Abstract
Although a whole arsenal of mechanisms are potentially involved in metabolic regulation, it is largely uncertain when, under which conditions, and to which extent a particular mechanism actually controls network fluxes and thus cellular physiology. Based on 13C flux analysis of Escherichia coli mutants, we elucidated the relevance of global transcriptional regulation by ArcA, ArcB, Cra, CreB, CreC, Crp, Cya, Fnr, Hns, Mlc, OmpR, and UspA on aerobic glucose catabolism in glucose-limited chemostat cultures at a growth rate of 0.1 h−1. The by far most relevant control mechanism was cyclic AMP (cAMP)-dependent catabolite repression as the inducer of the phosphoenolpyruvate (PEP)-glyoxylate cycle and thus low tricarboxylic acid cycle fluxes. While all other mutants and the reference E. coli strain exhibited high glyoxylate shunt and PEP carboxykinase fluxes, and thus high PEP-glyoxylate cycle flux, this cycle was essentially abolished in both the Crp and Cya mutants, which lack the cAMP-cAMP receptor protein complex. Most other mutations were phenotypically silent, and only the Cra and Hns mutants exhibited slightly altered flux distributions through PEP carboxykinase and the tricarboxylic acid cycle, respectively. The Cra effect on PEP carboxykinase was probably the consequence of a specific control mechanism, while the Hns effect appears to be unspecific. For central metabolism, the available data thus suggest that a single transcriptional regulation process exerts the dominant control under a given condition and this control is highly specific for a single pathway or cycle within the network.
Some parts of metabolic networks are organism specific, but the core network is highly conserved. Almost all aerobic bacteria have a similar set of about 100 enzymes that catalyze the formation of biosynthetic building blocks, energy, and cofactors. This core network is ubiquitous because all specialized catabolic pathways finally merge into one or more of the common intermediates. Obviously, not all core reactions are simultaneously active and the evolved regulatory structure of an organism ensures appropriate and flexible activity of the various enzymes under the conditions normally encountered. One key regulatory task is to direct carbon fluxes such that all of the necessary biomass components are synthesized at the appropriate stoichiometry and rate from a wide range of substrates.
Transcriptional regulation is generally considered the main microbial control mechanism, and a complicated network of global and specific transcription factors could potentially manage this distribution of fluxes (31). Such transcriptional control of metabolic activity is firmly established for the degradation and biosynthesis branches of the network. A typical example is aromatic amino acid biosynthesis with fine tuning of flux into the various branches by allosteric feedback inhibition of key enzymes but transcriptional regulation as the general control mechanism for absence or presence of the pathway (41). The situation is much less clear, however, for the central metabolic network that catalyzes the major flows of carbon under all conditions, but often in opposite directions. In this study, we attempted to quantify the relevance of global transcriptional regulation for the control of intracellular carbon fluxes.
Previously, we investigated the global transcriptional control of glucose metabolism in aerobic batch cultures of Escherichia coli, with the main result that only one of seven global regulators exerted specific control on any central metabolic flux; i.e., ArcA repressed the tricarboxylic acid (TCA) cycle flux by more than 60% (40). Under these conditions, high extracellular glucose concentrations invoke the catabolite repression response. This is a general microbial response in which a number of mechanistically distinct transcriptional regulation processes enable organisms to feast on their preferred substrate and minimize the metabolic load for expressing the uptake and catabolic machinery of less preferred substrates (5, 27, 46). Since the dominance of catabolite repression overrides many other regulation processes, we focus on the transcriptional control of glucose metabolism at low or absent catabolite repression. Among the few conditions that allow the study of this situation are glucose-limited chemostat cultures are low dilution rates (10, 11, 32, 37), with the additional benefit that such steady-state cultivation avoids growth rate-dependent effects of mutants that may obscure results in batch cultures (21).
Here, we systematically quantify the control of 12 global regulators of the distribution of metabolic fluxes in otherwise isogenic E. coli mutants (4) at a dilution rate of 0.1 h−1 in mini-scale (35), glucose-limited chemostat cultures. These regulators are involved in oxygen sensing (ArcA, ArcB, Fnr) (29, 53), catabolite repression (Cra, CreB, CreC Crp, Cya) (3, 45, 46), regulation of carbohydrate utilization (Mlc) (42), modulation of carbon flow at growth arrest (UspA) (39), global gene regulation and chromosome organization (Hns) (2), and osmotic regulation (OmpR) (33). To identify regulation mechanisms that actually control the distribution of fluxes, we quantified in vivo enzyme activity through 13C-based metabolic flux analysis (47, 48). A particular focus was on the newly discovered phosphoenolpyruvate (PEP)-glyoxylate cycle that is active under glucose hunger conditions in slow-growing E. coli chemostat cultures (12, 25, 35, 43) but whose transcriptional regulation remained elusive.
MATERIALS AND METHODS
Strains and growth conditions.
All mutants were obtained from the KEIO knockout collection in the E. coli BW25113 background (4, 26), a close relative of MG1655. For clarity, we use a mutant nomenclature that reflects the deleted genes (Table 1). For complementation, the crp gene was amplified, from the start codon to the stop codon, by PCR from E. coli BW25113 genomic DNA and cloned behind the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter of plasmid pTrc99A (Pharmacia). Expression was induced by cultivation in the presence of 1 mM IPTG.
TABLE 1.
E. coli strains used in this study
| Straina | Relevant genotype | Reference |
|---|---|---|
| BW25113 | lacIqrrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | 7 |
| ArcA (JW4364) | BW25113 ΔarcA::kan | 26 |
| ArcB (JW3177) | BW25113 ΔarcB::kan | 26 |
| Cra (JW0078) | BW25113 Δcra::kan | 26 |
| CreB (JW4361) | BW25113 ΔcreB::kan | 26 |
| CreC (JW4362) | BW25113 ΔcreC::kan | 26 |
| Crp (JW3320) | BW25113 Δcrp::kan | 26 |
| Cya (JW3778) | BW25113 Δcya::kan | 26 |
| Fnr (JW1328) | BW25113 Δfnr::kan | 26 |
| Hns (JW1225) | BW25113 Δhns::kan | 26 |
| Mlc (JW1586) | BW25113 Δmlc::kan | 26 |
| OmpR (JW3368) | BW25113 ΔompR::kan | 26 |
| UspA (JW3462) | BW25113 ΔuspA::kan | 26 |
The original strain designation is given in parentheses.
For all cultivations, we used M9 minimal medium that contained (per liter of deionized water) 0.8 g NH4Cl, 0.5 g NaCl, 7.5 g Na2HPO4 · 2H2O, and 3.0 g KH2PO4. The following components were sterilized separately and then added (per liter final medium): 2 ml of 1 M MgSO4, 1 ml of 0.1 M CaCl2, 0.3 ml of 1 mM filter-sterilized thiamine HCl, and 10 ml of a trace element solution containing (per liter) 1 g of FeCl3 · 6H2O, 0.18 g of ZnSO4 · 7H2O, 0.12 g of CuCl2 · 2H2O, 0.12 g of MnSO4 · H2O, and 0.18 g of CoCl2 · 6H2O. Sterilized glucose was added to a final concentration of 1 g/liter as the limiting nutrient. For labeling experiments, either a mixture of 50% (wt/wt) [1-13C]glucose (99%; Cambridge Isotope Laboratories, Andover, MA) and 50% (wt/wt) natural glucose or a mixture of 20% (wt/wt) [U-13C]glucose (99%; Cambridge Isotope Laboratories, Andover, MA) and 80% (wt/wt) natural glucose was used.
Frozen glycerol stock cultures were first grown in M9 medium that was supplemented with 5% (vol/vol) Luria-Bertani complex medium. Upon overnight incubation, 1 ml culture broth was used to inoculate a 10-ml-scale bioreactor (35) with M9 medium. Depending on mutant growth rates, the medium feed for glucose-limited chemostat operation was initiated after 4 to 8 h of batch growth. Eight parallel chemostat experiments were done in the previously described miniature bioreactors (35). Briefly, aeration was achieved with water-saturated air at a flow rate of 20 ml/min, a constant temperature was assured through incubation in a 37°C water bath, and a constant pH was achieved through appropriately buffered medium. To avoid the selection of high-affinity mutants (55) and to minimize the selection of subpopulations (38), a new starter culture was prepared for each chemostat experiment.
Analytical procedures and physiological parameters.
Cell growth was monitored as the optical density at 600 nm (OD600). Glucose and acetate concentrations were determined enzymatically with commercial kits (Beckman-Coulter, Zurich, Switzerland, or Dispolab, Dielsdorf, Switzerland). All physiological parameters were determined from cultures in a steady state that was typically achieved after seven culture volume changes. Correlation factors for cellular dry weight and OD600 were predetermined from batch cultures of each mutant for the determination of biomass yields and specific consumption and production rates. Chemostat aliquots for 13C analyses were withdrawn from cultures that exhibited constant OD600 readings for at least two volume changes, i.e., typically after seven volume changes. These cultures were also in an isotopic steady state because growth occurred on a labeled substrate for the entire bioreactor batch and chemostat cultivation.
Crude cell extracts for in vitro enzyme assays were prepared from pellets of 10-ml culture aliquots. For control, batch cultures were grown in 50 ml of M9 minimal medium supplemented with 5 g/liter glucose in 500-ml shake flasks on a rotary shaker at 37°C. Pellets were resuspended in 3 ml of lysis buffer and disrupted with a French press. Isocitrate dehydrogenase and PEP carboxykinase activities were monitored spectrophotometrically by following the rate of NADPH production or NADH consumption at 349 nm, assuming an extinction coefficient of 6.2 mM−1 cm−1 (1, 24). Isocitrate lyase activity was monitored by following the formation of glyoxylic acid phenylhydrazone at 324 nm with an extinction coefficient of 17 mM−1 cm−1 (8).
Metabolic flux ratio analysis by GC-MS.
Samples for gas chromatography-mass spectrometry (GC-MS) analysis were prepared as described previously (13). Briefly, cell pellets were hydrolyzed in 6 M HCl at 105°C for 24 h in sealed Eppendorf microtubes. Hydrolysates were dried under a stream of air at around 60°C and subsequently derivatized at 85°C in 30 μl dimethylformamide (Fluka, Switzerland) and 30 μl N-(tert-butyldimethylsilyl)-N-methyl-trifluoroacetamide with 1% (vol/vol) tert-butyldimethylchlorosilane (Fluka, Switzerland) for 60 min (14). Derivatized amino acids were analyzed on a series 8000 gas chromatograph combined with an MD 800 mass spectrometer (Fisons Instruments, Beverly, MA). The GC-MS-derived mass isotope distributions of proteinogenic amino acids were then corrected for naturally occurring isotopes (13). The corrected mass distributions were related to the in vivo metabolic activities with previously described algebraic equations and statistical data treatment, which quantified 10 ratios of fluxes through converging reactions and pathways to the synthesis of five intracellular metabolites (13) by using the software Fiat Flux (56).
In the standard network for metabolic flux ratio calculations of Fiat Flux (56), the glyoxylate shunt is considered inactive and the fraction of oxaloacetate originating from PEP can be determined as described before (13). The activity of the glyoxylate shunt can be diagnosed from the calculated CO2 labeling content from [U-13C]glucose experiments. If the calculated value falls outside its theoretical boundaries (0% and the degree of fractional label in the input glucose), the glyoxylate shunt is active. In these cases, the fraction of oxaloacetate derived through the glyoxylate shunt was also considered (35). Even when considering an active glyoxylate shunt, the calculated fraction of 13C-labeled CO2 can fall outside of its theoretical boundaries. When this happened, the fraction of 13C-labeled CO2 was estimated on the basis of a linear correlation with the dilution rate, which was determined from data sets with well-determined 13CO2 fractions (data not shown) (35).
13C-constrained metabolic net flux analysis.
Intracellular net fluxes were estimated with the previously described (14) stoichiometric model that contained all major pathways of central carbon metabolism, including the glyoxylate shunt and the Entner-Doudoroff (ED) pathway, by using the software Fiat Flux (56). The reaction matrix consisted of 26 unknown fluxes and 21 metabolite balances (including the three experimentally determined rates of glucose uptake and acetate and biomass production). To solve this underdetermined system of equations with five degrees of freedom, eight of the flux ratios calculated as described above were used as additional constraints as described before (14, 35, 50), i.e., serine derived through the Embden-Meyerhoff-Parnas (EMP) pathway, pyruvate derived through the ED pathway, oxaloacetate originating from PEP, PEP originating from oxaloacetate, pyruvate originating from malate (upper and lower boundaries), oxaloacetate derived through the glyoxylate shunt (upper boundary), and PEP derived through the pentose phosphate pathway (upper bound). The first four ratios were used as equality constraints, while the latter four were used as boundary constraints.
Fluxes into biomass were calculated from the known metabolite requirements for macromolecular compounds and the growth rate-dependent RNA and protein contents (9). The sum of the weighted square residuals of the constraints from both metabolite balances and flux ratios was minimized by using the MATLAB function fmincon, and the residuals were weighted by dividing through the experimental error (14). The computation was repeated at least five times with randomly chosen initial flux distributions to ensure identification of the global minimum, and the system always converged to the same solution. For each metabolite that was used as a precursor for biomass synthesis, a proportional error to its requirements for biomass of 2%, rather than 4%, was assigned (14).
RESULTS
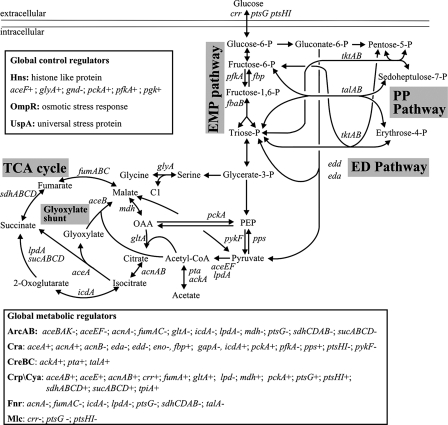
To identify transcriptional mechanisms that control the distribution of intracellular fluxes in the central metabolism of E. coli, we chose otherwise isogenic knockout mutants of the global metabolic regulators ArcA, ArcB, Cra, Crp, Cya, CreB, CreC, and Mlc and the more general global regulators Hns, OmpR, and UspA from the KEIO library (4). Except for UspA and OmpR, DNA binding sites upstream of central metabolic genes are known for the above regulators (Fig. 1) and UspA is a modulator of carbon flow during growth arrest that does not act on the genetic level (39). Since our particular interest was in aerobic glucose metabolism under conditions without or with strongly reduced glucose repression, we investigated glucose-limited chemostat cultures at a low dilution rate of 0.1 h−1, at which E. coli is generally considered to be derepressed (11, 32, 37). The large number of steady-state experiments with at least duplicate 13C experiments for each mutant (68 in total) was made manageable by the use of our recently described mini-scale chemostat system (35).
FIG. 1.
Central carbon metabolism in E. coli. Arrowheads indicate the assumed reaction reversibility. The insets provide an overview of central metabolic genes that are regulated by the investigated global regulators. Plus and minus signs indicate positive and negative transcriptional regulation, respectively. Only regulated genes in the central metabolism are shown for clarity. PP, pentose phosphate; CoA, coenzyme A; OAA, oxaloacetate; P, phosphate.
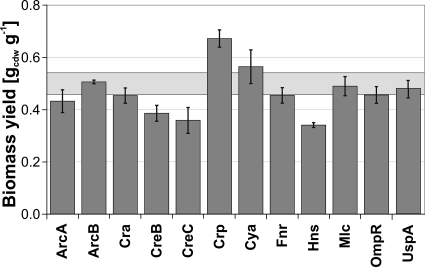
For most regulator knockouts, the steady-state growth physiology was not significantly affected. Generally, overflow metabolism in the form of acetate secretion did not occur in our slow-growing cultures. Only the ArcB, Crp, Cya, and Mlc mutants secreted acetate at rates below 0.1 mM g (dry weight)−1 h−1 (see Table S1 in the supplemental material). Most mutants exhibited a biomass yield similar to or slightly lower than that of the parent (Fig. 2). Such yield reductions are not unexpected given that global regulators with pleiotrophic phenotypes were deleted. Remarkable exceptions were the Crp and Cya mutants, with 13% and 34% higher biomass yields, respectively. Altered yields indicate that the interrupted regulation mechanism was active under the present condition and a relevant cellular process, metabolic or other, was affected. Whether or not such active mechanisms actually control the distribution of intracellular fluxes in central metabolism or whether the yield effect is simply an indirect consequence of other altered regulation of other biological processes will be investigated in the following.
FIG. 2.
Biomass yields of selected global regulator mutants in glucose-limited chemostat culture at a dilution rate of 0.1 h−1. Values are averages from two to six independent cultivations, and the error bars represent the standard deviations. The light gray box indicates the standard deviation from six reference strain experiments. Variations in the dilution rate between 0.09 and 0.13 h−1 were corrected by using linear regression on the glucose uptake rate for yield calculations (equation 3 and Table 1 in reference 35).
Is there global transcriptional control of intracellular flux distribution?
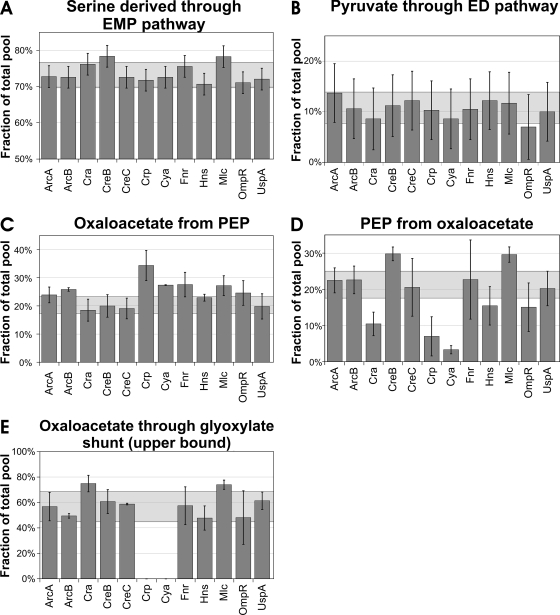
To identify potentially altered distributions of intracellular fluxes in the regulator knockouts, we performed 13C-labeling experiments with [U-13C]glucose and [1-13C]glucose in mini-scale chemostat cultures. Initial interpretation of data was done by metabolic flux ratio analysis (13), which directly quantifies intracellular ratios of converging fluxes in central metabolism (Fig. 3). Once inside the cell, glucose may be catabolized via the EMP, pentose phosphate, or ED pathway in E. coli (Fig. 1). The relative use of these initial pathways is quantified by the fraction of serine derived through the EMP pathway (Fig. 3A) and the fraction of pyruvate derived through the ED pathway (Fig. 3B). Although the entire carbon flux proceeds through these pathways, their relative contributions remained stable in all of the mutants, within the parental ranges of 73% ± 4% and 11% ± 3%, respectively.
FIG. 3.
Origin of key metabolic intermediates in E. coli global regulator mutants during continuous, glucose-limited growth. The fraction of serine derived through the EMP pathway and the fraction of pyruvate derived through the ED pathway were obtained from experiments with 50% [1-13C]glucose and 50% natural glucose. All other ratios were from experiments with 20% [U-13C]glucose and 80% natural glucose. For [U-13C]glucose experiments, the bars represent the average of two to four independent cultivations and the error is the standard deviation. For all [1-13C]glucose experiments, the bars represent a single experiment and error bars are the experimental error calculated from redundant mass distribution (13, 35). The light gray boxes indicate the range of values obtained from three experiments with the parent strain.
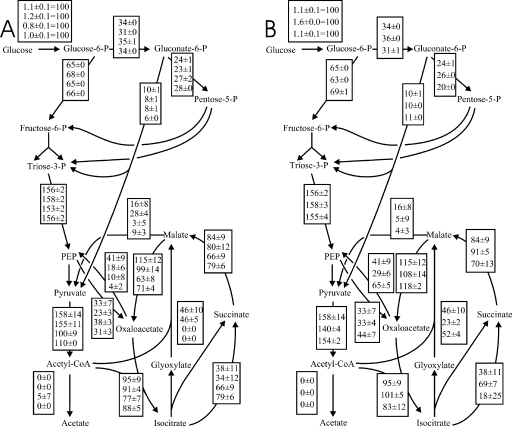
To place these local flux ratios into their network context, absolute fluxes were calculated from the physiological data with the flux ratios as 13C constraints (14). While the absolute glucose flux into the network varied by a factor of almost 2, as a consequences of the altered biomass yields, the relative distribution of fluxes in the upper part of the metabolism was essentially the same in all of the mutants (data partly shown in Fig. 4). Five mutants exhibited a significant, albeit weakly altered, fraction of oxaloacetate originating from PEP (Fig. 3C). Since this ratio quantifies the biosynthetic (anaplerotic) flux versus both the catabolic TCA cycle flux to oxaloacetate and the glyoxylate shunt to oxaloacetate, it is also sensitive to yield and growth rate differences and thus does not directly signify a specific regulation process. Although the flux ratios did not indicate obvious metabolic changes in the Hns mutant, net flux analysis revealed significantly increased TCA cycle flux between isocitrate and succinate and a concomitantly reduced glyoxylate shunt flux (Fig. 4B), which is masked by the upper boundary determination for the ratio of the glyoxylate shunt flux. In the absence of known gene targets in either the TCA cycle or the glyoxylate shunt, this effect might be indirect. The slightly reduced flux in the reverse direction from oxaloacetate to PEP (Fig. 4B) is consistent with the known inducing effect of HNS on pckA expression (28) (Fig. 1).
FIG. 4.
Metabolic net fluxes in (from top to bottom) the E. coli reference strain and the Cra, Crp, and Cya mutants (A) and for the reference strain and the Hns and Mlc mutants (B) in glucose-limited continuous cultures at a dilution rate of 0.1 h−1. Flux values are normalized to the specific glucose uptake rate (given in the first box). Values are the mean of two to four independent experiments, and the errors indicate the standard deviation for these independent experiments. Only selected fluxes are shown for clarity. P, phosphate; CoA, coenzyme A.
The by far strongest metabolic effects were seen for four mutants in two other flux ratios related to oxaloacetate, i.e., PEP originating from oxaloacetate through the gluconeogenic PEP carboxykinase reaction (Fig. 3D) and the upper boundary for oxaloacetate derived through the glyoxylate shunt (Fig. 3E). For the Mlc mutant, the effects in all three oxaloacetate-related flux ratios were consistently small and are probably related to perturbed PEP metabolism through inactivation of the negative, Mlc-dependent control of the PEP-driven phosphotransferase system for glucose uptake (51) (Fig. 1). For the Cra, Crp, and Cya mutants, however, at least one of the oxaloacetate ratios was very strongly affected, and collectively, the data suggest that these regulators are involved in the control of the PEP-glyoxylate cycle.
Transcriptional control of the PEP-glyoxylate cycle.
Recently identified as an alternative to the TCA cycle, the bifunctional anabolic and catabolic PEP-glyoxylate cycle is characterized by the conjoint activity of glyoxylate shunt and PEP carboxykinase for complete oxidation of PEP to CO2 (12). These key reactions are subject to several transcriptional regulation processes, in particular to catabolite repression (18, 19, 43, 45). In E. coli, catabolite repression is mediated by cyclic AMP (cAMP)-cAMP receptor protein (CRP) and cAMP-independent Cra, both capable of acting as activators and repressors of target gene expression (Fig. 1) (5, 45, 46). Completely inactive in glucose batch cultures, the cycle catalyzes substantial carbon fluxes in parallel to the well-known TCA cycle in slow-growing, strictly glucose-limited chemostat cultures (12, 17, 25, 35). These results are fully consistent with the 45 to 65% contribution of the glyoxylate shunt to oxaloacetate synthesis in almost all of the mutants investigated here (Fig. 3E).
In the Cya (encoding adenylate cyclase) and Crp mutants that lack the cAMP-CRP complex, in vivo glyoxylate shunt activity was essentially abolished (Fig. 3E). Additionally, both mutants catalyzed only basal fluxes through the second key reaction catalyzed by PEP carboxykinase (Fig. 3D). The overall distribution of flux in the network of both mutants demonstrates complete absence of the PEP-glyoxylate cycle because (i) the glyoxylate shunt flux was zero and (ii) the gluconeogenic fluxes from oxaloacetate to PEP and from malate to pyruvate were very low compared to those in the parent and much smaller than the anaplerotic flux from PEP to oxaloacetate (Fig. 4A). Consequently, both mutants exhibited almost doubled TCA cycle fluxes. Thus, cAMP-dependent catabolite repression appears to control the PEP-glyoxylate cycle flux under glucose limitation.
These in vivo flux data are qualitatively corroborated by in vitro enzyme activities in the Crp and Cya mutants, which exhibited significantly reduced activity of the glyoxylate shunt key enzyme isocitrate lyase while that of the competing TCA cycle enzyme isocitrate dehydrogenase was increased (Table 2). The in vitro PEP carboxykinase activity was not altered. To verify that the observed changes were not due to polar effects of the still present marker gene or secondary-site mutations, we complemented the Crp mutant with the plasmid-based Crp gene and cultivated the complemented mutant under the same chemostat conditions. In vitro enzyme activities demonstrated that both the isocitrate lyase and isocitrate dehydrogenase activities were restored to the wild-type level (Table 2). Consistently, the flux ratios around oxaloacetate were restored to the wild-type level (data not shown), indicating that the observed regulatory and flux phenotypes were indeed caused by the crp deletion.
TABLE 2.
In vitro enzyme activities in crude cell extracts of batch and glucose-limited chemostat cultures of the Cya and Crp mutants and the E. coli reference strain
| Enzyme | Avg enzyme sp act (μmol min−1 g protein−1) ± SDa
|
|||||
|---|---|---|---|---|---|---|
| Batch, BW2513 | Chemostat
|
|||||
| BW2513 | Cya− | Crp− | BW2513/pTrc99A | Crp− pT::Crp | ||
| Isocitrate lyase | 19 ± 7 | 95 ± 5 | 40 ± 5 | 35 ± 5 | 90 ± 5 | 75 ± 15 |
| Isocitrate dehydrogenase | 370 ± 30 | 35 ± 5 | 45 ± 5 | 85 ± 15 | 30 ± 5 | 20 ± 5 |
| PEP carboxykinase | 10 ± 1 | 24 ± 2 | 23 ± 3 | 15 ± 3 | NDb | ND |
Results of duplicate experiments are shown.
ND, not done.
In contrast to regulation by Cya and Crp, cAMP-independent catabolite repression through Cra (45) has no apparent control over the glyoxylate shunt under this condition (Fig. 3E and 4A). Since the flux from oxaloacetate to PEP was much lower in the Cra mutant compared to the parent (Fig. 3D and 4A), the known Cra-dependent transcriptional activation of pckA (45) appears to exert a partial control on the flux through the PEP carboxykinase reaction of the PEP-glyoxylate cycle. This effect was insufficient to abolish the PEP-glyoxylate cycle, however, because the combined gluconeogenic fluxes from malate to pyruvate and oxaloacetate to PEP still exceed the anabolic flux from PEP to oxaloacetate in the Cra mutant (Fig. 4A). From this result and from the unaltered PEP carboxykinase in vitro activity in the Cya and Crp mutants, we conclude that the PEP-glyoxylate cycle control appears to act primarily on the glyoxylate shunt portion of the cycle.
DISCUSSION
Under the severe glucose limitation at the low dilution rate used here, substantial fluxes through the PEP-glyoxylate cycle were expected (12, 25, 35). The precise regulation mechanism that effectively controlled this major flux rerouting from the TCA cycle to the PEP-glyoxylate cycle, however, was unclear. Based on in vivo flux and in vitro enzyme data on global regulator mutants, we demonstrate that PEP-glyoxylate cycle activity is strongly controlled by induction through the cAMP-CRP complex under the conditions applied. This finding is consistent with the reported increased mRNA and protein levels of PEP carboxykinase and glyoxylate shunt enzymes at a dilution rate of 0.1 h−1, relative to higher growth rates (25). Thus, growth rate-dependent PEP-glyoxylate cycle fluxes under glucose limitation (35) are apparently controlled by the intracellular cAMP level, which is elevated at dilution rates below 0.1 h−1 (32, 37). Strain-dependent differences in fluxes through the PEP-glyoxylate cycle and the glyoxylate shunt are therefore most likely explained by varying cAMP levels (9, 12, 36).
Conversely, ArcA repression of key PEP-glyoxylate cycle genes was not effective under the present conditions because the fluxes through the cycle were similar in the mutant and reference strains. Cra regulation appears to exert partial control of the PEP carboxykinase flux, which is 50% lower in the Cra mutant than in the reference strain. This control does not extent to the overall PEP-glyoxylate cycle though, because malic enzyme partly substitutes for PEP carboxykinase. Excess glyoxylate-to-malate fluxes are thus routed to pyruvate (rather than PEP), as was also shown previously for other mutants (16). Collectively, the above results illustrate that allosteric inhibition of PEP carboxykinase activity through PEP and ATP (49) is not a relevant control mechanism under the present conditions.
The remaining regulators investigated here had no apparent effects on the relative distribution of flux through the central metabolism. The sole exception was an increased TCA cycle flux in the Hns mutant, which is probably an unspecific response because genetic targets of HNS are currently not known to be involved in the TCA cycle. We cannot exclude the possibility that rapidly occurring, natural rpoS mutations (30, 38) were present in subpopulations of our cultures, but metabolic impacts are unlikely, as judged from negligible flux deviations between multiple experiments with all of the mutants and the reference strain. This view is consistent with reproducible but significantly different flux patterns in different rpoS deletion mutants (data not shown).
While transcriptional regulation is simultaneously active on many metabolic enzymes, only one or a few transcription processes appear to actually control central metabolic fluxes under a given condition. In batch cultures with abundant glucose, even under fully aerobic conditions, only ArcA repression was relevant with a negative control of TCA cycle fluxes by a factor of 2 (40). ArcA was irrelevant, however, in slow-growing chemostat cultures with their extremely low glucose concentrations. cAMP-CRP activation exhibits a pattern for the PEP-glyoxylate cycle that is the converse of that of ArcA for the TCA cycle but clearly does not exert flux control through all of its genetic targets. Although cAMP-CRP induction of the TCA cycle genes suc and sdh (Fig. 1) must be absent in the Cra and Cya mutants, both mutants actually have higher TCA cycle fluxes.
By in vivo monitoring of pathway activity within their network context, our results provide quantitative insights into the control of how fluxes are distributed through the network. This approach is complementary to metabolic control analysis that defines a quantitative link between flux through a particular pathway and metabolic or genetic control of its constituent enzymes (44, 52). As discussed above, accumulating evidence suggests that transcriptional flux control is often specific for a particular pathway or cycle in a highly condition-dependent fashion. Since fluxes are the integrated consequence of all regulatory and biochemical interactions within the network (20, 48), it is perhaps not overly surprising that our results deviate from or extend previous conclusions that were exclusively based on genetic evidence. One common misconception is that catabolite repression of sdhCDAB and other genes effectively splits the E. coli TCA cycle into a two-branched pathway during growth on readily fermentable substrates such as glucose (6, 34). Our and previous flux data from E. coli batch and chemostat cultures on glucose demonstrate clearly (15, 16, 22, 25, 40), however, that despite active catabolite repression, there is substantial cyclic operation. While transcriptional regulation often modulates the expression of metabolic genes, as shown by numerous DNA chip experiments (e.g., references 19, 23, and 54), apparently only a few such modulations directly control the in vivo flux through a given pathway.
Supplementary Material
Acknowledgments
This work was supported by a scholarship from the EPFL to A.N.
Footnotes
Published ahead of print on 25 January 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Al Zaid Siddiquee, K., M. J. Arauzo-Bravo, and K. Shimizu. 2004. Metabolic flux analysis of pykF gene knockout Escherichia coli based on 13C-labeling experiments together with measurements of enzyme activities and intracellular metabolite concentrations. Appl. Microbiol. Biotechnol. 63407-417. [DOI] [PubMed] [Google Scholar]
- 2.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 247-17. [DOI] [PubMed] [Google Scholar]
- 3.Avison, M. B., R. E. Horton, T. R. Walsh, and P. M. Bennett. 2001. Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal media. J. Biol. Chem. 27626955-26961. [DOI] [PubMed] [Google Scholar]
- 4.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the KEIO collection. Mol. Syst. Biol. 22006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brückner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209141-148. [DOI] [PubMed] [Google Scholar]
- 6.Cronan, J. E., Jr., and D. C. LaPorte. 1996. Tricarboxylic acid cycle and glyoxylate bypass, p. 206-216. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon, G. H., and H. L. Kornberg. 1959. Assay methods for key enzymes of the glyoxylate cycle. Biochem. J. 733-10. [Google Scholar]
- 9.Emmerling, M., M. Dauner, A. Ponti, J. Fiaux, M. Hochuli, T. Szyperski, K. Wüthrich, J. E. Bailey, and U. Sauer. 2002. Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J. Bacteriol. 184152-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferenci, T. 2008. Bacterial physiology, regulation and mutational adaptation in a chemostat environment. Adv. Microb. Physiol. 53169-229. [DOI] [PubMed] [Google Scholar]
- 11.Ferenci, T. 2001. Hungry bacteria—definition and properties of a nutritional state. Environ. Microbiol. 3605-611. [DOI] [PubMed] [Google Scholar]
- 12.Fischer, E., and U. Sauer. 2003. A novel metabolic cycle catalyzes glucose oxidation and anaplerosis in hungry Escherichia coli. J. Biol. Chem. 27846446-46451. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, E., and U. Sauer. 2003. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur. J. Biochem. 270880-891. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, E., N. Zamboni, and U. Sauer. 2004. High-throughput metabolic flux analysis based on gas chromatography-mass spectrometry derived 13C constraints. Anal. Biochem. 325308-316. [DOI] [PubMed] [Google Scholar]
- 15.Flores, S., G. Gosset, N. Flores, A. A. de Graaf, and F. Bolivar. 2002. Analysis of carbon metabolism in Escherichia coli strains with an inactive phosphotransferase system by 13C labeling and NMR spectroscopy. Metab. Eng. 4124-137. [DOI] [PubMed] [Google Scholar]
- 16.Fong, S. S., A. Nanchen, B. O. Palsson, and U. Sauer. 2006. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J. Biol. Chem. 2818024-8033. [DOI] [PubMed] [Google Scholar]
- 17.Franchini, A. G., and T. Egli. 2006. Global gene expression in Escherichia coli K-12 during short-term and long-term adaptation to glucose-limited continuous culture conditions. Microbiology 1522111-2127. [DOI] [PubMed] [Google Scholar]
- 18.Goldie, H. 1984. Regulation of transcription of the Escherichia coli phosphoenolpyruvate carboxykinase locus: studies with pck-lacZ operon fusions. J. Bacteriol. 159832-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gosset, G., Z. Zhang, S. Nayyar, W. A. Cuevas, and M. H. Saier, Jr. 2004. Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J. Bacteriol. 1863516-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hellerstein, M. K. 2003. In vivo measurement of fluxes through metabolic pathways: the missing link in functional genomics and pharmaceutical research. Annu. Rev. Nutr. 23379-402. [DOI] [PubMed] [Google Scholar]
- 21.Hoskisson, P. A., and G. Hobbs. 2005. Continuous culture—making a comeback? Microbiology 1513153-3159. [DOI] [PubMed] [Google Scholar]
- 22.Hua, Q., C. Yang, T. Baba, H. Mori, and K. Shimizu. 2003. Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J. Bacteriol. 1857053-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hua, Q., C. Yang, T. Oshima, H. Mori, and K. Shimizu. 2004. Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures. Appl. Environ. Microbiol. 702354-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Igamberdiev, A. U., and P. Gardeström. 2003. Regulation of NAD- and NADP-dependent isocitrate dehydrogenases by reduction levels of pyridine nucleotides in mitochondria and cytosol of pea leaves. Biochim. Biophys. Acta 1606117-125. [DOI] [PubMed] [Google Scholar]
- 25.Ishii, N., K. Nakahigashi, T. Baba, M. Robert, T. Soga, A. Kanai, T. Hirasawa, M. Naba, K. Hirai, A. Hoque, P. Y. Ho, Y. Kakazu, K. Sugawara, S. Igarashi, S. Harada, T. Masuda, N. Sugiyama, T. Togashi, M. Hasegawa, Y. Takai, K. Yugi, K. Arakawa, N. Iwata, Y. Toya, Y. Nakayama, T. Nishioka, K. Shimizu, H. Mori, and M. Tomita. 2007. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316593-597. [DOI] [PubMed] [Google Scholar]
- 26.Ito, M., T. Baba, H. Mori, and H. Mori. 2005. Functional analysis of 1440 Escherichia coli genes using the combination of knock-out library and phenotype microarrays. Metab. Eng. 7318-327. [DOI] [PubMed] [Google Scholar]
- 27.Körner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27559-592. [DOI] [PubMed] [Google Scholar]
- 28.Laurent-Winter, C., S. Ngo, A. Danchin, and P. Bertin. 1997. Role of Escherichia coli histone-like nucleoid-structuring protein in bacterial metabolism and stress response—identification of targets by two-dimensional electrophoresis. Eur. J. Biochem. 244767-773. [DOI] [PubMed] [Google Scholar]
- 29.Liu, X., and P. De Wulf. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 27912588-12597. [DOI] [PubMed] [Google Scholar]
- 30.Maharjan, R., S. Seeto, L. Notley-McRobb, and T. Ferenci. 2006. Clonal radiation in a constant environment. Science 313514-517. [DOI] [PubMed] [Google Scholar]
- 31.Martínez-Antonio, A., and J. Collado-Vides. 2003. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 6482-489. [DOI] [PubMed] [Google Scholar]
- 32.Matin, A., and M. K. Matin. 1982. Cellular levels, excretion, and synthesis rates of cyclic AMP in Escherichia coli grown in continuous culture. J. Bacteriol. 149801-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuno, T., and S. Mizushima. 1987. Isolation and characterization of deletion mutants of ompR and envZ, regulatory genes for expression of the outer membrane proteins OmpC and OmpF in Escherichia coli. J. Biochem. (Tokyo) 101387-396. [DOI] [PubMed] [Google Scholar]
- 34.Nam, T.-W., Y.-H. Park, H.-J. Jeong, S. Ryu, and Y.-J. Seok. 2005. Glucose repression of the Escherichia coli sdhCDAB operon, revisited: regulation by the CRP-cAMP complex. Nucleic Acids Res. 336712-6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nanchen, A., A. Schicker, and U. Sauer. 2006. Nonlinear dependency of intracellular fluxes on growth rate in miniaturized continuous cultures of Escherichia coli. Appl. Environ. Microbiol. 721164-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noronha, S. B., H. J. C. Yeh, T. F. Spande, and J. Shiloach. 2000. Investigation of the TCA cycle and the glyoxylate shunt in Escherichia coli BL21 and JM109 using 13C-NMRMS. Biotechnol. Bioeng. 68316-327. [PubMed] [Google Scholar]
- 37.Notley-McRobb, L., A. Death, and T. Ferenci. 1997. The relationship between external glucose concentration and cAMP levels inside Escherichia coli: implications for models of phosphotransferase-mediated regulation of adenylate cyclase. Microbiology 1431909-1918. [DOI] [PubMed] [Google Scholar]
- 38.Notley-McRobb, L., T. King, and T. Ferenci. 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 184806-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nyström, T., and F. C. Neidhardt. 1994. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 11537-544. [DOI] [PubMed] [Google Scholar]
- 40.Perrenoud, A., and U. Sauer. 2005. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J. Bacteriol. 1873171-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pittard, A. J. 1996. Biosynthesis of the aromatic amino acids, p. 458-484. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 42.Plumbridge, J. 2002. Regulation of gene expression in the PTS in Escherichia coli: the role and interactions of Mlc. Curr. Opin. Microbiol. 5187-193. [DOI] [PubMed] [Google Scholar]
- 43.Prasad Maharjan, R., P. L. Yu, S. Seeto, and T. Ferenci. 2005. The role of isocitrate lyase and the glyoxylate cycle in Escherichia coli growing under glucose limitation. Res. Microbiol. 156178-183. [DOI] [PubMed] [Google Scholar]
- 44.Rossell, S., C. C. van der Weijden, A. Lindenbergh, A. van Tuijl, C. Francke, B. M. Bakker, and H. V. Westerhoff. 2006. Unraveling the complexity of flux regulation: a new method demonstrated for nutrient starvation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1032166-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saier, M. H., Jr., and T. M. Ramseier. 1996. The catabolite repressor/activator (Cra) protein of enteric bacteria. J. Bacteriol. 1783411-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saier, M. H., Jr., T. M. Ramseier, and J. Reizer. 1996. Regulation of carbon utilization, p. 1325-1343. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 47.Sauer, U. 2004. High-throughput phenomics: experimental methods for mapping fluxomes. Curr. Opin. Biotechnol. 1558-63. [DOI] [PubMed] [Google Scholar]
- 48.Sauer, U. 2006. Metabolic networks in motion: 13C-based flux analysis. Mol. Syst. Biol. 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sauer, U., and B. J. Eikmanns. 2005. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29765-794. [DOI] [PubMed] [Google Scholar]
- 50.Sauer, U., V. Hatzimanikatis, J. E. Bailey, M. Hochuli, T. Szyperski, and K. Wuthrich. 1997. Metabolic fluxes in riboflavin-producing Bacillus subtilis. Nat. Biotechnol. 15448-452. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka, Y., K. Kimata, T. Inada, H. Tagami, and H. Aiba. 1999. Negative regulation of the pts operon by Mlc: mechanism underlying glucose induction in Escherichia coli. Genes Cells 4391-399. [DOI] [PubMed] [Google Scholar]
- 52.ter Kuile, B. H., and H. V. Westerhoff. 2001. Transcriptome meets metabolome: hierarchical and metabolic regulation of the glycolytic pathway. FEBS Lett. 500169-171. [DOI] [PubMed] [Google Scholar]
- 53.Unden, G., S. Achebach, G. Holighaus, H. G. Tran, B. Wackwitz, and Y. Zeuner. 2002. Control of FNR function of Escherichia coli by O2 and reducing conditions. J. Mol. Microbiol. Biotechnol. 4263-268. [PubMed] [Google Scholar]
- 54.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 1871591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wick, L. M., H. Weilenmann, and T. Egli. 2002. The apparent clock-like evolution of Escherichia coli in glucose-limited chemostats is reproducible at large but not at small population sizes and can be explained with monod kinetics. Microbiology 1482889-2902. [DOI] [PubMed] [Google Scholar]
- 56.Zamboni, N., E. Fischer, and U. Sauer. 2005. FiatFlux—a software for metabolic flux analysis from 13C-glucose experiments. BMC Bioinformatics 6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.