Abstract
The Escherichia coli guaB promoter (PguaB) regulates the transcription of two genes, guaB and guaA, that are required for de novo synthesis of GMP, a precursor for the synthesis of guanine nucleoside triphosphates. The activity of PguaB is subject to growth rate-dependent control (GRDC). Here we show that the A+T-rich sequence located between positions −59 and −38 relative to the guaB transcription start site stimulates transcription from PguaB ∼8- to 10-fold and, in common with other UP elements, requires the C-terminal domain of the RNA polymerase α subunit for activity. Like the rrnB P1 UP element, the PguaB UP element contains two independently acting subsites located at positions −59 to −47 and −46 to −38 and can stimulate transcription when placed upstream of the lacP1 promoter. We reveal a novel role for the PguaB UP element by demonstrating that it is required for GRDC. The involvement of the UP element in GRDC also requires the participation of sequences located at least 100 bp upstream of the guaB transcription start site. These sequences are required for down-regulation of PguaB activity at lower growth rates.
Escherichia coli RNA polymerase (RNAP) is a multisubunit complex consisting of an α subunit homodimer, single β, β′, and ω subunits, and one of several σ subunits (14). The α subunit contains an N-terminal domain (αNTD; residues 8 to 232) and a C-terminal domain (αCTD; residues 249 to 329) (5, 21, 44). αNTD and αCTD are separated by a flexible linker that is at least 13 amino acids long (5, 20, 25). αCTD plays important roles in transcription activation by contacting a number of transcription factors (6, 18) or through interacting with an A+T-rich DNA sequence referred to as the UP element (16).
UP elements stimulate transcription by recruiting αCTD to DNA (32, 43). The best-studied UP element spans the region from −59 to −38 at the rrnB P1 promoter (16). The rrnB P1 UP element contains two domains, a ∼9-bp proximal UP element subsite located at positions −46 to −38 and a ∼13-bp distal UP element subsite located at positions −59 to −47. Each subsite functions independently by recruiting one αCTD (11). The amino acid residues in αCTD that comprise the 265 determinant, including residues V264, R265, N268, N294, G296, K298, and S299, interact with UP element and A-tract sequences by contacting the DNA backbone, and R265 makes additional contacts with bases in the minor groove (4, 32, 43). These residues are the most important for rrnB P1 UP element utilization both in vivo and in vitro (13, 35). In the absence of a strong distal UP element subsite, αCTD bound to a consensus or near-consensus proximal UP element subsite also makes productive contacts with region 4.2 of σ70 that serve to stimulate transcription (7, 35).
The E. coli guaB promoter (PguaB) regulates the transcription of two genes, guaB and guaA, which together constitute the guaBA operon. The guaB and guaA genes encode IMP dehydrogenase and GMP synthetase, respectively, and are required for the biosynthesis of GMP from IMP (22, 42). PguaB is regulated by the cyclic AMP (cAMP) receptor protein (CRP), and CRP binds to a ∼300-bp DNA fragment containing PguaB that includes a predicted CRP site centered at position −117.5 relative to the guaB transcription start site (19). PguaB also contains putative binding sites for PurR and DnaA overlapping the core promoter region (23, 41). In addition, an A+T-rich sequence located between positions −59 and −38 at PguaB matches the consensus 22-bp UP element sequence [5′-nnAAA(A/T)(A/T)T(A/T)TTTTnnAAAAnnn-3′; n represents a nonconserved position]) at 13 out of 15 conserved positions (Fig. 1), but its role as an UP element has not been confirmed (10, 19).
FIG. 1.
Comparison of A+T-rich sequences used in this work with the consensus sequence for UP elements. Consensus UP element sequences and the putative PguaB UP element (positions −59 to −38 relative to the guaB transcription start site) are aligned with the 22-bp general consensus sequence for UP elements (10). Nonconserved positions are indicated by a lowercase “n.” Mismatches to the general consensus UP element sequence are shown in boldface. The degree of the match to the general consensus is shown for each sequence. The consensus 4541 UP element was made by combining optimal proximal and distal UP element subsites (11). The consensus 4192 UP element was selected by a SELEX procedure (10). The predicted PguaB proximal UP element subsite contains PguaB sequences from position −46 to −38, whereas the predicted PguaB distal UP element subsite contains sequences from −59 to −47. The SUB sequence is nonspecific for αCTD and contributes minimally to transcription (10, 30).
The term growth rate-dependent control (GRDC) is used to refer to situations where the abundance of a particular transcript or protein product represents an increasing fraction of the cell mass at higher growth rates. For example, in E. coli, the synthesis rates of rRNA and tRNA increase in proportion to the square of the growth rate, with the result that stable RNA accounts for an increased fraction of the cellular mass with increasing growth rate (9, 15, 27). One consequence of GRDC of stable RNA synthesis is that the consumption of nucleoside triphosphates also increases in a growth rate-dependent manner (28). Because GMP is a precursor for the synthesis of GTP, it follows that the rate of synthesis of GMP should be regulated according to the growth rate. Consistent with this, it has been shown that PguaB activity is subject to GRDC (8). The best-characterized promoter subject to GRDC that also contains an UP element is the rrnB P1 promoter, although the UP element plays no role in GRDC of this promoter (2, 28). In this work, we show that the A+T-rich sequence located upstream of the PguaB −35 region does indeed function as an UP element and that, like the rrnB P1 UP element, it contains promoter-proximal and -distal subsites that are able to activate transcription independently. However, we demonstrate that, in contrast to the situation at the rrnB P1 promoter, the PguaB UP element is required for GRDC of PguaB and this regulation requires the participation of sequences further upstream.
MATERIALS AND METHODS
Strains.
The bacterial strains and plasmids used in this study are listed in Table 1. For promoter measurements in vivo, strains containing single-copy promoter-lacZ transcriptional fusions were employed. All transcriptional fusions were carried on λ prophages and were constructed using a system based on λimm21 (30, 40). All PguaB-lacZ fusions were made in the VH1000 genetic background and had downstream endpoints at +36 with respect to the guaB transcription start site. All lacP1-lacZ transcriptional fusions were made in strain NK5031. Apart from strain RLG4288, lacP1 derivatives each contain the core lacP1 promoter (positions −37 to +52) with non-lac sequences located upstream of position −37 relative to the lacP1 transcription start site. Strain NKGLAC contains a lacP1-lacZ fusion in which the predicted PguaB UP element is located at positions −59 to −38.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype/descriptiona | Promoter designationb | Source or reference |
|---|---|---|---|
| Bacterial strains | |||
| NK5031 | F′ lacZΔM5262 supF Nalr | 17 | |
| VH1000 | MG1655 lacZ lacI pyrE+ | 12 | |
| NKGLAC | NK5031/λ [SUB(−66 to −60)-PguaB(−59 to −38)-lacP1(−37 to +52)]-lacZ | This study | |
| RLG4208 | NK5031/λ [rrnB P1(−66 to −60)-4192(−59 to −38)-lacP1(−37 to +52)]-lacZ | 10 | |
| RLG4282 | NK5031/λ [rrnB P1(−88 to −38, Δ72)-lacP1(−37 to +52)]-lacZ | 31 | |
| RLG4288 | NK5031/λ [SUB(−58 to −41)-lacP1(−40 to +52)]-lacZ | Core lacP1 | 31 |
| VH1000G-253 | VH1000/λ [PguaB(−253 to +36)]-lacZ | PguaB(−253 to +36) | This study |
| VH1000G-133 | VH1000/λ [PguaB(−133 to +36)]-lacZ | PguaB(−133 to +36) | This study |
| VH1000G-133SUB | VH1000/λ [PguaB(−133 to −60)-SUB(−59 to −38)-PguaB(−37 to +36)]-lacZ | PguaB(−133 to +36) (no UP) | This study |
| VH1000G-100 | VH1000/λ [PguaB(−100 to +36)]-lacZ | PguaB(−100 to +36) | This study |
| VH1000G-84 | VH1000/λ [PguaB(−84 to +36)]-lacZ | PguaB(−84 to +36) | This study |
| VH1000G-69 | VH1000/λ [PguaB(−69 to +36)]-lacZ | PguaB(−69 to +36) | This study |
| VH1000G-59 | VH1000/λ [SUB(−66 to −60)-PguaB(−59 to +36)]-lacZ | PguaB(−59 to +36) | This study |
| VH1000G-PROX | VH1000/λ [SUB(−66 to −47)-PguaB(−46 to +36)]-lacZ | This study | |
| VH1000G-DIS | VH1000/λ [SUB(−66 to −60)-PguaB(−59 to −47)-SUB(−46 to −38)-PguaB(−37 to +36)]-lacZ | This study | |
| VH1000G-CONS | VH1000/λ [4541(−59 to −38)-PguaB(−37 to +36)]-lacZ | This study | |
| VH1000G-37 | VH1000/λ [SUB(−59 to −38)-PguaB(−37 to +36)]-lacZ | Core PguaB | This study |
| Plasmids | |||
| pBluescript II KS | E. coli-specific cloning vector (Apr) | 1 | |
| pMSB1 | Derivative of promoter cloning plasmid, pRS1553, for construction of lacZ fusions (Apr) | 30 | |
| pRLG770 | Derivative of pKM2 for in vitro transcription reactions | 36 |
Apr, ampicillin resistance. The architecture of each promoter is shown in brackets. The SUB sequence is a DNA sequence that is nonspecific for αCTD. 4192 is the consensus 4192 UP element (10); 4541 is the consensus 4541 UP element (11). For SUB and UP element sequences, see Fig. 1. The SUB sequence inserted at positions −66 to −60 (5′-GACTGCA-3′) separates an UP element from the upstream A+T-rich EcoRI site. At other promoters, the corresponding region of SUB is used to replace the UP element sequences shown in Fig. 1. Δ72 indicates a deletion of an A:T base pair at position −72 at rrnB P1, which results in loss of binding and activation by the factor for inversion stimulation (36).
Designations of promoter derivatives referred to in the text are given where appropriate.
Media and growth conditions.
Bacteria were grown at 37°C. NK5031 strain derivatives, each containing a lacP1-lacZ transcriptional fusion, were grown in Luria-Bertani medium. VH1000 strain derivatives, each containing a PguaB-lacZ transcriptional fusion, were grown in M9 minimal medium supplemented with 0.2% (wt/vol) glucose and 5 μg/ml thiamine. Strains containing plasmids encoding wild-type (WT) α or mutant α derivatives were grown in M9 minimal medium with 0.4% (wt/vol) glucose, 5 μg/ml thiamine, 0.8% (wt/vol) Casamino Acids, and 100 μg/ml ampicillin. To achieve different cellular growth rates, cells were grown in M9 minimal medium containing 5 μg/ml thiamine and one of the following (in order of increasing growth rate supported): 0.4% (wt/vol) glycerol, 0.4% succinate plus 20 amino acids, 0.4% (wt/vol) glycerol plus 20 amino acids, 0.4% (wt/vol) glycerol plus 1% (wt/vol) Casamino Acids, 0.4% (wt/vol) glycerol plus 2% (wt/vol) Casamino Acids, 0.4% (wt/vol) glucose plus 20 amino acids, or 0.4% (wt/vol) glucose plus 0.8% (wt/vol) Casamino Acids (“20 amino acids” comprised each amino acid at a final concentration of 20 μg/ml).
Recombinant DNA techniques.
Standard methods for plasmid isolation and DNA manipulation were used throughout (37). All promoter derivatives were made by PCR and contained an upstream EcoRI restriction site and a downstream HindIII restriction site. PCR products were digested with EcoRI and HindIII, ligated to pMSB1 for use in construction of single-copy lacZ fusions (30), and also ligated to pRLG770 for in vitro transcription assays. All amplified sequences were checked by DNA sequencing.
Measurement of transcription in vivo.
Dense starter cultures of bacteria containing single-copy promoter-lacZ fusions were diluted 100-fold into the same growth medium and grown at 37°C to an optical density at 600 nm (OD600) of 0.3 to 0.5. For GRDC experiments, starter cultures were grown overnight in medium supporting the lowest growth rate (i.e., using glycerol as the sole carbon source). Strains containing plasmids encoding WT α and mutant α derivatives were inoculated from fresh colonies into minimal medium at an OD600 of ∼0.007 and grown at 37°C to an OD600 of ∼0.335 to 0.45 in the presence of 100 μg/ml ampicillin. Cells were permeabilized using chloroform-sodium dodecyl sulfate, and the β-galactosidase activity was determined using a published protocol (26). For GRDC experiments, cells were disrupted by sonication, and the β-galactosidase activity was normalized to the activity corresponding to a growth rate of 1 doubling per h, which was calculated from a linear regression curve fitted to a plot of β-galactosidase activity versus growth rate.
Measurement of transcription in vitro.
Multiple-round transcription reactions were performed at 30°C for 20 min using a previously described method (36). Each 25-μl reaction mixture contained 40 mM Tris-acetate (pH 7.9), 150 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml acetylated bovine serum albumin (Promega), 200 μM ATP, 200 μM GTP, 200 μM CTP, 10 μM UTP, and 5 μCi of [α-32P]UTP (800 Ci/mmol; Perkin-Elmer). Each reaction mixture also contained 0.2 nM supercoiled template DNA (pRLG770 derivative) and either a commercially available WT E. coli RNAP holoenzyme (Epicenter) or E. coli core RNAP purified as described elsewhere (E. Campbell, S. Darst, S. I. Husnain, B. Nickels, M. S. Thomas, K.-A. Twist, and L. Westblade, unpublished data) and subsequently reconstituted with σ70. The latter form of RNAP contained either WT α or a derivative lacking αCTD (RNAPΔαCTD), and RNAP activity was standardized as described previously (Campbell et al., unpublished). Reactions were initiated with RNAP and terminated with 25 μl of stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol). Samples, containing equal amounts of template DNA, were fractionated on a 5.5% acrylamide gel containing 7 M urea, and transcript abundance was quantified using a FujiFilm FLA3000 phosphorimager. The amount of transcript was determined by deducting the background activity in the lane from the value for the transcript abundance.
DNase I footprinting.
DNase I footprinting was performed according to a published method (38). Derivatives of pBluescript II KS containing EcoRI-XhoI DNA fragments extending from position −133 to +36 of the guaB promoter were purified following electrophoresis in 6% acrylamide gels (25), labeled at the downstream (XhoI) end with [γ-32P]ATP (6,000 Ci/mmol; Perkin-Elmer) using T4 polynucleotide kinase, and purified using QIAQuick columns (Qiagen). Labeled DNA fragment (4 nM) was incubated at room temperature for 30 min in a buffer containing 20 mM HEPES (pH 8.0), 5 mM MgCl2, 50 mM potassium glutamate, 1 mM dithiothreitol, 500 μg/ml acetylated bovine serum albumin, and 7% glycerol in the presence or absence of either 55 nM WT reconstituted RNAP or 119 nM reconstituted RNAPΔαCTD, in a final volume of 20 μl, after which DNA fragments were digested with DNase I. Reactions were stopped using a buffer containing 0.3 M sodium acetate (pH 5.2) and 10 mM EDTA. Samples were purified by phenol-chloroform extraction and ethanol precipitation, and DNA fragments were separated in a 6% acrylamide-7 M urea sequencing gel. A Maxam-Gilbert G+A sequencing ladder was run alongside. Footprints were visualized using a FujiFilm FLA3000 phosphorimager.
RESULTS
Activation of PguaB in vivo by sequences located upstream of the core promoter elements.
To analyze the contribution to transcription of sequences present upstream of PguaB, promoter derivatives with different upstream endpoints were used to generate transcriptional fusions to the lacZ gene and introduced into the E. coli chromosome as a single copy. The full-length WT PguaB derivative used in this study contained promoter sequences extending upstream to position −253 relative to the guaB transcription start site. This promoter and a promoter derivative containing sequences up to −133 both harbored the putative CRP site. In the other promoter derivatives, the upstream deletion removed the putative CRP site. All promoter derivatives had a downstream endpoint located at +36 (Fig. 2A). The promoter activity was determined by measuring the β-galactosidase activity in exponentially growing cells.
FIG. 2.
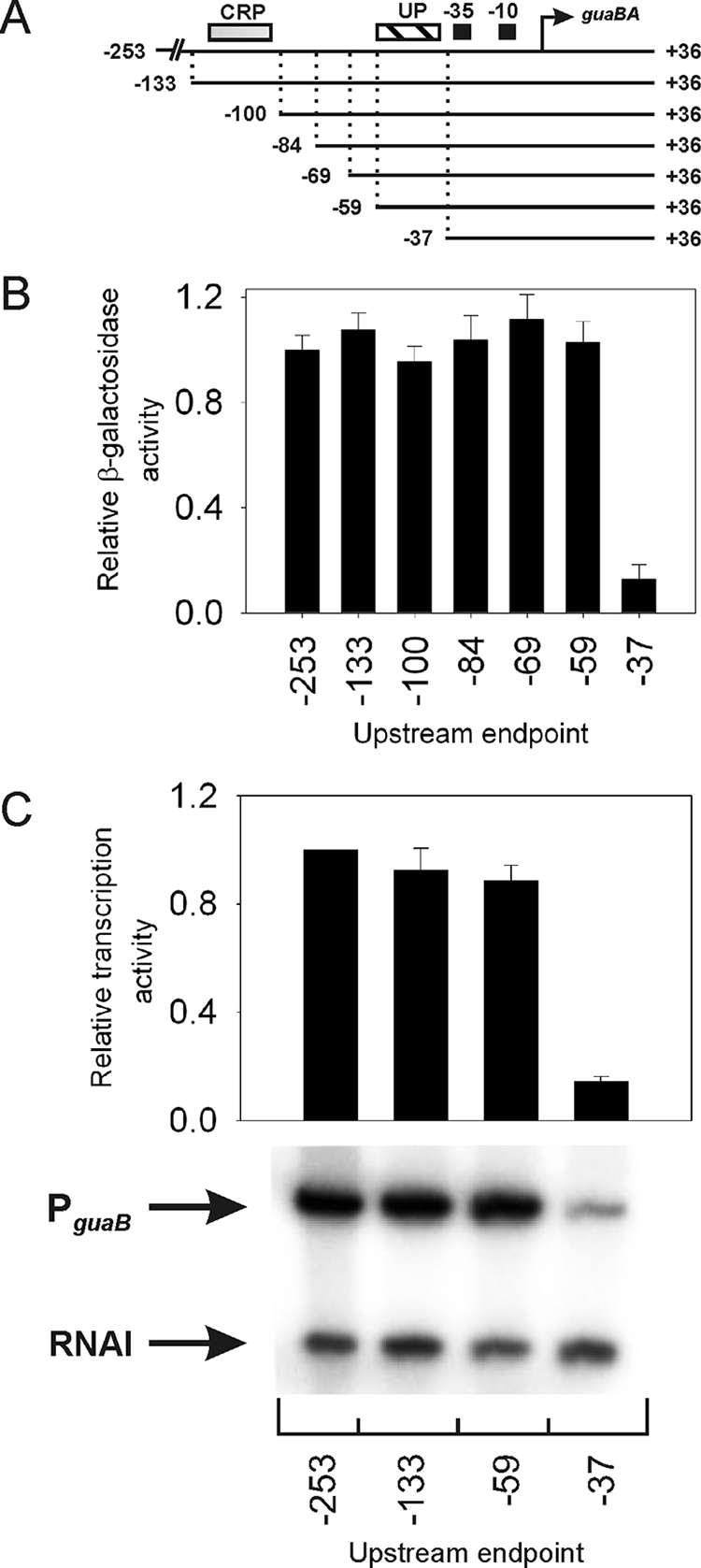
Contributions of sequences upstream of the guaB promoter to transcription. (A) The WT guaB promoter (−253 to +36) and derivatives each having a different upstream endpoint were constructed. Deletion endpoints are indicated relative to the guaB transcription start site. The predicted CRP binding site centered at position −117.5, the putative PguaB UP element (designated “UP”), the PguaB −35 and −10 hexamers, and the guaB transcription start site are shown (diagram not to scale). PguaB(−37 to +36) is referred to as core PguaB and contains the SUB sequence from positions −59 to −38 (Fig. 1). (B) The β-galactosidase activity (in Miller units) specified by each promoter-lacZ fusion in strain VH1000 growing logarithmically in M9-glucose minimal medium is expressed relative to the activity of PguaB(−253 to +36) (activity, 3,003 Miller units; relative activity, 1). (C) Multiple-round in vitro transcription assays were performed using a subset of guaB promoter derivatives with different upstream endpoints cloned into plasmid pRLG770, in the presence of 10 nM RNAP. The PguaB-directed transcript produced from this plasmid contains ∼180 nucleotides (RNA-I contains ∼110 nucleotides). Each promoter activity was determined by measuring the transcript abundance and is expressed relative to the activity of PguaB(−253 to +36), set at 1. In panels B and C, values are means and standard deviations from three independent experiments.
The results from the deletion analysis revealed that removal of the sequence from position −253 to −60 does not lead to any significant changes in the efficiency of transcription from PguaB (Fig. 2B). This suggests that sequences upstream of position −60 do not contribute significantly to transcription from PguaB in cells growing under the conditions employed. Transcription was also measured from a PguaB derivative with an upstream endpoint at position −37, which is referred to as the core guaB promoter (see the legend to Fig. 2). The core guaB promoter contains a “SUB” sequence in place of the putative PguaB UP element at positions −59 to −38 (Fig. 1). The SUB sequence has been shown to minimize the contribution to transcription from the recruitment of αCTD to DNA (10, 30). Comparison of the activity of PguaB(−59 to +36) with core PguaB indicated that the putative PguaB UP element stimulates the core promoter approximately eightfold in vivo (Fig. 2B).
Activation of PguaB in vitro by sequences located upstream of the core promoter elements.
To determine whether upstream guaB promoter sequences stimulate transcription in the absence of additional regulatory factors, multiple-round in vitro transcription reactions were performed using a subset of the promoter derivatives employed for the in vivo study (see above). Reactions were initiated with 10 nM RNAP. The results showed that, in the absence of other factors, sequences located upstream of position −59 do not contribute significantly to the transcriptional activity of PguaB. In contrast, deletion of the putative PguaB UP element gave rise to a ∼6-fold decrease in transcription activity compared to that of the full-length guaB promoter (Fig. 2C). These results are in good agreement with the findings from the in vivo analysis (see above) and demonstrate that the putative PguaB UP element can function in the absence of additional regulatory factors.
The putative PguaB UP element contains independently acting subsites.
We measured transcription both in vivo and in vitro from PguaB(−59 to +36), which contained the putative PguaB UP element sequence, and PguaB derivatives containing only the predicted PguaB proximal UP element subsite, only the predicted PguaB distal UP element subsite, or a consensus UP element (Fig. 1). The locations of the PguaB UP element subsites used here correspond to the proximal and distal UP element subsites defined in a previous study (11). The consensus UP element used in this analysis (the 4541 UP element [Fig. 1]) has been described previously (11). The results were compared to the transcription activity of core PguaB.
The results showed that the contributions of each tested sequence to transcription in vivo and in vitro are comparable (Fig. 3A and B). Whereas the putative PguaB UP element afforded an ∼8- to 10-fold increase in transcription from PguaB, the putative PguaB proximal UP element subsite stimulated transcription ∼5- to 6-fold and the putative PguaB distal UP element subsite afforded a ∼3-fold increase in transcription. These results demonstrate that the putative PguaB UP element contains subsites that can function independently in stimulating transcription from PguaB and that together they function as coactivators of transcription. The consensus UP element stimulated transcription from PguaB ∼11-fold, i.e., similar to the level afforded by the putative PguaB UP element.
FIG. 3.
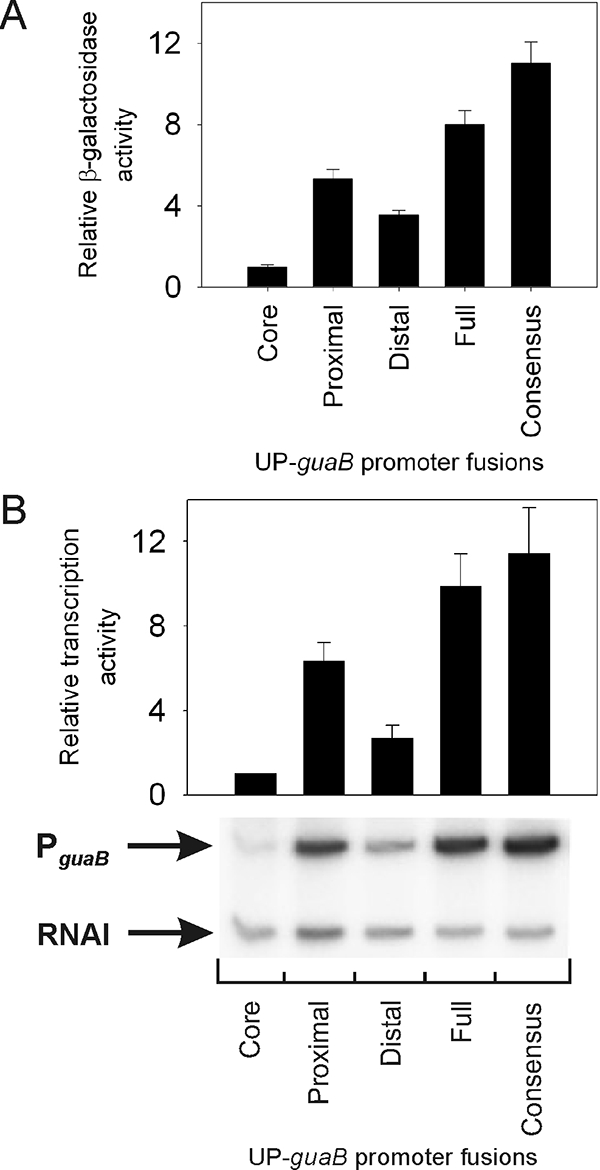
Contribution of subsites within the PguaB A+T-rich sequence to transcription. guaB promoter derivatives containing the consensus 4541 UP element located at positions −59 to −38, the complete 22-bp PguaB A+T-rich element located at −59 to −38 (full), the predicted PguaB proximal UP element subsite (positions −46 to −38), or the predicted PguaB distal UP element subsite (positions −59 to −47), or lacking an UP element (core PguaB), were constructed. Core PguaB contains a SUB sequence from position −59 to −38. PguaB containing the proximal UP element subsite also contains a SUB sequence from position −59 to −47, and PguaB containing the distal UP element subsite contains a SUB sequence from position −46 to −38. (A) The β-galactosidase activity (in Miller units) specified by each promoter-lacZ fusion in strain VH1000 growing logarithmically in M9-glucose minimal medium is expressed relative to the activity of core PguaB (activity, 385 Miller units, relative activity, 1). (B) Multiple-round in vitro transcription assays were performed using each guaB promoter construct in plasmid pRLG770 in the presence of 5 nM RNAP. The activity of each promoter is expressed relative to that of core PguaB, set at 1. In both panels, values are means and standard deviations from three independent experiments.
Activation of the lac promoter in vivo by the putative PguaB UP element.
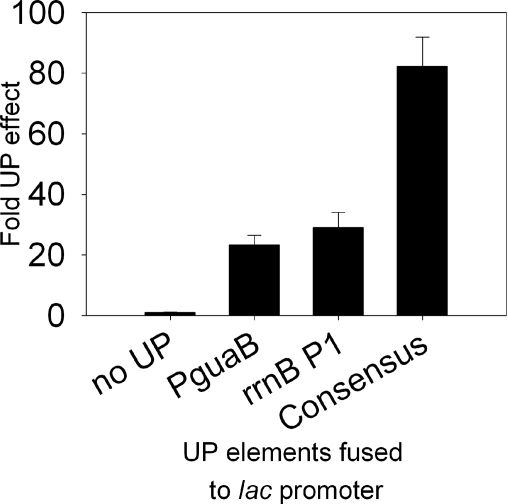
The rrnB P1 UP element is capable of stimulating transcription from other promoter sequences (31). To determine whether this also holds true for the putative PguaB UP element, transcription was measured in vivo from the core lac promoter (core lacP1) containing the putative PguaB UP element located at positions −59 to −38. To compare the strength of activation due to the putative PguaB UP element with that from other UP element sequences, transcription from core lacP1 fused to either the rrnB P1 UP element or a consensus UP element was also quantified. The consensus UP element used here (the 4192 UP element [Fig. 1]) has been described previously (10). The results were compared to the transcription activity of the core lac promoter (i.e., lacP1 without an UP element). The results show that the putative PguaB UP element stimulated transcription from core lacP1 ∼23-fold, i.e., at a much higher level than stimulation of the guaB promoter (Fig. 4). By comparison, the rrnB P1 UP element stimulated transcription from the core lac promoter ∼30-fold, whereas the consensus UP element afforded an ∼80-fold increase in promoter activity.
FIG. 4.
Activation of the lacP1 promoter by the PguaB A+T-rich element. lacP promoter derivatives contain WT lacP1 sequences from −37 to + 52 and one of the following: the consensus 4192 UP element (“Consensus”), the rrnB P1 UP element, the PguaB UP element, or no UP element, as indicated. UP element sequences are positioned from −59 to −38 relative to the lacP1 transcription start site. The core lac promoter contains lac promoter sequences as far upstream as position −40 and a SUB sequence from positions −58 to −41 (31). The β-galactosidase activity (in Miller units) specified by each promoter-lacZ fusion is the mean (and standard deviation) from three independent experiments and is expressed as a percentage of the activity of the core lac promoter (activity, 36 Miller units; relative activity, 1).
Role of αCTD in transcription from PguaB in vivo.
To investigate whether αCTD plays a role in transcription at PguaB, transcription from PguaB(−59 to +36) and core PguaB was measured in merodiploid cells containing the WT rpoA allele on the chromosome and a plasmid encoding either WT α or a mutant derivative harboring an alanine substitution at position 265 (α265A). The results show that the activity of PguaB(−59 to +36) did not differ significantly in the presence or absence of α265A (see Fig. S1A in the supplemental material). However, in the presence of α265A, transcription from core PguaB increased ∼2.9-fold over that for cells containing only WT α. This can be explained by the rationale that a decrease in the activity of the putative UP element in the presence of α265A leads to a compensatory increase in transcription from core PguaB. The effect of the mutant α on UP element function was therefore quantified as described previously (13, 24, 33). Thus, in the presence of α265A, the stimulation of PguaB due to the putative UP element was reduced by a factor of ∼2.5 (from 9.5-fold to 3.6-fold) (see Fig. S1B in the supplemental material).
Role of αCTD in transcription from PguaB in vitro.
To demonstrate that αCTD is required for stimulation of PguaB by the A+T-rich element, transcription was also measured in vitro from PguaB(−59 to +36) and from core PguaB in the presence of the RNAP holoenzyme containing WT α or α lacking αCTD (RNAPΔαCTD). The results show that transcription from PguaB(−59 to +36) decreased ∼10-fold in the presence of RNAPΔαCTD in comparision to the activity measured in the presence of WT RNAP. On the other hand, the activity of core PguaB did not differ significantly in the presence of either RNAP, and in each case the promoter activity was comparable to the activity of PguaB(−59 to +36) in the presence of RNAPΔαCTD (Fig. 5A). This indicates that the PguaB A+T-rich element requires αCTD to function in transcription activation.
FIG. 5.
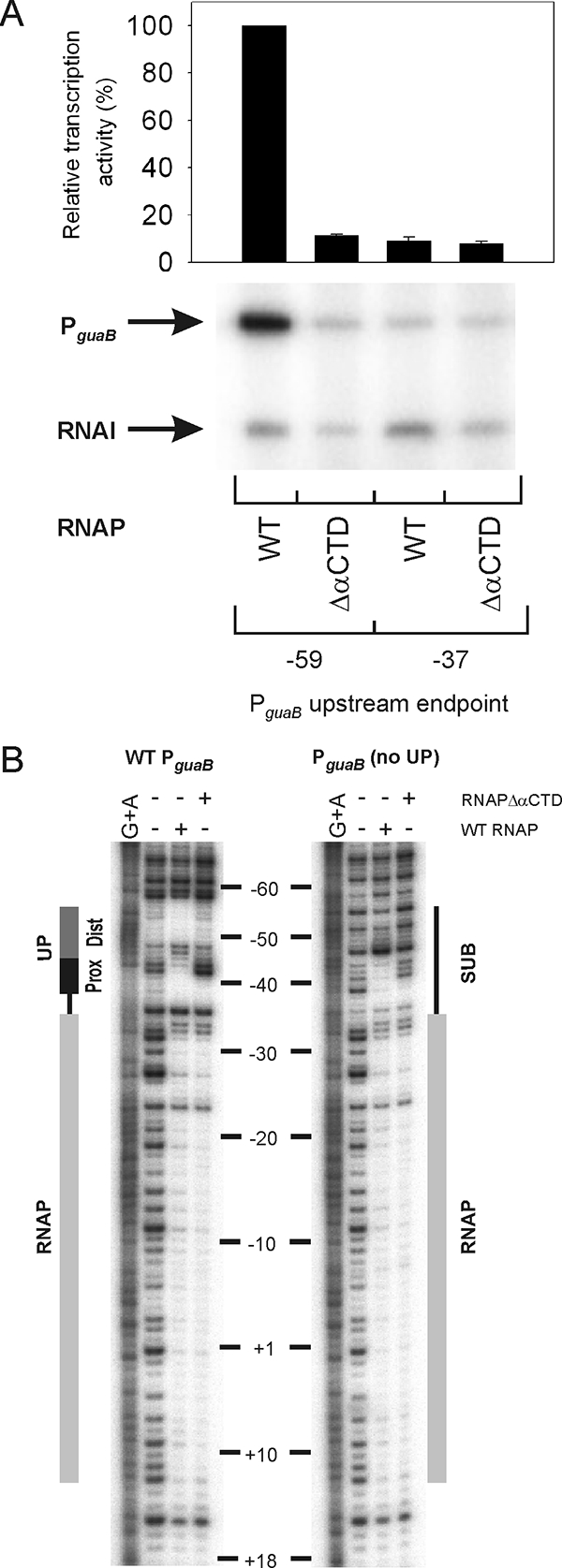
Role of αCTD in transcription from PguaB in vitro. (A) Multiple-round transcription reactions were performed with the core guaB promoter and PguaB(−59 to +36) in the presence of either 5 nM WT reconstituted RNAP or 11 nM reconstituted RNAPΔαCTD. The concentrations of the two RNAPs employed showed comparable activities, as measured by multiple-round transcription from the lacUV5 promoter (data not shown). The activity of each promoter is expressed as a percentage of that of PguaB(−59 to +36) in the presence of WT RNAP, set at 100%. Each promoter activity is the mean and standard deviation from three independent experiments. (B) DNase I footprinting was performed with the PguaB(−133 to +36) DNA fragment (WT) and with PguaB(−133 to +36) (no UP). Experiments were performed in the presence of either WT reconstituted RNAP or RNAPΔαCTD. The region protected by RNAP and the locations of the UP element and SUB sequence are indicated. Nucleotide positions are shown relative to the guaB transcription start site, and lanes containing the G+A ladder are indicated.
DNase I footprinting analysis was employed to probe for αCTD-DNA interactions at PguaB. The putative PguaB UP element contains a GC dinucleotide at positions −46 and −45 (Fig. 1) that is susceptible to cleavage by DNase I. In the presence of WT RNAP, cleavage at this position in PguaB(−133 to +36) is abolished, but this protection is lost in the presence of RNAPΔαCTD, indicating that αCTD interacts with this region of the guaB promoter (Fig. 5B). Replacement of the putative PguaB UP element with a SUB sequence [i.e., PguaB(−133 to +36) (no UP)] did not abolish protection of the region corresponding to the proximal UP element subsite by WT RNAP (i.e., the SUB sequence was still bound by αCTD).
The PguaB UP element is required for GRDC.
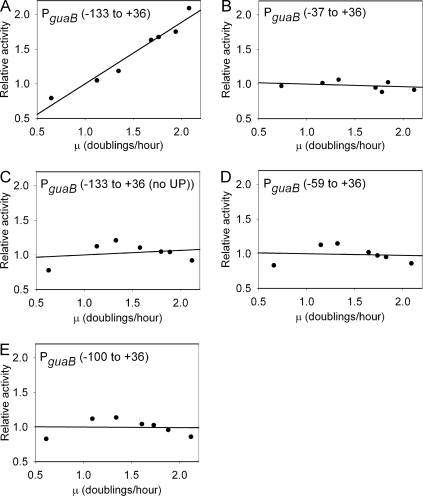
GRDC of PguaB was examined by growing cells harboring single-copy PguaB-lacZ fusions in media that supported different cellular growth rates and measuring the β-galactosidase activity. The activity of PguaB(−133 to +36) was found to increase in direct proportion to an increasing growth rate, i.e., a twofold increase in the growth rate was accompanied by a twofold increase in promoter activity (Fig. 6A). As expected, deletion of upstream sequences to position −37 (core PguaB) resulted in a decrease in guaB promoter activity at all growth rates (Fig. 6B). However, the decrease was progressively more marked at higher growth rates, and this resulted in a complete loss of GRDC. To investigate whether the UP element contributes to GRDC, the UP element in PguaB(−133 to +36) was replaced by a SUB sequence [i.e., PguaB(−133 to +36) (no UP)]. This manipulation also resulted in a loss of GRDC (as well as causing a sharp decrease in guaB promoter activity), suggesting that the UP element is required for GRDC (Fig. 6C). To determine whether sequences located upstream of the UP element are also required for GRDC, the activity of PguaB(−59 to +36) was measured at different cellular growth rates. The results demonstrated that sequences located upstream of the UP element are also required for GRDC (Fig. 6D). Moreover, at the lower growth rates (i.e., μ ≤ 1.0), this promoter derivative is at least 2.3-fold more active than PguaB(−133 to +36), suggesting that sequences located upstream of the UP element are required for the repression of PguaB in slowly growing cells. To localize these sequences more precisely, the activity of the PguaB(−100 to +36) promoter was also measured at different growth rates. The results show that this promoter derivative behaves similarly to PguaB(−59 to +36) (Fig. 6E), implying that sequences located between −133 and −100 participate in the GRDC of PguaB.
FIG. 6.
Requirement of the PguaB UP element for GRDC. The effects of growth rate on the activities of the indicated PguaB derivatives were determined by growing strain VH1000 harboring fusions of these promoters to lacZ in different media and determining the β-galactosidase activities. The activity of each promoter was normalized to a value of 1.0 at a growth rate of 1 doubling per h. The β-galactosidase activity specified by each promoter (in Miller units) at a growth rate of 1 doubling per h was as follows: PguaB(−133 to +36), 1,588; PguaB(−37 to +36), 418; PguaB(−133 to +36) (no UP), 419; PguaB(−59 to +36), 3,573; PguaB(−100 to +36), 3,009. Each data point represents the mean promoter activity and the mean growth rate. Means were calculated using data obtained from three independent experiments.
DISCUSSION
Previously, a 22-bp UP element-like sequence extending from position −59 to −38 relative to the guaB transcription start site was identified by comparison to the consensus sequence for UP elements (10, 19). We demonstrate here that this sequence does function as an UP element. First, this sequence stimulates transcription from PguaB ∼8- to 10-fold both in vivo and in vitro. Moreover, it can activate other promoters such as lacP1, which it can stimulate at least 20-fold. The variation in transcription enhancement at different promoters is a feature of other UP elements (for example, a consensus UP element stimulates transcription from PguaB ∼11-fold, from rrnB P1 ∼340-fold, and from lacP1 ∼80-fold in vivo) (11, 31). The lower contribution of the PguaB UP element to transcription from PguaB relative to that for lacP1 is likely to be due to differences in promoter architecture that result in a higher activity of the core guaB promoter in comparison to lacP1 (i.e., core PguaB is ∼10-fold more active than core lacP1). In particular, PguaB has a consensus −10 sequence that is separated from the −35 region by 17 bp, whereas at lacP1 the −10 sequence (which has a poorer match to the consensus sequence) is separated from the −35 region by an 18-bp spacer.
Second, the PguaB UP element appears to contain independently functioning subsites that correspond in location to the previously identified proximal and distal subsites in the rrnB P1 and consensus UP elements (11). Accordingly, we assume that each PguaB UP element subsite functions by contacting a single αCTD. As with the rrnB P1 UP element, the PguaB-proximal UP element subsite stimulates transcription to a greater degree than the distal subsite (11). It has been shown that αCTD bound to consensus or near-consensus proximal UP element subsites functionally interacts with region 4 of σ70 and that such interactions may account for the observation that proximal UP element subsites contribute more to transcription activation than distal UP element subsites (7, 35). Since the PguaB-proximal UP element subsite matches the consensus sequence, it is possible that αCTD-σ70 interactions may be important for PguaB UP element function.
Third, the activity of the PguaB UP element requires αCTD. Thus, deletion of αCTD abolishes UP element-dependent transcription from PguaB in vitro and is accompanied by a loss of protection of the UP element by RNAP. Although protection of non-UP element sequences located upstream of the core guaB promoter by WT RNAP was observed, these interactions do not contribute a strong stimulatory effect on transcription. This observation mirrors findings from a previous study that demonstrated protection of the region between −35 and −70 at the lacUV5 promoter by αCTD, although these sequences do not contain an UP element (34). Incorporation into RNAP of α subunits containing an amino acid substitution in the DNA binding region (α265A) was also shown to decrease UP element-dependent transcription from PguaB in vivo. However, in this situation, UP element-independent transcription from the core guaB promoter fully compensated for the reduction in UP element function. This suggests that core PguaB is subject to a feedback mechanism that can counter regulatory inputs acting outside of the core promoter region. Although a compensatory mechanism (involving the initiating nucleotide for transcription) operates under similar conditions at the rrnB P1 promoter, it does not restore promoter activity completely in the presence of α265A (13, 39). This is probably due to the much larger negative effect that impairment of the αCTD-UP element interaction exerts on rrnB P1 activity.
The PguaB UP element is absolutely required for GRDC of PguaB. This contrasts with the situation at two other growth rate-regulated promoters (rrnB P1 and the leuV promoter), where GRDC does not require the UP element (2, 3, 29). Our results also show that, in addition to the PguaB UP element, sequences located between positions −133 and −100 are required for GRDC, and their role is to facilitate the inhibition of PguaB at lower growth rates. This region contains a sequence exhibiting a good match to the CRP site consensus sequence, suggesting that CRP may participate in GRDC of PguaB. These observations do not rule out the possibility that an additional factor(s) that binds to the region between −100 and the UP element is involved. Therefore, GRDC of PguaB may be explained by one of the following two general models. (i) Binding of one or more regulatory proteins to sequences located upstream of, or overlapping, position −100 (for example, CRP) inhibits UP element function. This could occur directly or indirectly, by facilitating occupancy or remodeling of the region upstream of (and overlapping) the UP element by other DNA-binding proteins. (ii) The PguaB UP element, with or without the participation of other DNA-binding proteins, may alter the orientation of the upstream DNA so as to allow a regulator bound near position −100 to inhibit RNAP, perhaps by bringing it into close proximity to the core promoter region. In both models, DNA occupancy by at least one of the DNA-binding proteins, although not necessarily the factor binding upstream of position −100, would increase at lower growth rates. Work is currently under way to explore these possibilities.
Supplementary Material
Acknowledgments
This work was supported by a family Ph.D. sponsorship awarded to S.I.H., kindly provided by S. M. Husnain, and a project research grant awarded to M.S.T. by the Wellcome Trust (grant 073917).
We are indebted to W. Ross, T. Gaal, H. Murray, and R. L. Gourse (University of Wisconsin—Madison) for technical advice, strains, plasmids and helpful discussions. We are grateful to T. Belyaeva (University of Leeds) and D. Browning (University of Birmingham) for advice on DNase I footprinting. We thank N. Savery (University of Bristol) for technical advice. We also thank L. Westblade for the generous gift of RNAPs containing WT and mutant α subunits and for providing σ70.
Footnotes
Published ahead of print on 18 January 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alting-Mees, M. A., and J. M. Short. 1989. pBluescript II: gene mapping vectors. Nucleic Acids Res. 179494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartlett, M. S., and R. L. Gourse. 1994. Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J. Bacteriol. 1765560-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer, B. F., R. M. Elford, and W. M. Holmes. 1993. Mutagenesis and functional analysis of the Escherichia coli tRNA (Leu 1) promoter. Mol. Microbiol. 7265-273. [DOI] [PubMed] [Google Scholar]
- 4.Benoff, B., H. Yang, C. L. Lawson, G. Parkinson, J. Liu, E. Blatter, Y. W. Ebright, H. M. Berman, and R. H. Ebright. 2002. Structural basis of transcription activation: the CAP-αCTD-DNA complex. Science 2971562-1566. [DOI] [PubMed] [Google Scholar]
- 5.Blatter, E. E., W. Ross, H. Tang, R. L. Gourse, and R. H. Ebright. 1994. Domain organization of RNA polymerase α subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell 78889-896. [DOI] [PubMed] [Google Scholar]
- 6.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293199-213. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H., H. Tang, and R. H. Ebright. 2003. Functional interaction between RNA polymerase alpha subunit C-terminal domain and σ70 in UP-element- and activator-dependent transcription. Mol. Cell 111621-1633. [DOI] [PubMed] [Google Scholar]
- 8.Davies, I. J., and W. T. Drabble. 1996. Stringent and growth-rate-dependent control of the gua operon of Escherichia coli K-12. Microbiology 1422429-2437. [DOI] [PubMed] [Google Scholar]
- 9.Dennis, P. P., M. Ehrenber, and H. Bremer. 2004. Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol. Mol. Biol. Rev. 68639-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estrem, S. T., T. Gaal, W. Ross, and R. L. Gourse. 1998. Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl. Acad. Sci. USA 959761-9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estrem, S. T., W. Ross, T. Gaal, Z. W. Chen, W. Niu, R. H. Ebright, and R. L. Gourse. 1999. Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase α subunit. Genes Dev. 132134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaal, T., M. S. Bartlett, W. Ross, C. L. Turnbough, Jr., and R. L. Gourse. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in Bacteria. Science 2782092-2097. [DOI] [PubMed] [Google Scholar]
- 13.Gaal, T., W. Ross, E. E. Blatter, H. Tang, X. Jia, V. V. Krishnan, N. Assa-Munt, R. H. Ebright, and R. L. Gourse. 1996. DNA-binding determinants of the α subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev. 1016-26. [DOI] [PubMed] [Google Scholar]
- 14.Geszvain, K. M., and R. Landick. 2004. The structure of bacterial RNA polymerase, p. 283-296. In N. P. Higgins (ed.), The bacterial chromosome. American Society for Microbiology, Washington, DC.
- 15.Gourse, R. L., T. Gaal, M. S. Bartlett, J. A. Appleman, and W. Ross. 1996. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu. Rev. Microbiol. 50645-677. [DOI] [PubMed] [Google Scholar]
- 16.Gourse, R. L., W. Ross, and T. Gaal. 2000. UPs and downs in bacterial transcription initiation: the role of the α subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 37687-695. [DOI] [PubMed] [Google Scholar]
- 17.Guarente, L., G. Lauer, T. M. Robert, and M. Ptashne. 1980. Improved methods for maximising expression of a cloned gene: a bacterium that synthesizes rabbit β-globin. Cell 20543-553. [DOI] [PubMed] [Google Scholar]
- 18.Hochschild, A., and S. L. Dove. 1998. Protein-protein contacts that activate and repress prokaryotic transcription. Cell 92597-600. [DOI] [PubMed] [Google Scholar]
- 19.Hutchings, M. I., and W. T. Drabble. 2000. Regulation of the divergent guaBA and xseA promoters of Escherichia coli by the cyclic AMP receptor protein. FEMS Microbiol. Lett. 187115-122. [DOI] [PubMed] [Google Scholar]
- 20.Jeon, Y. H., T. Yamazaki, T. Otomo, A. Ishihama, and Y. Kyogoku. 1997. Flexible linker in the RNA polymerase α subunit facilitates the independent motion of the C-terminal activator contact domain. J. Mol. Biol. 267953-962. [DOI] [PubMed] [Google Scholar]
- 21.Kimura, M., N. Fujita, and A. Ishihama. 1994. Functional map of the α subunit of Escherichia coli RNA polymerase. Deletion analysis of the amino-terminal assembly domain. J. Mol. Biol. 242107-115. [DOI] [PubMed] [Google Scholar]
- 22.Mehra, R. K., and W. T. Drabble. 1981. Dual control of the gua operon of Escherichia coli K12 by adenine and guanine nucleotides. J. Gen. Microbiol. 12327-37. [DOI] [PubMed] [Google Scholar]
- 23.Meng, L. M., M. Kilstrup, and P. Nygaard. 1990. Autoregulation of PurR repressor synthesis and involvement of purR in the regulation of purB, purC, purL, purMN and guaBA expression in Escherichia coli. Eur. J. Biochem. 187373-379. [DOI] [PubMed] [Google Scholar]
- 24.Meng, W., T. Belyaeva, N. J. Savery, S. J. Busby, W. E. Ross, T. Gaal, R. L. Gourse, and M. S. Thomas. 2001. UP element-dependent transcription at the Escherichia coli rrnB P1 promoter: positional requirements and role of the RNA polymerase alpha subunit linker. Nucleic Acids Res. 294166-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng, W., N. J. Savery, S. J. W. Busby, and M. S. Thomas. 2000. The Escherichia coli RNA polymerase α subunit linker: length requirements for transcription activation at CRP-dependent promoters. EMBO J. 191555-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Nomura, M., R. Gourse, and G. Baughman. 1984. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 5375-117. [DOI] [PubMed] [Google Scholar]
- 28.Paul, B. J., W. Ross, T. Gaal, and R. L. Gourse. 2004. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 38749-770. [DOI] [PubMed] [Google Scholar]
- 29.Pokholok, D. K., M. Redlak, C. L. Turnbough, Jr., S. Dylla, and W. M. Holmes. 1999. Multiple mechanisms are used for growth rate and stringent control of leuV transcriptional initiation in Escherichia coli. J. Bacteriol. 1815771-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao, L., W. Ross, J. A. Appleman, T. Gaal, S. Leirmo, P. J. Schlax, M. T. Record, Jr., and R. L. Gourse. 1994. Factor independent activation of rrnB P1. An “extended” promoter with an upstream element that dramatically increases promoter strength. J. Mol. Biol. 2351421-1435. [DOI] [PubMed] [Google Scholar]
- 31.Ross, W., S. E. Aiyar, J. Salomon, and R. L. Gourse. 1998. Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J. Bacteriol. 1805375-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross, W., A. Ernst, and R. L. Gourse. 2001. Fine structure of E. coli RNA polymerase-promoter interactions: α subunit binding to the UP element minor groove. Genes Dev. 15491-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross, W., K. K. Gosink, J. Salomon, K. Igarashi, C. Zou, A. Ishihama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science 2621407-1413. [DOI] [PubMed] [Google Scholar]
- 34.Ross, W., and R. L. Gourse. 2005. Sequence-independent upstream DNA-αCTD interactions strongly stimulate Escherichia coli RNA polymerase-lacUV5 promoter association. Proc. Natl. Acad. Sci. USA 102291-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross, W., D. A. Schneider, B. J. Paul, A. Mertens, and R. L. Gourse. 2003. An intersubunit contact stimulating transcription initiation by E. coli RNA polymerase: interaction of the α C-terminal domain and σ region 4. Genes Dev. 171293-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross, W., J. F. Thompson, J. T. Newlands, and R. L. Gourse. 1990. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 93733-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Savery, N. J., T. Belyaeva, and S. J. W. Busby. 1996. Protein-DNA interactions, p. 1-43. In K. Docherty (ed.), Gene transcription: DNA binding proteins, essential techniques. Wiley/BIOS Press, Chichester, United Kingdom.
- 39.Schneider, D. A., and R. L. Gourse. 2003. Changes in Escherichia coli rRNA promoter activity correlate with changes in initiating nucleoside triphosphate and guanosine 5′-diphosphate 3′-diphosphate concentrations after induction of feedback control of ribosome synthesis. J. Bacteriol. 1856185-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 5385-96. [DOI] [PubMed] [Google Scholar]
- 41.Tesfa-Selase, F., and W. T. Drabble. 1996. Specific binding of DnaA protein to a DnaA box in the guaB gene of Escherichia coli K12. Eur. J. Biochem. 241411-416. [DOI] [PubMed] [Google Scholar]
- 42.Tiedeman, A. A., and J. M. Smith. 1984. Isolation and characterization of regulatory mutations affecting the expression of the guaBA operon of Escherichia coli K-12. Mol. Gen. Genet. 19577-82. [DOI] [PubMed] [Google Scholar]
- 43.Yasuno, K., T. Yamazaki, Y. Tanaka, T. S. Kodama, A. Matsugami, M. Katahira, A. Ishihama, and Y. Kyogoku. 2001. Interaction of the C-terminal domain of the E. coli RNA polymerase α subunit with the UP element: recognizing the backbone structure in the minor groove surface. J. Mol. Biol. 306213-225. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, G., and S. A. Darst. 1998. Structure of the Escherichia coli RNA polymerase α subunit amino-terminal domain. Science 281262-266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.