Abstract
Escherichia coli DH10B was designed for the propagation of large insert DNA library clones. It is used extensively, taking advantage of properties such as high DNA transformation efficiency and maintenance of large plasmids. The strain was constructed by serial genetic recombination steps, but the underlying sequence changes remained unverified. We report the complete genomic sequence of DH10B by using reads accumulated from the bovine sequencing project at Baylor College of Medicine and assembled with DNAStar's SeqMan genome assembler. The DH10B genome is largely colinear with that of the wild-type K-12 strain MG1655, although it is substantially more complex than previously appreciated, allowing DH10B biology to be further explored. The 226 mutated genes in DH10B relative to MG1655 are mostly attributable to the extensive genetic manipulations the strain has undergone. However, we demonstrate that DH10B has a 13.5-fold higher mutation rate than MG1655, resulting from a dramatic increase in insertion sequence (IS) transposition, especially IS150. IS elements appear to have remodeled genome architecture, providing homologous recombination sites for a 113,260-bp tandem duplication and an inversion. DH10B requires leucine for growth on minimal medium due to the deletion of leuLABCD and harbors both the relA1 and spoT1 alleles causing both sensitivity to nutritional downshifts and slightly lower growth rates relative to the wild type. Finally, while the sequence confirms most of the reported alleles, the sequence of deoR is wild type, necessitating reexamination of the assumed basis for the high transformability of DH10B.
Molecular biology studies rely heavily on Escherichia coli for essential operations, ranging from the simple propagation of plasmid DNA to the creation of large clone libraries for whole-genome sequence determination. Among the strains developed as hosts for these everyday applications, DH10B (17) is commonly used across the research community, taking advantage of particularly useful properties exhibited by the strain. These include high transformation efficiency, the ability to take up and stably maintain large plasmids, the lack of methylation-dependent restriction systems (MDRS), and colony screening via lacZ-based α-complementation. However, analysis of sequenced bacterial artificial chromosome (BAC) clones derived from DH10B shows a high incidence of insertion sequence (IS) transposition from the chromosome into the cloned fragment (25).
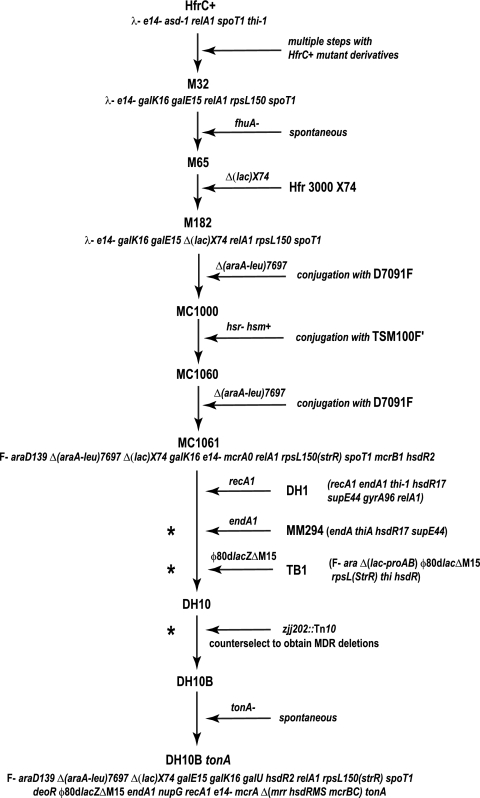
The genome of DH10B was constructed before the modern era of molecular biology, through a series of genetic manipulations (Fig. 1). The progenitors were all K-12 strains, with the exception of D7091F, in which a region surrounding the Δ(araA-leu)7697 deletion had been derived from E. coli B SB3118 by P1 transduction (John Wertz, personal communication). Ultimately, MC1061 (9) served as a starting point for Hanahan and coworkers to replace alleles by using a series of P1 transductions that resulted in DH10B (17). Among the engineered gene replacements were recA1 to improve clone stability by inhibiting the homologous recombination system; endA1, which inactivates the encoded periplasmic DNA-specific endonuclease, thereby enhancing DNA stability during transformation; and a φ80 derivative containing the lacZΔM15 mutation for screening by α-complementation. Recombination functions for the latter two steps were provided by expressing RecA from a plasmid that was subsequently cured by treatment with coumermycin (Frederic Bloom, personal communication). The resultant strain, DH10, was also reported to contain an unspecified mutation in deoR which was hypothesized to increase transformation efficiency, though no explanation was offered (19). A DH10 derivative containing the allele mdoB::Tn10 (zjj202::Tn10 in reference 22) was subjected to fusaric acid treatment to counterselect against the tetracycline resistance gene in the transposon, again in the presence of the RecA-expressing plasmid. DH10B was one isolate from this selection with a deletion spanning the marker and the flanking region, including the MDRS loci (mrr, mcrA, mcrB, and mcrC). Deletion of the MDRS loci was shown to improve the cloning efficiency of mammalian DNA, in which cytosine is commonly methylated (17). Later, a strain with a spontaneous mutation in tonA that confers resistance to the bacteriophages T1, T5, and φ80 was isolated, and that strain, DH10B tonA, was also commercialized (Invitrogen Corp.).
FIG. 1.
Construction of DH10B. The steps leading to the creation of MC1061 from HfrC+ are outlined, followed by the steps described by Grant et al. (17) to generate DH10B from MC1061. Genotypes of selected key strains are indicated. The DH10B tonA genotype is a composite compiled from multiple sources. Steps involving a pBR322-recA plasmid are indicated with an asterisk. The branched pathway consisting of 25 steps leading from wild-type K-12 to HfrC+ (3) has been omitted for clarity.
Given the number of steps involving transfer or deletion of undefined DNA fragments in the genesis of DH10B, an examination of the relationship between the stated DH10B genotype and the actual genome sequence was warranted. To do so, we took advantage of sequence reads corresponding to the DH10B genome that had been gathered as “contaminant” sequences during the bovine genome sequencing project carried out at the Human Genome Sequencing Center (HGSC) at Baylor College of Medicine. These reads were assembled into the complete DH10B sequence using a new assembly engine, the SeqMan genome assembler (SMGA). The sequence confirms that most of the stated genotype is correct but also uncovers a plethora of additional changes that were not known. Together, these alterations have important implications for the biology of the strain and indicate that the assumed genetic basis for some of the phenotypes should be reevaluated.
MATERIALS AND METHODS
DNA sequencing and contaminant screening.
DH10B reads were obtained from a set of about 4 million reads produced by low-coverage sequencing of bovine BACs as part of the bovine genome project. Each BAC DNA preparation is contaminated by a small (<1%) amount of DNA from the DH10B host. These E. coli reads were identified and distinguished from the bovine BAC reads by comparison to the E. coli MG1655 sequence. This procedure gave 180,727 reads after additional screening against cloning and sequencing vector and trimming sequences to high-quality bases (Phred 20). An additional 3,076 reads were isolated by this procedure using the E. coli IS10 (GenBank accession no. AY319289.1) and φ80 (unpublished data) sequences, in order to obtain regions present in DH10B but absent in MG1655.
During the finishing phase, gaps between contigs were closed by the direct sequencing of PCR products generated from primers flanking the gaps, and for larger gaps, small insert (SMIL) libraries were generated from the PCR products spanning the gaps. Appropriate primer pairs were generated from the assembly using SeqMan software (DNAStar, Inc.).
Sequence assembly.
Genome assembly was done primarily using the SMGA (DNAStar, Inc.). Sequences were preprocessed within SMGA to trim vector and low-quality sequences and to check for more than 900 known repeats. Once the sequences are processed, assembly consists of three steps. First, each sequence read is parsed into overlapping 25-base oligomers and a running count for each oligomer is tabulated. Oligomers occurring more than once in a sequence or more than 1.5 times the expected coverage are marked as repeat oligomers. Sequences are then scanned against the oligomer table, and one nonrepeat oligomer for each 20-base window is chosen (oligomer tag). Oligomer tags are placed in a bucket-and-chain table that includes the position and orientation for the oligomer in each sequence (1). Second, oligomer tags are reorganized into a sequence-sequence overlap table. Each sequence read is scanned against the overlap table to generate a list of sequences that have compatible oligomer tags. Third, contigs are built by sequential addition of overlapping reads. Initial alignments are started using pairs of nonrepeated, overlapping sequences which have mate pair reads that also overlap. Additional constrained sequence pairs are then added, followed by nonrepeat sequences lacking mate pair information. Next, sequences with one nonrepeated end and one repeated end are added. Fully repeated sequences are added to an existing contig only if they have a nonrepeated mate pair within that contig. Finally, repeat sequences that cannot be unambiguously placed into existing contigs are combined into repeat contigs and incorporated manually.
Following the initial assembly, contigs were ordered within SeqMan Pro (DNAStar, Inc.), first using available mate pair data and then alignment information with MG1655 from the Mauve genome aligner (10). When possible, adjacent contigs were merged, resulting in a 36-contig scaffold spanning the genome. Gaps were filled by sequencing of directed PCR products and, where necessary, the corresponding MG1655 sequence. The draft sequence was then used as a template for a complete reassembly by SMGA to maximize coverage and quality. Repeat regions were manually incorporated to ensure accuracy. A second round of PCR-based directed sequencing was used to complete the sequence.
Annotation.
The consensus sequence of the first complete (single-contig) assembly was imported into the Web-based ASAP database (15, 16) to aid analysis and annotation by multiple users in different locations. Annotated Feature coordinates were updated when refinements to the assembly produced new genome versions. Genes identical to those in MG1655 were identified using genome alignment data generated by Mauve (10); single-nucleotide polymorphisms (SNPs) and other sequence differences from MG1655 were also identified from Mauve alignments. Pseudogenes were identified by manual inspection and carefully verified. IS elements were identified using RepeatMasker (A. F. A. Smit, R. Hubley, and P. Green, unpublished data; http://repeatmasker.org) searching against the ISFinder database (40). The genome and annotations are freely accessible for viewing in ASAP (https://asap.ahabs.wisc.edu), and annotations may be added or updated by members of the community upon registration.
Growth measurements.
Cultures were grown in MOPS (morpholinethanesulfonic acid) minimal medium (32) supplemented with 0.1% glucose and amino acids (40 μg/ml each) as described previously. For each growth curve, a single colony was used to inoculate 50 ml fresh medium in a 250-ml baffled flask and grown at 37°C with shaking in an orbital water bath. Optical density measurements at 600 nm (OD600) were taken every minute using an automated system.
Analysis of mutation rates.
d-Cycloserine resistance assays were performed as previously described (12, 33). Briefly, in a fluctuation assay, 20 tubes of 1 ml mineral salts medium (17a) supplemented with glucose and thiamine were inoculated with 104 cells each, and cultures were grown to early stationary phase. Fifty-microliter aliquots from each tube were then spread on minimal plates containing d-cycloserine (0.04 mM). The estimated number of mutations per tube (m) was calculated from the number of colonies by using the Ma-Sandri-Sarkar maximum-likelihood method (39). Equation 41 from the report of Stewart et al. (41) was used to extrapolate the obtained m value, valid for 50 μl, to 1 ml. Statistical comparisons of m values were made only when the difference in total cell number was negligible (<3%, P ≥ 0.6, with a two-tailed, unpaired t test). The total number of cells in a tube was calculated by spreading dilutions from three tubes onto nonselective plates. Dividing the number of mutations per tube by the average total number of cells in a tube gives the mutational rate (mutation/cell/generation).
In a second protocol, appropriate for detection of base substitutions, cells resistant to rifampin were selected. Resistant cells carry base substitutions in rpoB (24). Twenty tubes of 1 ml LB were inoculated with 104 cells each, and cultures were grown to early stationary phase. Appropriate dilutions were seeded onto LB agar plates and LB agar plates containing rifampin (100 μg/ml), and colony counts were performed after 24 or 48 h, respectively. Mutation frequencies are reported as a proportion of the number of rifampin-resistant colonies relative to the total viable count. The results correspond to the mean value obtained in two independent experiments.
Analysis of mutation types.
Analysis of the mutational spectrum of the cycA gene has been described previously (12). Briefly, a 1,877-bp genomic segment encompassing the entire gene was amplified from mutant cells using the primer pair cycA1/cycA2. A representative sample was obtained by analyzing 5 colonies from each parallel plate, yielding a total of 96 samples per experiment. The amplified fragments were resolved on an agarose gel and compared to a fragment generated from the wild-type template. Identical sizes indicated a mutation affecting only one or a few nucleotides, a decrease in size or failure of amplification indicated a deletion, and a detectable increase in size indicated an IS insertion. Where further analysis of the insertion mutants was desired, the identity of the ISs was determined by PCR using combinations of oppositely oriented IS-specific primers and primers flanking cycA.
Primers for PCR analysis of mutations.
The primers used for PCR analysis of mutations (33) included the following: cycA1, 5′-CTGATGCCGGTAGGTTCT-3′; cycA2, 5′-GCGCCATCCAGCATGATA-3′; for IS1, IS1A1 (5′-TCGCTGTCGTTCTCA-3′) and IS1A2 (5′-AAGCCACTGGAGCAC-3′); for IS2, UK1R (5′-TCGCAGGCATACCATCAA-3′) and UK2R (5′-CAGACGGGTTAACGGCA-3′); for IS5, IS5ki3 (5′-ATAGGCTGATTCAAGGCA-3′) and IS5ki2 (5′-GCTCGATGACTTCCACCA-3′); and for IS150, IS150ki1 (5′-ACGTGCCGAGATGATCCT-3′) and IS150ki2 (5′-GCGCCATCCAGCATGATA-3′).
RESULTS
Sequence assembly of DH10B.
DH10B commonly serves as the host for the construction and propagation of large-insert genomic DNA libraries used in whole-genome sequencing efforts. These libraries generally consist of hundreds of thousands to millions of independent clones, each containing a large genomic DNA fragment (∼150 kb) ligated into a BAC vector. During the construction of sublibraries from BAC clones, small amounts of DH10B genomic DNA are present that are occasionally cloned and sequenced. Sequence reads from such “contaminating” clones are then identified in silico and filtered out by comparison against a database containing the MG1655 reference genome (7), known repeated bacterial sequence elements, phages, and other elements. In this way, almost 200,000 sequence reads were collected either entirely from DH10B or from chimeras of bacterial and bovine DNA. All reads were generated by Sanger sequencing technology, and mate pair data were available for most reads. Based on an average read length of 700 bp and a DH10B genome size of 4.6 million bases, these data sets represent an average depth of coverage of approximately 25.
The complete genome sequence was then assembled de novo using a new desktop sequence assembly package, the SMGA. The core of SMGA consists of an efficient sequence analyzer that breaks down sequences into a bucket-and-chain hash table of overlapping subsequences, or oligomers (1), and a segmented banded aligner that can rapidly align sequences of any length with nearly constant memory usage (see Materials and Methods). The assembler makes full use of mate pair information, when available, and handles repeat elements in a conservative manner. SMGA is also capable of assembling data sets, either individually or in combination, from traditional Sanger sequencing or massively parallel, next-generation technologies, such as pyrosequencing (29).
The Sanger sequence data set for DH10B was assembled into contigs using SMGA. Scaffolds spanning the genome were organized first using mate pair information and, second, by alignment with the MG1655 genome (7, 35) using the Mauve genome aligner (10). Following gap filling, the resulting draft consensus sequence was then used as a template to reassemble the entire data set using SMGA. Remaining low-coverage areas and consensus ambiguities were resolved using directed sequencing of PCR products corresponding to the areas in question. Finally, the sequence was independently verified with SOLiD (Sequencing by Oligonucleotide Ligation and Detection) technology using the final consensus as a template for assembly (K. McKernan, J. Malek, H. Peckham, F. R. Blattner, and G. M. Weinstock, unpublished results).
Overview of the DH10B genome.
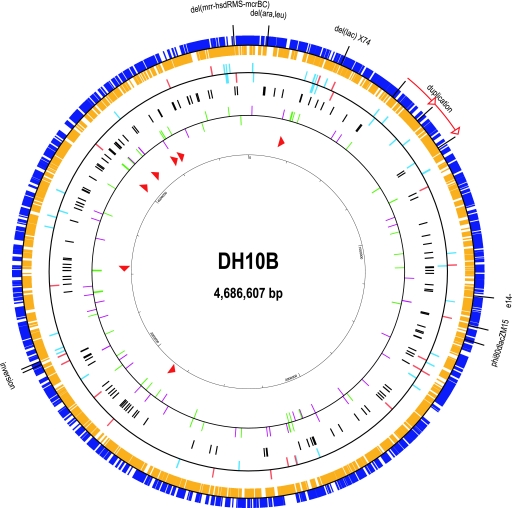
The circular genome of DH10B is 4,686,137 base pairs in length (Fig. 2) and can be readily aligned end to end with the wild-type MG1655 sequence with a genome aligner such as Mauve (Fig. 3). As expected, the extensive colinearity between DH10B and MG1655 is also reflected in the gene content. Among the 4,305 protein-encoding genes present in MG1655, 4,058 have identical counterparts in DH10B and another 30 genes contain one or more synonymous SNPs. The remaining 217 genes each differ from their MG1655 counterpart at the protein level, ranging from single-amino-acid substitutions to complete deletions. Other than IS and phage elements, we have not detected any protein-encoding genes that are not also present in MG1655.
FIG. 2.
Circle diagram of the DH10B genome. Protein-encoding genes are shown in the outer ring, with blue boxes representing those genes coded on the forward strand and gold boxes representing those on the complementary strand. Large deletions and insertions are indicated outside of the circle. In the next inner ring, IS elements that cause gene disruptions or are intergenic are indicated with red or aqua tick marks, respectively, with the positions above or below the line indicating which strand is transcribed. SNPs are then indicated with black tick marks. tRNAs (green tick marks) and small regulatory RNAs (purple tick marks) are shown, with those on the forward strand above the line and those on the complementary strand below it. Finally, rRNA operons are shown as red arrowheads.
FIG. 3.
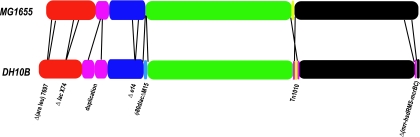
Comparison of the DH10B and MG1655 genomes. Schematic of the two genomes showing colinear blocks of sequence. The endpoints of each block are defined by an adjacent sequence that is present only in one genome. The major structural rearrangements in DH10B are indicated below the genome representation.
Among the stable and regulatory RNA genes, all seven rRNA operons are intact, although 15 SNPs are observed in five different rRNA genes. All but three of these SNPs are sequence variants found in other copies of the rRNA gene within the DH10B genome and are thus attributable to recombinational gene-conversion events. The exceptions (G191A in rrsB and C1161Tand C1162T in rrsE) are all transitions; the consequence of these changes, if any, is unknown. Eighty-five of the 86 tRNA-encoding genes are entirely conserved between the two strains. The one variant tRNA, encoded by argQ, contains a C-to-T transition at position 11. The mutation is of unknown functional consequence, and furthermore, the other three paralogous tRNAArg genes are wild type. However, of the 60 genes encoding small regulatory RNAs that have been accurately defined for MG1655, four have deletions ranging from one nucleotide to complete removal in DH10B.
Large-scale alterations of the DH10B genome.
DH10B contains several large structural differences relative to MG1655 (Fig. 3; Table 1). Four known large-scale deletions, Δ(ara leu)7697, ΔlacX74, Δ(mrr-hsdRMS-mcrBC), and precise excision of the e14 prophage, together remove 135,044 bp and 121 genes either completely or partially (Table 1). Excision of e14 also restores a wild-type icd (b1136), although the gene produces a functional protein in the presence of the phage as well (21). These deletions remove multiple operons encoding diverse cellular functions in addition to those targeted for a desired phenotype (see Table S1 in the supplemental material).
TABLE 1.
Select genetic alterations in DH10B
| Allele | Alteration typea | Gene(s) affected | DH10B sequenceb | Amino acid change(s)c |
|---|---|---|---|---|
| Δ(ara leu)7697 | Deletion | (b0059) b0060-b0079 (b0080) | After 62378 (ΔMG 62379-88274) | HepA Δ1-295; FruR Δ1-82, L83K |
| araD139 | Deletion | b0061 | Part of Δ(ara leu)7697 | Null |
| tonA | Wild type and missense | b0150 | G142348R | W254W or W254STOP |
| ΔlacX74 | Deletion | (b0321) b0322-b0352 | After 313972 (ΔMG 338422-3746342) | YahG Δ292-472 |
| DH10Bdup | Duplication | b0553-b0655 | Tandem duplication of 514341-627601 | Wild type |
| galK16 | IS2 disruption | b0757 | IS2 insertion from 728375-729703 | Δ57-383 |
| galE15 | Missense | b0759 | G844839A | S122F |
| deoR | Wild type | b0840 | No change | Wild type |
| e14 | Excision | (b1136) b1137-b1159 | After 1250887 (ΔMG 1195443-1210646) | Icd restored to wild type |
| mcrA | Deletion | b1159 | Deleted by e14 excision | Null |
| galU | Wild type | b1236 | No change | Wild type |
| φ80dlacZΔM15 | Insertion | b0340-b0349 | Insert from 1349613-1396987 | LacZ (b0344) Δ11-42 |
| recA1 | Missense | b2699 | G2913852A | G160D |
| relA1 | IS2 disruption | b2784 | IS2 insertion from 3003960-3005291 | Δ87-744 |
| endA1 | Missense | b2945 | G3184059A | E208K |
| Tn10.10d | Inversion | (b2964) b2965-b2972 (b4466) | 3200798-3211928 | NupG Δ222-418; YghJ Δ1-541 |
| nupG | IS10 disruption | b2964 | IS10R insertion at 3199443 | Δ222-418 |
| rpsL | Missense | b3342 | T3570191C | K43R |
| rph | Frameshift | b3643 | G inserted at 3911483 | Corrects rph-1 |
| spoT1 | Insertion | b3650 | ATCAGG insertion from 3918250-3918255; C3918768T | QD insertion after D84; H255Ye |
| Δ(mrr-hsdRMS-mcrBC) | Deletion | b4312-b4358 (b4359) | Partial Tn10 from 4640587-4641916 (ΔMG 4538777-4595456) | MdoB Δ668-750 |
Relative to MG1655.
Numbers are genomic coordinates. Base changes all refer to the forward strand. MG represents MG1655.
Changes relative to the MG1655 sequence. Protein names and deleted amino acids refer to affected genes on the flanks of large deletions, except for LacZ, which is an internal deletion.
Coordinates refer to the internal segment of the transposon that is inverted relative to MG1655.
Residue 257 of SpoT1 due to two-amino-acid insertion after position 84.
There are two major insertions in DH10B relative to MG1655. First, the φ80dlacZΔM15 insertion is a mosaic element described in detail below. Second, a 113-kb region of genome (genomic coordinates 514341 to 627601) is precisely duplicated in tandem. IS5 elements immediately flanking this segment presumably provided the homology necessary for creating the duplication. The 106 duplicated genes encode functions ranging from membrane components and transporters to transcriptional regulators.
Finally, an 11-kb segment at coordinates 3087540 to 3098670 is inverted relative to MG1655. This region is flanked by IS10 elements and can function as a transposon, as evidenced by its identification in the mouse BAC clone AC125523 (GenBank GI:28626891). We confirmed the authenticity of the inversion by PCR.
Polymorphisms.
In addition to the 30 genes carrying synonymous SNPs in DH10B, 66 genes have missense mutations relative to MG1655 and 5 genes have nonsense mutations (Fig. 4A; see also Table S2 in the supplemental material). Eleven genes with missense mutations also contain a synonymous SNP. In addition, 42 SNPs are in intergenic regions. While some clustering of SNPs is apparent, particularly in regions of directed strain construction, polymorphisms are found across the genome.
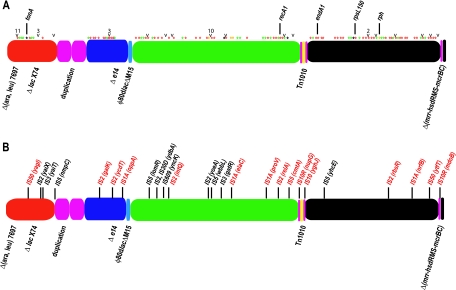
FIG. 4.
Distribution of small-scale mutations in the DH10B genome. A schematic of the genome illustrated in Fig. 3 shows the distribution of SNPs and frameshifts (A) and positioning of IS elements (B). (A) Differences in the DH10B sequence affecting conserved domain sequences are indicated by asterisks, with different colors indicating the types of change. Green, silent SNP; red, missense SNP; black, nonsense SNP; yellow, frameshift. Genes in which the mutation has a known phenotypic consequence are indicated. Note that the frameshift in rph corrects the rph-1 allele present in MG1655. SNPs in intergenic regions are indicated with a caret, and at sites where more than a SNP is present, the number of changes is given. (B) IS-disrupted genes in common with MG1655 are indicated in black, while those unique to DH10B are in red. The specific IS element at each location is indicated; the disrupted gene is in parentheses. For clarity, only differences affecting gene regions are shown.
Among the genes with missense mutations, three were expected, recA1, endA1, and rpsL150 (Table 1). DH10B contains seven genes, which have single-amino-acid changes, classified as essential by Baba et al. (2), including rpsL. The rpsL150 allele is known to confer streptomycin resistance, but the functional consequences for the other essential genes (dnaA, glmS, glyQ, lpxK, mreC, and murA) are unknown.
The five nonsense mutations (in chiA, gatZ, tonA, yigA, and ygcG), all result in substantially truncated protein products. Approximately half the tonA sequence reads are wild type and half are mutant, indicating that the bovine BAC library was made with competent cells from both the original DH10B strain and DH10B tonA. There is also a sequence ambiguity in the rpsT gene (causing a synonymous CAG-to-CAA change at genomic coordinate 20897) that may represent a mixed population as well.
Four genes are affected by frameshift mutations relative to MG1655: flhC, mglA, fruB, and rph. The 1-bp insertion in rph restores the wild-type reading frame from the rph-1 frameshift found in both MG1655 and W3110, thus alleviating the partial requirement for pyrimidine that is characteristic of both wild-type strains (23).
IS elements and phage elements.
The DH10B genome contains significantly more IS elements than MG1655 (63 elements compared to 43) (see Table S3 in the supplemental material). Among the 63 elements in DH10B, 26 are located within coding regions, including all 11 (disrupting 10 genes) found in MG1655 (Fig. 4B). Similarly, all 32 IS elements in MG1655 intergenic regions are the same elements in the same intervals in DH10B, except that the IS5 element located near the flhD promoter in DH10B is replaced by IS2 in MG1655. The DH10B duplication results in an extra copy of both IS5 and IS186B. Three IS elements in DH10B intergenic regions are not present at those sites in MG1655.
Six of the DH10B disruptions, including the previously known galK16 allele, affect carbon source uptake and metabolic pathways. The relA1 allele, which was present in the early progenitor strains (Fig. 1), inactivates the major GDP/GTP pyrophosphokinase responsible for producing (p)ppGpp during a stringent response (30). DH10B also harbors the spoT1 allele (Table 1) which increases basal (p)ppGpp levels by reducing the hydrolase activity of the bifunctional enzyme (14).
MG1655 contains 10 defective prophages, of which 8 are identical in DH10B with the exception of a synonymous SNP in the intR gene of Rac. As noted above, e14 is excised from DH10B; prophage Qin has two alterations, a missense mutant-encoding SNP in ydfU and an IS2 disruption of intQ. The φ80lacZΔM15 prophage was added during construction and is not present in MG1655.
In the final sequence assembly, we observed a contig that corresponds to a precisely excised, circular episome of the Rac defective phage element. The ability of Rac to exist as the oriJ plasmid has been documented previously (11). As oriJ plasmids do not replicate in the presence of a Rac prophage (11), the integrated and plasmid forms of the element must be derived from different DH10B cell subpopulations. The oriJ plasmid copy number has not been reported, but the depth of coverage of the episomal contig is twice that of the prophage, arguing that Rac excision and maintenance as a plasmid occur with significant frequency.
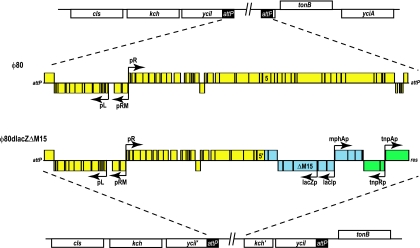
φ80lacZΔM15.
φ80 is a temperate lambdoid phage and shares a common genetic organization with λ (38), although there is limited homology at the nucleotide level (13). Integration of the phage occurs at the attP site in the 5′ end of yciI (27). Defective φ80 phages have also been used as transfer vectors to move desired alleles between strains (5, 6, 8). Using the complete φ80 sequence (G. Plunkett III, unpublished data) as a reference, we evaluated the structure of the φ80lacZΔM15 locus in DH10B (Fig. 5). The defective prophage consists of the three following segments: (i) 28,648 bp of the φ80 prophage ending within gene 5, (ii) a 12,744-bp piece of the E. coli chromosome extending from the middle of cynS to mhpD which also contains the lac region including the lacZΔM15 allele, and (iii) the 5,983-bp Tn1000 segment of the F plasmid. The 5′ end of the element is properly recombined at the attP site, whereas the 3′ end consists of the Tn1000 3′-end terminal repeat res, integrated into a partial copy of kch. Downstream of the disrupted kch gene is an intact copy of yciI including an attP site. This organization is consistent with the aberrant excision and recombination events necessary both to generate the defective phage element and integrate it into the recipient genome (5).
FIG. 5.
Comparison of φ80 and φ80dlacZΔM15 prophage structures. Diagrams of the two prophages show their chromosomal contexts and more-detailed schematics of the open reading frames (boxes) and major promoters within the two elements. Open reading frames above the center horizontal lines of the diagrams are transcribed in the forward direction, while those below the lines are coded on the complementary strand. Color boxes in the prophages indicate the source of the genes: yellow, φ80; blue, E. coli chromosome; green, Tn1000 from the F plasmid. The two types of terminal sequences, attP and res, are indicated, as is the lacZΔM15 allele. In φ80dlacZΔM15, the φ80 sequences end within gene 5 (indicated as 5′).
Comparison with MG1655 and W3110.
The two sequenced wild-type K-12 strains, MG1655 and W3110 (7, 20), display extensive sequence conservation across the entire lengths of their genomes (20). Differences are limited to a large inversion in W3110 (22), 13 IS and defective prophage insertions in only one of the strains, and nine base pair changes in eight genes (20).
Comparison of the DH10B sequence with the two wild-type strains argues that DH10B is more closely related to MG1655. DH10B does not contain the W3110 inversion and is identical to MG1655 at 16 out of 21 sites of divergence between the two wild-type strains (Table 2). The exception is the IS5 insertion in the flhD promoter in both DH10B and W3110 rather than the IS2 found in MG1655. Interestingly, DH10B lacks the unique 23S rRNA SNPs of the two other strains [rrlE(G2256A) in W3110, and rrlD(A2547G T2548A G2549T) in MG1655].
TABLE 2.
Comparison of DH10B, MG1655, and W3110 sequences
| Region affecteda | Typeb | Polymorphism inc:
|
DH10B correspondenced | |
|---|---|---|---|---|
| MG1655 | W3110 | |||
| alsK | Insertion | wt | IS5 | MG1655 |
| dcuC | Insertion | wt | IS5 | MG1655 |
| gatA | Insertion | wt | IS5 | MG1655 |
| rcsC | Insertion | wt | IS2 | MG1655 |
| tdcD | Insertion | wt | IS5 | MG1655 |
| tnaB | Insertion | wt | IS5 | MG1655 |
| flhD promoter | Insertion | IS1 | IS5 | W3110 |
| csgC-IG-ymdA | Insertion | wt | IS2 | MG1655 |
| lrhA-IG-yfhQ | Insertion | wt | IS1 | MG1655 |
| tnaC-IG-tnaA | Insertion | wt | IS5 | MG1655 |
| ychE-IG-oppA | Insertion | wt | IS2 | IS1A.2 |
| yieE-IG-yidZ | Insertion | wt | IS2 | MG1655 |
| eutA-IG-eutB | Insertion | CPZ-55 | wt | MG1655 |
| acnA | Missense | A (A522) | G (G522) | MG1655 |
| crp | Missense | C (T29) | A (K29) | MG1655 |
| intQ | Missense | T (F274) | C (L274) | IS2 |
| ycdT | Missense | C (A130) | T (V130) | IS2 |
| rpoS | Nonsense | C (Q33) | T (STOP33) | MG1655 |
| yedJ | Silent | C (V219) | T (V219) | MG1655 |
| rrlE | SNP | A2256 | G2256 | MG1655 |
| dcuA | Frameshift | wt | TT after nt 182 | MG1655 |
Mutations within genes are indicated by the gene name only. Mutations in intergenic (IG) regions are indicated along with flanking gene names.
Type of mutation in W3110 relative to MG1655.
SNPs in conserved domain sequences are indicated by the base present and the corresponding amino acid in parentheses. For rrlE, the base change and position within the gene are given. For dcuA, TT is inserted after nucleotide 182 in the gene. CPZ-55 is a prophage. wt, wild type.
DH10B correspondence with MG1655 or W3110 is indicated. Where DH10B differs from both, the associated change is noted.
Growth requirements.
DH10B is unable to grow on synthetic minimal medium. The strain lacks the leuLABCD operon [part of Δ(ara leu)7697], and indeed, when leucine is added to MOPS minimal medium with 0.1% glucose, DH10B grows aerobically with a doubling time of 76 min while MG1655 has a doubling time of 69 min in the same medium. The disparity in doubling times between DH10B and MG1655 was also observed with additional amino acid supplements. These results demonstrate that DH10B is strictly auxotrophic only for leucine and suggest that the lower growth rates are not due to partial requirements for other amino acids.
DH10B mutation rate.
The observations of Kovarik and coworkers regarding the high incidence of IS transposition (25), and the number and distribution of SNPs described here, raised the possibility that mutation frequencies are elevated in DH10B compared to those in the wild type. To test this hypothesis, the spontaneous mutation rates of both DH10B and MG1655 were determined using two different assays, namely, (i) a d-cycloserine resistance assay, detecting all types of mutations in the cycA gene (37, 43), and (ii) a rifampin resistance assay, detecting point mutations in the essential rpoB gene (24).
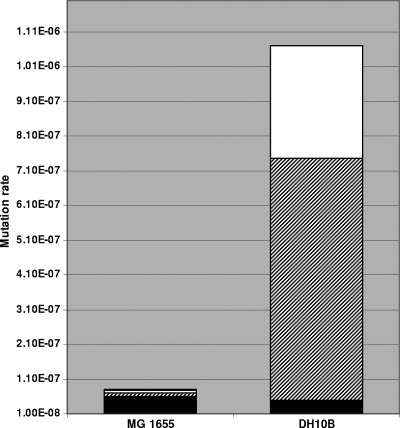
Mutation rate measurements based on d-cycloserine resistance were significantly different between DH10B and MG1655. Data from four independent experiments showed that the total spontaneous mutation rate of cycA in growing DH10B cultures was 13.5 times higher than that of MG1655 (1.07 × 10−6 compared to 7.90 × 10−8, respectively). The mutation spectra obtained from the two strains are also dramatically different (Fig. 6). In MG1655, 74% of the mutations obtained were point mutations, 24% were IS insertions, and 2% were deletions. About half of the IS-mediated disruptions were caused by IS150. In contrast, in DH10B, 95% of cycA mutations were IS insertions, 72% of which were IS150 mediated. Although the proportions of point mutations were vastly different (74% in MG1655 versus 5% in DH10B), the actual rates of point mutations were similar in the two strains (5.86 × 10−8 for MG1655 versus 5.02 × 10−8 in DH10B). No deletions were found among the cycA alleles in DH10B.
FIG. 6.
Comparison of the mutational spectra in DH10B and MG1655. A bar graph shows the distribution of cycA mutations obtained in MG1655 and DH10B. Rectangles within each bar represent point mutations (black), IS150 insertions (hatched), and other IS insertions (IS1, IS2, IS5, and not determined) (white). The rectangle representing deletions in MG1655 is not visible, and no deletions were detected in DH10B.
The similar point mutation frequencies of DH10B and MG1655 were confirmed by the rifampin resistance assay. Based on two independent experiments, the frequencies of rpoB point mutations were 2.43 × 10−9 for DH10B and 2.24 × 10−9 for MG1655. These rates are lower than those for cycA because only noninactivating rpoB mutations are viable.
Together, the results are consistent with the high level of IS transposition observed previously for DH10B (25) and imply that the numerous SNPs found in DH10B are a result of the extensive genetic manipulation that went into the construction of the strain rather than an increased point mutation rate.
DISCUSSION
We have taken advantage of sequence reads collected at HGSC during the bovine genome sequencing project to construct the complete DH10B genomic sequence by using a new desktop sequence assembler, SMGA. The sequence is informative in the understanding of both the biology of DH10B and the intended and unintended changes that can result during extensive strain construction using classic genetic methods.
Although the alleles targeted during the various construction steps are basically as expected, the deoR gene is a significant exception. The mutation of deoR was thought to be responsible for the enhanced transformation efficiency of DH10B (17, 19), but its sequence is unambiguously wild type. Mutations of deoR were originally isolated by selecting for mutants that grew rapidly on inosine but not uridine, due to the constitutive activation of the deoCABD operon (31). Using a similar selection scheme, Hanahan observed that the fast-growing strains also had higher transformation efficiencies of large plasmids and assumed that this was also caused by mutation of deoR (19a). The wild-type deoR locus indicates either that the two phenotypes (fast growth on inosine and high transformation efficiency) are completely separable or that another undefined locus (or loci) is responsible. In favor of the latter possibility, the same selection scheme was used to independently isolate the highly transformable DH5 strain (19a) which has now also been shown to contain a wild-type deoR gene (Invitrogen Corp., unpublished results). Interestingly, the multiple-deletion strain, MDS42, has transformation properties similar to those of DH10B (33), raising the possibility that they may share a common subset of mutated genes accounting for the phenotype. Even when pseudogenes and phage elements are excluded, there are still 52 mutated genes in common to be investigated. A systematic investigation of this set and its effect on transformation efficiency is now possible.
The 13.5-fold-higher mutation rate in DH10B than in MG1655 is entirely due to increased IS transposition. This is consistent with previous findings showing a high incidence of IS transposition into eukaryotic BAC library clones (25). In those studies, IS10 was the most frequently observed element in the BAC clones, while IS150 transposition predominated in our study. It is important to note that cycA does not contain a preferred IS10 target site (18, 25), so that IS10 transpositions are expected to be rare. No target site specificity has yet been reported for IS150 or other IS3 family members, but this is under investigation in a separate project. IS150 transposase levels and/or activity could be elevated in DH10B. IS150 transposase production is regulated by a highly efficient programmed translational frameshifting mechanism (42), although precise details of the frameshifting mechanism are only now emerging. No obvious connections with the DH10B genotype are evident.
Conserved IS5 elements were likely involved in creating the large tandem duplication that doubles the gene dosage of 106 genes. Such duplications are quite common in E. coli and Salmonella enterica serovar Typhimurium, probably due to RecA-dependent unequal-sister-strand exchanges between repeated sequences (36), although they are lost at high frequency unless they confer some selective advantage under the given growth conditions. While recA1 in DH10B would allow fixation of the duplication even in the absence of a selective advantage, the three construction steps following introduction of recA1 (Fig. 1) employed a wild-type RecA-expressing plasmid that was subsequently cured from the strain. This implies either that the duplication arose very late in the construction and was fixed by curing of the recA plasmid or that it confers a selective advantage for growth on complex media (e.g., Luria-Bertani or tryptone broth) that are generally used for culturing DH10B. One candidate operon for positive selection is gltLKJI, which encodes the glutamate-aspartate ABC family transporter (28). Cells growing in complex media consume available amino acids in a sequential fashion, with serine and aspartate being used during exponential growth and others such as glutamate used in the transition to stationary phase (4, 34). Doubling the expression of gltLKJI could enhance the uptake of these amino acids, providing a growth advantage.
The range of nutrients that DH10B can utilize is limited by the deletion of numerous metabolic pathways. Nevertheless, DH10B requires only leucine as a supplement for growth on minimal media with a suitable carbon source. The consistently lower growth rates observed with DH10B cultures compared to MG1655 are likely a consequence of elevated basal (p)ppGpp levels caused by the spoT1 allele as seen in different backgrounds (14, 26). Consistent with the “relaxed” phenotype imparted by relA1, however, DH10B does exhibit extensive growth lags during nutrient downshifts (data not shown).
Classical genetic strain construction has been an invaluable tool in elucidating much basic molecular biology. DH10B can be considered an extreme case by the extent of manipulations and the resulting changes to the genome. While most of the changes do not impact the strain itself, IS transposition into cloned fragments propagated in this host sends a strong cautionary signal regarding uncharacterized genomes. Recent advances in sequencing technology and strain construction are now allowing such issues to be eliminated (33).
Supplementary Material
Acknowledgments
We are grateful to Joel Malek and Heather Peckham at Applied Biosystems for performing the sequencing of DH10B on the SOLiD system. We thank John Wertz (Yale Stock Center) and Frederic Bloom for providing detailed information on the construction of MC1061 and DH10B, respectively. We also thank Dmitry Shevchenko and John Campbell from Scarab Genomics for PCR verification of the inversion within Tn10.10 and stimulating discussions. We also thank Benjamin German, a high school interm at Scarab Genomics, for excellent technical assistance. The finishing phase was performed by Lisa Hemphill and the production staff at the Baylor College of Medicine HGSC.
This work was supported by NIH grant U54 HG003273 at the Baylor College of Medicine HGSC and by an OTKA grant to G.P.
Footnotes
Published ahead of print on 1 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allex, C. F., S. F. Baldwin, J. W. Shavlik, and F. R. Blattner. 1997. Increasing consensus accuracy in DNA fragment assemblies by incorporating fluorescent trace representations. Proc. Int. Conf. Intell. Syst. Mol. Biol. 53-14. [PubMed] [Google Scholar]
- 2.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 21 February 2006, posting date. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed]
- 3.Bachmann, B. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 4.Baev, M. V., D. Baev, A. J. Radek, and J. W. Campbell. 2006. Growth of Escherichia coli MG1655 on LB medium: monitoring utilization of amino acids, peptides, and nucleotides with transcriptional microarrays. Appl. Microbiol. Biotechnol. 71317-322. [DOI] [PubMed] [Google Scholar]
- 5.Beckwith, J. R., and E. R. Signer. 1966. Transposition of the lac region of Escherichia coli. I. Inversion of the lac operon and transduction of lac by phi80. J. Mol. Biol. 19254-265. [DOI] [PubMed] [Google Scholar]
- 6.Beckwith, J. R., E. R. Signer, and W. Epstein. 1966. Transposition of the lac region of E. coli. Cold Spring Harbor Symp. Quant. Biol. 31393-401. [DOI] [PubMed] [Google Scholar]
- 7.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 8.Brickman, E., and J. Beckwith. 1975. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J. Mol. Biol. 96307-316. [DOI] [PubMed] [Google Scholar]
- 9.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138179-207. [DOI] [PubMed] [Google Scholar]
- 10.Darling, A. C., B. Mau, F. R. Blattner, and N. T. Perna. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 141394-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz, R., P. Barnsley, and R. H. Pritchard. 1979. Location and characterisation of a new replication origin in the E. coli K12 chromosome. Mol. Gen. Genet. 175151-157. [DOI] [PubMed] [Google Scholar]
- 12.Feher, T., B. Cseh, K. Umenhoffer, I. Karcagi, and G. Posfai. 2006. Characterization of cycA mutants of Escherichia coli. An assay for measuring in vivo mutation rates. Mutat. Res. 595184-190. [DOI] [PubMed] [Google Scholar]
- 13.Fiandt, M., Z. Hradecna, H. A. Lozeron, and W. Szybalski. 1971. Electron micrographic mapping of deletions, insertions, inversions and homologies in the DNAs of coliphages lambda and pHi80, p. 329-354. In A. D. Hershey (ed.), The bacteriophage lambda. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 14.Fiil, N. P., B. M. Willumsen, J. D. Friesen, and K. von Meyenburg. 1977. Interaction of alleles of the relA, relC, and spoT genes in Escherichia coli: analysis of the interconversion of GTP, ppGpp and pppGpp. Mol. Gen. Genet. 15087-101. [DOI] [PubMed] [Google Scholar]
- 15.Glasner, J. D., P. Liss, G. Plunkett III, A. Darling, T. Prasad, M. Rusch, A. Byrnes, M. Gilson, B. Biehl, F. R. Blattner, and N. T. Perna. 2003. ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 31147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glasner, J. D., M. Rusch, P. Liss, G. Plunkett III, E. L. Cabot, A. Darling, B. D. Anderson, P. Infield-Harm, M. C. Gilson, and N. T. Perna. 2006. ASAP: a resource for annotating, curating, comparing, and disseminating genomic data. Nucleic Acids Res. 34D41-D45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 874645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Hall, B. G. 1998. Activation of the bgl operon by adaptive mutation. Mol. Biol. Evol. 151-5. [DOI] [PubMed] [Google Scholar]
- 18.Halling, S. M., and N. Kleckner. 1982. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell 28155-163. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan, D., J. Jessee, and F. R. Bloom. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 20463-113. [DOI] [PubMed] [Google Scholar]
- 19a.Hanahan, D. July 1989. Biologically pure Escherichia coli cell line which is a deoR− mutant and which is more transformation efficient with foreign plasmids than deoR+ Escherichia coli cell lines, process for obtaining these cell lines, methods of use. U.S. patent 4,851,348.
- 20.Hayashi, K., N. Morooka, Y. Yamamoto, K. Fujita, K. Isono, S. Choi, E. Ohtsubo, T. Baba, B. L. Wanner, H. Mori, and T. Horiuchi. 21 February 2006, posting date. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. doi: 10.1038/msb4100049. [DOI] [PMC free article] [PubMed]
- 21.Hill, C. W., J. A. Gray, and H. Brody. 1989. Use of the isocitrate dehydrogenase structural gene for attachment of e14 in Escherichia coli K-12. J. Bacteriol. 1714083-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill, C. W., and B. W. Harnish. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. USA 787069-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen, K. F. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 1753401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin, D. J., and C. A. Gross. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 20245-58. [DOI] [PubMed] [Google Scholar]
- 25.Kovarik, A., M. A. Matzke, A. J. Matzke, and B. Koulakova. 2001. Transposition of IS10 from the host Escherichia coli genome to a plasmid may lead to cloning artefacts. Mol. Genet. Genomics 266216-222. [DOI] [PubMed] [Google Scholar]
- 26.Laffler, T., and J. A. Gallant. 1974. Stringent control of protein synthesis in E. coli. Cell 347-49. [DOI] [PubMed] [Google Scholar]
- 27.Leong, J. M., S. Nunes-Duby, C. F. Lesser, P. Youderian, M. M. Susskind, and A. Landy. 1985. The phi 80 and P22 attachment sites. Primary structure and interaction with Escherichia coli integration host factor. J. Biol. Chem. 2604468-4477. [PubMed] [Google Scholar]
- 28.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 285-13. [DOI] [PubMed] [Google Scholar]
- 29.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y. J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. Alenquer, T. P. Jarvie, K. B. Jirage, J. B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metzger, S., G. Schreiber, E. Aizenman, M. Cashel, and G. Glaser. 1989. Characterization of the relA1 mutation and a comparison of relA1 with new relA null alleles in Escherichia coli. J. Biol. Chem. 26421146-21152. [PubMed] [Google Scholar]
- 31.Munch-Petersen, A., P. Nygaard, K. Hammer-Jespersen, and N. Fiil. 1972. Mutants constitutive for nucleoside-catabolizing enzymes in Escherichia coli K12. Isolation, charactrization and mapping. Eur. J. Biochem. 27208-215. [DOI] [PubMed] [Google Scholar]
- 32.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Posfai, G., G. Plunkett III, T. Feher, D. Frisch, G. M. Keil, K. Umenhoffer, V. Kolisnychenko, B. Stahl, S. S. Sharma, M. de Arruda, V. Burland, S. W. Harcum, and F. R. Blattner. 2006. Emergent properties of reduced-genome Escherichia coli. Science 3121044-1046. [DOI] [PubMed] [Google Scholar]
- 34.Pruss, B. M., J. M. Nelms, C. Park, and A. J. Wolfe. 1994. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J. Bacteriol. 1762143-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riley, M., T. Abe, M. B. Arnaud, M. K. Berlyn, F. R. Blattner, R. R. Chaudhuri, J. D. Glasner, T. Horiuchi, I. M. Keseler, T. Kosuge, H. Mori, N. T. Perna, G. Plunkett, 3rd, K. E. Rudd, M. H. Serres, G. H. Thomas, N. R. Thomson, D. Wishart, and B. L. Wanner. 2006. Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth, J. R., N. Benson, T. Galitski, K. Haack, J. Lawrence, and L. Miesel. 1996. Rearrangements of the bacterial chromosome—formation and applications, p. 2256-2276. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 37.Russell, R. R. 1972. Mapping of a d-cycloserine resistance locus in Escherichia coli K-12. J. Bacteriol. 111622-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rybchin, V. N. 1984. Genetics of bacteriophage phi 80—a review. Gene 273-11. [DOI] [PubMed] [Google Scholar]
- 39.Sarkar, S., W. T. Ma, and G. H. Sandri. 1992. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica 85173-179. [DOI] [PubMed] [Google Scholar]
- 40.Siguier, P., J. Perochon, L. Lestrade, J. Mahillon, and M. Chandler. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34D32-D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart, F. M., D. M. Gordon, and B. R. Levin. 1990. Fluctuation analysis: the probability distribution of the number of mutants under different conditions. Genetics 124175-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogele, K., E. Schwartz, C. Welz, E. Schiltz, and B. Rak. 1991. High-level ribosomal frameshifting directs the synthesis of IS150 gene products. Nucleic Acids Res. 194377-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wargel, R. J., C. A. Hadur, and F. C. Neuhaus. 1971. Mechanism of d-cycloserine action: transport mutants for d-alanine, d-cycloserine, and glycine. J. Bacteriol. 1051028-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.