Abstract
The heptapeptide-nucleotide microcin C (McC) targets aspartyl-tRNA synthetase. Upon its entry into a susceptible cell, McC is processed to release a nonhydrolyzable aspartyl-adenylate that inhibits aspartyl-tRNA synthetase, leading to the cessation of translation and cell growth. Here, we surveyed Escherichia coli cells with singly, doubly, and triply disrupted broad-specificity peptidase genes to show that any of three nonspecific oligopeptidases (PepA, PepB, or PepN) can effectively process McC. We also show that the rate-limiting step of McC processing in vitro is deformylation of the first methionine residue of McC.
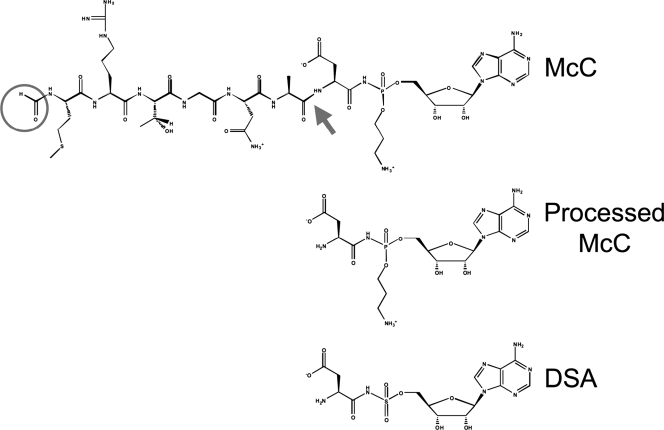
Microcins are a class of antibacterial agents produced by Escherichia coli and its close relatives (1, 2, 14). Microcins are characterized by their small size (<10 kDa) and narrow specificity of antibacterial action. Microcins are produced from ribosomally synthesized peptide precursors. Microcins B, C, and J are heavily modified by dedicated maturation enzymes (6, 14). The object of this study, microcin C (McC), is a heptapeptide with a covalently attached C-terminal-modified AMP (9, 12). The peptide moiety of McC is encoded by the 21-bp-long mccA gene, the shortest bacterial gene known (7, 8). Mature McC (Fig. 1) has a molecular mass of 1,178 Da and contains formylated N-terminal methionine, a C-terminal aspartate instead of an asparagine encoded by the mccA gene, and an AMP residue attached to the α carboxyl group of the C-terminal aspartate through an N-acyl phosphoramidate linkage. The phosphate group is additionally modified by a propylamine group.
FIG. 1.
The structure of McC. The chemical structure of McC, a peptide-nucleotide, is shown at the top. The N-terminal formyl group is circled, and the peptide bond that must be cleaved to yield processed McC is shown by an arrow. Processed McC, an inhibitor of Asp-RS, is shown below. The synthetic Asp-RS inhibitor DSA is shown at the bottom of the figure.
McC is processed inside sensitive cells, and the product of such processing, a nonhydrolyzable analog of aspartyl-adenylate (Fig. 1), strongly inhibits translation by preventing the synthesis of aminoacylated tRNAAsp by aspartyl-tRNA synthetase (Asp-RS) (11). Whereas unprocessed McC has no effect on the aminoacylation reaction, processed McC has no effect on the growth of sensitive cells at concentrations at which intact McC efficiently inhibits growth (11). Thus, McC is a Trojan horse inhibitor (15): the peptide moiety is required for the entry of unprocessed McC into sensitive cells, where it must be processed by peptidases to release the inhibitory aminoacyl nucleotide part of the drug.
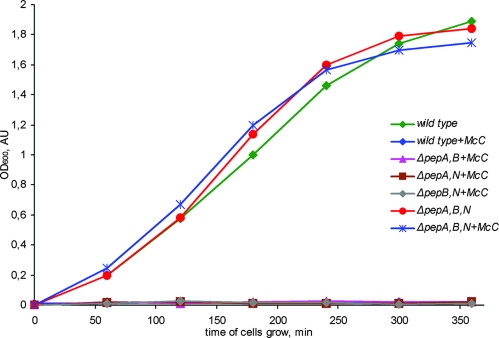
Cells that are unable to process McC should become resistant to it. E. coli K-12 encodes three broad-specificity oligopeptidases, peptidase A, B, and N (13). Since any one of these peptidases is potentially involved in McC processing, resistance mutations due to defective processing are expected to be difficult to find. Indeed, a recent analysis of McC-resistant E. coli generated by random transposon-mediated mutagenesis failed to reveal mutants defective in processing. Instead, numerous intake mutations due to lesions in the YejABEF transporter were identified (15). To identify peptidases that are involved in McC processing, we therefore systematically created mutants with single, double, and triple disruptions of the pepA, pepB, and pepN genes, coding for peptidases A, B, and N, respectively, of the McC-sensitive E. coli K-12 strain BW28357 cells (5), and tested the resultant mutants for their ability to grow in the presence of McC. We found that all singly and doubly disrupted mutants were viable, that they showed no visible growth anomalies on rich medium at 37°C and that they were as sensitive to McC as the parental strain was (data not shown and Fig. 2). Thus, none of the three broad-specificity peptidases appears to be solely responsible for McC processing inside the cell. The ΔpepA ΔpepB ΔpepN triple mutant was also viable, with no visible growth anomalies on rich medium at 37°C (Fig. 2). However, this strain was fully resistant to McC (Fig. 2), suggesting that no processing takes place when all three broad-specificity peptidases are inactivated.
FIG. 2.
Cells lacking peptidases A, B, and N are McC resistant. The growth of the indicated cells in the presence or in the absence of 10 μg/ml of McC is shown. The results presented are representative of those obtained from three independent experiments.
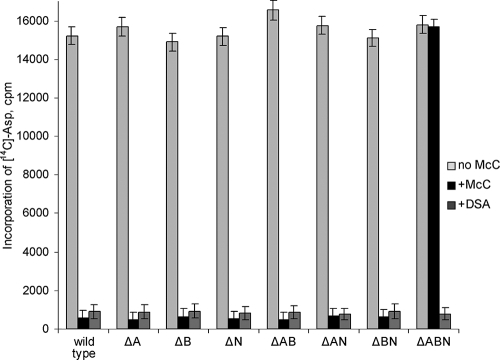
To prove that McC processing is affected in the ΔpepA ΔpepB ΔpepN cells, cytoplasmic extracts from the wild-type and the various mutant cells were prepared, and their abilities to perform tRNAAsp aminoacylation in the presence of McC were tested. As expected, the wild-type cell extracts were highly active in tRNAAsp aminoacylation, which is catalyzed by Asp-RS, but the reaction was blocked upon addition of McC (Fig. 3). Similar results were obtained with extracts prepared from cells lacking one or two of the three broad-specificity peptidases (Fig. 3). However, extracts prepared from the triple mutant cells were unaffected by McC (Fig. 3). At the same time, the aminoacylation reaction in triple-mutation cell extracts was readily inhibited by a processed McC analog, synthetic aspartyl-sulfamoyl-adenosine (DSA [3]), indicating that Asp-RS in these enzymes is still subject to inhibition by nonhydrolyzable aspartyl-adenylates. We therefore conclude that the lack of inhibition seen with reaction mixtures containing extracts of triple-mutation cells and intact McC is due to the lack of processing of McC in these extracts.
FIG. 3.
Asp-RS inhibition in extracts of peptidase mutants. S30 extracts of the indicated cells were prepared and incubated with or without 2 μg/ml DSA or McC for 20 min to allow processing of the latter, and the tRNAAsp aminoacylation reaction was carried out. The amounts of aminoacylated tRNAAsp (measured as the incorporation of [C14]Asp in trichloroacetic acid-precipitable material) are shown (see reference 12 for experimental details).
The results presented above argue strongly that any one of the three broad-specificity peptidases, peptidase A, B, or N, is able to cleave the ultimate peptide bond of McC, which is a prerequisite for processed McC production. Moreover, because all three peptidases are known to cleave only N-terminal residues of the peptides (13), peptidase A, B, or N will also be able to remove other McC amino acids. The N-terminal amino acid in mature McC is a formylated methionine found in all ribosomally synthesized polypeptides encoded by open reading frames with the initiating codon ATG. Neither of the three peptidases is able to remove formylated methionine (13). The formyl group of Met1 is removed by a specialized essential enzyme, peptide deformylase (PDF) (17). Deformylated Met1 can next be removed by methionine aminopeptidase (MAP), also an essential enzyme (17). The activity of MAP depends on the nature of the penultimate amino acid (the so-called N-end rule [18, 19]). In the case of McC, deformylated methionine can conceivably also be removed by broad-specificity oligopeptidases.
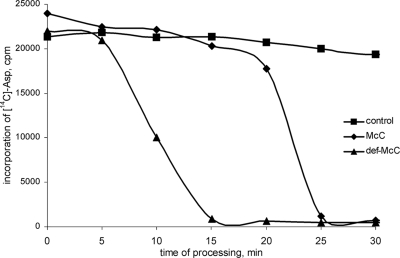
Mass spectrometric analysis of McC produced by wild-type E. coli cells reveals a major peak with a molecular mass of 1,178 Da that corresponds to the formylated molecule shown in Fig. 1B. However, a minor peak of 1,150 Da is also present. The 28-atomic-unit difference likely corresponds to the loss of the formyl group from the N-terminal methionine of McC (Fig. 1). Nuclear magnetic resonance analysis confirmed this notion (data not shown). Interestingly, the mass spectrometric analysis of McC samples purified from the cultured medium of cells containing the McC overproduction plasmid (7) but lacking peptidases A, B, and N revealed a single mass peak of 1,150 Da, corresponding to that of deformylated McC; no 1,178-Da peak was detected. The reason for the apparently very efficient deformylation of McC in triple-mutation cells is unknown; perhaps there exists a feedback loop that links the activity of PDF with the activity of peptidases in the cell. To determine the effect of the formyl group on McC processing, the reaction of tRNAAsp aminoacylation was repeated (using the wild-type cell extracts) in the presence of equal amounts of formylated or deformylated McC (Fig. 4). In contrast to reactions shown in Fig. 3, residual aminoacylation activity was monitored over time, which allows one to determine the appearance of processed McC in the extract. Inhibition of tRNAAsp aminoacylation by formylated McC occurred after a distinctive lag of ∼20 min, the time required for processing (10). In contrast, reactions containing deformylated McC were inhibited much sooner (with a lag time of less than 10 min). Thus, the results are consistent with the idea that a delay in the processing time of formylated McC, compared to that of deformylated McC, is due to the action of PDF.
FIG. 4.
The rate of McC processing depends on the presence of the N-terminal formyl group. S30 extracts of the indicated cells were prepared and incubated with or without 2 μg/ml formylated (McC) or deformylated (def-McC) McC to allow processing for the indicated amounts of time, and the tRNAAsp aminoacylation reaction was carried out. The amounts of aminoacylated tRNAAsp (measured as the incorporation of [C14]Asp in trichloroacetic acid-precipitable material) are shown (see reference 11 for experimental details). Control reaction mixtures contained water instead of McC.
To determine if MAP is involved in the removal of deformylated Met1 from McC, we incubated the wild-type and deformylated microcins with the wild-type or triple-mutation cell extracts for a time sufficient to allow complete processing and then followed their fate with mass spectrometry. The results showed, as expected (11), that both the formylated and the deformylated microcin peaks disappeared in wild-type cell extracts. In contrast, in the mutant cell extracts, deformylated McC remained intact, and no additional peaks (for example, corresponding to a product of methionine removal by MAP) were seen. On the other hand, incubation of the formylated McC led to the accumulation of a 1,150-Da peak, the product of deformylation. On the basis of these results, we conclude that MAP is not involved in McC processing, presumably because the Arg residue in the second position of McC makes the removal of Met1 inefficient (19). Instead, the removal of deformylated Met1 and further processing of the peptide moiety of McC are carried out by peptidases A, B, and N.
Since deformylated McC is processed faster, we were interested in determining whether it would be a more potent cell growth inhibitor than formylated McC produced by wild-type cells. Surprisingly, deformylated McC (at concentrations up to 100 μM) failed to produce growth inhibition zones on the lawns of McC-sensitive cells. In contrast, formylated McC produced clear inhibition zones at concentrations of 10 μM or less. We therefore conclude that the N-terminal formyl group has a dramatic effect on McC uptake.
In this work, we demonstrate that any one of the three broad-specificity peptidases encoded by the E. coli genome can perform the suicidal act of McC processing. We also show that deformylation of the N-terminal methionine by PDF is a rate-limiting step in McC processing in wild-type cell extracts. On the other hand, it appears that facilitated transport of McC in the cell requires the N-terminal formyl group, since the in vivo activity of DSA was comparable to that of deformylated McC (data not shown). The results therefore suggest that YejABEF, the ABC transporter responsible for the transport of McC inside the cell, is able to specifically recognize N-terminal formyl groups of the peptides it transports.
From the point of view of using McC as a platform for the generation of antibacterial compounds, the observed lack of specificity in processing is welcome news, for it appears that if McC can find its way into a bacterial cell cytoplasm, it will be processed with the release of inhibitory aspartyl-adenylate, for almost every bacterium known encodes at least one nonspecific peptidase (and every bacterium encodes Asp-RS, which is the target of processed McC).
The availability of peptidase mutants described in this paper makes it possible to screen rapidly for the involvement of various peptidases in processing Trojan horse inhibitors whose activation requires the hydrolysis of peptide bonds (4, 16). For example, we used the panel of our mutants to show that only the ΔpepA ΔpepN and ΔpepA ΔpepB ΔpepN strains are resistant to albomycin, a Trojan horse inhibitor that targets Ser-RS (4). Thus, PepA and PepN are jointly required for albomycin processing. Curiously, previous work suggested that PepN alone is sufficient for albomycin processing (4). The reasons for this apparent discrepancy are unknown.
Acknowledgments
We thank Charles Miller for help and insight at the initial stages of this work. We thank Pieter Van de Vijver and A. Van Aerschot, Rega Institute for Medical Research (K. U. Leuven) for the gift of DSA and discussions. We thank Mikhail S. Gelfand for help with bioinformatic analysis.
This work was supported by the Northeast Biodefense Center U54-AI057158-Lipkin grant, a Burroughs Wellcome Career Award, a Russian Academy of Sciences Presidium program in molecular and cellular biology new groups grant, a Rutgers University Technology Commercialization Fund Grant (to K.S.), and Russian Foundation for Basic Research grant 06-04-48865 (to A.M.).
Footnotes
Published ahead of print on 25 January 2008.
REFERENCES
- 1.Baquero, F., and F. Moreno. 1984. The microcins. FEMS Microbiol. Lett. 23117-124. [Google Scholar]
- 2.Baquero, F., D. Bouanchaud, M. C. Martinez-Perez, and C. Fernandez. 1978. Microcin plasmids: a group of extrachromosomal elements coding for low-molecular-weight antibiotics in Escherichia coli. J. Bacteriol. 135342-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernier, S., P. M. Akochy, J. Lapointe, and R. Chenevert. 2005. Synthesis and aminoacyl-tRNA synthetase inhibitory activity of aspartyl adenylate analogs. Bioorg. Med. Chem. 1369-75. [DOI] [PubMed] [Google Scholar]
- 4.Braun, V., K. Günthner, K. Hantke, and L. Zimmermann. 1983. Intracellular activation of albomycin in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 156308-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Destoumieux-Garzon, D., J. Peduzzi, J., and S. Rebuffat. 2002. Focus on modified microcins: structural features and mechanisms of action. Biochimie 84511-519. [DOI] [PubMed] [Google Scholar]
- 7.Fomenko, D. E., A. Z. Metlitskaya, J. Peduzzi, C. Goulard, G. Katrukha, L. V. Gening, S. Rebuffat, and I. A. Khmel. 2003. Microcin C51 plasmid genes: possible source of horizontal gene transfer. Antimicrob. Agents Chemother. 472868-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Pastor, J. E., J. L. San Millan, and F. Moreno. 1994. The smallest known gene. Nature 369281. [DOI] [PubMed] [Google Scholar]
- 9.Guijarro, J. I., J. E. Gonzalez-Pastor, F. Baleux, J. L. San Millan, M. A. Castilla, M. Rico, F. Moreno, and M. Delepierre. 1995. Chemical structure and translation inhibition studies of the antibiotic microcin C7. J. Biol. Chem. 27023520-23532. [DOI] [PubMed] [Google Scholar]
- 10.Kazakov, T., A. Metlitskaya, and K. Severinov. 2007. Structure-activity analysis of translation inhibitor microcin C. J. Bacteriol. 1892114-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metlitskaya, A., T. Kazakov, A. Kommer, O. Pavlova, I. Krashenninikov, V. Kolb, I. Khmel', and K. Severinov. 2006. Aspartyl-tRNA synthetase is the target of peptidenucleotide antibiotic Microcin C. J. Biol. Chem. 28118033-18042. [DOI] [PubMed] [Google Scholar]
- 12.Metlitskaya, A. Z., G. S. Katrukha, A. S. Shashkov, D. A. Zaitsev, T. A. Egorov, and I. A. Khmel. 1995. Structure of microcin C51, a new antibiotic with a broad spectrum of activity. FEBS Lett. 357235-238. [DOI] [PubMed] [Google Scholar]
- 13.Miller, C. G. 1996. Protein degradation and proteolytic modification, p. 938-954. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 14.Moreno, F., J. E. Gonzalez-Pastor, M. R. Baquero, and D. Bravo. 2002. The regulation of microcin B, C and J. operons. Biochimie 84521-529. [DOI] [PubMed] [Google Scholar]
- 15.Novikova, M., A. Metlitskaya, K. Datsenko, T. Kazakov, A. Kazakov, B. Wanner, and K. Severinov. 2007. The Escherichia coli Yej transporter is required for the uptake of translation inhibitor microcin C. J. Bacteriol. 1898361-8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reader, J. S., P. T. Ordoukhanian, J. G. Kim, V. de Crecy-Lagard, I. Hwang, S. Farrand, and P. Schimmel. 2005. Major biocontrol of plant tumors targets tRNA synthetase. Science 3091533. [DOI] [PubMed] [Google Scholar]
- 17.Severinov, K., E. Semenova, A. Kazakov, T. Kazakov, and M. S. Gelfand. 2007. The post-translationally modified microcins. Mol. Microbiol. 61380-1394. [DOI] [PubMed] [Google Scholar]
- 18.Solbiati J., A. Chapman-Smith, J. L. Miller, C. G. Miller, and J. E. Cronan, Jr. 1999. Processing of the N termini of nascent polypeptide chains requires deformylation prior to methionine removal. J. Mol. Biol. 290607-614. [DOI] [PubMed] [Google Scholar]
- 19.Varshavsky, A. 1995. The N-end rule. Cold Spring Harbor Symp. Quant. Biol. 60461-478. [DOI] [PubMed] [Google Scholar]