Abstract
Salmonella enterica serovar Typhimurium delivers a variety of proteins via the Salmonella pathogenicity island 1 (SPI1)-encoded type III secretion system into host cells, where they elicit several physiological changes, including bacterial invasion, macrophage apoptosis, and enteropathogenesis. Once Salmonella has established a systemic infection, excess macrophage apoptosis would be detrimental to the pathogen, as it utilizes macrophages as vectors for systemic dissemination throughout the host. Therefore, SPI1 expression must be restricted to one or a few specific locations in the host. In the present study, we have demonstrated that the expression of this complex of genes is repressed by the ATP-dependent ClpXP protease, which therefore suppresses macrophage apoptosis. Depletion of ClpXP caused significant increases in the amounts of two SPI1-encoded transcriptional regulators, HilC and HilD, leading to the stimulation of hilA induction and therefore activation of SPI1 expression. Our evidence shows that ClpXP regulates cellular levels of HilC and HilD via the control of flagellar gene expression. Subsequent experiments demonstrated that the flagellum-related gene product FliZ controls HilD posttranscriptionally, and this in turn activates HilC. These findings suggest that the ClpXP protease coregulates SPI1-related virulence phenotypes and motility. ClpXP is a member of the stress protein family induced in bacteria exposed to hostile environments such as macrophages.
Salmonella enterica serovar Typhimurium is a facultative intracellular pathogen that causes gastroenteritis in humans and systemic diseases similar to typhoid fever in mice. Specific virulence factors encoded within the Salmonella pathogenicity islands (SPIs) are required for the development of salmonellosis (20, 24, 37, 43). One SPI, SPI1, encodes a type III secretion system with a multiprotein secretion apparatus, termed a needle complex, as well as effector proteins that are injected into the host cell cytoplasm via this apparatus (19, 32, 33, 56, 67). Various functions have been attributed to the SPI1 type III secretion system, including actin rearrangement, which causes the bacterium to be engulfed (9, 18, 47); macrophage apoptosis (7, 26, 30); and enteropathogenesis (65). After invasion of intestinal epithelial cells, Salmonella is able to disseminate to any tissue. It propagates within macrophages, a process that requires a second type III secretion system encoded on SPI2. Once Salmonella has established a systemic infection, excess macrophage apoptosis would be detrimental. During this systemic phase, the organisms need to repress and delay the onset of apoptosis to allow sufficient time for them to replicate, escape, and invade new macrophages. Thus, SPI1 gene expression must be suppressed during the systemic phase of infection. We have previously reported that a member of the ATP-dependent protease family, Lon, negatively regulates SPI1 gene expression (58, 60) and that this down-regulation is quite important for suppressing SPI1-dependent apoptosis sufficiently to allow time for Salmonella to replicate within the macrophages (57). It has been demonstrated that Lon is also important for down-regulating SPI1 gene expression in epithelial cells (5).
ATP-dependent proteolysis functions as a precise regulatory mechanism that limits the availability of key enzymes or critical regulatory proteins controlling gene expression. In addition to Lon, four ATP-dependent proteases, i.e., ClpAP, ClpXP, HslVU, and FtsH, have been found in gram-negative bacteria (for a review, see references 22 and 42). These are collectively called AAA+ (ATPases associated with diverse cellular activities plus) proteases (46). ClpXP is a bipartite protease responsible for degrading certain key regulatory proteins and aberrant translation products bearing the SsrA degradation tag, which is added cotranslationally to nascent polypeptides when ribosomes stall (16, 21, 23, 61). The ClpP component of ClpXP consists of two stacked heptameric rings, which enclose a central chamber containing the proteolytic active site. The ClpX component is a hexameric-ring ATPase that binds substrate proteins, denatures them, and translocates the unfolded polypeptides into the ClpP degradation chamber (48).
In the present study, we demonstrate that ClpXP negatively regulates the expression of SPI1 and hence suppresses macrophage apoptosis and the invasion of epithelial cells. The control of expression of the SPI1 type III secretion system is complex: several transcriptional regulators are present within the island. Among these, the master regulator is HilA, a member of the OmpR/ToxR family of transcriptional regulators, which can activate expression either directly or indirectly by increasing the expression of another regulator, InvF (2, 31, 37, 39). InvF, in a complex with the chaperone protein SicA, induces expression of the sic/sip operon (11). Two homologous proteins, HilC and HilD, both members of the AraC/XylS family of transcriptional regulators, induce hilA expression (12, 51, 53). HilC and HilD can each individually bind to the DNA immediately upstream of hilA, and it is believed that this binding leads to hilA expression (12, 52, 53). It is further suggested that HilD acts as a direct activator of hilA (6), as opposed to a derepressor as previously assumed (52, 53). It has also been demonstrated that HilC and HilD each induce the expression of the other as well as inducing their own promoters (40, 47). Recent data have shown that loss of HilD decreases hilC transcription but that loss of HilC does not significantly alter hilD expression (13, 15), suggesting that HilD may be at the top of the hierarchy of regulation of SPI1 expression. In addition to these SPI1-encoded regulators, genetic studies have identified many regulatory proteins encoded outside the island. One such regulator, Lon, has been shown to degrade both HilC and HilD, thus down-regulating SPI1 gene expression (58).
The structure of the SPI1 type III secretion apparatus is similar to the basal structure of the flagellum (4). Assembly of a flagellum requires the export of protein subunits from the cytoplasm to the outer surface of the cell by a mechanism that resembles those of secretion systems (4, 27). Furthermore, many components of the flagellar export apparatus share a high degree of homology with those of the type III secretion machinery. These findings suggest that these two type III machineries have a common evolutionary origin. We have previously reported that ClpXP negatively regulates flagellum biogenesis by controlling the turnover of the FlhD/FlhC complex, which functions as a master regulator at the apex of the transcription hierarchy of the flagellar regulon (61, 62).
Here, we have studied the mechanism by which ClpXP negatively controls SPI1 gene expression. The present results, coupled with our previous results (61, 62), indicate that ClpXP coregulates the expression of two different type III secretion systems, SPI1 related and flagellum related.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains used in this study are derivatives of S. enterica serovar Typhimurium χ3306 and are listed in Table 1. Plasmids pTKY559 and pTKY562 carry a hilC promoter-lacZ fusion and a hilD promoter-lacZ fusion, respectively (60). Plasmid pTKY651 carries a PA1lacO-1 promoter-hilD fusion (60).
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant propertiesa | Reference |
|---|---|---|
| χ3306 | Virulent wild type | 25 |
| CS2007 | clpP::Cm in χ3306 | 66 |
| CS2110 | hilA::Tn5 lacZY in χ3306 | 60 |
| CS2120 | hilA::Tn5 lacZY in CS2007 | This study |
| CS2462 | fliZ::Km in χ3306 | This study |
| CS2464 | fliZ::Km in CS2007 | This study |
| CS2609 | flhD::Tn10 in χ3306 | 61 |
| CS2610 | flhD::Tn10 in CS2007 | 61 |
| CS2724 | ΔhilC in χ3306 | 60 |
| CS2725 | ΔhilD in χ3306 | 60 |
| CS2732 | hilA::Tn5 lacZY in CS2724 | 60 |
| CS2733 | hilA::Tn5 lacZY in CS2725 | 60 |
| CS2802 | ΔhilC ΔhilD in χ3306 | 60 |
| CS2815 | hilA::Tn5 lacZY in CS2802 | 60 |
| CS3222 | hilA::Km in χ3306 | This study |
| CS3319 | clpP::Cm in CS2732 | This study |
| CS3320 | clpP::Cm in CS2733 | This study |
| CS3321 | clpP::Cm in CS2815 | This study |
| CS3322 | clpP::Cm in CS2724 | This study |
| CS3325 | clpP::Cm in CS2725 | This study |
| CS3328 | clpP::Cm in CS2802 | This study |
| CS3329 | fliZ::Km in CS2802 | This study |
Cm, chloramphenicol resistance; Km, kanamycin resistance.
To construct the fliZ disruption mutants, P22 phages were propagated on KK1397 (34) to transduce the fliZ::Km mutation. The lysate was used for infection of S. enterica serovar Typhimurium strains, and the transductants were selected by kanamycin resistance. Bacterial cells were routinely grown in L broth (1% Bacto tryptone [Difco, Detroit, MI], 0.5% Bacto yeast extract [Difco], 0.5% sodium chloride [pH 7.4]) or on L agar. The media were supplemented with chloramphenicol (25 μg ml−1), ampicillin (25 μg ml−1), kanamycin (25 μg ml−1), and/or nalidixic acid (25 μg ml−1) when necessary. Bacterial cells were grown aerobically in L broth.
DNA techniques.
DNA purification, ligation, restriction analysis, PCR amplification, DNA sequencing, and agarose gel electrophoresis were carried out as previously described (66).
Macrophage infection.
Cultured RAW264.7 macrophages were seeded onto a 24-well plate at a density of 2 × 105cells per well and incubated overnight at 37°C. Prior to infection, the macrophages were washed with Hanks balanced salt solution (HBSS) and challenged with Salmonella strains at a multiplicity of infection of 10. The plates were centrifuged for 5 min at 500 × g to enhance and synchronize infection, followed by incubation for 30 min at 37°C to permit phagocytosis. The free bacteria were removed by three washes with HBSS. Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum and 100 μg of gentamicin per ml was added, and the cells were incubated for 1.5 h at 37°C. The cells were washed with HBSS three times, followed by incubation with DMEM containing 10% fetal bovine serum and 10 μg of gentamicin per ml at 37°C.
Detection of histone-associated cytoplasmic DNA fragments after Salmonella-induced macrophage cell death.
At 6 h after infection, the macrophages were washed with HBSS and treated with 0.5 ml of lysis buffer for 30 min at room temperature. The lysates were centrifuged at 200 × g for 10 min at 4°C, and a 0.3-ml portion of the supernatant was taken for further assay. The samples were diluted 1:100, and the cytoplasmically located histones bound to fragmented DNA were quantified colorimetrically using Cell Death Detection ELISAPLUS (Roche Diagnostics).
Assay for invasion of epithelial cells.
Intestine-407 cells were invaded by Salmonella in 24-well tissue culture plates as described previously (58, 59) with some modifications. Bacterial cells were washed with HBSS and used to inoculate monolayers previously washed with HBSS, at a multiplicity of infection of 10. The monolayers were incubated for 2 h at 37°C and then washed thoroughly with HBSS and lysed with 0.2% Triton X-100 in phosphate-buffered saline to determine the total number of bacteria associated with the cultured cells. Alternatively, to assess the number of intracellular bacteria, the infected tissue culture cells were further incubated for 3 h in DMEM containing 100 μg of gentamicin per ml to eliminate extracellular bacteria before lysis with the Triton X-100 solution. Bacterial numbers were determined by plating the lysates on L agar plates after appropriate dilution.
Collection of whole-cell proteins and secreted proteins.
Bacterial cells were grown aerobically in 10 ml of L broth to an optical density at 600 nm of 1.0 at 37°C. To prepare whole-cell proteins, bacterial cells were harvested by centrifuging 1 ml of the culture and then suspended in 200 μl of sample buffer (36). Cell lysates (20 μl) were applied to each lane of a sodium dodecyl sulfate (SDS)-polyacrylamide gel. To prepare the proteins secreted into the medium, 7 ml of the same culture was centrifuged to remove cells. The filtered supernatant was mixed with prechilled trichloroacetic acid (final concentration, 10%), chilled on ice for 15 min, and centrifuged at 10,000 × g for 20 min. The pellet was washed once with acetone and then solubilized in 70 μl sample buffer (36). The sample (15 μl) was loaded on each lane and subjected to immunoblotting analysis.
Immunoblot analysis.
Equivalent numbers of bacterial cells were suspended in sample buffer (36), boiled for 5 min, and subjected to SDS-10% polyacrylamide gel electrophoresis. Separated proteins on the gels were transferred onto Immobilon-P polyvinylidene difluoride membranes (Millipore, Bedford, MA). Proteins were reacted with rabbit anti-SipB serum (1:12,500), anti-SipC serum (1:12,500), anti-HilA serum (1:12,500), anti-HilC serum (1:12,500), and anti-HilD serum (1:12,500), followed by alkaline phosphatase-conjugated anti-rabbit immunoglobulin G as the secondary antibody. The enzymatic reactions were performed in the presence of 300 μg ml−1 of nitroblue tetrazolium (Dojin, Kumamoto, Japan) and 150 μg ml−1 of bromochloroindolylphosphate (Amresco, Solon, OH). Anti-HilA, -HilC, -HilD, and -SipC antisera were previously established in our laboratory (60).
β-galactosidase assay.
The activity of β-galactosidase was determined by the method of Platt et al. (49). The enzyme units presented here are the averages from at least three independent assays.
RESULTS
Depletion of the ClpXP protease in S. enterica serovar Typhimurium stimulates the induction of apoptosis in macrophages.
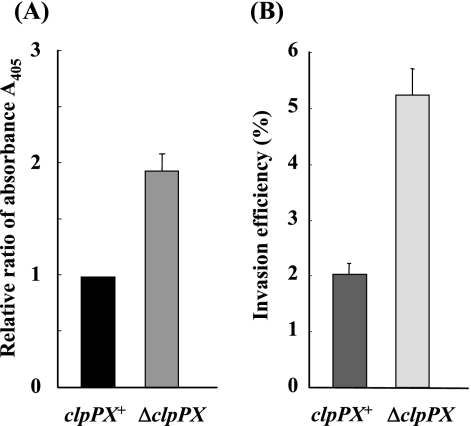
In a previous study, we characterized the genomic organization of the clpP region of S. enterica serovar Typhimurium pathogenic strain χ3306, clpP-clpX-lon, and constructed an insertional mutation in the clpP gene (66). The insertion of a chloramphenicol resistance cassette into clpP resulted in a polar mutation in clpX, which together with clpP constitutes an operon. However, the insertion evidently did not block the expression of lon preceded by its own promoter, which is downstream of clpX, because anti-Lon antiserum detected the corresponding protein in ΔclpPX mutant cells (66). Disruption of the clpP gene did not affect the cell growth rate (62, 66). During characterization of the virulence phenotype of the ΔclpPX mutant, we found cytoplasmic histone-associated DNA fragments, which constitute a marker for apoptotic cells (Fig. 1A), indicating that the mutant induces rapid and excessive macrophage apoptosis (6 h after infection). Rapid macrophage apoptosis by Salmonella infection is known to be mediated by an SPI1-encoded protein, SipB (26). Although Salmonella also induces SPI2-dependent, SPI1-independent apoptosis of macrophages, the execution of SPI2-dependent apoptosis is considerably delayed (18 to 24 h after infection) and is masked when the SPI1 genes are expressed (44, 64). Another aspect of SPI1-related virulence, the invasion of epithelial cells, was assessed in both the ΔclpPX mutant and the parental cells using cultured Intestine-407 cells (Fig. 1B). The level of invasion by ΔclpPX was significantly higher than that of invasion by the parental clpPX+ strain, suggesting that ClpXP is involved in the control of Salmonella invasiveness.
FIG. 1.
(A) Levels of cytoplasmic nucleosomal fragments in macrophages infected with S. enterica serovar Typhimurium strains χ3306 (clpPX+) and CS2007 (ΔclpPX). RAW264.7 cells were infected with bacteria at a multiplicity of infection of 10. At 6 h after infection, the levels of histone-associated cytoplasmic DNA fragments were measured by quantitative enzyme-linked immunosorbent assay using an antihistone antibody as described in Materials and Methods. The data are the means and standard derivations for each strain tested in triplicate. (B) Invasion efficiency of strains χ3306 (clpPX+) and CS2007 (ΔclpPX). Cultured Intestine-407 cells were used to assess invasiveness as described in Materials and Methods. The data are the means and standard derivations for each strain tested in triplicate. Invasion efficiency was expressed as the ratio of the number of intracellular bacteria to the number of bacteria adhering to the cultured cells.
ClpXP negatively regulates the expression of the SPI1 regulon.
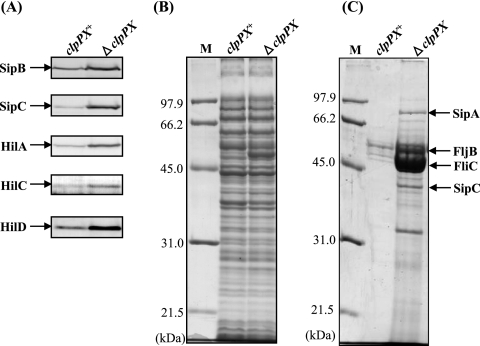
The enhanced ability of ΔclpPX to induce macrophage apoptosis and to invade epithelial cells suggested that clpPX deletion results in increased expression of SPI1 proteins. To examine this possibility, we initially used immunoblotting to compare the cellular levels of SipB and SipC, which are known to translocate into epithelial cells, in ΔclpPX cells and the isogenic clpPX + cells. SipB is necessary and sufficient for the induction of rapid macrophage apoptosis (26). SipC itself is known to cause bundling of actin filaments directly (19). The amounts of SipB and SipC in ΔclpPX cells were apparently higher than those in the clpPX+ cells (Fig. 2A). We have previously identified the SPI1 proteins SipA and SipC by mass spectrometry of secreted proteins separated by SDS-polyacrylamide gel electrophoresis (58). The Coomassie blue-stained gel patterns of the secreted proteins indicate that significantly higher levels of SipA and SipC are secreted in the ΔclpPX mutant (Fig. 2C). The 50-kDa band that accumulates in the ΔclpPX strain (Fig. 2B) is a flagellin protein, FliC, as previously identified (62). S. enterica serovar Typhimurium expresses two antigenically distinct flagellins encoded by fliC and fljB, and the alternative expression of these two genes results in an oscillation of phenotype known as phase variation, which occurs with frequencies ranging from 10−3 to 10−5 per bacterium per generation (55). Mass spectrometry demonstrated that cultures of strain χ3306 (clpPX+) include both the FliC and FljB proteins (62). The profile of secreted proteins shows that FliC and FljB are greatly increased by the disruption of clpPX (Fig. 2C). We then examined the cellular levels of HilA, a transcriptional activator with a central role in the regulatory hierarchy of SPI1 gene expression. The results show that the level of HilA is greatly increased in ΔclpPX cells. Two SPI1 gene products, HilC and HilD, have been shown to bind to the upstream repressing sequence of hilA to induce its transcription (12, 52, 53). In addition, overproduction of HilC and HilD has been shown to increase hilA expression significantly (13, 40). It is suggested that HilD directly activates hilA transcription (6). Therefore, the amounts of HilC and HilD in ΔclpPX cells were compared with those in the clpPX+ cells. The levels of both HilC and HilD were significantly increased by disruption of clpPX, suggesting that the ClpXP protease may down-regulate hilA transcription by a mechanism dependent on both HilC and HilD.
FIG. 2.
Effect of depletion of ClpXP protease on cellular levels of various SPI1 proteins. Bacterial cells of strains χ3306 (clpPX+) and CS2007 (ΔclpPX) were used. (A) Immunoblotting analysis of the cellular lysates using anti-SipB, anti-SipC, anti-HilA, anti-HilC, and anti-HilD sera. (B) Coomassie brilliant blue-stained SDS-10% polyacrylamide gel electrophoretic patterns of the same samples used for immunoblotting. Lane M contains the molecular mass standard. (C) Coomassie brilliant blue-stained gel patterns of the proteins secreted into the medium. The proteins were prepared as described in Materials and Methods.
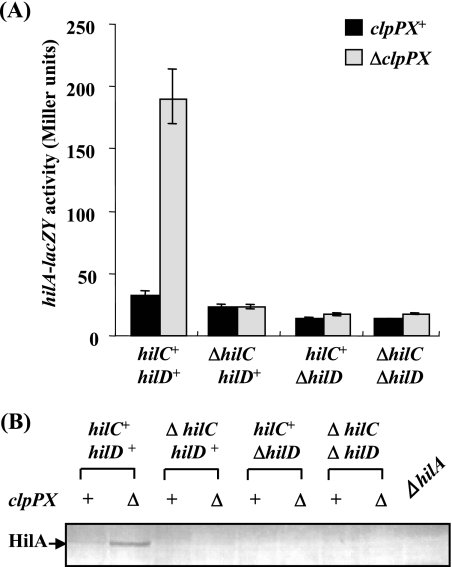
To examine this possibility, the effect of clpPX disruption on the expression of hilA was examined by use of a chromosomal lacZY fusion in strains with ΔhilC and/or ΔhilD backgrounds. As shown in Fig. 3A, disruption of clpPX increased hilA transcription approximately sixfold in hilC+ hilD+ cells. In contrast, the stimulation of hilA expression in ΔclpPX cells was completely suppressed by introducing the ΔhilC and/or ΔhilD mutation. The results of immunoblotting of cell lysates prepared from various strains with the mutation backgrounds used for transcriptional analysis (Fig. 3A) are shown in Fig. 3B. The apparent levels of HilA are consistent with the transcriptional analysis results. That is, the amount of HilA in the ΔclpPX mutant cells was larger than that in clpPX+ cells, and the accumulation of HilA in ΔclpPX cells was suppressed by introducing the hilC and/or hilD mutation. These results strongly suggest that ClpXP regulates hilA transcription through a pathway dependent on both HilC and HilD.
FIG. 3.
Effect of depletion of ClpXP protease on the expression of the hilA gene in the absence of the hilC and/or hilD gene. (A) The expression levels of a lacZ fusion to the hilA promoter were assayed. The values represent the means and standard deviations for samples tested at least in triplicate. The strains used were CS2110 (clpPX+ hilC+ hilD+), CS2120 (ΔclpPX hilC+ hilD+), CS2732 (clpPX+ ΔhilC hilD+), CS3319 (ΔclpPX ΔhilC hilD+), CS2733 (clpPX+ hilC+ ΔhilD), CS3320 (ΔclpPX hilC+ ΔhilD), CS2815 (clpPX+ ΔhilC ΔhilD), and CS3321 (ΔclpPX ΔhilC ΔhilD). (B) Immunoblotting analysis of the cellular lysates using anti-HilA serum. The strains used were χ3306 (clpPX+ hilC+ hilD+), CS2007 (ΔclpPX hilC+ hilD+), CS2724 (clpPX+ ΔhilC hilD+), CS3322 (ΔclpPX ΔhilC hilD+), CS2725 (clpPX+ hilC+ ΔhilD), CS3325 (ΔclpPX hilC+ ΔhilD), CS2802 (clpPX+ ΔhilC ΔhilD), CS3328 (ΔclpPX ΔhilC ΔhilD), and CS2110 (ΔhilA).
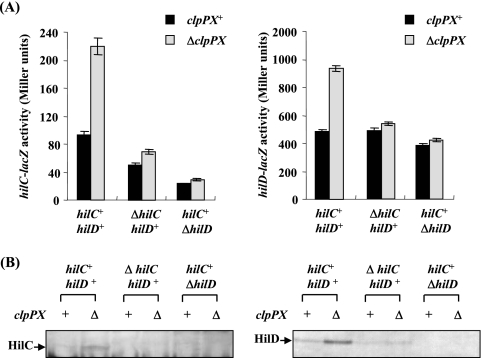
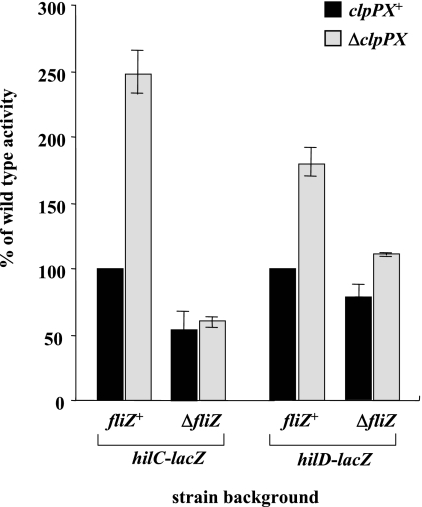
We then examined the effect of clpPX disruption on the transcription from hilC and hilD promoters using lacZ fusion on plasmids. As shown in Fig. 4A, disruption of clpPX increased hilC transcription 2.6-fold and hilD transcription 1.9-fold. HilC and HilD each activate the expression of the other as well as inducing their own promoters (39, 47), so we then measured transcription from the hilC and hilD promoters on plasmids in hilC- or hilD-disrupted backgrounds on the chromosome. The results show that the enhanced clpPX disruption effect on the transcription from hilC and hilD promoters was abolished by introducing either ΔhilC or ΔhilD mutations on the chromosome. The immunoblotting analyses show that the increased clpPX disruption effect on cellular levels of HilC and HilD disappeared when either ΔhilD or ΔhilC was introduced (Fig. 4B). Together, these findings suggest that the both HilC and HilD need to be accumulated to cause a significant stimulation of hilA expression by clpPX disruption.
FIG. 4.
Effect of depletion of ClpXP protease on the expression of the hilC and hilD genes in the absence of hilC or hilD gene. (A) The expression levels of lacZ fusions to hilC and hilD promoters were assayed. The values represent the means and standard derivations for samples tested at least in triplicate. Plasmid pTKY559 (hilC promoter-lacZ fusion) or pTKY562 (hilD promoter-lacZ fusion) was introduced into bacterial strains χ3306 (clpPX+ hilC+ hilD+), CS2007 (ΔclpPX hilC+ hilD+), CS2724 (clpPX+ ΔhilC hilD+), CS3322 (ΔclpPX ΔhilC hilD+), CS2725 (clpPX+ hilC+ ΔhilD), and CS3325 (ΔclpPX hilC+ ΔhilD). The resultant strains were used for determination of β-galactosidase activity. (B) Immunoblotting of the cellular lysates using anti-HilC and anti-HilD sera. The strains used were χ3306, CS2007, CS2724, CS3322, CS2725, and CS3325.
ClpXP regulates SPI1 gene expression via control of flagellar gene expression.
How does ClpXP control the levels of HilC and HilD? Our previous finding that it negatively regulates the expression of the flagellar regulon by controlling the turnover of the master regulator, FlhD/FlhC (62), allowed us to hypothesize that it may regulate HilC and HilD by controlling flagellar gene expression. In S. enterica serovar Typhimurium, the transcription of more than 50 genes related to flagellum biogenesis forms a highly ordered cascade divided into three classes, 1, 2, and 3 (35). At the apex of the transcription hierarchy lies the sole operon flhDC. Its products, FlhD and FlhC, act together in a FlhD/FlhC hetero-oligomer to activate the promoter of class 2 genes (28, 38). Our previous study demonstrated that ClpXP recognizes and degrades the FlhD/FlhC hetero-oligomer, resulting in down-regulation of the flagellar regulon (61). One of seven operons in class 2, fliAZY, encodes an alternative sigma factor, FliA (σ28), that is required for the transcription of class 3 genes, producing FliZ and FliY. Previous studies have reported that the fliA::Tn5 mutation significantly decreases expression of the hilA-lacZ transcriptional fusion and that expression is restored by a trans-complementing plasmid expressing fliZ, suggesting that FliZ is required for hilA expression (29, 41). Therefore, ClpXP may possibly control the cellular levels of the transcriptional regulators HilC and HilD by a pathway strongly dependent on FliZ. To examine this possibility, we measured the levels of HilC and HilD in clpPX+ and ΔclpPX cells after introducing a ΔfliZ mutation. The results (Fig. 5) show that the enhancement of cellular HilC and HilD levels by clpPX disruption disappeared in the ΔfliZ mutant, suggesting that the control of HilC and HilD levels by ClpXP depends on the function of FliZ. Owing to the effect of the ΔfliZ mutation on HilC and HilD levels, the enhancement of HilA by clpPX disruption was also extinguished. As expected, disruption of the flhDC operon, encoding the master regulator for expression of the flagellar regulon, blocked the increase of HilC and HilD levels caused by clpPX disruption. Taking these results together, it is suggested that SPI1 expression is regulated by ClpXP through a mechanism by which FliZ controls the cellular levels of the transcriptional regulators HilC and HilD.
FIG. 5.
Effects of disruption of fliZ or flhDC on enhancement of cellular levels of SPI1 regulator proteins by depletion of ClpXP. Whole-cell lysates were separated on SDS-10% polyacrylamide gels and then subjected to immunoblotting using anti-HilA, anti-HilC, or anti-HilD serum. The bacterial strains used were χ3306 (clpPX+ fliZ+ flhDC+), CS2007 (ΔclpPX fliZ+ flhDC+), CS2462 (clpPX+ ΔfliZ flhDC+), CS2464 (ΔclpPX ΔfliZ flhDC+), CS2609 (clpPX+ fliZ+ ΔflhDC), CS2610 (ΔclpPX fliZ+ ΔflhDC), CS2802 (ΔhilC ΔhilD), and CS3222 (ΔhilA).
Transcriptional analysis of hilC and hilD in fliZ+ and ΔfliZ cells.
To examine whether FliZ controls the transcription of hilC and hilD, we measured the efficiency of transcription from hilC and hilD promoters using lacZ fusions in cells with different genetic backgrounds. The results (Fig. 6) show that the ΔfliZ mutation moderately decreased the transcription of hilD in the clpPX+ background, notwithstanding the marked decrease in the amount of HilD protein caused by this mutation (Fig. 5), suggesting that FliZ may regulate HilD production at a posttranscriptional and/or posttranslational level.
FIG. 6.
Levels of hilC and hilD transcription in the absence of ClpXP and/or FliZ in cells. The levels of expression of a lacZ fusion to the hilC or hilD promoter in cells with different genetic backgrounds were assayed. Plasmid pTKY559 (hilC promoter-lacZ fusion) or pTKY562 (hilD promoter-lacZ fusion) was introduced into bacterial strains χ3306 (clpPX+ fliZ+), CS2007 (ΔclpPX fliZ+), CS2462 (clpPX+ ΔfliZ), and CS2464 (ΔclpPX ΔfliZ). The resultant stains were used for determination of β-galactosidase activity. The values represent the means and standard deviations for samples tested at least in triplicate.
In contrast, the ΔfliZ mutation significantly decreased the transcription of hilC in cells with a clpPX+ background. It has been demonstrated that HilC and HilD each significantly activate the expression of the other when they are overproduced (13, 40). It has also recently been reported that loss of HilD decreases hilC transcription but that loss of HilC does not significantly alter hilD expression, so it is proposed that HilD is the apex of regulation of SPI1 expression (13, 15). Therefore, the decrease of hilC transcription caused by the ΔfliZ mutation in cells with a clpPX+ background could be explained by the decreased amount of HilD.
FliZ modulates HilD at the posttranscriptional level.
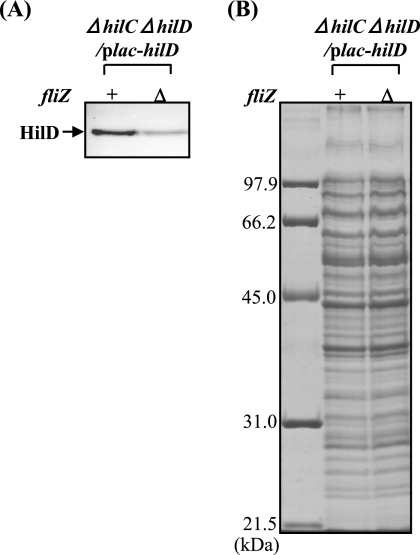
To examine whether FliZ is involved in the posttranscriptional control of HilD, we decided to compare the levels of HilD protein in fliZ+ and ΔfliZ cells where hilD is expressed under the regulation of the PA1lacO-1 promoter system. For this purpose, strains CS2802 (fliZ+ ΔhilC ΔhilD) and CS3329 (ΔfliZ ΔhilC ΔhilD) were transformed with the plasmid pTKY651, in which the hilD promoter is replaced by the PA1lacO-1 promoter. The immunoblotting results in Fig. 7 show that HilD was detectable in the absence of IPTG (isopropyl-β-d-thiogalactopyranoside), a condition in which hilD was expressed by read-through from the PA1lacO-1 promoter. The results show that disruption of fliZ significantly decreased the cellular level of HilD even when the corresponding gene was expressed by the PA1lacO-1 promoter system, suggesting that HilD is controlled posttranscriptionally by FliZ. To test this inference, we analyzed the in vivo stability of HilD. There was no difference in HilD half-life between the fliZ+ and ΔfliZ cells, suggesting that HilD is not controlled posttranslationally by FliZ (data not shown).
FIG. 7.
Cellular levels of HilD protein expressed by a PA1lacO-1 promoter-hilD fusion in wild type and FliZ-depleted cells. Plasmid pTKY651 (PA1lacO-1 promoter-hilD fusion) was introduced into bacterial strains CS2802 (fliZ+ ΔhilC ΔhilD) and CS3329 (ΔfliZ ΔhilC ΔhilD). Cells of the resultant strains were grown to an optical density at 600 nm of 1.0, collected, lysed, and run on SDS-10% polyacrylamide gels. The separated proteins were transferred to a membrane and then immunostained with anti-HilD antiserum (A). Coomassie brilliant blue-stained gel electrophoretic patterns of the same samples used for immunoblotting are also shown (B). The leftmost lane in panel B contains molecular mass standards.
These findings, coupled with our previous results (61, 62), suggest that the overproduction of FliZ due to accumulation of the FlhD/FlhC master regulator of the flagellar regulon in clpPX-deleted cells results in the accumulation of HilD by posttranscriptional control and consequently leads to increased SPI1 expression.
DISCUSSION
Expression of SPI1 genes is tightly regulated at several stages in a complex manner by regulators within and outside the island. As summarized in Fig. 8, HilA is the central regulator in the overall scheme of SPI1 regulation and is known to activate expression of the prg/org and inv/spa operons. Read-through from inv/spa leads to the activation of sic/spa. InvF, in a complex with the chaperone protein SicA, also induces expression of the sic/sip operon, so HilA activates genes encoding all the components necessary for a functional SPI1 type III secretion system (2, 11, 12, 15, 37). The products of other genes within SPI1, hilC and hilD, have been shown to activate hilA expression (12, 50, 53). HilC and HilD are also capable of activating hilC and hilD expression independently of each other, and they act in a complex regulatory loop to control hilA expression (13, 50, 53).
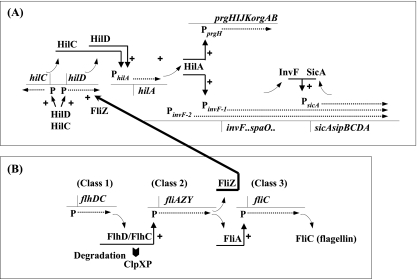
FIG. 8.
A model of the coordinated regulation of SPI1 gene expression and flagellar gene expression by ClpXP protease. (A) Regulatory cascade of SPI1 gene expression. HilC and HilD bind directly upstream of the master regulator gene hilA to induce its expression. HilA directly activates the SPI1-encoded prg/org and inv/spa operons by binding just upstream of PprgH and PinvF-1. The inv/spa transcript reads through the sic/sip operon. Activation of PinvF-1 leads to production of InvF, in a complex with the chaperone protein SicA, and then induces expression of the sic/sip operon. (B) Regulatory cascade of flagellar gene expression. The products, FlhD and FlhC, act together in a FlhD/FlhC complex as the master regulator at the apex of a transcription hierarchy comprising three classes of genes. FliA, which is an alternative sigma factor, σ28, is required to transcribe the class 3 genes. ClpXP degrades the FlhD/FlhC complex, leading to the down-regulation of flagellar regulon expression. FliZ, which is encoded by the fliAZY operon of the flagellar regulon, activates the expression of hilD at the posttranscriptional level, leading to the stimulation of SPI1 gene expression. ClpXP negatively regulates the expression of SPI1 genes through the repression of flagellar regulon expression. See the text for details and references.
In the present study, we have demonstrated that ClpXP negatively regulates SPI1 gene expression. We observed that depletion of ClpXP caused a significant increase in the amount of HilC and HilD proteins (Fig. 2), which is associated with increased expression of all SPI1 genes. Immunoblotting actually revealed that HilA and proteins encoded by genes in the sic/sip operon, SipB and SipC, are greatly increased in the ΔclpPX mutant. We hypothesized that ClpXP may regulate HilC and HilD through the control of the flagellar regulon, since ClpXP has been shown to regulate its expression negatively by degrading the FlhD/FlhC master regulator, which functions at the apex of the transcription hierarchy of the regulon (61, 62). Subsequent experiments showed that the enhancement of cellular levels of HilC and HilD by clpPX disruption seemed to be caused by an increase in the flagellar protein FliZ, encoded by the class 2 fliAZY operon. That is to say, disruption of fliZ abolished the increase of HilC and HilD caused by deleting clpPX (Fig. 5). Transcriptional analysis of hilC and hilD in the fliZ+ and ΔfliZ backgrounds suggested that FliZ controls HilC at the transcriptional level and may regulate HilD production at the posttranscriptional level (Fig. 6). Immunoblotting demonstrated that the ΔfliZ mutation decreased the cellular level of HilD even when hilD was transcribed from the PA1lacO-1 promoter (Fig. 7), suggesting that hilD is controlled posttranscriptionally. Furthermore, it seems unlikely that FliZ controls HilD at the posttranslational level (our unpublished data). Therefore, it can be concluded that FliZ controls HilD at the posttranscriptional level. The function of FliZ is unknown, and its predicted product shows no significant homology to any known protein or structural motif.
When the levels of transcription from the hilC and hilD promoters are compared between clpPX+ cells and ΔclpPX cells in the fliZ+ background, it is evident that clpPX disruption increases the expression of both hilC and hilD (Fig. 4A and Fig. 6). It has been demonstrated that HilD is at the top of the hierarchy of the SPI1 regulatory loop and has a predominant role, though apparently it is not sufficient on its own to activate SPI1 (13, 15). Production of HilD leads to transcriptional activation of HilC, and each activates the expression of both (13, 40). Therefore, the increased transcription of hilC and hilD in the ΔclpPX cells could be explained by amplification of the regulatory loop in which FliZ triggered an increase in the amount of HilD via posttranscriptional control. On the other hand, the stimulation of hilA expression by clpPX disruption was abrogated by introducing the ΔhilC mutation (Fig. 3). This could be because HilD did not accumulate in the ΔhilC ΔclpPX double mutant cells (Fig. 4). As stated above, it is known that HilC and HilD can activate the expression of hilC and hilD independently of each other as well as inducing their own promoters (40, 47). Therefore, it is likely that the moderate effect on HilD production by FliZ needs to be amplified in the regulatory loop to cause significant stimulation of hilA expression in the ΔclpPX mutant cells.
In addition to HilC and HilD, RtsA, which belongs to the AraC/XylS family of transcriptional regulators, has been shown to activate expression of SPI1 genes by binding upstream of hilA to induce its expression (13, 14). HilC, HilD, and RtsA are all also capable of activating expression of hilC, hilD, and rtsA independently of each other, constituting a complex feed-forward regulatory loop that controls hilA expression (13, 14, 15). Therefore, it can be speculated that RtsA acts as an amplifier, leading to the overexpression of all SPI1 genes in the ΔclpPX mutant cells.
SPI1 is a complex regulatory system, and many different signals have been shown to feed into the network. The current report suggests that all the global regulators seem to control hilA expression in a HilD-dependent manner (15). One such regulator is the posttranscriptional regulatory protein CsrA, which binds to the messages of its targets and alters mRNA stability (51). In the ΔcsrA mutant, the expression of SPI1 genes is greatly reduced (1). SirA in the BarA/SirA two-component regulatory system acts by inducing the expression of two small RNAs, the CsrB and CsrC RNAs, which are antagonistic to CsrA. Overproduction of SirA induces the expression of a hilA-lacZ transcriptional fusion only when HilD is present (13). Thus, SirA induction of csrBC prevents CsrA action, indirectly activating hilD expression posttranscriptionally (17). The RNA chaperone Hfq has recently been recognized as a major posttranscriptional regulator of bacterial gene expression that participates in numerous regulatory pathways (63). A recent report demonstrates that the Δhfq mutation drastically reduces Salmonella invasiveness (54). It has also demonstrated that Δhfq cells have sevenfold-reduced levels of hilC, hilD, and rtsA mRNAs, suggesting that Hfq affects signal transmission further upstream in the SPI1-activating cascade.
HilE is a major negative regulator of SPI1 expression. Deletion of hilE increases expression of hilA only when HilD is present. Although the mechanism of hilE action is not well understood, the results from bacterial two-hybrid studies suggest that HilE binds directly to HilD, preventing its action (3). hilE is regulated by several systems that feed into the SPI1 regulation circuit. The systems that negatively regulate SPI1 expression, e.g., the two-component PhoP/PhoQ and PhoR/PhoB regulatory systems and FimZY for type 1 fimbrial expression, seem to function primarily through HilE (3, 15). Lon protease is a powerful negative regulator of SPI1 expression (58, 60). Depletion of Lon increases hilA expression 40-fold and causes a 10-fold increase in invasiveness. Lon has been shown to regulate HilD posttranslationally, specifically by degradation. Taking these findings together, we suggest that HilD production is controlled largely at both the posttranscriptional and posttranslational levels and that this is the key to SPI1 regulation. To understand SPI1 regulation completely, the molecular mechanism by which hilD is regulated at the posttranscriptional level must be studied in detail.
ClpXP is also known to be a member of the family of stress proteins, which are induced in response to hostile environments (for a review, see reference 45). It has been demonstrated that stress proteins are selectively induced in S. enterica serovar Typhimurium growing within macrophages, where the bacteria are exposed to a variety of bactericidal mechanisms (8). We previously reported that hilA expression was repressed in macrophages after phagocytosis but that this gene was continuously expressed in a S. enterica serovar Typhimurium strain with a defect in Lon, another member of the stress protein family, growing within macrophages (57). Furthermore, we demonstrated that this derepression of SPI1 genes by a Lon-depleted mutant led to rapid and massive macrophage apoptosis by a mechanism involving caspases 1 and 3 (57). Once a systemic Salmonella infection has been established, excess apoptosis of macrophages, upon which the organism is reliant, would be detrimental to the bacteria. Thus, SPI1 gene expression must be controlled in bacteria growing within macrophages to suppress apoptosis sufficiently to allow time for the bacterium to replicate, escape, and invade new macrophages, leading to systemic infection. To ensure this, Salmonella can take advantage of a unique condition in the macrophage to repress SPI1. The ClpXP and Lon proteases may contribute to the down-regulation of SPI1 expression within macrophages. A recent report has demonstrated that expression of both fliC, encoding a flagellar filament, and fliA, encoding σ28, is repressed during intracellular growth and that this repression depends on functional ClpXP (10). The increase in ClpXP in S. enterica serovar Typhimurium by its response to the hostile environment within macrophages after phagocytosis would ensure the repression of both SPI1 genes and the flagellar regulon.
Acknowledgments
We thank T. Tomoyasu, A. Tokumitsu, and A. Suzuki for technical assistance.
This work was supported by grants-in-aid for scientific research (17390125) and research on priority areas (19041015) from the Ministry for Education, Culture, Sports, Sciences and Technology of the Japanese Government.
Footnotes
Published ahead of print on 1 February 2008.
REFERENCES
- 1.Altier, C., M. Suemoto, and S. D. Lawhon. 2000. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect. Immun. 686790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bajaj, V., C. Hwang, and C. A. Lee. 1995. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18715-727. [DOI] [PubMed] [Google Scholar]
- 3.Baxter, M. A., and B. D. Jones. 2005. The fimYZ genes regulate Salmonella enterica serovar Typhimurium invasion in addition to type 1 fimbrial expression and bacterial motility. Infect. Immun. 731377-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blocker, A., K. Komiyama, and S. Aizawa. 2003. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl. Acad. Sci. USA 1003027-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boddicker, J. D., and B. D. Jones. 2004. Lon protease activity causes down-regulation of Salmonella pathogenicity island 1 invasion gene expression after infection of epithelial cells. Infect. Immun. 722002-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boddicker, J. D., B. M. Knosp, and B. D. Jones. 2003. Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators. J. Bacteriol. 185525-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan, M. A., and B. T. Cookson. 2000. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol. Microbiol. 3831-40. [DOI] [PubMed] [Google Scholar]
- 8.Buchmeier, N. A., and F. Heffron. 1990. Induction of Salmonella stress proteins upon infection of macrophages. Science 248730-732. [DOI] [PubMed] [Google Scholar]
- 9.Collazo, C. M., and J. E. Galán. 1997. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol. Microbiol. 24747-756. [DOI] [PubMed] [Google Scholar]
- 10.Cummings, L. A., W. D. Wilkerson, T. Bergsbaker, and B. T. Cookson. 2006. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol. Microbiol. 61795-809. [DOI] [PubMed] [Google Scholar]
- 11.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 201850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichelberg, K., W. D. Hardt, and J. E. Galán. 1999. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol. Microbiol. 33139-152. [DOI] [PubMed] [Google Scholar]
- 13.Ellermeier, C. D., J. R. Ellermeier, and J. M. Slauch. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57691-705. [DOI] [PubMed] [Google Scholar]
- 14.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1855096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellermeier, J. R., and J. M. Slauch. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 1024-29. [DOI] [PubMed] [Google Scholar]
- 16.Flynn, J. M., S. B. Neher, Y. I. Kim, R. T. Sauer, and T. A. Baker. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11671-683. [DOI] [PubMed] [Google Scholar]
- 17.Fortune, D. R., M. Suemoto, and C. Alter. 2006. Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect. Immun. 74331-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu, Y., and J. E. Galán. 1998. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol. Microbiol. 27359-368. [DOI] [PubMed] [Google Scholar]
- 19.Galán, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 2841322-1328. [DOI] [PubMed] [Google Scholar]
- 20.Galán, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 866383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez, M., F. Rasulova, M. R. Maurizi, and R. Woodgate. 2000. Subunit-specific degradation of the UmuD/D′ heterodimer by the ClpXP protease: the role of trans recognition in UmuD′ stability. EMBO J. 195251-5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottesman, S. 2003. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell Dev. Biol. 19565-587. [DOI] [PubMed] [Google Scholar]
- 23.Gottesman, S., E. Roche, Y.-N. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 121338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groisman, E. A., and H. Ochman. 1993. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 123779-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gulig, P. A., and R. Curtiss III. 1987. Plasmid-associated virulence of Salmonella typhimurium. Infect. Immun. 552891-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hersh, D., D. M. Monack, M. R. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA 962396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hueck, C. J. 1998. The type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikebe, T., S. Iyoda, and K. Kutsukake. 1999. Structure and expression of the fliA operon of Salmonella typhimurium. Microbiology 1451389-1396. [DOI] [PubMed] [Google Scholar]
- 29.Iyoda, S., T. Kamidoi, K. Hirose, K. Kutsukake, and H. Watanabe. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 3081-90. [DOI] [PubMed] [Google Scholar]
- 30.Jesenberger, V., K. J. Procyk, J. Yuan, S. Reipert, and M. Baccarini. 2000. Salmonella-induced caspase-2 activation in macrophages: a novel mechanism in pathogen-mediated apoptosis. J. Exp. Med. 1921035-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaniga, K., J. C. Bossio, and J. E. Galán. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13555-568. [DOI] [PubMed] [Google Scholar]
- 32.Kimbrough, T. G., and S. I. Miller. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. USA 9711008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galán, and S. I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280602-605. [DOI] [PubMed] [Google Scholar]
- 34.Kutsukake, K., T. Ikebe, and S. Yamamoto. 1999. Two novel regulatory genes, fliT and fliZ, in the flagellar regulon of Salmonella. Genes Genet. Syst. 74287-292. [DOI] [PubMed] [Google Scholar]
- 35.Kutsukake, K., Y. Ohya, and T. Iino. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laemmli, U. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 37.Lee, C. A., B. D. Jones, and S. Falkow. 1992. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc. Natl. Acad. Sci. USA 891847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, X., and P. Matsumura. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 1767345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lostroh, C. P., and C. A. Lee. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 31281-1291. [DOI] [PubMed] [Google Scholar]
- 40.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1832733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1821872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maurizi, M. R. 1992. Proteases and protein degradation in Escherichia coli. Experientia 48178-200. [DOI] [PubMed] [Google Scholar]
- 43.Mills, D. M., V. Bajaj, and C. A. Lee. 1995. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol. Microbiol. 15749-759. [DOI] [PubMed] [Google Scholar]
- 44.Monack, D. M., W. W. Navarre, and S. Falkow. 2001. Salmonella-induced macrophage death: the role of caspase-1 in death and inflammation. Microbes Infect. 31201-1212. [DOI] [PubMed] [Google Scholar]
- 45.Morimoto, R. I., A. Tissieres, and C. Georgopoulos. 1994. Progress and perspectives on the biology of heat shock proteins and molecular chaperone, p. 1-30. In R. I. Morimoto, A. Tissieres, and C. Georgopoulos (ed.), The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Neuwald, A. F., L. Aravind, J. L. Spouge, and E. V. Koonin. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 927-43. [PubMed] [Google Scholar]
- 47.Olekhnovich, I. N., and R. J. Kadner. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 1844148-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ortega, J., S. K. Singh, T. Ishikawa, M. R. Maurizi, and A. C. Steven. 2000. Visualization of substrate binding and translocation by the ATP-dependent protease, ClpXP. Mol. Cell 61515-1521. [DOI] [PubMed] [Google Scholar]
- 49.Platt, T., B. Mueller-Hill, and J. H. Miller. 1972. Assay of β-galactosidase, p. 325-355. In J. H. Miller (ed.), Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 50.Rakeman, J. L., H. R. Bonifield, and S. I. Miller. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 1813096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romeo, T., M. Gong, M. Y. Liu, and A. M. Brun-Zinkernagel. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 1754744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schechter, L. M., and C. A. Lee. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 401289-1299. [DOI] [PubMed] [Google Scholar]
- 53.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32629-642. [DOI] [PubMed] [Google Scholar]
- 54.Sittka, A., V. Pfeiffer, K. Tedin, and J. Vogel. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 63193-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stocker, B. A. D. 1949. Measurement of the rate of mutation of flagellar antigenic phase in Salmonella typhimurium. J. Hyg. 47398-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sukhan, A., T. Kubori, J. Wilson, and J. E. Galan. 2001. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J. Bacteriol. 1831159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takaya, A., A. Suzuki, Y. Kikuchi, M. Eguchi, E. Isogai, T. Tomoyasu, and T. Yamamoto. 2005. Derepression of Salmonella pathogenicity island 1 genes within macrophages leads to rapid apoptosis via caspase-1- and caspase-3-dependent pathways. Cell. Microbiol. 779-90. [DOI] [PubMed] [Google Scholar]
- 58.Takaya, A., T. Tomoyasu, A. Tokumitsu, M. Morioka, and T. Yamamoto. 2002. The ATP-dependent Lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J. Bacteriol. 184224-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takaya, A., T. Tomoyasu, H. Matsui, and T. Yamamoto. 2004. The DnaK/DnaJ chaperone machinery of Salmonella enterica serovar Typhimurium is essential for invasion of epithelial cells and survival within macrophages, leading to systemic infection. Infect. Immun. 72690-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takaya, A., Y. Kubota, E. Isogai, and T. Yamamoto. 2005. Degradation of the HilC and HilD regulator proteins by ATP-dependent Lon protease leads to downregulation of Salmonella pathogenicity island 1 gene expression. Mol. Microbiol. 55839-852. [DOI] [PubMed] [Google Scholar]
- 61.Tomoyasu, T., A. Takaya, E. Isogai, and T. Yamamoto. 2003. Turnover of FlhD and FlhC, master regulator proteins for Salmonella flagellum biogenesis, by the ATP-dependent ClpXP protease. Mol. Microbiol. 48443-452. [DOI] [PubMed] [Google Scholar]
- 62.Tomoyasu, T., T. Ohkishi, Y. Ukyo, A. Tokumitsu, A. Takaya, M. Suzuki, K. Sekiya, H. Matsui, K. Kutsukake, and T. Yamamoto. 2002. The ClpXP ATP-dependent protease regulates flagellum synthesis in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valentin-Hansen, P., M. Erickson, and C. Udesen. 2004. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 511525-1533. [DOI] [PubMed] [Google Scholar]
- 64.van der Velden, A. W., S. W. Lindgren, M. J. Worley, and F. Heffron. 2000. Salmonella pathogenicity island 1-independent induction of apoptosis in infected macrophages by Salmonella enterica serotype Typhimurium. Infect. Immun. 685702-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36997-1005. [DOI] [PubMed] [Google Scholar]
- 66.Yamamoto, T., H. Sashinami, A. Takaya, T. Tomoyasu, H. Matsui, T. Hanawa, S. Kamiya, and A. Nakane. 2001. Disruption of the genes for ClpXP protease in Salmonella enterica serovar Typhimurium results in a persistent infection in mouse, and development of the persistence requires endogenous gamma interferon and tumor necrosis factor alpha. Infect. Immun. 693164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou, D., and J. E. Galán. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 31293-1298. [DOI] [PubMed] [Google Scholar]