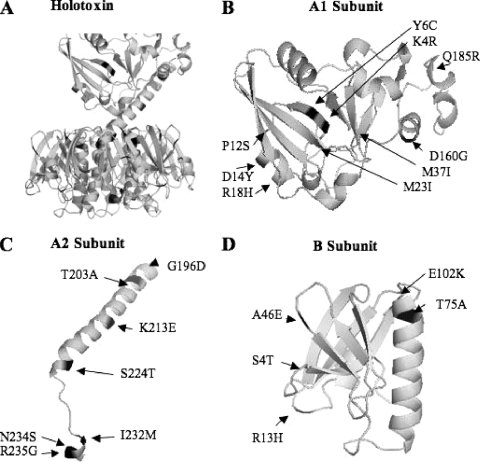
The previous RFLP analyses indicated that a considerable genetic diversity existed among the elt operons carried by the tested ETEC strains, particularly among strains producing LT only. To further evaluate the extent of the genetic diversity of the LT-encoding genes, we sequenced LT-encoding regions of the elt operons of 51 ETEC strains, including the H10407 and the 258909-3 strains, originally isolated from patients in Bangladesh (9, 13). Taken together, the sequence analysis revealed 50 base changes at 48 different polymorphic sites in the elt operon. Thirty-four polymorphic sites were located in eltA, including 4 sites in the sequence encoding the signal peptide, 17 sites in the sequence encoding the A1 subunit, and 13 sites in the sequence encoding the A2 subunit. The eltB cistron showed 16 polymorphic sites, including 5 sites in the sequence encoding the signal peptide and 11 sites in the structural gene. Thirty polymorphic sites resulted in changes in the amino acid sequences, including 21 amino acid changes in eltA and 9 in eltB (Fig. 2). The replacement of asparagin by aspartic acid in position 238 was observed only in the elt operon of H10407 and thus was not considered for the definition of an additional LT type. Of the remaining 20 polymorphic sites in LTA, 2 were located in the signal peptide (F-10V and F-10A), 10 in the A1 subunit (K4R, Y6C, P12S, D14Y, R18H, M23I, M37I, D160G, Q185R, and S190L), and 8 in the A2 subunit (T193A, G196D, T203A, K213E, S224T, I232M, N234S, and R235G). In the LTB subunit, four polymorphic sites were located in the signal peptide (C-16F, A-10V, C-4Y, and Y-2H) and five sites in the mature protein amino acid sequence (S4T, R13H, A46E, T75A, and E102K). In Fig. 2, the positions of the detected polymorphic sites are indicated in the structures of LT subunits, except two sites in the A1 subunit (Q185R and S190L) and one in the A2 subunit (T193A) located in loops with unresolved structures.
FIG. 2.
LT structural model with locations of the polymorphic sites detected in the present study. The LT structural coordinates were retrieved from the PDB (code 1LTS). (A) Structural model of the whole toxin (A and B subunits). (B) Structural model of the A1 subunit with positions (arrows) of nine polymorphic sites with amino acid residues present in the available PDB file. (C) Structural model of the A2 subunit with positions (arrows) of seven polymorphic sites with amino acid residues present in the available PDB file. (D) Structural model of the B subunit with positions (arrows) of the five polymorphic sites with amino acid residues present in the available PDB file. Polymorphic sites are shaded dark. Three polymorphic sites (Q185R, S190L, and T193A) were located on a loop encompassing the disulfide bridge linking A1 and A2 subunits, which has not been structurally solved in the PDB file.
Based on the sequence analysis of the elt operon, 16 LT types were detected with regard to the elt sequence of the reference H10407 strain. The polymorphic site at position 238 was not considered for definition of LT types, since it was restricted to the H10407 strain. As indicated in Table 3, the most frequently found LT type (LT1) was shared by 24 of the 51 tested ETEC strains, including the reference H10407 strain and all tested O6:H16 strains. The second most frequently expressed LT type (LT2) was characterized by four polymorphic sites in the A subunit (S190L, G196D, K213E, and S224T) and one polymorphic site in the B subunit (T75A). This LT type seems to have a widespread distribution; it was detected among both LT+ and LT+/ST+ strains and in an ETEC strain (258909-3) derived from a subject in Bangladesh. This LT allele was also independently reported 10 years ago in a Japanese tourist returning from Israel (20). LT3, displayed by two O88:H25 strains, showed two polymorphic sites in the A subunit (K213E and R235G), while LT4 showed the same amino acid sequence as the previously described LTp (20, 50) with two polymorphic sites (K4R and K213E) in the A and three polymorphic sites (S4T, A46E, and E102K) in the B subunit. The remaining 12 LT types differ from LTs produced by the H10407 strain by the presence of from one (such as the LT9 and LT12 types) to six (such as the LT7 and LT14 types) polymorphic sites (Table 3). In total, six different LT types (LT1, LT3, LT5, LT9, LT12, and LT13) were grouped in the RFLP-I group, five (LT2, LT7, LT14, LT15, and LT16) in RFLP-V, and two (LT1 and LT10) in RFLP-IV, reflecting the higher discriminatory power of the sequencing analysis with regard to RFLP typing (Table 2). Similarly, RFLP-VI, previously recognized as indicative of LTp, encompassed two LT types (LT4 and LT6). The remaining RFLP types encompassed a single LT type each (Table 2).
TABLE 3.
Deduced amino acid compositions of variant A and B subunits obtained from sequenced elt operons of all ETEC strains analyzed in this study
| LT allelic type | ETEC strain serotype (no. of samples) | Positions of the mutated amino acids in subunit:
|
|
|---|---|---|---|
| A | B | ||
| LT1 | H10407 (O78:H11)a | ||
| O6:H16 (15) | |||
| O8:H9 (2) | |||
| O7:H18 (1) | |||
| O114:H− (1) | |||
| O23:H16 (2) | |||
| O23:H33/H45 (1) | |||
| O159:H34 (1) | |||
| LT2 | O78:H12 (4) | S190L, G196D, K213E, S224T | T75A |
| O159:H21 (3) | |||
| O128:H− (1) | |||
| Ont:H8 (1) | |||
| Ont:H21 (1) | |||
| O159:H4 (1) | |||
| LT3 | O88:H25 (2) | K213E, R235G | |
| LT4 | O23:H28 (1) | K4R, K213E | S4T, A46E, E102K |
| LT5 | O48:H21 (1) | K213E, R235G | R13H |
| O88:H25 (1) | |||
| LT6 | O148:H28 (1) | K4R, K213E | S4T, A46E |
| LT7 | O106:H40 (1) | P12S, S190L, G196D, K213E, S224T | T75A |
| LT8 | O152:H2 (1) | T203A, K213E | R13H |
| LT9 | O167:H5 (1) | D14Y | |
| LT10 | Ont:Hnt (1) | Q185R, N234S | |
| LT11 | Ont:H− (1) | M37I, T193A, K213E, I232M | |
| LT12 | Ont:H− (1) | M37I | |
| LT13 | Ont:H9 (1) | R18H, M23I | |
| LT14 | O159:H17 (1) | D160G, S190L, G196D, K213E, S224T | T75A |
| LT15 | O78:H12 (1) | S190L, K213E | T75A |
| LT16 | O78:H12 (1) | Y6C, S190L, G196D, K213E | T75A |
Based on the corrected amino acid sequence of the LT encoded by strain H10407 (6). The polymorphic site at position 238 (N238D) found in the elt operon of strain H10407 was not considered for the definition of a new LT type.
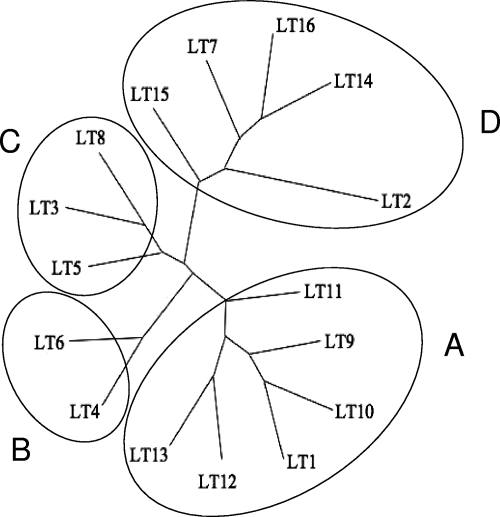
The LT polymorphism observed among the tested ETEC strains showed that some of the natural variants are related. To illustrate the similarity relationships of the LT variants detected in the present study, the amino acids of the complete structural sequences of the two subunits were aligned and an unrooted phylogram was constructed based on the neighbor-joining algorithm (Fig. 3). The 16 LT types were clustered in four major groups with six (LT1, LT9, LT10, LT12, LT13, and the most distantly related, LT11), two (LT4 and LT6), three (LT3, LT5, and LT8), and five (LT2, LT7, LT14, LT15, and LT16) members, referred to as A, B, C, and D, respectively. Notably, the most frequently found LT types, LT1 and LT2, were clustered in the two most distantly related groups, while group C encompassed LT types sharing complete identity or close similarity to the previously described LTp (Fig. 3).
FIG. 3.
Unrooted phylogram constructed by the neighbor-joining method showing the sequence relationships of the concatenated amino acid sequences of the mature A and B subunits of the detected LT variants.
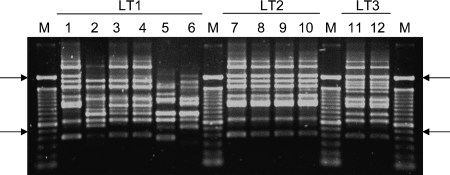
Clonally distinct ETEC strains may express the same LT type.
Our previous analyses have demonstrated that ETEC strains sharing the same serotype and virulence-associated factors may be clonally related, as demonstrated by RAPD analysis (29, 31, 32). In order to evaluate whether ETEC strains sharing a specific LT type were clonally related, we determined the RAPD band profiles of strains expressing either the LT1, LT2, or LT3 type (Fig. 4) . Indeed, ETEC strains of serotype O6:H16 (15 strains), O78:H12 (6 strains), and O88:H25 (2 strains) belonged to different clonal groups and expressed different LT types (LT1, LT2, and LT3, respectively). On the other hand, O78:H11, O8:H9, O6:H16, O7:H18. and O114:H− ETEC strains belong to different clonal groups but share the same LT type (LT1). These results indicate that, based on the strain set analyzed, clonally distinct ETEC strains may share the same LT allele, suggesting lateral transfer of the elt operon among clonally unrelated strains.
FIG. 4.
RAPD profiles of representative ETEC strains expressing the prevailing LT types reported in this study. RAPD profiles were obtained by amplification of genomic DNA with the 1254 primer. LT1, RAPD profiles of ETEC strains sharing the LT1 type: lane 1, reference strain H10407 (O78:H11); lane 2, strain PE0260 (O8:H9); lane 3, strain 4291-1 (O6:H16); lane 4, strain 4292-1 (O6:H16); lane 5, strain PE0215 (O7:H18); lane 6, strain 4101-1 (O114:H−). LT2, RAPD profiles of ETEC strains sharing the LT2 type: lane 7, strain PE0347 (O78:H12); lane 8, strain PE0379 (O78:H12); lane 9, strain 25A-1 (O78:H12); lane 10, strain 2781-5 (O78:H12). LT3, RAPD profiles of ETEC strains sharing the LT3 type: lane 11, strain 136-I (O88:H25); lane 12, strain 136-II (O88:H25). Lane M, 100-bp molecular-size ladder (Invitrogen). The arrows indicate the positions of 500 (bottom) and 2,072 (top) bp.
Functional characterization of LT variants expressed by ETEC strains.
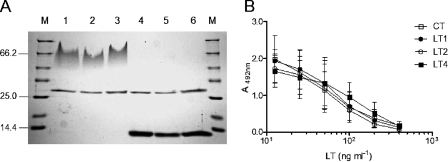
Although the observed polymorphism of LT expressed by human-derived ETEC strains did not involve amino acid residues known to participate directly either in ADP ribosylation or GM1 receptor binding, conformational changes may indirectly affect the biological activities of variant LT types with regard to the reference LT derived from the H10407 strain. Based on the widespread occurrence of the polymorphic sites and their close proximity to amino acids residues known to affect the activity or stability of the toxin, we selected seven LT types (LT1, LT2, LT4, LT5, LT9, LT13, and LT16) to be tested, either as purified toxins (LT1, LT2, and LT4) or as sonically disrupted whole-cell extracts, both in vitro, by means of the cytotonic effect on cultured Y-1 cells, and in vivo, based on fluid accumulation in rabbit ligated ileal loops. LT1, LT4, LT5, and LT2 were representative of the A, B, C, and D groups. On the other hand, LT9, LT13, and LT16 were tested due to the close proximity of the polymorphic site to the Arg7 residue, previously shown to dramatically affect the ADP-ribosyltransferase activity of the toxin (26a). Polyacrylamide gel analyses of the three toxins purified by galactose affinity chromatography showed that all detected B subunits were assembled into stable pentamers in unboiled samples (Fig. 5). Additionally, densitometric analysis of stained gels indicated that all three toxins presented similar A and B subunit distribution ratios (1:5) (Fig. 5 and data not shown). In vitro analysis of the purified LT1, LT2, and LT4 showed no significant difference with regard to binding affinity to the ganglioside in competitive GM1-binding assays (Fig. 5). Nonetheless, the toxic effects of LT4 proved to be significantly less than those detected with LT1, LT2, LT5, LT9, LT13, and LT16, either with the purified proteins or with whole-cell extracts, both in vitro with Y-1 cells and in vivo with fluid accumulation in the rabbit ligated ileal loop assays (Table 4) . These results indicate that LT4, which is identical to the previously described LTp, has reduced toxicity to eukaryotic cells, while the other tested LT variants showed behavior similar to that of the reference toxin produced by the H10407 ETEC strain.
FIG. 5.
Purification of three LT types and competitive binding assays carried out with the GM1 ganglioside. (A) Representatives of LT1, LT2, and LT4 purified by galactose affinity chromatography and sorted in a Coomassie blue-stained polyacrylamide gel. Samples of purified toxins were applied in the polyacrylamide gels before (lanes 1 to 3) or after (lanes 4 to 6) 5 min of boiling. The samples of purified LT were as follows: LT1, lanes 1 and 4; LT2, lanes 2 and 5; and LT4, lanes 3 and 6. (B) GM1-binding curves of different LT variants determined in competitive ELISA with peroxidase-labeled CT. Shown are Scatchard plots for the binding of wild-type LT (LT1), LT from ETEC 25A-1 (LT2), LT from ETEC 1372-1 (LT4), and CT. The data are means ± standard deviations of duplicate samples and are representative of three independent experiments.
TABLE 4.
Toxic activities of different LT types measured both in vitro, by the cytotonic action on cultivated Y-1 cells, and in vivo, by fluid accumulation in rabbit ligated ileal loops
| LT typea | Cytotonic effect on Y-1 cells (ng ml−1)b | Fluid accumulation in rabbit ligated ileal loop (ml 5 cm−1)c |
|---|---|---|
| Purified toxins | ||
| LT1 (H10407) | 3.5 ± 2.0 | 10.0 ± 0.5 |
| LT2 (25A-1) | 1.64 ± 0.6 | 12.75 ± 3.18 |
| LT4 (1372-1) | 65.65 ± 18.75 | ≤0.5 |
| Cell lysates | ||
| LT− (4611-4)d | ≤0.5 | |
| LT1 (H10407) | 0.89 ± 0.43 | 6.0 ± 3.0 |
| LT2 (25A-1) | 0.62 ± 0.41 | 12.5 ± 1.5 |
| LT4 (1372-1) | 15.67 ± 5.43 | ≤0.5 |
| LT5 (136-III) | 0.29 ± 0.05 | 11.5 ± 3.54 |
| LT9 (4702-1) | 0.59 ± 0.08 | 7.85 ± 1.63 |
| LT13 (PE0615) | 0.29 ± 0.07 | 10.53 ± 0.92 |
| LT16 (2781-5) | 0.44 ± 0.2 | 9.75 ± 2.47 |
Different LT types, as either purified toxins or sonically disrupted whole-cell extracts. The LT-producing ETEC strains are indicated in parentheses.
The values indicate the concentrations of LT that induced visible cytotonic effects in 50% of the Y-1 cells examined. The concentrations of LT were determined by capture ELISA using LT-specific antibodies, as described in Materials and Methods.
Volume (ml) of liquid accumulated in rabbit ligated ileal loops following inoculation of 150 ng of toxin, either purified or in whole-cell extracts.
LT− ETEC strain.
DISCUSSION
The present report is the first systematic study of the natural genetic diversity of LT-Is produced by ETEC strains isolated from humans. Based on a set of 51 strains, we detected a rather high variability of LT, which was disclosed by RFLP and DNA-sequencing methods. The detected polymorphism represents a considerable increase in the available knowledge of the diversity of the elt operon. Indeed, six out of the seven RFLP types and 16 of the 31 reported polymorphic sites are described for the first time and clearly demonstrate that the natural diversity of LTs produced by human-derived ETEC strains has been underestimated during recent decades. Moreover, the restricted number of strains tested and the fact that most of them were isolated in Brazil suggest that LT diversity, as well as the possible impacts of such diversity on the epidemiology of the ETEC-associated disease, remains underestimated on a worldwide scale.
The detection of high natural diversity of LTs in a rather small set of ETEC strains may be ascribed to two major facts. First, we carried out a systematic analysis of LT genetic diversity, employing two powerful molecular typing methods (RFLP and DNA sequencing). Second, the nature of the selected strain set was designed to encompass ETEC strains previously shown to be genetically heterogeneous, as inferred from their serotypes (20 different serotypes and six nontypeable strains) and toxinogenic profiles. Half (25 strains) of the tested strains were strains producing LT only, usually recognized by their heterogeneous nature, lack of an identifiable CF, and frequent recovery from asymptomatic subjects (33). Indeed, among the 25 tested LT+ ETEC strains, we found all seven detected RFLP types and 14 of the 16 LT sequence types. On the other hand, LTs encoded by 26 LT+/ST+ ETEC strains were classified in three RFLP types and four LT types. These results suggest that LTs produced by LT-only ETEC strains, particularly among strains isolated from asymptomatic subjects, are more genetically variable than LTs produced by LT+/ST+ strains. This finding raises interesting questions concerning the high incidence of ETEC strains producing only LT among nondiarrheic children in epidemiological studies carried out in different regions of endemicity (32). Additionally, such findings might indicate that in areas of endemicity, ETEC strains producing LT only are subject to selective forces, leading to a more diverse repertoire of LT sequences that could contribute to immunological escape mechanisms. Future studies aimed at the determination of LT types produced by ETEC strains isolated from different geographical regions should shed more light on the relevance of LT typing and the epidemiology of ETEC-associated disease. RFLP typing has been the only method employed for many years to differentiate LTh and LTp (25, 45, 47). Although restricted, such differentiation was the most convincing demonstration of the natural diversity of LT-Is produced by wild-type ETEC strains. In the present study, seven RFLP types were identified among the 51 ETEC strains, including the HhaI polymorphism at the eltB cistron, previously employed to differentiate LTh and LTp, and a new HhaI HphI polymorphism in which the loss of the HhaI site was due to a distinct base replacement. The finding of six new polymorphic sites in the elt operon reinforces the use of such molecular typing methods as simple and fast tools to disclose LT variants produced by wild-type ETEC strains isolated from humans or other vertebrate hosts. On the other hand, our results demonstrate that ascribing a specific RFLP type to an ETEC group, such as those derived from different mammalian hosts or based on strains sharing a specific serotype or virulence-associated markers, may not disclose the real LT diversity, as demonstrated by RFLP types I, IV, V, and VI, which encompass two to six different LT types identified by DNA sequencing. The lower resolution of the RFLP analysis is clearly demonstrated by the artificial differentiation of LTp and LTh. Among the four ETEC strains sharing the loss of the HhaI restriction site, three were genetically distinct and belonged to different LT types.
The location of the elt operon in high-molecular-weight plasmids and the frequent finding of flanking palindromic sequences strongly suggest that lateral gene transfer mechanisms contributed to the dissemination of the LT-encoding genes among wild-type ETEC strains (37, 39, 49). The dynamic nature of ETEC-associated genes has been confirmed by multilocus sequence-typing analyses of clonally distinct E. coli strains (43). Indeed, the acquisition of enterotoxin genes may be sufficient to generate an ETEC strain, which may belong to many phylogenetically distinct lineages. In the present study, we identified 16 LT types, taking into account 31 amino acid replacements with regard to the toxin encoded by the reference H10407 strain. These results further demonstrate the dynamic nature of the elt operon based on the widespread distribution of two LT types (LT1 and LT2), which are shared by clonally unrelated ETEC strains, as demonstrated by RAPD analysis. The same conclusion may be drawn with strains sharing the same phenotypic and genotypic profiles but expressing different LT types, as illustrated by the O88:H25 and O78:H12 strains. On the other hand, we observed some close relationships between some LT types and the phenotypic and genotypic features of certain ETEC groups. For example, all tested O6:H16 (16) and O78:H12 (4) strains carried the same LT types, LT1 and LT2, respectively. Taken together, the present observations show that much still has to be done in order to achieve a better understanding of the dynamics of the elt operon transfer among wild-type ETEC strains with similar or different phenotypic traits and genetic compositions.
The LT2 type, characterized by four mutations in the A subunit (S190L, G196D, K213E, and S224T) and one mutation in the B subunit (T75A), was shown to be the second most widely distributed LT type among wild-type ETEC strains. This LT type was detected in six different serotypes and was recovered from subjects in different regions in Brazil and Bangladesh (represented by the 258909-3 strain). The same LT has also been detected in an ETEC strain isolated from a Japanese tourist, further demonstrating the distribution of the type on a worldwide scale (20). Interestingly, the same group reported that, in contrast to other ETEC strains, the gene encoding this variant LT was integrated into the chromosomal DNA of the strain (20). In our strains, preliminary data indicated that, at least in some strains, the elt operon encoding the LT2 type is located on episomal plasmids, but further analyses should evaluate in more detail the locations of the replicons encoding different LT types.
The close proximity of the reported sequences of the LT types allowed us to establish putative evolutionary relationships among them, which resulted in the definition of four major similarity groups. The close proximity of the LT variant amino acid sequences, which differ in a maximum of seven residues, restrict precise phylogenetic inferences but suggest that the two most widely distributed LT types (LT1 and LT2) are representative of two divergent LT groups, while the third and fourth groups (B and C) represent different groups encompassing LT4 (LTp) and the closely related LT6 type (group B), as well as LT3, LT5, and LT8 in a fourth group (C). These data led us to conclude that the differentiation of LTh and LTp is artificial and should be reconsidered based on a more detailed analysis of the natural diversity of LTs produced by ETEC strains isolated from different mammalian hosts. Thus, the finding of an LT4-producing ETEC strain in a human host would not necessarily be indicative of a zoonotic infection. On the other hand, the definition of four LT groups based on sequence similarity suggests that the LT variants are subject to evolutionary forces that may affect biochemical and biological features of the toxins encoded by different ETEC strains. Indeed, a clear correlation between genetic diversity and altered biological functions has already been established for Shiga-like toxin (Stx) variants produced by enterohemorragic E. coli strains (8, 41).
In our initial characterization of the biological functions of natural LT variants expressed by wild-type ETEC strains, toxins representing seven LT types were selected for in vitro and in vivo testing with either purified toxins (LT1, LT2, and LT4) or whole-cell extracts. The purified toxins revealed no significant difference with regard to receptor (GM1) binding affinity, but a probe of the toxic effects of the LT variants indicated that LT4, which is identical to the previously reported LTp, has reduced cytotonic effects on Y-1 cells and did not lead to fluid accumulation in rabbit ligated ileal loops. Previous evidence, based on whole-cell lysates or culture supernatants, indicated that LTp has toxic effects, including the activation of adenylate cyclase, similar to the toxin produced by the human-derived ETEC strains (22). In contrast to such results, the purified LT4, as well as whole bacterial lysate of the wild-type ETEC strain, showed reduced toxic effects under in vitro and in vivo conditions. The purified toxin was also evaluated for purity, the presence of an altered A and B ratio distribution, and susceptibility to protease attack without any evidence for altered behavior with regard to other tested LT variants (unpublished observation). Indeed, the closely related LT6 type also showed reduced toxicity when tested with Y-1 cells and whole bacterial lysates, suggesting that replacement of lysine by arginine at position 4 of the A1 subunit affects the biological activities of these LT variants (unpublished observations). The close proximity of this polymorphic site to the arginine residue at position 7 suggests that the K4R substitution would change the structural organization of the toxin active site in a fashion similar to that of the R7K mutant generated by site-directed mutagenesis (44). Further functional characterization of LT4, together with biochemical and structural analyses, is currently under investigation and will help us to evaluate the impact of this polymorphism on the activities of LTs produced by these wild-type ETEC strains.
As recently demonstrated by us, wild-type ETEC strains may differ in the amounts of LT produced and/or secreted under in vitro and in vivo growth conditions (24). Indeed, the inherent variability of ETEC strains in secreting LTs may correlate with the incidence and severity of diarrheal episodes in either humans or other mammalian hosts (2, 4, 40). The findings that LTs produced by ETEC strains isolated from humans show considerable genetic diversity add a further aspect to be evaluated concerning the contribution of these features to the complex epidemiology of the disease and may contribute to the design and development of more rational prophylactic and therapeutic approaches against ETEC-associated diarrhea.
Acknowledgments
This work was supported by CNPq and FAPESP grants.
We gratefully acknowledge the contributions of M. Magalhães (Federal University of Pernambuco, Recife, Brazil), A. Régua-Mangia (Oswaldo Cruz Foundation, Rio de Janeiro, Brazil), and A. M. Svennerholm (University of Göteborg, Göteborg, Sweden) for providing some of the strains used in the present study. We gratefully acknowledge the contributions of M. O. Lasaro (The Wistar Institute) in critically reading the manuscript, J. Cabrera-Crespo in the purification of LT, and A. J. Piantino Ferreira in the rabbit ligated ileal loop assay. We also gratefully acknowledge the invaluable technical assistance of C. Calderon and L. C. da Silva.
Footnotes
Published ahead of print on 25 January 2008.
REFERENCES
- 1.Bäckström, M., V. Shahabi, S. Johansson, S. Teneberg, A. Kjellberg, H. Miller-Podraza, J. Holmgren, and M. Lebens. 1997. Structural basis for differential receptor binding of cholera and Escherichia coli heat-labile toxins: influence of heterologous amino acid substitutions in the cholera B-subunit. Mol. Microbiol. 24489-497. [DOI] [PubMed] [Google Scholar]
- 2.Berberov, E. M., Y. Zhou, D. H. Francis, M. A. Scott, S. D. Kachman, and R. A. Moxley. 2004. Relative importance of heat-labile enterotoxin in the causation of severe diarrheal disease in the gnotobiotic piglet model by a strain of enterotoxigenic Escherichia coli that produces multiple enterotoxins. Infect. Immun. 723914-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, R. E., S. S. Morris, and J. Bryce. 2003. Where and why are 10 million children dying every year? Lancet 3612226-2234. [DOI] [PubMed] [Google Scholar]
- 4.Coster, T. S., M. K. Wolf, E. R. Hall, F. J. Cassels, D. N. Taylor, C. T. Liu, F. C. Trespalacios, A. DeLorimier, D. R. Angleberger, and C. E. McQueen. 2007. Immune response, ciprofloxacin activity, and gender differences after human experimental challenge by two strains of enterotoxigenic Escherichia coli. Infect. Immun. 75252-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dallas, W. S. 1983. Conformity between heat-labile toxin genes from human and porcine enterotoxigenic Escherichia coli. Infect. Immun. 40647-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domenighini, M., M. Pizza, M. G. Jobling, R. K. Holmes, and R. Rappuoli. 1995. Identification of errors among database sequence entries and comparison of correct amino acid sequences for the heat-labile enterotoxins of Escherichia coli and Vibrio cholerae. Mol. Microbiol. 151165-1167. [DOI] [PubMed] [Google Scholar]
- 7.Dubreuil, J. D. 1997. Escherichia coli STb enterotoxin. Microbiology 1431783-1795. [DOI] [PubMed] [Google Scholar]
- 8.Eklund, M., K. Leino, and A. Siitonen. 2002. Clinical Escherichia coli strains carrying stx genes: stx variants and stx-positive virulence profiles. J. Clin. Microbiol. 404585-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, D. G., R. P. Silver, D. J. Evans, Jr., D. G. Chase, and S. L. Gorbach. 1975. Plasmid-controlled colonization factor associated with virulence in Escherichia coli enterotoxigenic for humans. Infect. Immun. 12656-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaastra, W., and A. M. Svennerholm. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4444-452. [DOI] [PubMed] [Google Scholar]
- 11.Giannella, R. A. 1983. Escherichia coli heat-stable enterotoxin: biochemical and physiological effects on the intestine. Prog. Food Nutr. Sci. 7157-165. [PubMed] [Google Scholar]
- 12.Gomes, T. A. T., P. M. Griffin, C. Ivey, L. R. Trabulsi, and S. R. T. S. Ramos. 1996. EPEC infections in São Paulo. Rev. Microbiol. 2725-33. [Google Scholar]
- 13.Gothefors, L., C. Ahren, B. Stoll, D. K. Barua, F. Orskov, M. A. Salek, and A. M. Svennerholm. 1985. Presence of colonization factor antigens on fresh isolates of fecal Escherichia coli: a prospective study. J. Infect. Dis. 1521128-1133. [DOI] [PubMed] [Google Scholar]
- 14.Green, B. A., R. J. Neill, W. T. Ruyechan, and R. K. Holmes. 1983. Evidence that a new enterotoxin of Escherichia coli which activates adenylate cyclase in eucaryotic target cells is not plasmid mediated. Infect. Immun. 41383-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guth, B. E., E. G. Aguiar, P. M. Griffin, S. R. Ramos, and T. A. Gomes. 1994. Prevalence of colonization factor antigens (CFAs) and adherence to HeLa cells in enterotoxigenic Escherichia coli isolated from feces of children in Sao Paulo. Microbiol. Immunol. 38695-701. [DOI] [PubMed] [Google Scholar]
- 16.Guth, B. E., C. L. Pickett, E. M. Twiddy, R. K. Holmes, T. A. Gomes, A. A. Lima, R. L. Guerrant, B. D. Franco, and L. R. Trabulsi. 1986. Production of type II heat-labile enterotoxin by Escherichia coli isolated from food and human feces. Infect. Immun. 54587-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirst, T. 1999. Cholera toxin and Escherichia coli heat-labile enterotoxin. Academic Press, New York, NY.
- 18.Holmgren, J., J. Adamsson, F. Anjuere, J. Clemens, C. Czerkinsky, K. Eriksson, C. F. Flach, A. George-Chandy, A. M. Harandi, M. Lebens, T. Lehner, M. Lindblad, E. Nygren, S. Raghavan, J. Sanchez, M. Stanford, J. B. Sun, A. M. Svennerholm, and S. Tengvall. 2005. Mucosal adjuvants and anti-infection and anti-immunopathology vaccines based on cholera toxin, cholera toxin B subunit and CpG DNA. Immunol. Lett. 97181-188. [DOI] [PubMed] [Google Scholar]
- 19.Honda, T., T. Tsuji, Y. Takeda, and T. Miwatani. 1981. Immunological nonidentity of heat-labile enterotoxins from human and porcine enterotoxigenic Escherichia coli. Infect. Immun. 34337-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imamura, S., N. Kido, M. Kato, H. Kawase, A. Miyama, and T. Tsuji. 1997. A unique DNA sequence of human enterotoxigenic Escherichia coli enterotoxin encoded by chromosomal DNA. FEMS Microbiol. Lett. 146241-245. [DOI] [PubMed] [Google Scholar]
- 21.Inoue, T., T. Tsuji, M. Koto, S. Imamura, and A. Miyama. 1993. Amino acid sequence of heat-labile enterotoxin from chicken enterotoxigenic Escherichia coli is identical to that of human strain H 10407. FEMS Microbiol. Lett. 108157-161. [DOI] [PubMed] [Google Scholar]
- 22.Kantor, H. S., P. Tao, and C. Wisdom. 1974. Action of Escherichia coli enterotoxin: adenylate cyclase behavior of intestinal epithelial cells in culture. Infect. Immun. 91003-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karmali, M. A., M. Petric, C. Lim, R. Cheung, and G. S. Arbus. 1985. Sensitive method for detecting low numbers of verotoxin-producing Escherichia coli in mixed cultures by use of colony sweeps and polymyxin extraction of verotoxin. J. Clin. Microbiol. 22614-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lasaro, M. A., J. F. Rodrigues, C. Mathias-Santos, B. E. Guth, A. Regua-Mangia, A. J. Piantino Ferreira, M. Takagi, J. Cabrera-Crespo, M. E. Sbrogio-Almeida, and L. C. de Souza Ferreira. 2006. Production and release of heat-labile toxin by wild-type human-derived enterotoxigenic Escherichia coli. FEMS Immunol. Med. Microbiol. 48123-131. [DOI] [PubMed] [Google Scholar]
- 25.Leong, J., A. C. Vinal, and W. S. Dallas. 1985. Nucleotide sequence comparison between heat-labile toxin B-subunit cistrons from Escherichia coli of human and porcine origin. Infect. Immun. 4873-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llop, P., P. Caruso, J. Cubero, C. Morente, and M. M. Lopez. 1999. A simple extraction procedure for efficient routine detection of pathogenic bacteria in plant material by polymerase chain reaction. J. Microbiol. Methods. 3723-31. [DOI] [PubMed] [Google Scholar]
- 26a.• Lobbet. 1991. •.
- 27.Maas, R. 1983. An improved colony hybridization method with significantly increased sensitivity for detection of single genes. Plasmid 10296-298. [DOI] [PubMed] [Google Scholar]
- 28.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pacheco, A. B., L. C. Ferreira, M. G. Pichel, D. F. Almeida, N. Binsztein, and G. I. Viboud. 2001. Beyond serotypes and virulence-associated factors: detection of genetic diversity among O153:H45 CFA/I heat-stable enterotoxigenic Escherichia coli strains. J. Clin. Microbiol. 394500-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pacheco, A. B., B. E. Guth, D. F. de Almeida, and L. C. Ferreira. 1996. Characterization of enterotoxigenic Escherichia coli by random amplification of polymorphic DNA. Res. Microbiol. 147175-182. [DOI] [PubMed] [Google Scholar]
- 31.Pacheco, A. B., B. E. Guth, K. C. Soares, L. Nishimura, D. F. de Almeida, and L. C. Ferreira. 1997. Random amplification of polymorphic DNA reveals serotype-specific clonal clusters among enterotoxigenic Escherichia coli strains isolated from humans. J. Clin. Microbiol. 351521-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pacheco, A. B., K. C. Soares, D. F. de Almeida, G. I. Viboud, N. Binsztein, and L. C. Ferreira. 1998. Clonal nature of enterotoxigenic Escherichia coli serotype O6:H16 revealed by randomly amplified polymorphic DNA analysis. J. Clin. Microbiol. 362099-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qadri, F., A. M. Svennerholm, A. S. Faruque, and R. B. Sack. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18465-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regua-Mangia, A. H., B. C. Guth, J. R. da Costa Andrade, K. Irino, A. B. Pacheco, L. C. Ferreira, V. Zahner, and L. M. Teixeira. 2004. Genotypic and phenotypic characterization of enterotoxigenic Escherichia coli (ETEC) strains isolated in Rio de Janeiro city, Brazil. FEMS Immunol. Med. Microbiol. 40155-162. [DOI] [PubMed] [Google Scholar]
- 35.Reis, M. H., B. E. Guth, T. A. Gomes, J. Murahovschi, and L. R. Trabulsi. 1982. Frequency of Escherichia coli strains producing heat-labile toxin or heat-stable toxin or both in children with and without diarrhea in Sao Paulo. J. Clin. Microbiol. 151062-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez, J., and J. Holmgren. 2005. Virulence factors, pathogenesis and vaccine protection in cholera and ETEC diarrhea. Curr. Opin. Immunol. 17388-398. [DOI] [PubMed] [Google Scholar]
- 37.Schlor, S., S. Riedl, J. Blass, and J. Reidl. 2000. Genetic rearrangements of the regions adjacent to genes encoding heat-labile enterotoxins (eltAB) of enterotoxigenic Escherichia coli strains. Appl. Environ. Microbiol. 66352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schweizer, H. P. 2001. Vectors to express foreign genes and techniques to monitor gene expression in Pseudomonads. Curr. Opin. Biotechnol. 12439-445. [DOI] [PubMed] [Google Scholar]
- 39.So, M., W. S. Dallas, and S. Falkow. 1978. Characterization of an Escherichia coli plasmid encoding for synthesis of heat-labile toxin: molecular cloning of the toxin determinant. Infect. Immun. 21405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinsland, H., P. Valentiner-Branth, H. K. Gjessing, P. Aaby, K. Molbak, and H. Sommerfelt. 2003. Protection from natural infections with enterotoxigenic Escherichia coli: longitudinal study. Lancet 362286-291. [DOI] [PubMed] [Google Scholar]
- 41.Stephan, R., and L. E. Hoelzle. 2000. Characterization of shiga toxin type 2 variant B-subunit in Escherichia coli strains from asymptomatic human carriers by PCR-RFLP. Lett. Appl. Microbiol. 31139-142. [DOI] [PubMed] [Google Scholar]
- 42.Tsuji, T., S. Taga, T. Honda, Y. Takeda, and T. Miwatani. 1982. Molecular heterogeneity of heat-labile enterotoxins from human and porcine enterotoxigenic Escherichia coli. Infect. Immun. 38444-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner, S. M., R. R. Chaudhuri, Z. D. Jiang, H. DuPont, C. Gyles, C. W. Penn, M. J. Pallen, and I. R. Henderson. 2006. Phylogenetic comparisons reveal multiple acquisitions of the toxin genes by enterotoxigenic Escherichia coli strains of different evolutionary lineages. J. Clin. Microbiol. 444528-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Akker, F., E. A. Merritt, M. Pizza, M. Domenighini, R. Rappuoli, and W. G. Hol. 1995. The Arg7Lys mutant of heat-labile enterotoxin exhibits great flexibility of active site loop 47-56 of the A subunit. Biochemistry 3410996-11004. [DOI] [PubMed] [Google Scholar]
- 45.Vinal, A. C., and W. S. Dallas. 1987. Partition of heat-labile-enterotoxin genes between human and animal Escherichia coli isolates. Infect. Immun. 551329-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolf, M. K. 1997. Occurrence, distribution, and associations of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin. Microbiol. Rev. 10569-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto, T., T. Gojobori, and T. Yokota. 1987. Evolutionary origin of pathogenic determinants in enterotoxigenic Escherichia coli and Vibrio cholerae O1. J. Bacteriol. 1691352-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto, T., T. Tamura, and T. Yokota. 1984. Primary structure of heat-labile enterotoxin produced by Escherichia coli pathogenic for humans. J. Biol. Chem. 2595037-5044. [PubMed] [Google Scholar]
- 49.Yamamoto, T., and T. Yokota. 1981. Escherichia coli heat-labile enterotoxin genes are flanked by repeated deoxyribonucleic acid sequences. J. Bacteriol. 145850-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto, T., and T. Yokota. 1983. Sequence of heat-labile enterotoxin of Escherichia coli pathogenic for humans. J. Bacteriol. 155728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]