Abstract
Stimulus perception by the KdpD/KdpE two-component system of Escherichia coli is still controversial with respect to the nature of the stimulus that is perceived by the sensor kinase KdpD. Limiting potassium concentrations in the medium or high osmolality leads to KdpD/KdpE signal transduction, resulting in kdpFABC expression. It has been hypothesized that changes in turgor are sensed by KdpD through alterations in the physical state of the cytoplasmic membrane. However, in this study the quantitative determination of expression levels of the kdpFABC operon revealed that the system responds very effectively to K+-limiting conditions in the medium but barely and to various degrees to salt and sugar stress. Since the current view of stimulus perception calls for mainly intracellular parameters, which might be sensed by KdpD, we set out to test the cytoplasmic concentrations of ATP, K+, Na+, glutamate, proline, glycine, trehalose, putrescine, and spermidine under K+-limiting conditions. As a first result, the determination of the cytoplasmic volume, which is a prerequisite for such measurements, revealed that a transient shrinkage of the cytoplasmic volume, which is indicative of a reduction in turgor, occurred only under osmotic upshift but not under K+-limiting conditions. Furthermore, the intracellular ATP concentration significantly increased under osmotic upshift, whereas only a slight increase occurred after a potassium downshift. Finally, the cytoplasmic K+ concentration rose severalfold only after an osmotic upshock. For the first time, these data indicate that stimulus perception by KdpD correlates neither with changes in the cytoplasmic volume nor with changes in the intracellular ATP or K+ concentration or those of the other solutes tested. In conclusion, we propose that a reduction in turgor cannot be the stimulus for KdpD.
A typical bacterial two-component system (35, 23), consisting of the membrane-bound sensor kinase KdpD and the cytoplasmic response regulator KdpE, controls the expression of the kdpFABC operon (29, 40), which codes for the high-affinity potassium uptake system KdpFABC, a four-subunit K+-transporting P-type ATPase (2). This complex is synthesized only when the constitutive K+ transporters, Kup and Trk, do not provide sufficient amounts of potassium for the maintenance of turgor (10). The sensor kinase KdpD is a homodimeric integral membrane protein of 99 kDa (43, 15). It can be functionally divided into an input domain (residues 1 to 660) and a transmitter domain (residues 661 to 894). The input domain comprises an unusually large cytoplasmic N-terminal domain (residues 1 to 395), four narrow-spaced transmembrane helices followed by an arginine-rich region, and an additional 140 residues of the cytoplasmic C-terminal part. The C-terminal cytoplasmic transmitter region consists of a catalytic domain carrying all features characteristic of bacterial histidine kinases, including a conserved histidine residue (His 673), a dimerization domain, and several conserved motifs involved in binding and hydrolysis of ATP (27). The input and transmitter domains are linked by a coiled-coil region. The 395-amino- acid-long cytoplasmic N-terminal domain is unique in size among all bacterial sensor kinases. It contains a slightly modified Walker A and B motif (18) that was shown to bind 8-azido-ATP (13) and a Usp domain of the ATP-binding type (33). The function of the N-terminal domain has not been clarified in every detail, but it has been shown to play a role in stabilizing the ternary complex of KdpD, KdpE, and the kdpFABC promoter region of the chromosome (14) and to regulate KdpD phosphatase activity through ATP binding (19).
At potassium-limiting conditions or high osmolality, KdpD is autophosphorylated at the conserved histidine residue of the C-terminal catalytic domain, whereby one monomer translocates a γ-phosphate of ATP to the other (39). Subsequently, the phosphoryl group is transferred in a kinase reaction to the cytoplasmic response regulator KdpE. KdpE∼P dimerizes and serves as a positive transcriptional regulator for the genes of the kdpFABC operon (38). At normal growth conditions, the phospho-KdpE-specific phosphatase activity of KdpD prevails over its kinase activity (3), and as a consequence, KdpE∼P is dephosphorylated and dissociates from the promoter binding region, thereby terminating the expression of the kdpFABC operon.
It has been hypothesized that KdpD senses changes in turgor or some effect thereof (8, 21). This was mainly due to the discovery of kdpFABC being an osmotically upregulated operon under the control of KdpD/KdpE. In addition, an integral membrane protein, such as KdpD, could mechanistically be affected by changes in the physicochemical state of the membrane such as, e.g., strain or shape, which in turn are closely related to turgor. At first glance, the induction of kdpFABC expression under potassium-limiting conditions fitted well into this picture, since the level of intracellular potassium plays an important role in the maintenance of turgor. However, whether the turgor level changes under potassium-limiting conditions has so far not been demonstrated, since turgor can only be estimated but cannot be measured directly in gram-negative bacteria. The idea that KdpD senses changes in turgor was challenged by the fact that osmotic upshifts using iso-osmolal concentrations of nonionic solutes, such as sugars, did not exhibit the same kdpFABC expression level that was seen with salts (1). Therefore, it was suggested that KdpD somehow senses the cells’ need for potassium to maintain turgor. Sugiura et al. (37) suggested mechanistically different sensing mechanisms for potassium-limiting conditions and osmotic stress. However, Zimmann et al. (44) showed that the extension of the fourth transmembrane helix encompassing the arginine cluster is mainly involved in sensing both stimuli, which may not be separable. Furthermore, Heermann et al. (14) proposed an additional, more-complex regulatory network for kdpFABC expression. Although the cytoplasmic potassium concentration was already shown not to be the stimulus for KdpD (22), recent work postulated that intracellular potassium is sensed by the C-terminal domain of KdpD (31). Since kdpFABC expression does not correlate with the outside potassium concentration (10, 22) and since a KdpD derivative lacking the four transmembrane helices does not fully abolish kdpFABC expression (16), mainly intracellular parameters were suggested to be sensed by KdpD. ATP could be a candidate, since a regulatory role of binding of ATP to the N-terminal domain in the phosphatase activity of KdpD was shown (19). Furthermore, a significant increase in the ATP level occurs in osmotically stressed cells (26). However, other solutes, such as, e.g., glutamate or compatible solutes (proline and trehalose), which are involved in the osmoresponse of Escherichia coli (reviewed in references 6 and 41), also might play a role in signal transduction elicited by potassium-limiting conditions. To determine the concentration of cytoplasmic solutes, the measurement of the cytoplasmic volume is a prerequisite. Therefore, we set out to determine the cytoplasmic volume of growing E. coli cells that were shifted either to nominally potassium-free medium or, as a control, to higher osmolality. The results presented here provide clear evidence that neither the cytoplasmic volume (and hence changes in turgor) nor the concentration of several cytoplasmic solutes, such as K+, ATP, putrescine, spermidine, trehalose, glutamate, and proline, can be the stimulus for KdpD.
MATERIALS AND METHODS
Strains, media, and growth conditions.
E. coli K12 and E. coli M2701 (metB mel-7 galK rpsL) (32) were used throughout this study. Minimal phosphate-buffered K115 and K0 media have been described (9). Briefly, K115 contains 115 mM K+, and K0 is identical to K115 except that K+ is replaced by Na+. Every mixture of these two media is isosmotic and allows adjustment of any potassium concentration in the range of 0.02 to 115 mM. Osmotic solutes were added directly to the medium at the desired concentration. In the case of E. coli M2701, methionine (20 mg/liter) and streptomycin (200 mg/liter) were added to the growth medium. Cells were incubated aerobically at 37°C.
Medium shift.
Cells were grown to exponential phase, and 20 to 40 ml of the culture was vacuum filtered through a 0.45-μm nitrocellulose membrane. Cells were washed once and resuspended in the same volume of prewarmed (37°C) growth medium of the new composition, and growth was continued with shaking. Depending on the optical density of the culture, this procedure took 30 to 60 s. The moment when filter-collected cells were placed in the new culture flask was considered time zero. Depending on the parameter to be measured, the first sample was taken between 10 and 60 s after time zero. As a control, cells were also shifted into prewarmed medium (37°C) of the same composition in order to identify disturbances due to the filtration procedure. In a typical experiment, cells were grown to mid-exponential growth phase without osmotic or potassium-limiting stress and were then transferred into a medium of the same osmolality but lacking potassium. In the case of osmotic upshift, the osmostressor was added directly to the growth medium. If not stated otherwise, the term osmotic upshift is used as a synonym only for salt stress elicited by NaCl.
Determination of kdpFABC expression by Q-RT-PCR.
RNA was extracted from cells of exponential growth phase using the RNeasy minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. After DNase treatment, the cDNA was synthesized with the RevertAid First Strand cDNA synthesis kit. The sequences of the primer pair kdpAfor2/kdpArev2, used for the quantitative reverse transcription-PCR (Q-RT-PCR), were GCC GCC AGC GGG ATT GCG G and CTT CAA CGG TAT TCA CAG CCT G, respectively. As an internal standard, the primer pair gapAfor1/gapArev1 (CTC CAC TCA CGG CCG TTT CG/CTT CGC ACC AGC GGT GAT GTG) for the E. coli housekeeping gene gap was used. Binding of the primers for the Q-RT-PCR for all strains and amplification of the appropriate single PCR product corresponding to the kdpA gene were controlled by PCR. The run protocol for all primer pairs began with 2 min at 95°C, followed by a 40-fold cycle of 15 s at 95°C, 30 s at 62°C, and 30 s at 72°C, and ended with 5 min at 72°C in an iCycler thermal cycler (Bio-Rad). To correct for sampling errors, the levels of expression of kdpA as determined from the cycle threshold (CT) values were normalized to the level of the housekeeping gene. For statistical analysis, the data from different measurements were averaged. The induction of kdpFABC expression was then usually given as the ratio of the values of the normalized expression rate for the induced state (<0.1 mM K+ or high osmolality) divided by the values of the normalized expression for the noninduced state (20 mM K+ or low osmolality). This indicates the extent of kdpFABC expression after a shift of cells from noninducing to inducing conditions.
Measurements of solute distribution in growing cells.
The size of the cytoplasmic water space as an indicator of the cytoplasmic volume was determined for growing cells by a modification of the method of Stock et al. (36). 3H2O (1 mCi/ml) and 0.1 mCi/ml [14C]sucrose (ubiquitously labeled) were added simultaneously to 1.3 ml of growing cells of E. coli that were placed in a 1.5-ml reaction tube. The [14C]sucrose was purified prior to use by thin-layer chromatography in order to remove contaminating traces of [14C]glucose. Since E. coli K12 exhibited sucrose uptake activity (data not shown), the E. coli strain M2701, which does not show such properties but otherwise resembles the wild type (32), was used. After 60 s, cells were centrifuged for 120 s through a 200-μl silicone oil layer of appropriate density (usually 1.04 g/cm3 and up to 1.07 g/cm3 in the case of medium of high osmolality), which was sufficient to collect all of the cells. The bottom of the reaction tube was carefully separated with a strong razor blade, cutting through the oil layer in order to avoid contamination of the cell pellet by traces of the supernatant. Cell lysis was achieved by resuspending the cell pellet in 1 ml 0.4 M NaOH at 62°C and shaking for 1 h. The amounts of radioactivity of the two nuclides in the cell pellet were determined simultaneously by liquid scintillation counting using a dual-label protocol that converts the original counts per minute of each nuclide to disintegrations per minute (dpm) to account for the overlap of the energy distribution spectra of the two nuclides. For quantification, the radioactivity of 50 μl of the supernatant was also counted. Since [3H]water enters every part of the cell, the total cellular water space including the extracellular water layer can be calculated as follows: (3H dpm of the pellet) × 50 μl/(3H dpm of 50 μl of the supernatant).
Since [14C]sucrose enters every part of the cell except for the cytoplasm, the extracellular water layer plus the periplasmic space can be calculated as follows: (14C dpm of the pellet) × 50 μl/(14C dpm of 50 μl of the supernatant).
The difference between the space occupied by tritiated water and the space occupied by sucrose results in the water space of the cytoplasm, which includes all water layers bound to biological surfaces, the so-called bound water (11, 5), and the free diffusing water, the so-called bulk water. In the following, the water space of the cytoplasm will be referred to as the cytoplasmic volume, which does not encompass the volume of all macromolecules in the cytoplasm. The concentrations of cytoplasmic solutes will be calculated on the basis of the cytoplasmic volume.
ATP measurements.
The ATP concentration of the cytoplasm of growing cells was determined using the Enliten ATP assay (Promega). Briefly, cells of 0.2 ml cell culture were collected by centrifugation for 30 s, resuspended in 50 μl of ice-cold 2% trichloroacetic acid, and immediately frozen at −18°C. After thawing, 1 ml of 50 mM Tris buffer (pH 7.7) was added and cell debris was pelleted by centrifugation for 60 s.
Determination of dry weight, total protein, cell count, and viability of the cells.
Dry weight was determined by filtration of 40 ml cell culture through a 0.45-μm nitrocellulose membrane, drying of the pellet at 105°C for 1 h, and determination of its weight using a calibrated Sartorius 2006MP microbalance. Total protein was determined according to the protocol of Peterson (28), using bovine serum albumin (Pierce) as a standard. The number of cells per ml cell culture was determined manually in a Thoma chamber according to the manufacturer's instructions using light microscopy at magnification ×1,000. The viability of cells was determined by the spread plate method. Colonies were counted manually.
Determination of cellular and cytoplasmic potassium and sodium.
Cellular potassium and sodium contents were determined by flame photometry of extracts of cells that were collected by centrifugation through silicon oil. Shortly, cells of 1 ml cell culture were collected by centrifugation through a 200-μl silicon oil layer and extracted by using 5% trichloroacetic acid, freeze-thawing at −18°C, and heating to 100°C for 10 min. Cell debris was pelleted, and potassium and/or sodium in the supernatant were measured by using flame photometry in an Eppendorf ELEX 6361 flame photometer. Extracellular (i.e., medium) concentrations were directly measured after appropriate dilution.
Metabolite analysis.
Before or after a medium shift, 40 to 50 ml of cells were collected, resuspended in 5 ml of the same medium lacking glucose, and immediately frozen with shaking in liquid N2. After lyophilization, polyamines, sugars, and amino acids were determined by derivatization with 9-fluorenylmethyl chloroformate prior to high-performance liquid chromatography and subsequent UV detection as described previously (17).
Microscopy and image analysis.
Cells were visualized by phase-contrast microscopy (1,000-fold) and photographed by using a Spot digital camera, model 2.3.0 (Diagnostic Instruments, Inc.), using Spot Advanced Software, version 3.1 (Diagnostic Instruments, Inc.). Photos were taken without any fixation of the cells and as shortly as possible (approximately 10 s) after an osmotic upshift by the addition of 0.8 M NaCl or KCl to the side of the coverslip. To obtain reliable results, we have constantly observed the same cell throughout the entire procedure. The two-dimensional size of hundreds of cells was quantified by image analysis using the ImageJ Java software (http://rsb.info.nih.gov/ij/). This software tool distinguishes cells from other particles in the growth medium and allows the determination of the area occupied by every single cell.
RESULTS
Induction profile of kdpFABC expression following potassium downshift or osmotic upshift.
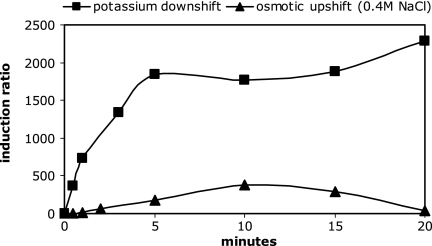
The ratio of the normalized kdpFABC expression rates of E. coli cells before and after a shift in the composition of the medium revealed that KdpD/KdpE respond very effectively to a potassium downshift. Thirty seconds after the shift, the induction of kdpFABC expression was already 400-fold, and it reached almost 2,000-fold after 5 min (Fig. 1). This high level not only was maintained but also increased further, up to 7,000-fold at 3 h after the shift (data not shown), due to a further decrease in the K+ concentration in the medium. In contrast, after imposition of salt stress by 0.4 M NaCl, kdpFABC expression was only transiently induced, reaching 3-fold induction after 30 s, 300-fold induction after 10 min, and only 8-fold induction after 60 min (data not shown). If osmotic stress was imposed by 0.6 M sucrose (iso-osmolar with 0.4 M NaCl), kdpFABC expression was also transiently induced, but only a 20-fold induction was observed after 10 min (data not shown). It is important to mention that salt stress from the addition of 0.4 M potassium chloride did not result in the expression of the kdpFABC operon (data not shown). In summary, KdpD responds to salt stress or the addition of sugar in various degrees that fall far short of the response to potassium depletion (Table 1). In a control experiment, no induction was observed when cells were transferred into a growth medium of the same composition as that before the shift, demonstrating that the handling procedure was not responsible for any induction of the kdpFABC operon. It should be mentioned that although variations of the data set are caused by the extremely low noninduced-state values, the overall magnitude of the induction ratio stayed quite constant (Table 1).
FIG. 1.
Induction of the kdpFABC operon following a potassium downshift or an osmotic upshift. Cells of E. coli K12 were cultivated to exponential growth in K20 minimal medium and then shifted to high osmolality by 0.4 M NaCl (triangles) or potassium-free medium (squares). At time points indicated, normalized mRNA levels for kdpFABC expression were quantified by Q-RT-PCR as described in Materials and Methods. The induction ratio was calculated by division of the amount of normalized mRNA after the shift by the amount before the shift. Each data point represents values from three independent measurements.
TABLE 1.
Response of kdpFABC expression to various stimuli
| Stimulusa | kdpFABC induction ratio (fold)c | Time of onset of expression | Duration |
|---|---|---|---|
| K+ limitation | 1,000 | Seconds | Permanent |
| Salt (NaCl) stressb | 100 | Minutes | Transient |
| Salt (KCl) stressb | None | NAd | NA |
| Sugar stressb | 10 | Minutes | Transient |
Applied by different medium conditions. Details are given in the text.
Iso-osmolar medium conditions.
The induction ratio was determined by Q-RT-PCR as described in the text and in the legend to Fig. 1. Only the orders of magnitude are indicated.
NA, not applicable.
Changes in cytoplasmic volume following a potassium downshift or an osmotic upshift.
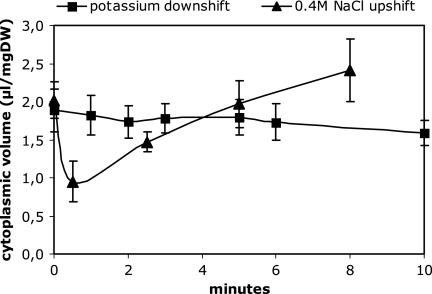
The cytoplasmic volume of exponentially growing cells with an optical density of 0.5 was 1.89 (± 0.28) μl/mg dry weight in minimal medium containing 20 mM potassium. This value was significantly lower at a low optical density and higher at a high optical density (data not shown). Therefore, for all experiments, the cytoplasmic volume was determined. After a potassium downshift, the external potassium concentration immediately decreased from 20 mM to 74 (± 6) μM and reached 8 (± 4) μM after 60 min, clearly indicating the potassium-limiting state of the growth medium. After the shift into nominally potassium-free medium, there was hardly any change in the cytoplasmic volume (Fig. 2). The space occupied by [14C]sucrose and the total water space were the same before and after the potassium downshift. As a result, no significant change in the concentration of any cytoplasmic solute, based on the cytoplasmic volume, occurred during the first minutes after the shift. In cells that were shifted into a medium of the same composition, none of the cell compartments were affected except that the size of the cytoplasmic and total volumes slowly increased during long-term growth. This was not the case after a K+ downshift.
FIG. 2.
Cytoplasmic volume of E. coli M2701 after a potassium downshift or an osmotic upshift. Cells were grown to exponential phase in K20 minimal medium and then transferred to K0 minimal medium (squares) or to high osmolality by the addition of 0.4 M NaCl (triangles). At time points indicated, the cytoplasmic volume was determined as described in Materials and Methods. The data result from six (potassium downshift) or three (osmotic upshift) independent measurements. Values are given with standard deviations.
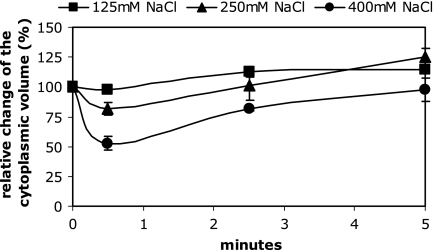
As reported previously (6), osmotically stressed cells showed an immediate and drastic decrease in the cytoplasmic volume after the addition of osmotic solutes to growing cells (Fig. 2). Since there is no change in the cytoplasmic volume upon K+ limitation, the decrease in the cytoplasmic volume of osmotically stressed cells also served as a control for the reliability of the method. The cytoplasmic volume already was reduced from 1.96 to 1.02 μl/mg dry weight 30 s after the addition of 0.4 M NaCl, corresponding to a 48% decrease, and was restored approximately 5 min after the medium shift. It was also observed that the water content of the cytoplasm increased up to 120% of the initial value 8 min after the osmotic upshock. It is important to mention that salts or sugars were comparably effective (data not shown). Interestingly, comparable values were obtained when NaCl was replaced by KCl. We also confirmed that the decrease in the cytoplasmic volume depends on the extent of the osmotic upshift (Fig. 3). This also served as a control for the sensitivity of cytoplasmic volume determination. In all cases, a shrinkage and subsequent restoration of the cytoplasmic volume originated from changes in the total water space of the cell. The space occupied by sucrose, namely, the periplasmic space plus the extracellular water layer, stayed largely unchanged (data not shown). The resulting conclusion that osmotically stressed cells immediately reduce their overall size to the same extent that they reduce their cytoplasmic volume could also be verified by phase-contrast microscopy and image analysis (see Fig. 6).
FIG. 3.
Changes in the cytoplasmic volume of E. coli M2701 after osmotic upshock of different strengths. The indicated amounts of NaCl were added to cells growing exponentially in K20 minimal medium. At time points indicated, the cytoplasmic volumes were measured as described in Materials and Methods and are given as percentages of the initial value. Measurements were done in duplicate.
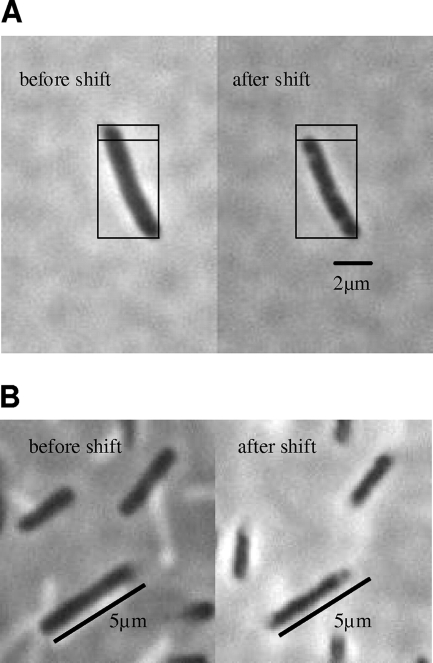
FIG. 6.
Cells of E. coli M2701 before and after an osmotic upshift. Cells of E. coli M2701 in K20 minimal medium that were attached to the microscope slide without fixation were photographed before and 10 s after an osmotic upshift by the addition of 0.8 M NaCl (A) or 0.8 M KCl (B) to the side of the coverslip. Pictures were taken at 1,000-fold magnification under a phase-contrast microscope. For easier size estimation, the subdivided squares of identical size or metering bars were introduced in the figure.
Changes in cytoplasmic ATP concentrations following a potassium downshift or an osmotic upshift.
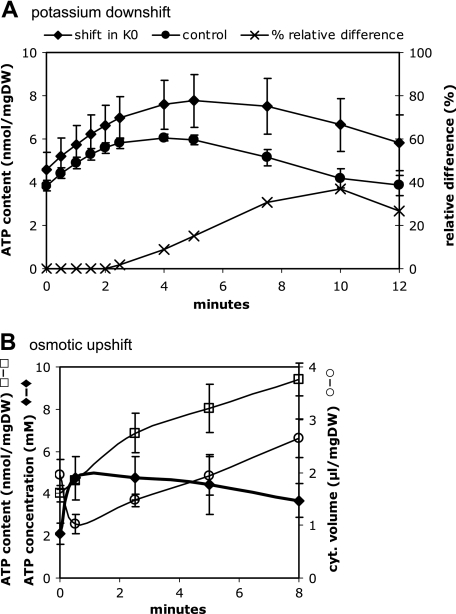
The cytoplasmic ATP content increased from 4.5 (±0.8) nmol/mg dry weight to 7.76 (±1.2) nmol/mg dry weight during the first 5 min after a potassium downshift and decreased to 6.65 (±1.2) nmol/mg dry weight 10 min after the shift (Fig. 4A). However, cells that were shifted to the same medium also showed an increase in the cytoplasmic ATP content from 3.84 (± 0.3) nmol/mg dry weight to 5.96 (± 0.2) nmol/mg dry weight 5 min after the shift and a decrease to nearly initial values after 10 min. To arrive at the changes in the ATP level, all values were normalized to percentages of the initial value, and for each time point, the corrected values for the experiment and the control were then subtracted from each other. As can clearly be seen, 5 min after the potassium downshift, the intracellular ATP content is only 15% higher than that in control cells, and it is 37% higher after 10 min before it decreases again to values comparable to the situation before the shift (data not shown). Since the cytoplasmic volume showed only a slight decrease, by 5 to 10%, under these conditions (see above), the change in the ATP concentration is approximately proportional to the change in the ATP content. On the basis of the cytoplasmic volume (1.89 [± 0.28] μl/mg dry weight before and 1.80 [± 0.23] μl/mg dry weight 5 min after the shift), the cytoplasmic ATP concentration was calculated to be 2.26 (± 0.52) mM before and 2.66 (± 0.52) mM 5 min after the shift, corresponding to a 17% increase during the first 5 min after the shift.
FIG. 4.
ATP content of E. coli M2701 following a potassium downshift or an osmotic upshift. Cells were grown to exponential growth in K20 minimal medium and then transferred to K0 minimal medium (A) or shifted to higher osmolality by the addition of 0.4 M NaCl (B). At time points indicated, intracellular ATP was measured as described in Materials and Methods. As a control, ATP was also measured in control cells that were shifted into medium of the same composition (circles in panel A). Values were normalized to percentages of the initial value and subtracted from each other (panel A, ×). The ATP concentration (mM) was calculated on the basis of the ATP content in nmol/mg dry weight (DW) divided by the cytoplasmic (cyt.) volume in μl/mg dry weight (B).
The ATP concentration immediately increased from 2.1 mM to 4.7 mM after imposition of salt stress by 0.4 M NaCl (Fig. 4B). This was due to both a moderate increase in the cytoplasmic ATP content and the immediate shrinkage of the cytoplasmic volume (see above). The ATP content of the cytoplasm increased constantly over a time period of 8 min from 4 to 9.4 nmol/mg dry weight, whereas the cytoplasmic volume decreased by 50% after 30 s and was restored after 5 min. Eight minutes after the upshift, the cytoplasmic ATP concentration was 3.6 mM.
Changes in cytoplasmic potassium concentrations following a potassium downshift or an osmotic upshift.
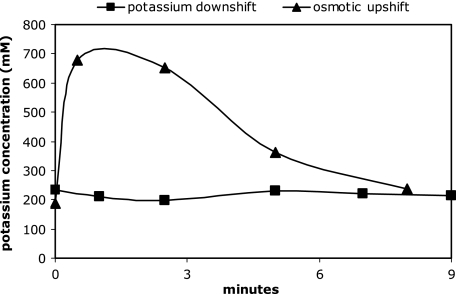
After a potassium downshift, the cytoplasmic potassium concentration hardly changed at all (Fig. 5). However, the cytoplasmic potassium concentration immediately increased transiently after salt stress elicited by 0.4 M NaCl. Since the increase in the cytoplasmic potassium content from 352 nmol/mg dry weight to 941 nmol/mg within the first 3 min was accompanied by a steep decrease in the cytoplasmic water space, the cytosolic potassium concentration increased from 186 mM to 676 mM during the first minute after an osmotic upshift, corresponding to a 3.6-fold increase (Fig. 5). Subsequently, it slowly decreased again to 238 mM, which was slightly higher than the initial value. This is in agreement with the observation by Epstein and Schultz (10), who showed that the intracellular potassium content of growing cells depends on medium osmolality.
FIG. 5.
Cytoplasmic potassium concentration for E. coli M2701 after a potassium downshift or an osmotic upshift. Cells of E. coli M2701 were cultivated to exponential growth and then transferred to potassium-free medium (squares), or at time point zero, 0.4 M NaCl was added (triangles), as described in Materials and Methods. The cytoplasmic potassium content was measured by using flame photometry at time points indicated, and then the cytoplasmic potassium concentration was calculated based on the results of the cytoplasmic volume determinations. The potassium content was corrected for the amounts of potassium in the periplasmic and extracellular water space. Each data point represents values from three independent measurements.
Other cytoplasmic solutes.
The cytoplasmic sodium concentration remained unchanged. The concentrations of putrescine, spermidine, trehalose, glutamate, and glycine were not significantly altered after a potassium downshift. However, as already reported previously, an increase in the glutamate and trehalose concentrations was observed under osmotic upshift (data not shown) (7).
Viability, cell number, total protein, and dry weight.
The viability, total cell count, total protein, and dry weight per ml cell culture did not change after a potassium downshift compared to the situation before the shift (data not shown). In addition, the ratio of the cell dry weight per ml cell culture to the optical density remained constant over the range used for the measurements in this study (data not shown). This indicates that the dry weight of the cells is a valid reference for the cytoplasmic volume.
Microscopy and image analysis following a potassium downshift or an osmotic upshift.
Out of 6 pictures of E. coli M2701 cells in K20 medium, 202 cells were depicted and the average pixel area was determined to be 556 (± 30) using the ImageJ analysis tool. Three minutes after a shift to potassium-free medium, the average pixel area of 408 cells out of 6 pictures was 528 (± 17), corresponding to a 5% decrease in the two-dimensional cell size, which is in the range of statistical error (4.3% ± 1.1%). Thus, no significant immediate change of the cell size after a potassium downshift could be observed by optical means. Interestingly, it could be optically confirmed that a potassium downshift prevented cells from slowly increasing their volume during long-term growth as they do at potassium concentrations of 20 mM (data not shown).
Three minutes after the addition of NaCl (salt stress), the average pixel area of 176 cells out of 6 pictures was 445 (± 25), corresponding to a 20% decrease in the two-dimensional cell size, with a statistical error of 5.6% (± 0.1%). Calculating the volume and the area of a rod-shaped bacterium with a ratio of its total length to its diameter of 3.75 (e.g., a length of 4.5 μm and a diameter of 1.2 μm) reveals that a 20% decrease in its two-dimensional size corresponds to a 30% decrease in its three-dimensional size. This is comparable to the values of the cytoplasmic volume obtained by solute distribution. In Fig. 6A, two pictures of the same cell before and 10 s after a nonquantitative but maximally 0.8 M NaCl upshift reveal the significant and spontaneous reduction in cell size in vivo. By approximation of the cell dimensions using a 2-μm metering bar (approximately 6.45 μm in length and 1.2 μm in diameter before the shift and 5.6 μm in length and 1 μm in diameter after the shift), the cellular volume was calculated to be 6.84 μm3 before and 4.13 μm3 after the shift, corresponding to a 40% volume reduction. The beginning of plasmolysis can be seen at the slightly brighter spots appearing in the osmostressed cell. A similar volume decrease can also be obtained by the addition of KCl instead of NaCl (Fig. 6B).
DISCUSSION
In order to search for cellular parameters which might serve as the primary stimulus in triggering the signaling cascade of the KdpD/KdpE system, two situations were carefully analyzed under which kdpFABC expression is known to be induced: first, a sudden decrease in the potassium concentration; and second, a sudden increase in medium osmolality. It has already been observed, by measuring β-galactosidase activities of a kdpFABC promoter-lacZ fusion, though not often noted, that salt or sugar stress is less effective than potassium-limiting conditions in causing kdpFABC expression (e.g., see reference 44). In this study, it turned out for the first time from mRNA measurements that the kdpFABC operon is indeed much more effectively expressed following a potassium downshift than with salt or sugar stress (see Table 1). Since RT-PCR data are normalized to mRNA levels of other housekeeping genes, in our case gap, a general stress effect on the transcription apparatus can be excluded. However, when a system responds to high concentrations of NaCl and hardly or not at all to isotonic media of different compositions (e.g., sucrose or KCl; see Table 1), it does not really respond to osmotic upshock per se. It is more likely that synthesis of the KdpFABC system is induced by salt or sugar stress but in various degrees that fall far short of the response to a potassium downshift. Therefore, it seems either that the stimulus perceived under potassium-limiting conditions is stronger itself or that the sensitivity of KdpD toward this stimulus is higher compared than under salt or sugar stress. In the latter case, two different stimuli might exist or the response to elevated osmolality is a secondary effect (see below) of a system that usually responds to potassium-limiting conditions. This could possibly explain why absolutely no kdpFABC induction occurs if KCl is used for imposing salt stress. As expected, under these conditions an excess of potassium prevents kdpFABC induction. If both stresses are communicated via, e.g., the same intracellular parameter, the change in this parameter is then expected to be greater after a potassium downshift than after an osmotic upshift. Since many cellular parameters are known to respond to an osmotic upshift, the focus of this work was on the physiological state of cells after a potassium downshift. As a control, the same experiments were carried out with cells subjected to an osmotic upshift.
The measurements of the cytoplasmic volume were chosen mainly to address two cellular aspects: first, the relation of changes in turgor to kdpFABC expression; and second, the determination of the cytoplasmic concentrations of potential solutes, which might function as stimuli for KdpD. It has previously been hypothesized that KdpD is a potential sensor of changes in turgor through the detection of alterations in membrane strain (see reference 22 and references therein). Since turgor or membrane strain cannot be measured directly for E. coli, we took advantage of the fact that a reduction in the cytoplasmic volume correlates with a reduction in turgor (4). As shown in this report, osmotically stressed cells exhibit a shrinkage of the cytoplasm which did not occur under potassium downshift. Consequently, under the latter conditions, cells did not suffer from a loss of water, at least not to such an extent as to change the cytoplasmic volume or the shape of the membrane. It is well established that many bacteria maintain turgor that is crucial for growth and that has been estimated to be 0.5 to 5 atmospheres in gram-negative species and up to 10 to 30 atmospheres in gram-positive ones (6, 4, 30). It has not been fully clarified whether turgor in gram-negative bacteria is exerted across the inner membrane or the entire cell envelope. In the first case, the cytoplasmic membrane would be pressed against the cell wall; in the latter, the periplasm would be isosmotic with the cytoplasm and both the cytoplasmic membrane and the murein sacculus would be pressed against the outer membrane, as proposed by Stock et al. (36) and Cayley et al. (4). The surrounding cell wall was shown to function as an elastic sacculus (20). As long as the murein sacculus is in its elastic state, turgor directly correlates with the expansion of the cell. Only if the elasticity has reached its maximum can increases in turgor occur without expanding the cell any further, and vice versa: only beyond maximum elasticity can a reduction of turgor happen without shrinkage of the cell. However, Koch and Woeste (20) showed that the murein sacculus of growing cells of E. coli is far from being maximally extended. A reduction in turgor can therefore be excluded when cells of E. coli do not decrease their cellular or cytoplasmic volume. Furthermore, if the shrinkage of the cytoplasm after an osmotic upshock, with its concomitant effect on the inner membrane, was the stimulus for KdpD, then one would expect an at least equal if not stronger shrinkage under potassium-limiting conditions. However, under osmotic upshift, the cytoplasmic volume was transiently reduced and kdpFABC expression was low and transient, whereas after a potassium downshift, the cytoplasmic volume was unaffected and kdpFABC expression was high. Therefore, a reduction in turgor cannot be a stimulus for KdpD. This view is further supported by the observation that osmotic stress elicited by potassium chloride exhibits the expected transient decrease in the cytoplasmic volume but does not elicit kdpFABC expression. Therefore, the low level of kdpFABC expression under salt stress elicited by sodium chloride might be a “secondary effect” due to the inhibitory effect of sodium ions on the potassium uptake systems (W. Epstein, personal communication), thereby mimicking K+ limitation.
Since it is known from previous observations (26) and confirmed in our studies that osmotically stressed cells exhibit an elevated level of ATP and since the input domain of KdpD possesses a regulatory ATP binding site, it was proposed that changes in the ATP concentration could be a stimulus for KdpD (19). However, after a potassium downshift, the ATP concentration is hardly affected compared to that in osmotically stressed cells, which show a more than twofold-higher ATP concentration. In the study by Ohwada and Sagisaka (26) using 0.8 M NaCl, the decrease in the cytoplasmic volume was not taken into account, which leads to an even higher ATP level. Therefore, it is quite unlikely that changes in the ATP concentration are the stimulus for KdpD.
Under conditions of potassium downshift leading to potassium concentrations in the medium below 80 μM, the cytoplasmic potassium concentration was not significantly affected compared to the massive accumulation after an osmotic upshift. Nevertheless, kdpFABC expression was maximally induced under K+-limiting conditions. It is interesting to note that the cytoplasmic potassium concentration had already returned to initial values 8 min after an osmotic upshift, which is much faster than reported by others (7, 25). This discrepancy can be partly explained by the fact that in our case the concentration of potassium was calculated on the basis of the cytoplasmic volume, which already was higher 8 min after an osmotic upshock than the initial values.
The conditions under which kdpFABC expression is induced have often been referred to as the cells’ need for potassium (21, 22). Inadequate potassium accumulation prevents water accumulation, which in turn does not allow an increase in cell size sufficient for subsequent cell division, and as a consequence, growth ceases. Potassium is the only ion that enters the cytoplasm after an osmotic upshift in concentrations high enough to restore turgor. As far as the cytoplasmic volume is concerned, we showed that the cells’ need for potassium is differently satisfied by the two kdpFABC inducing conditions. Under osmotic stress conditions, the intracellular potassium concentration has to be adjusted to much higher values to restore water content and turgor. Under potassium-limiting conditions, the cells showed neither a loss of cytoplasmic water nor a significant reduction in the potassium concentration. Consequently, cells do not necessarily have to restore either of the two. The cells’ need for potassium is then reflected by the challenge of maintaining a constant cytoplasmic level of potassium against a more than 2,000-fold concentration gradient across the membrane (inside, e.g., 200 mM; outside, e.g., 80 μM) compared to only 10-fold at, e.g., 20 mM K+ outside. However, the hypothesis that KdpD could be a sensor of this concentration gradient is not correct, because the expression levels of kdpFABC can be different at constant concentration gradients. An elevated intracellular potassium concentration of, e.g., 400 mM, a response to high osmolality of the medium, results in a 20-fold gradient of K+ across the membrane at 20 mM K+ outside. Under these conditions, kdpFABC expression is turned on. However, the same 20-fold gradient across the membrane can be established (inside, 200 mM; outside, 10 mM K+), but kdpFABC is not expressed. In addition, kdpFABC expression is already high in cells grown with 20 mM potassium that are deleted for the trk genes, whereas it stays uninduced in wild-type cells grown at the same external potassium concentration (22). Thus, the gradient of K+ is also not a very likely candidate to act as a stimulus for KdpD. As a consequence, although potassium-limiting conditions exhibit high kdpFABC induction ratios, neither the outside nor the inside potassium concentration nor its gradient across the membrane can explain the kdpFABC expression profile over the range of conditions that have been tested so far.
In previous studies, Asha and Gowrishankar (1) already have postulated on the basis of kdp-lacZ fusions and corresponding β-galactosidase activities that kdpFABC expression was not turgor regulated. They put forward the hypothesis that a decrease in the specific rate of K+ fluxes through the constitutive K+ transport systems would represent the stimulus for KdpD, without providing a suitable mechanism by which that could occur. In our view, this is not too far away from the descriptive term of the cells’ need for potassium used by Epstein and coworkers. However, the question remains how the cells’ need for potassium is satisfied. One possibility would be the interaction of KdpD with another protein (an accessory sensor), which would put KdpD in the position to sense the cells’ need for potassium. That histidine kinases do interact with, e.g., transporters has been shown for DcuS (sensor for fumarate) and DcuB (fumarate/succinate antiporter) (G. Unden, personal communication). Furthermore, Steyn et al. (34) could show that in Mycobacterium tuberculosis the N-terminal domain of KdpD interacts with the lipoproteins LprF and LprJ. Although no homologous proteins have been found in E. coli, it is still conceivable that due to low homology they have so far not been identified. In addition, in vitro transphosphorylation experiments between noncognate sensor kinase/response regulator pairs in E. coli revealed no cross talk partner for the C-terminal domain of KdpD (42). Consequently, the cells’ need for potassium must be recognized by other factors. In our view, this could be a so-far-unaddressed question related to processes that heavily depend not only on the presence of K+ but also on the continuing uptake of potassium to generate sufficient turgor during cell elongation. The larger the cell size as a consequence of cell elongation, the more net potassium is needed per cell to maintain the same turgor. Since KdpD is an integral membrane protein, it could, e.g., be affected by the incorporation of phospholipids into the cytoplasmic membrane, comparable to the constant incorporation of murein into the sacculus. Our observation that the cytoplasmic volume of cells slowly and constantly increases during long-term growth at a sufficient potassium concentration while it remains constant during growth after a potassium downshift points in such a direction. In this context, it is interesting to note that the kdpFABC induction is transient in the case of salt stress (only NaCl) or sugar stress but permanent in the case of a potassium downshift, which can be switched off only by the addition of sufficient potassium.
These considerations would call for a hypothesis in which KdpD senses the insertion of fatty acids into the inner-membrane phospholipids. The fatty acids are incorporated by two membrane-bound enzymes, the sn-glycerol-3-phosphate acyltransferase and the 1-acyl-sn-glycerol-3-phosphate acyltransferase, encoded by the genes plsB and plsC, respectively (12). It is conceivable that these enzymes exhibit lower transferase activity in cases where membrane extension does not occur (K+ limitation). The low activity state of these or other enzymes of the lipid synthesis complex (24) might allow autophosphorylation of KdpD by direct protein-protein interaction or via a mediator protein, like the lipoproteins LprF and LprJ, found in M. tuberculosis (34). To explore this possibility, experiments are under way in our laboratory to look for direct interactions between these enzymes and KdpD and to search for mediator proteins.
Acknowledgments
We thank Monika Nietschke for expert technical assistance, E. Galinski (University of Bonn) for the determination of the various solutes, K. Schmid (University of Osnabrueck) for providing E. coli strain M2701, and W. Epstein (University of Chicago) for helpful discussions and critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 431), the Fonds der Chemischen Industrie, and the Ministerium für Wissenschaft und Kultur of the State of Lower Saxony.
Footnotes
Published ahead of print on 1 February 2008.
REFERENCES
- 1.Asha, H., and J. Gowrishankar. 1993. Regulation of kdp operon expression in Escherichia coli: evidence against turgor as signal for transcriptional control. J. Bacteriol. 1754528-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bramkamp, M., K. Altendorf, and J. C. Greie. 2007. Common patterns and unique features of P-type ATPases: a comparative view on the KdpFABC complex from Escherichia coli. Mol. Membr. Biol. 24375-386. [DOI] [PubMed] [Google Scholar]
- 3.Brandon, L., S. Dorus, M. Epstein, K. Altendorf, and K. Jung. 2000. Modulation of KdpD phosphatase implicated in the physiological expression of the Kdp ATPase of Escherichia coli. Mol. Microbiol. 381086-1092. [DOI] [PubMed] [Google Scholar]
- 4.Cayley, S., H. J. Guttman, and M. T. Record, Jr. 2000. Biophysical characterization of changes in amounts and activity of Escherichia coli cell and compartment water and turgor pressure in response to osmotic stress. Biophys. J. 781748-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cayley, S., B. A. Lewis, H. J. Guttman, and M. T. Record, Jr. 1991. Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J. Mol. Biol. 222281-300. [DOI] [PubMed] [Google Scholar]
- 6.Csonka, L. N. 1989. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53121-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinnbier, U., E. Limpinsel, R. Schmid, and E. P. Bakker. 1988. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch. Microbiol. 150348-357. [DOI] [PubMed] [Google Scholar]
- 8.Epstein, W. 1992. Kdp, a bacterial P-type ATPase whose expression and activity are regulated by turgor pressure. Acta Physiol. Scand. Suppl. 607193-199. [PubMed] [Google Scholar]
- 9.Epstein, W., and B. S. Kim. 1971. Potassium transport loci in Escherichia coli K-12. J. Bacteriol. 108639-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epstein, W., and S. G. Schultz. 1965. Cation transport in Escherichia coli. V. Regulation of cation content. J. Gen. Physiol. 49221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulton, A. B. 1982. How crowded is the cytoplasm? Cell 30345-347. [DOI] [PubMed] [Google Scholar]
- 12.Heath, R. J., S. Jackowski, and C. O. Rock. 2002. Fatty acid and phospholipid metabolism in prokaryotes, p. 55-92. In D. E. Vance and J. E. Vance (ed.), Biochemistry of lipids, lipoproteins and membranes, 4th ed. Elsevier, Amsterdam, The Netherlands.
- 13.Heermann, R., K. Altendorf, and K. Jung. 1998. The turgor sensor KdpD of Escherichia coli is a homodimer. Biochim. Biophys. Acta 1415114-124. [DOI] [PubMed] [Google Scholar]
- 14.Heermann, R., K. Altendorf, and K. Jung. 2000. The hydrophilic N-terminal domain complements the membrane-anchored C-terminal domain of the sensor kinase KdpD of Escherichia coli. J. Biol. Chem. 27517080-17085. [DOI] [PubMed] [Google Scholar]
- 15.Heermann, R., K. Altendorf, and K. Jung. 2003. The N-terminal input domain of the sensor kinase KdpD of Escherichia coli stabilizes the interaction between the cognate response regulator KdpE and the corresponding DNA-binding site. J. Biol. Chem. 27851277-51284. [DOI] [PubMed] [Google Scholar]
- 16.Heermann, R., A. Fohrmann, K. Altendorf, and K. Jung. 2003. The transmembrane domains of the sensor kinase KdpD of Escherichia coli are not essential for sensing K+ limitation. Mol. Microbiol. 47839-848. [DOI] [PubMed] [Google Scholar]
- 17.Huhn, G., J. Mattusch, and H. Schulz. 1995. Determination of polyamines in biological materials by HPLC with 9-fluorenylmethyl chloroformate precolumn derivatization. Fresenius J. Anal. Chem. 351563-566. [Google Scholar]
- 18.Jung, K., and K. Altendorf. 1998. Individual substitutions of clustered arginine residues of the sensor kinase KdpD of Escherichia coli modulate the ratio of kinase to phosphatase activity. J. Biol. Chem. 27326415-26420. [DOI] [PubMed] [Google Scholar]
- 19.Jung, K., and K. Altendorf. 1998. Truncation of amino acids 12-228 causes deregulation of the phosphatase activity of the sensor kinase KdpD of Escherichia coli. J. Biol. Chem. 27317406-17410. [DOI] [PubMed] [Google Scholar]
- 20.Koch, A. L., and S. Woeste. 1992. Elasticity of the sacculus of Escherichia coli. J. Bacteriol. 1744811-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laimins, L. A., D. B. Rhoads, and W. Epstein. 1981. Osmotic control of kdp operon expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 78464-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malli, R., and W. Epstein. 1998. Expression of the Kdp ATPase is consistent with regulation by turgor pressure. J. Bacteriol. 1805102-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mascher, T., J. D. Helmann, and G. Unden. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70910-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto, K., J. Kusaka, A. Nishibori, and H. Hara. 2006. Lipid domains in bacterial membranes. Mol. Microbiol. 611110-1117. [DOI] [PubMed] [Google Scholar]
- 25.McLaggan, D., J. Naprstek, E. T. Buurman, and W. Epstein. 1994. Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. J. Biol. Chem. 2691911-1917. [PubMed] [Google Scholar]
- 26.Ohwada, T., and S. Sagisaka. 1987. An immediate and steep increase in ATP concentration in response to reduced turgor pressure in Escherichia coli B. Arch. Biochem. Biophys. 259157-163. [DOI] [PubMed] [Google Scholar]
- 27.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 2671-112. [DOI] [PubMed] [Google Scholar]
- 28.Peterson, G. L. 1977. A simplification of the protein assay method of Lowry et al. Anal. Biochem. 83346-356. [DOI] [PubMed] [Google Scholar]
- 29.Polarek, J. W., G. Williams, and W. Epstein. 1992. The products of the kdpDE operon are required for expression of the Kdp ATPase of Escherichia coli. J. Bacteriol. 1742145-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poolman, B., J. J. Spitzer, and J. M. Wood. 2004. Bacterial osmosensing: roles of membrane structure and electrostatics in lipid-protein and protein-protein interactions. Biochim. Biophys. Acta 166688-104. [DOI] [PubMed] [Google Scholar]
- 31.Rothenbücher, M. C., S. J. Facey, D. Kiefer, M. Kossmann, and A. Kuhn. 2006. The cytoplasmic C-terminal domain of the Escherichia coli KdpD protein functions as a K+ sensor. J. Bacteriol. 1881950-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt, K. 1974. Ph.D. thesis. University of Regensburg, Regensburg, Germany.
- 33.Siegele, D. A. 2005. Universal stress proteins in Escherichia coli. J. Bacteriol. 1876253-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steyn, A. J. C., J. Joseph, and B. R. Bloom. 2003. Interaction of the sensor module of Mycobacterium tuberculosis H37Rv KdpD with members of the Lpr family. Mol. Microbiol. 471075-1089. [DOI] [PubMed] [Google Scholar]
- 35.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 36.Stock, J. B., B. Rauch, and S. Roseman. 1977. Periplasmic space in Salmonella typhimurium and Escherichia coli. J. Biol. Chem. 2527850-7861. [PubMed] [Google Scholar]
- 37.Sugiura, A., K. Hirokawa, K. Nakashima, and T. Mizuno. 1994. Signal-sensing mechanisms of the putative osmosensor KdpD in Escherichia coli. Mol. Microbiol. 14929-938. [DOI] [PubMed] [Google Scholar]
- 38.Sugiura, A., K. Nakashima, K. Tanaka, and T. Mizuno. 1992. Clarification of the structural and functional features of the osmoregulated kdp operon of Escherichia coli. Mol. Microbiol. 61769-1776. [DOI] [PubMed] [Google Scholar]
- 39.Voelkner, P., W. Puppe, and K. Altendorf. 1993. Characterization of the KdpD protein, the sensor kinase of the K+-translocating Kdp system of Escherichia coli. Eur. J. Biochem. 2171019-1026. [DOI] [PubMed] [Google Scholar]
- 40.Walderhaug, M. O., J. W. Polarek, P. Voelkner, J. M. Daniel, J. E. Hesse, K. Altendorf, and W. Epstein. 1992. KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. J. Bacteriol. 1742152-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wood, J. M. 1999. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol. Mol. Biol. Rev. 63230-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto, K., K. Hirao, T. Oshima, H. Aiba, R. Utsumi, and A. Ishihama. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 2801448-1456. [DOI] [PubMed] [Google Scholar]
- 43.Zimmann, P., W. Puppe, and K. Altendorf. 1995. Membrane topology analysis of the sensor kinase KdpD of Escherichia coli. J. Biol. Chem. 27028282-28288. [DOI] [PubMed] [Google Scholar]
- 44.Zimmann, P., A. Steinbrügge, M. Schniederberend, K. Jung, and K. Altendorf. 2007. The extension of the fourth transmembrane helix of the sensor kinase KdpD of Escherichia coli is involved in sensing. J. Bacteriol. 1897326-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]