Abstract
Tryptophan residues can possess a multitude of functions within a multidrug transport protein, e.g., mediating interactions with substrates or distal parts of the protein, or fulfilling a structural requirement, such as guiding the depth of membrane insertion. In this study, the nine tryptophan residues of the staphylococcal QacA multidrug efflux protein were individually mutated to alanine and phenylalanine, and the functional consequences of these changes were determined. Phenylalanine substitutions for each tryptophan residue were functionally tolerated. However, alanine modifications revealed an important functional role for three tryptophan residues, W58, W149, and W173, each of which is well conserved among QacA-related transport proteins in the major facilitator superfamily. The most functionally compromising mutation, an alanine substitution for W58, likely to be located at the extracellular interface of transmembrane segment 2, abolished all detectable QacA-mediated resistance and transport function. Second-site suppressor analyses identified several mutations that rescued the function of the W58A QacA mutant. Remarkably, all of these suppressor mutations were shown to be located in cytoplasmic loops between transmembrane helices 2 and 3 or 12 and 13, demonstrating novel functional associations between amino acid positions on opposite sides of the membrane and in distal N- and C-terminal regions of the QacA protein.
The qacA multidrug resistance determinant has been frequently identified on plasmids carried by clinical isolates of Staphylococcus aureus (21, 23, 30). The qacA gene encodes a multidrug transport protein, QacA (see Fig. 1), which displays exceptionally broad substrate specificity, conferring resistance to both monovalent and bivalent cationic antimicrobials from at least 12 different chemical families (5). Competition studies of the QacA transporter have determined that monovalent and bivalent cationic substrates interact with QacA at distinct sites (22); however, recent results have suggested that these sites are in physical proximity, possibly within the same extended binding region (32). Additionally, as observed for other multidrug transporters, the spatial organization of amino acid side chains that mediate stabilizing interactions with drugs within the QacA binding region appears to be flexible (3, 11). Transport mediated by QacA is powered by the proton motive force and can be described using the Michaelis-Menten kinetic parameters Km and Vmax (22).
FIG. 1.
Primary and secondary structures of the QacA multidrug transport protein. Amino acid residues known or predicted from hydropathy analysis to comprise the 14 TMS of QacA are shown within numbered boxes. The positions of mutated residues are given. The nine tryptophan residues of QacA are highlighted in black circles; residues mutated in W58A suppressor mutations are circled; and P309, used as a control for solvent accessibility analysis, is boxed. The locations and sequences of the MFS sequence motifs A and H (DHA2 specific), which were modified in these studies by mutagenesis of D79 and W173, respectively, are shown. Capitalized residues are 70% conserved, and lowercased residues are 30% conserved (25).
The QacA transport protein is classified as being within the drug:H+ antiporter 2 (DHA2) family of the major facilitator superfamily (MFS) of transport proteins (25) (www.tcdb.org). A defining feature of DHA2 family transporters, including QacA, is their organization into 14 α-helical transmembrane segments (TMS) (Fig. 1) (24, 25). This is in contrast to the majority of MFS transporters, which are composed of 12 TMS, such as the Escherichia coli lactose permease, glycerol-3-phosphate transporter, and putative multidrug transporter EmrD, which have been structurally characterized using 3-dimensional crystallography (2, 13, 34). Interestingly, these and other 12-TMS MFS transporters possess similar tertiary folds, with respect to the packing of their TMS, irrespective of their substrate specificities, vectorial mechanisms, or transporter subfamily classifications (12). The 12 TMS of these proteins are organized into two pseudosymmetrical domains consisting of the first and last 6 TMS, respectively. These domains interface primarily between TMS 2 and 11 and between TMS 5 and 8 and surround a central translocation region. Given the high structural conservation among 12-TMS MFS transporters, it has been suggested that these proteins operate using similar alternating-access functional mechanisms, where a centrally bound substrate(s) is provided access to the opposite side of the membrane by rocker switch-type motions between the N- and C-terminal domains (1, 16, 19, 20).
Although significant structural data are available for 12-TMS MFS transporters, the structure of a 14-TMS MFS transporter has yet to be determined. Nonetheless, given that 14-TMS MFS transporters are believed to have evolved from a 12-TMS precursor with the acquisition of two additional central TMS (27), the possibility that functionally related 12- and 14-TMS MFS transport proteins display similar tertiary organizations is an attractive hypothesis (15). Indeed, recent biochemical data have demonstrated that the DHA2 family tetracycline transporter, TetA(K), contains a solvent-accessible region along the length of TMS 10 (10), similar to the corresponding TMS 8 of 12-TMS MFS tetracycline transporters, which lines the central translocation region along its entire length (29).
The present study investigated the role(s) of the nine tryptophan residues in the QacA transport protein (Fig. 1). Tryptophan residues potentiate a range of molecular interactions, including cation-π, π-π, or aromatic stacking interactions, which could contribute to the stable binding of aromatic, cationic QacA substrates or form the basis for interactions between distal parts of the QacA protein (6, 7). Additionally, a number of tryptophan residues are predicted to be located at the periphery of the TMS in QacA, where they may function to guide the depth of TMS membrane insertion (Fig. 1) (4, 31). Several tryptophan residues within QacA are well conserved among DHA2 family transporters, and some have been found in previously recognized amino acid sequence motifs, suggesting that they function in a role common to these transporters (Fig. 1) (9, 25). Second-site suppressor studies were used to investigate the most functionally debilitating tryptophan substitution and revealed interesting functional complementation between residues in distal positions of the QacA polypeptide.
MATERIALS AND METHODS
Bacterial strains, plasmids, reagents, and media.
E. coli strain DH5α [supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] (8) was used as the host for all cloning procedures and functional analyses. The plasmids used were pBluescript II SK (Stratagene); the pBluescript II SK-based qacA clone pSK4322 (32), which encodes QacA protein with a C-terminal hexahistidine tag; and a pSK4322 derivative, pSK7400, that encodes a QacA W58A mutant (this study). Bacterial strains were cultured in Luria-Bertani broth and on Luria-Bertani agar plates, unless otherwise stated. Ampicillin was used at a concentration of 100 μg/ml for plasmid selection where required. Ampicillin, benzalkonium chloride, carbonyl cyanide m-chlorophenylhydrazone, chlorhexidine diacetate, dequalinium chloride, 4′,6′-diamidino-2-phenindole (DAPI), n-dodecyl-β-d-maltoside, ethidium bromide, N-ethylmaleimide (NEM), hydroxylamine hydrochloride, pentamidine isothionate, pyronin Y, and rhodamine 6G were purchased from Sigma. Fluorescein maleimide was obtained from Pierce. All other reagents were of analytical grade and were obtained through commercial sources.
DNA manipulations and random and site-directed mutagenesis.
Plasmid isolation and subcloning were conducted using standard molecular biological techniques (28). Alternatively, plasmids were isolated using the Quantum Prep plasmid miniprep kit (Bio-Rad). Restriction endonucleases were purchased from New England Biolabs and were used according to the manufacturer's instructions. Oligonucleotides were synthesized by GeneWorks (Australia). DNA sequencing was conducted at the Australian Genome Research Facility (Brisbane), and gene sequence data were assembled, stored, and analyzed using Sequencher, version 4.2.2 (Gene Codes Corp.).
Site-directed mutagenesis was conducted by the QuikChange (Stratagene) method using pairs of complementary oligonucleotide primers, which incorporated the desired amino acid change along with a silent mutation to add or remove a restriction endonuclease site in order to facilitate identification of mutant plasmids.
Second-site suppressor mutants of the nonfunctional QacA W58A mutant were generated by random hydroxylamine mutagenesis. Since hydroxylamine is a strong mutagen, appropriate safety procedures were used. The plasmid encoding the QacA W58A mutant, pSK7400, was treated in a buffer containing 50 mM potassium phosphate (pH 6.8), 100 mM EDTA, and 400 mM hydroxylamine hydrochloride for either (i) 45 min at 60°C or (ii) 1 h at 60°C and then 16 h at 37°C. Mutated pSK7400 DNA was purified using the GFX PCR DNA and gel band purification kit (Amersham Biosciences) and was transformed into E. coli DH5α cells. Functional W58A second-site suppressor mutants were selected by plating onto medium containing an inhibitory concentration (200 μg/ml) of ethidium bromide. To ensure that the ethidium resistance was conferred specifically by the qacA W58A mutant derivatives, the qacA genes carried by resistant colonies were isolated, cloned into fresh pBluescript II SK plasmids, and transformed into E. coli DH5α cells, and their ethidium resistance capacities were tested. Plasmids from resistant colonies were isolated and sequenced to identify the locations of the functionally restorative secondary mutations prior to further functional analyses.
Genes encoding QacA G400D or A403T mutants were constructed by subcloning the W58A G400D and W58A A403T mutant genes with wild-type qacA. The 534-bp BspEI/XbaI fragment of plasmid pSK4322 was replaced with the corresponding fragments of the pSK7400-derived plasmids encoding W58A G400D and W58A A403T QacA mutants to produce qacA derivatives encoding only the G400D and A403T mutations.
Western blot analyses.
Western blotting was conducted on lysates of E. coli cells expressing wild-type QacA or variants by using an anti-QacA rabbit polyclonal antiserum as described previously (32). Western blot membranes were scanned using a GS-710 calibrated imaging densitometer (Bio-Rad), and results were analyzed using Quantity One software (Bio-Rad) to determine the level of expression of each QacA variant relative to that of the wild-type protein.
MIC analyses.
Cultures of E. coli DH5α cells expressing each QacA mutant protein were grown from single freshly transformed colonies, diluted 1:20 in fresh medium, and grown to an optical density at 600 nm (OD600) of 0.6. After the cell density was adjusted to an OD600 of 0.1, the cells were diluted 1:10 and spotted using a multipoint replicator onto Mueller-Hinton agar medium containing either ethidium (25 to 1,000 μg/ml), rhodamine 6G (25 to 1,500 μg/ml), pyronin Y (25 to 500 μg/ml), benzalkonium (10 to 70 μg/ml), dequalinium (25 to 1,100 μg/ml), pentamidine (25 to 300 μg/ml), or chlorhexidine (0.5 to 6 μg/ml) at increasing concentrations.
Fluorescent transport assays.
Transport of the monovalent substrate ethidium and transport of the bivalent substrate DAPI were measured fluorimetrically in whole E. coli cells as previously described (22, 32). Briefly, 1-ml volumes of cells expressing QacA or a QacA derivative, suspended in 20 mM HEPES (pH 7.0) with an OD600 of 0.6, were loaded with either 15 μM ethidium or 8 μM DAPI in the presence of 10 mM carbonyl cyanide m-chlorophenylhydrazone. Substrate-loaded cells were washed in 20 mM HEPES (pH 7.0) and suspended in 1 ml of the same buffer. Transport was initiated by the addition of 125 mM sodium formate and was monitored fluorimetrically for at least 3 min using a Hitachi 4500 fluorescence spectrophotometer with excitation and emission wavelengths of 530 nm and 610 nm, respectively, for ethidium and 364 nm and 454 nm, respectively, for DAPI. Since ethidium transport by wild-type QacA and the QacA mutants was rapid, only the first minute of ethidium efflux was depicted so as to better resolve differences in the initial rates of transport mediated by these proteins. To determine the effect of maleimide modification, cells expressing cysteine-substituted QacA mutants were treated with 5 mM NEM for 20 min at 37°C prior to substrate loading.
Fluorescein maleimide reactivity analysis.
The fluorescein maleimide reactivities of QacA derivatives with single cysteine substitutions were determined both in whole cells and in disrupted cell membranes. The sample preparation and fluorescein maleimide labeling of QacA protein within disrupted cell membranes have been described previously (32). For labeling of QacA protein within whole cells, cells were collected from 16-h cultures of E. coli DH5α carrying pSK4322-based plasmids encoding a QacA derivative and were suspended in 15 mM Tris-HCl (pH 7.5)-10% glycerol. Fluorescein maleimide was added to a final concentration of 0.25 mM, and cells were incubated at 37°C for 25 min. Reactions were stopped by the addition of 10 volumes of 50 mM Tris-HCl (pH 8.0), 10% glycerol, and 20 mM NEM. QacA protein was solubilized using a mixture of 1% sodium dodecyl sulfate (SDS) and 0.5% n-dodecyl-β-d-maltoside. Nonlysed cells and insoluble debris were removed by centrifugation, and QacA protein was purified as outlined for cell membrane samples (32). Purified protein samples were fractionated on 10% SDS-polyacrylamide gels and scanned using a Molecular Imager FX (Bio-Rad) to reveal the associated fluorescein label. Total QacA protein was detected by staining gels with Coomassie blue R-250 for disrupted membrane samples or by Western blotting using an anti-QacA polyclonal antibody for samples purified from whole cells. Stained gels and Western blots were scanned using a GS-710 calibrated imaging densitometer (Bio-Rad), and all images were analyzed using Quantity One software (Bio-Rad) in order to determine the relative levels of fluorescein maleimide labeling.
RESULTS
Construction and expression of alanine- and phenylalanine-substituted tryptophan mutants.
The QacA export protein contains nine tryptophan residues, six of which are predicted to be membrane embedded and three of which are predicted to be within loop regions based on topological analyses (Fig. 1) (24, 26). Each of these nine tryptophan residues was individually mutated to both alanine and phenylalanine using site-directed mutagenesis of the qacA expression plasmid pSK4322. As described previously, this pBluescript-based plasmid allows constitutive low-level QacA protein expression in E. coli cells at a level ideal for functional studies (9). The expression level of each of the 18 QacA mutant proteins in E. coli DH5α cells, relative to that of wild-type QacA, was determined by semiquantitative Western blot analysis. None of the tryptophan mutations seriously compromised the expression potential of the QacA protein: the lowest level of expression determined, that for the W300A mutant, was 61% of that of wild-type QacA (Table 1). Notably, both the alanine and the phenylalanine mutations of W18 increased QacA protein expression levels significantly above those of the wild-type protein (Table 1).
TABLE 1.
Resistance profiles and expression levels of QacA tryptophan mutants
| QacA mutation | Relative expressiona | Relative MIC of mutantb
|
||||||
|---|---|---|---|---|---|---|---|---|
| Monovalent substrates
|
Bivalent substrates
|
|||||||
| Et (dye) | R6G (dye) | PyY (dye) | Bc (Qac) | Dq (Qac) | Pe (Dd) | Ch (Bg) | ||
| Wild-type QacA | 100 | 100 (400) | 100 (800) | 100 (200) | 100 (40) | 100 (900) | 100 (300) | 100 (4) |
| No QacA | N/A | 15 | 40 | 25 | 50 | 5 | 35 | 25 |
| Site-directed substitutions of native tryptophan residues | ||||||||
| W18A | 149 | 170 | 150 | 200 | ND | 110 | ND | 125 |
| W18F | 141 | 155 | 150 | 190 | ND | 110 | ND | 125 |
| W58A | 94 | 15 | 40 | 25 | 50 | 5 | 35 | 25 |
| W58F | 80 | 90 | 50 | 60 | ND | 55 | ND | 90 |
| W81A | 92 | 100 | 100 | ND | 100 | 100 | 100 | 100 |
| W81F | 79 | 90 | 95 | ND | 100 | 90 | 90 | 90 |
| W149A | 84 | 45 | 85 | 50 | ND | 55 | ND | 60 |
| W149F | 95 | 100 | 65 | 50 | ND | 65 | ND | 90 |
| W173A | 74 | 40 | 45 | 25 | 65 | 10 | 35 | 75 |
| W173F | 103 | 155 | 115 | 140 | ND | 110 | ND | 100 |
| W208A | 116 | 75 | 115 | 65 | ND | 60 | ND | 75 |
| W208F | 85 | 90 | 150 | 110 | ND | 110 | ND | 100 |
| W225A | 71 | 110 | 150 | 125 | ND | 110 | ND | 100 |
| W225F | 103 | 90 | 85 | 90 | ND | 100 | ND | 100 |
| W241A | 77 | 145 | 115 | 110 | ND | 110 | ND | 115 |
| W241F | 95 | 125 | 115 | 110 | ND | 110 | ND | 110 |
| W300A | 61 | 75 | 95 | ND | 100 | 100 | 85 | 25 |
| W300F | 95 | 100 | 95 | ND | 100 | 100 | 100 | 100 |
| W58A second-site suppressor mutants | ||||||||
| W58T | 98 | 45 | 50 | ND | 90 | 100 | 50 | 75 |
| W58A D79N | 70 | 40 | 65 | ND | 100 | 100 | 50 | 75 |
| W58A M391I | 48 | 50 | 55 | ND | 100 | 100 | 50 | 75 |
| W58A G400D | 38 | 140 | 80 | ND | 115 | 90 | 50 | 75 |
| W58A A403T | 22 | 100 | 80 | ND | 115 | 90 | 50 | 75 |
| G400D | 68 | 45 | 55 | ND | 75 | 5 | 40 | 25 |
| A403T | 80 | 30 | 70 | ND | 75 | 15 | 40 | 40 |
| Cysteine substitutions | ||||||||
| W58C | 91 | 15 | 40 | ND | 50 | 5 | 35 | 25 |
| D79C | 84 | 140 | 125 | ND | 100 | 80 | 100 | 100 |
| M391C | 69 | 80 | 125 | ND | 100 | 70 | 80 | 75 |
| G400C | 90 | 70 | 80 | ND | 100 | 90 | 60 | 75 |
| A403C | 79 | 55 | 95 | ND | 100 | 20 | 50 | 50 |
Expressed as a percentage of the wild-type QacA expression level. Values are averages from two Western blot analyses. N/A, not applicable.
MICs were determined for E. coli DH5α cells expressing QacA proteins. Each value is the percentage of the MIC for cells expressing wild-type QacA (given in parentheses [in micrograms per milliliter] in the first row) and is the average from at least two separate experiments. Et, ethidium; R6G, rhodamine 6G; PyY, pyronin Y; Bc, benzalkonium; Dq, dequalinium; Pe, pentamidine; Ch, chlorhexidine; Qac, quaternary ammonium compounds; Dd, diamidines; Bg, biguanidines; ND, not determined.
Resistance capacities of QacA tryptophan mutants.
MIC analyses were used to gauge the functional consequences of mutating the tryptophan residues in QacA, in terms of QacA-mediated resistance to a diverse range of representative drugs (Table 1). Remarkably, the W58A QacA mutant could not confer resistance to any of the compounds tested. Nonetheless, the W58F mutant maintained the capacity for at least intermediate-level resistance to all compounds, demonstrating that a tryptophan residue was not functionally essential at position 58. Serious reductions in the capacity for resistance to rhodamine 6G, pyronin Y, dequalinium, and pentamidine were observed after the W173A mutation in QacA, and the overall resistance capacity of the W149A mutant was reduced to intermediate levels (Table 1). Not surprisingly, W58, W149, and W173 are the tryptophan residues most highly conserved between QacA and other proteins in the DHA2 family of the MFS, as determined by amino acid sequence alignments (9, 25). Indeed, W173 is the first residue of amino acid sequence motif H, which is conserved in TMS 6 of DHA2 family transporters (Fig. 1) (25). In contrast to the W58A, W149A, and W173A mutants, cells expressing W18A, W18F, W173F, W225A, W241A, or W241F QacA mutant proteins were better equipped for growth on media containing QacA substrates than cells carrying wild-type QacA (Table 1). However, in some cases this may be a reflection of the higher expression levels of the mutant proteins (Table 1).
Transport mediated by QacA tryptophan mutants.
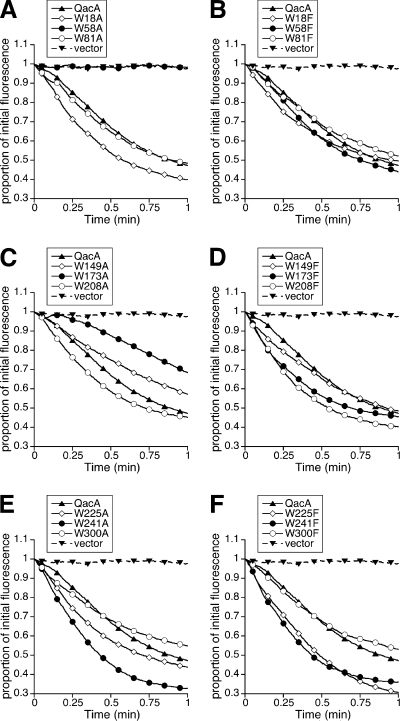
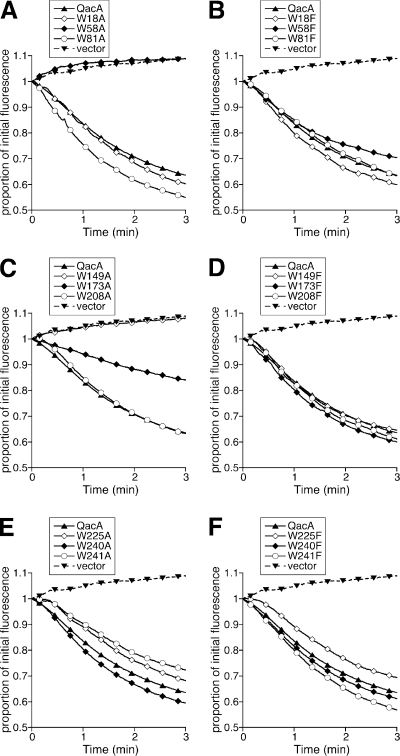
To further characterize the transport capabilities of mutant QacA proteins, each was examined for its capacity to induce efflux of ethidium and DAPI, representatives of monovalent and bivalent QacA substrates, respectively, by using fluorescence-based transport assays. In keeping with the results of MIC analyses, the W58A mutant was incapable of supporting either ethidium or DAPI transport (Fig. 2A and 3A). The capacity of the W173A mutant for substrate efflux was retarded, and the W149A mutant was functional in ethidium transport but failed to promote DAPI efflux at detectable levels (Fig. 2C and 3C). The remaining QacA mutants, including all those containing a mutation to phenylalanine, were able to induce the efflux of both ethidium and DAPI with initial transport rates close to or greater than those of wild-type QacA (Fig. 2 and 3).
FIG. 2.
Transport of the monovalent QacA substrate ethidium mediated by QacA mutants in which tryptophan has been replaced with alanine or phenylalanine. E. coli DH5α cells expressing each mutant (identified above each plot) were loaded with 15 μM ethidium bromide and monitored fluorimetrically after energization of transport. Transport reactions were conducted at least in duplicate, and the results of representative experiments are shown.
FIG. 3.
Transport of the bivalent QacA substrate DAPI mediated by QacA mutants in which tryptophan has been replaced with alanine or phenylalanine. E. coli DH5α cells expressing each mutant (identified above each plot) were loaded with 8 μM DAPI and monitored fluorimetrically after energization of transport. Transport reactions were conducted at least in duplicate, and the results of representative experiments are shown.
Isolation of W58A suppressor mutants.
The W58A amino acid substitution clearly affected the basic functional mechanism of QacA-mediated transport. To further explore this finding, second-site suppressor studies were conducted. Random mutants of the QacA W58A template were generated by hydroxylamine mutagenesis and transformed into E. coli DH5α cells in order to identify functional mutants on ethidium selective media. Five genes encoding qacA secondary mutants from the W58A derivative, each containing a distinct functionally restorative secondary mutation, were isolated. One mutation caused a primary-site substitution, changing the alanine at position 58 to a threonine residue. The remaining four mutations caused amino acid changes at positions distal to W58A, viz., D79N, M391I, G400D, and A403T. Western blot analyses were conducted to assess the level of expression of each suppressor mutant protein relative to those of wild-type QacA and the parental W58A mutant. The primary-site W58T mutant protein was expressed at a level close to those of wild-type and W58A mutant QacA; however, the four second-site mutants were expressed at lower levels (Table 1). The poor expression was particularly evident for the W58A G400D and W58A A403T QacA mutants, which were produced at 38% and 22% of wild-type levels, respectively (Table 1).
Functional analyses of W58A suppressor mutants.
The drug resistance profiles of the W58A suppressor mutants were determined using MIC analyses. Although these mutants were identified using only one QacA substrate, ethidium, all five mutations restored a degree of QacA-mediated resistance to all substrates tested (Table 1), supporting the idea that the deleterious effect of the W58A mutation was inherent to the basic QacA functional mechanism. At least intermediate levels of resistance to each substrate were mediated by the W58T, W58A D79N, and W58A M391I mutants (Table 1). Additionally, despite their poor expression, the levels of resistance conferred by the W58A G400D and W58A A403T QacA mutants were approximately equal to or, in the case of the monovalent substrates tested, greater than those of the other second-site mutants (Table 1). To determine their effects on QacA function in the absence of the W58A mutation, the most functionally restorative W58A second-site mutations, G400D and A403T, were isolated by replacing the portions of the W58A G400D and W58A A403T mutant plasmids encoding the W58A mutation with the wild-type sequence. The G400D and A403T QacA single-mutant proteins were expressed at levels below that of wild-type QacA but higher than those of the W58A G400D and W58A A403T double mutants. Nonetheless, these single mutants displayed poor overall drug resistance capacities that were close to or at background levels for some QacA substrates (Table 1). One possibility is that in the absence of the W58A mutation, these second-site suppressor mutations affect the QacA functional mechanism in a manner similar to that of the parental W58A substitution, albeit to a lesser extent.
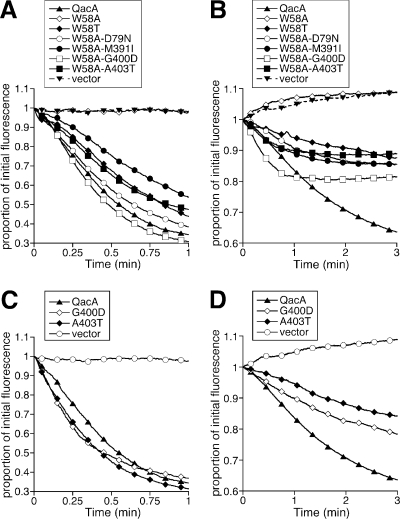
Ethidium and DAPI transport assays were conducted to extend the functional analysis of the suppressor mutants. These studies demonstrated that all five secondary mutations in the W58A template restored the capacity for QacA-mediated ethidium and DAPI transport (Fig. 4). Of note, whereas the suppressor mutations restored DAPI efflux capacity to the W58A QacA mutant and the initial rates of transport mediated by the suppressor mutants were significant, DAPI efflux mediated by a number of these mutants, particularly the W58A M391I, W58A G400D, and W58A A403T mutants, reached equilibrium earlier than that mediated by the wild-type protein or other single mutants (Fig. 4B). This could be indicative of a greater ability of DAPI to permeate cells containing these QacA W58A suppressor mutants. Interestingly, despite conferring only moderate ethidium resistance, the G400D and A403T single QacA mutants were able to mediate ethidium export at significant initial rates, and although these mutants facilitated moderate rates of DAPI efflux, they displayed significantly compromised capacities to confer resistance to another diamidine, pentamidine (Table 1; Fig. 4).
FIG. 4.
Transport mediated by W58A second-site suppressor mutants (A and B) and G400D and A403T QacA mutants (C and D). E. coli cells were loaded with either 15 μM ethidium bromide (A and C) or 8 μM DAPI (B and D), and transport was monitored fluorimetrically after energization. Transport reactions were conducted at least in duplicate, and the results of representative experiments are shown.
Membrane localization of suppressor positions.
The four distal amino acid substitutions that restored function to the W58A QacA mutant were in positions predicted from hydropathy to be in loops on the cytosolic side of the lipid bilayer, whereas the W58A mutation was predicted to be at the interface of TMS 2 and the extracellular loop between TMS 1 and 2 (loop 1-2) (Fig. 1) (24, 26). To test these positions experimentally, five cysteine replacement mutants, the W58C, D79C, M391C, G400C, and A403C mutants, were constructed and analyzed for their reactivities with the thiol-reactive, membrane-impermeant reagent fluorescein maleimide in both intact cells and disrupted membranes. The fluorescein maleimide reactivities of these cysteine mutants were compared to that of a previously constructed QacA mutant, the P309C mutant, which was shown to be moderately reactive with fluorescein maleimide, suggesting that position 309 is within extracellular loop 9-10, close to the boundary of TMS 10 (Fig. 1) (32). In disrupted membranes, the D79C, M391C, D400C, and A403C QacA mutants were far more reactive with fluorescein maleimide than the P309C QacA protein, suggesting that these positions are located within loop regions, as predicted (Fig. 5). When labeling was conducted in whole cells, the P309C QacA mutant remained moderately reactive with fluorescein maleimide. However, by contrast to the P309C protein, the D79C, M391C, G400C, and A403C QacA mutants were poorly reactive, suggesting that these positions are on the cytosolic side of the membrane, where they are less accessible to the membrane-impermeant fluorescein maleimide reagent. The fluorescein maleimide reactivities of the W58C QacA mutant were significantly lower than those of the P309C derivative but were similar in intact cells and disrupted membranes; therefore, position 58 of QacA is likely to be in its predicted membrane interfacial location, close to the extracellular side of the membrane. It is unlikely that the incorporated cysteine substitutions significantly affected the tertiary structure of QacA; therefore, these results support the predicted locations of the targeted amino acid positions and demonstrate that the second-site suppressor mutations of W58A occurred on the opposite side of the membrane from position 58.
FIG. 5.
Fluorescein maleimide reactivities of mutants containing cysteine substitutions at W58A suppressor positions in disrupted cell membranes (A) and whole cells (B). Fluorescein maleimide-treated QacA mutant proteins were purified and fractionated on SDS-polyacrylamide gels. (Top panels) Total QacA protein levels determined by Coomassie blue staining (A) or Western blotting with an anti-QacA antiserum (B). (Lower panels) Fluorescein maleimide label associated with mutant QacA proteins. The relative levels of fluorescein maleimide labeling as percentages of the labeling of the previously characterized QacA P309C mutant (32) are given below the gels and are averages from two repeat experiments.
Functional analyses of cysteine mutants.
Functional studies, including MIC analyses and fluorescent transport assays, of the W58C, D79C, M391C, G400C, and A403C QacA mutants were also conducted. Like the W58A mutation, the W58C substitution abolished QacA-mediated resistance to all substrates tested (Table 1). Nonetheless, compared to cells expressing no QacA protein, this mutant appeared to facilitate basal levels of drug efflux (Fig. 6A and B). Despite the conservation of D79 within the highly conserved signature motif, motif A of the MFS (Fig. 1) (25), its replacement with cysteine had little negative effect on QacA-mediated resistance to any substrate but resulted in increased resistance to ethidium and rhodamine 6G (Table 1). The D79C mutant was also able to mediate the efflux of ethidium and DAPI at initial rates greater than those mediated by wild-type QacA (Fig. 6A and B). The M391C, G400C, and A403C mutations within cytosolic loop 12-13 had modest overall effects on QacA-mediated resistance (Table 1). Of these QacA mutants, the most significant reductions in resistance capacity were seen with the A403C substitution, which caused an 80% reduction in the dequalinium resistance level from that conferred by the wild-type protein (Table 1). Additionally, the M391C, G400C, and A403C QacA mutants were able to facilitate the transport of ethidium and DAPI at initial rates approximately equal to or greater than those of wild-type QacA (Fig. 6A and B).
FIG. 6.
Ethidium and DAPI transport mediated by QacA mutants containing cysteine substitutions at W58A second-site suppressor positions. Transport was monitored fluorimetrically from cells loaded with ethidium bromide (A and C) or DAPI (B and D) after energization. The effects of maleimide modification at these positions were investigated by treating the cells with NEM prior to transport (C and D). Transport reactions were conducted at least in duplicate, and the results of representative experiments are shown.
Since each of the cysteine replacement mutants was at least partially reactive with fluorescein maleimide, duplicate transport studies were conducted after treatment of cells with the membrane-permeant thiol-reactive maleimide derivative NEM. This treatment did not affect the rate of transport from cells expressing wild-type QacA, which contains no native cysteine residues, nor did it affect transport from cells not expressing a QacA derivative (data not shown). Remarkably, however, the rates of substrate efflux mediated by most QacA cysteine mutants were modulated by preincubation with NEM. The rates of both ethidium and DAPI transport mediated by the W58C QacA mutant increased slightly upon reaction with NEM (Fig. 6C and D). Due to the low reactivity of this mutant with fluorescein maleimide, it may not be saturated with NEM; however, the increase in transport capacity suggests that a maleimide side chain is a better substitute for tryptophan at amino acid position 58 of QacA than the side chain carried by either alanine or cysteine. Additionally, NEM treatment had a negative impact on the rate of transport mediated by the M391C, G400C, and A403C mutants. This was most profound for the A403C QacA protein, where preincubation with NEM resulted in a complete loss of both ethidium and DAPI transport (Fig. 6C and D). Reaction with NEM only mildly reduced ethidium transport by the M391C and G400C proteins but showed a more significant effect on the DAPI transport mediated by these mutants (Fig. 6C and D).
DISCUSSION
The QacA efflux protein potentiates resistance to a broad range of lipophilic cationic antimicrobial compounds and is often carried by clinical isolates of S. aureus (5, 21, 23, 30). In this study, conservative and nonconservative amino acid mutations, to phenylalanine and alanine, respectively, were made for the nine tryptophan residues in QacA, and the functional effects of these changes were evaluated. In keeping with the high level of conservation of W58, W149, and W173 among transporters classified within the DHA2 family of the MFS (9, 25), nonconservative mutations of these residues to alanine were the most functionally damaging of the 18 tryptophan mutations examined (Table 1; Fig. 2 and 3). These derivatives were expressed at significant levels (Table 1); therefore, these residues are unlikely to mediate a role in protein stabilization or folding. Nonetheless, they may participate in functions common to all DHA2 family transporters, such as conformational transitions or proton translocation.
Single amino acid substitutions that abolish the transport of all substrates in QacA, such as the W58A mutation identified here, have been found to be rare. In our studies of the QacA transport protein, other substitutions with such a profound functional consequence have typically been for charged residues at intramembrane positions (5). Therefore, second-site suppressor studies of the W58A mutant were conducted to help gauge the basis of the functional defect within this mutant QacA protein. Substitutions at four distal amino acid positions were identified as restoring resistance and transport function to the W58A mutant, as was a threonine substitution at the primary site (W58T). Often, second-site suppressor mutations are in physical proximity to the primary deleterious mutation and therefore provide an indication of protein tertiary structure. However, the four amino acid substitutions that restored resistance and transport function to the compromised W58A QacA mutant, D79N, M391I, G400D, and A403T, were in positions predicted from hydropathy analysis to be in cytosolic loops, whereas the W58A mutation was predicted to be at the extracellular loop interface of TMS 2, i.e., on the opposite side of the lipid bilayer (Fig. 1). The results of fluorescein maleimide reactivity analysis (Fig. 5) support these predicted localizations, suggesting an interesting long-distance functional association of these distal positions.
Second-site suppressor mutations have been identified previously on opposite sides of the membrane in other MFS transporters. Indeed, suppressor mutations have been found in the loops flanking TMS 2 in both the E. coli TetA(B) tetracycline transporter (18, 33) and lactose permease (14), as with the W58A D79N QacA suppressor mutation described here. In the lactose permease, a T45R substitution in periplasmic loop 1-2 suppressed a deleterious D68S mutation in cytoplasmic loop 2-3 (14). Similarly, L30S and A40D mutations in loop 1-2 of the TetA(B) transporter have been shown to suppress deleterious amino acid substitutions for G62 and D66 in loop 2-3, respectively (18, 33). Therefore, a conformational association between amino acid residues on opposite sides of TMS 2 could be a common feature of all MFS transporters, including those containing 14 TMS.
The location of the M391I, G400D, and A403T second-site suppressor mutations of W58A in the loop connecting TMS 12 and 13 suggests a structural association between the N- and C-terminal regions of the QacA transporter (Fig. 1). To our knowledge, this is the first demonstration of such an association in a 14-TMS MFS protein; however, comparable observations have been made for 12-TMS MFS transporters (17). Loop 12-13 of QacA corresponds to cytoplasmic loop 10-11 of 12-TMS MFS transporters, according to the current model of 14-TMS MFS transporter evolution (6+2+6) (27). Interestingly, the L30S mutation in periplasmic loop 1-2 of TetA(B), identified as a suppressor of deleterious mutations in cytoplasmic loop 2-3 (18, 33), was also able to suppress a functionally damaging mutation, G332S, in cytoplasmic loop 10-11 of TetA(B) (17). Therefore, a conformational association of residues in periplasmic loop 1-2 of TetA(B) with residues in both cytoplasmic loops 2-3 and 10-11 has been shown. Of note, this correlates with the suppressor relationship of residues in cytoplasmic loops 2-3 (D79) and 12-13 (M391, G400, and A403) with a residue close to loop 1-2 (W58) in QacA (Fig. 1). Since in 12-TMS MFS transporters TMS 2 and 11 are known to interact at the interface of the N- and C-terminal domains (2, 13, 34), it is not surprising that amino acid residues in loops flanking these TMS are conformationally linked in TetA(B). Given that a comparable association between residues in corresponding regions of QacA was observed here, it is tempting to hypothesize that the tertiary fold around TMS 2 and 13 in QacA and perhaps other 14-TMS MFS transport proteins is similar to that around TMS 2 and 11 of 12-TMS MFS transporters.
Speculation on the functional role of W58 in QacA can be made based on the properties of functionally acceptable amino acid substitutions at this position. Since threonine constituted a suitable replacement for W58, it is clear that side chain aromaticity at position 58 is not essential for QacA-mediated transport function. Therefore, W58 is unlikely to participate in important stacking or cation-π bonds (6, 7, 35) with QacA substrates or adjacent amino acid side chains. Notably, although both the W58A and W58C QacA mutants displayed a complete lack of resistance function, the W58C mutant mediated basal levels of efflux, and treatment of this mutant with NEM, despite the fact that the mutant was only weakly reactive with maleimide derivatives (Fig. 5), resulted in an appreciable increase in efflux capacity, primarily for ethidium (Fig. 6D). Therefore, although cysteine and threonine are similar in size, there is an approximate relationship between QacA function and the bulkiness of the side chain at position 58. A possible scenario is that a bulky side chain is required at position 58 to promote the correct packing of TMS 2 with surrounding helices.
The G400D and A403T QacA mutants were expressed at higher levels than the W58A G400D and W58A A403T double mutants (Table 1), and despite catalyzing significant ethidium transport (Fig. 4C), they maintained low to moderate levels of resistance to each of the QacA substrates tested (Table 1). Therefore, whereas modifications to the QacA tertiary structure induced by the G400D and A403T mutations were functionally restorative in the W58A QacA background, they were detrimental to the wild-type protein. Cysteine substitutions for G400 and A403 were less functionally damaging (Table 1; Fig. 6A and B). However, modification of these cysteine side chains and that of the M391C mutant by NEM resulted in reduced transport function by these mutants, including a complete loss of transport function by the A403C mutant (Fig. 6C and D). Thus, in contrast to the apparent requirement for a bulky side chain at position 58, large side chains at positions 391, 400, and 403 may reduce the catalytic turnover of QacA by preventing conformation transitions required for substrate flux.
This study investigated the importance of tryptophan side chains for the drug efflux function of the 14-TMS MFS multidrug efflux protein QacA. An important functional role was observed for a number of these side chains, and an approximate size/function relationship was determined for the side chain at position 58. Suppressor mutations that rescued the nonfunctional W58A mutant demonstrated long-distance functional associations between residues on opposite sides of the membrane and in distal N- and C-terminal regions of the QacA polypeptide. The observation of suppressor relationships between residues in corresponding regions of 12-TMS MFS transporters is in keeping with the notion that 12- and 14-TMS MFS proteins may retain similar tertiary folds (15).
Acknowledgments
This work was supported by project grants 301938 and 457391 from the National Health and Medical Research Council (Australia).
Footnotes
Published ahead of print on 25 January 2008.
REFERENCES
- 1.Abramson, J., I. Smirnova, V. Kasho, G. Verner, S. Iwata, and H. R. Kaback. 2003. The lactose permease of Escherichia coli: overall structure, the sugar-binding site and the alternating access model for transport. FEBS Lett. 55596-101. [DOI] [PubMed] [Google Scholar]
- 2.Abramson, J., I. Smirnova, V. Kasho, G. Verner, H. R. Kaback, and S. Iwata. 2003. Structure and mechanism of the lactose permease of Escherichia coli. Science 301610-615. [DOI] [PubMed] [Google Scholar]
- 3.Adler, J., and E. Bibi. 2005. Promiscuity in the geometry of electrostatic interactions between the Escherichia coli multidrug resistance transporter MdfA and cationic substrates. J. Biol. Chem. 2802721-2729. [DOI] [PubMed] [Google Scholar]
- 4.Braun, P., and G. von Heijne. 1999. The aromatic residues Trp and Phe have different effects on the positioning of a transmembrane helix in the microsomal membrane. Biochemistry 389778-9782. [DOI] [PubMed] [Google Scholar]
- 5.Brown, M. H., and R. A. Skurray. 2001. Staphylococcal multidrug efflux protein QacA. J. Mol. Microbiol. Biotechnol. 3163-170. [PubMed] [Google Scholar]
- 6.Dougherty, D. A. 1996. Cation-π interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science 271163-168. [DOI] [PubMed] [Google Scholar]
- 7.Gallivan, J. P., and D. A. Dougherty. 1999. Cation-π interactions in structural biology. Proc. Natl. Acad. Sci. USA 969459-9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanahan, D. 1983. Studies on transformation in Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 9.Hassan, K. A., M. Galea, J. Wu, B. A. Mitchell, R. A. Skurray, and M. H. Brown. 2006. Functional effects of intramembranous proline substitutions in the staphylococcal multidrug transporter QacA. FEMS Microbiol. Lett. 26376-85. [DOI] [PubMed] [Google Scholar]
- 10.Hassan, K. A., K. L. Robinson, A. N. Smith, J. H. Gibson, R. A. Skurray, and M. H. Brown. 2006. Glycine-rich transmembrane helix 10 in the staphylococcal tetracycline transporter TetA(K) lines a solvent-accessible channel. Biochemistry 4515661-15669. [DOI] [PubMed] [Google Scholar]
- 11.Hassan, K. A., R. A. Skurray, and M. H. Brown. 2007. Transmembrane helix 12 of the Staphylococcus aureus multidrug transporter QacA lines the bivalent drug binding pocket. J. Bacteriol. 1899131-9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirai, T., J. A. Heymann, P. C. Maloney, and S. Subramaniam. 2003. Structural model for 12-helix transporters belonging to the major facilitator superfamily. J. Bacteriol. 1851712-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, Y., M. J. Lemieux, J. Song, M. Auer, and D.-N. Wang. 2003. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science 301616-620. [DOI] [PubMed] [Google Scholar]
- 14.Jessen-Marshall, A. E., and R. J. Brooker. 1996. Evidence that transmembrane segment 2 of the lactose permease is part of a conformationally sensitive interface between the two halves of the protein. J. Biol. Chem. 2711400-1404. [DOI] [PubMed] [Google Scholar]
- 15.Jin, J., A. A. Guffanti, C. Beck, and T. A. Krulwich. 2001. Twelve-transmembrane-segment (TMS) version (Δ TMS VII-VIII) of the 14-TMS Tet(L) antibiotic resistance protein retains monovalent cation transport modes but lacks tetracycline efflux capacity. J. Bacteriol. 1832667-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaback, H. R., R. Dunten, S. Frillingos, P. Venkatesan, I. Kwaw, W. Zhang, and N. Ermolova. 2007. Site-directed alkylation and the alternating access model for LacY. Proc. Natl. Acad. Sci. USA 104491-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawabe, T., and A. Yamaguchi. 1999. Transmembrane remote conformational suppression of the Gly-332 mutation of the Tn10-encoded metal-tetracycline/H+ antiporter. FEBS Lett. 457169-173. [DOI] [PubMed] [Google Scholar]
- 18.Kimura, T., T. Sawai, and A. Yamaguchi. 1997. Remote conformational effects of the Gly-62→Leu mutation of the Tn10-encoded metal-tetracycline/H+ antiporter of Escherichia coli and its second-site suppressor mutation. Biochemistry 366941-6946. [DOI] [PubMed] [Google Scholar]
- 19.Law, C. J., Q. Yang, C. Soudant, P. C. Maloney, and D. N. Wang. 2007. Kinetic evidence is consistent with the rocker-switch mechanism of membrane transport by GlpT. Biochemistry 4612190-12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemieux, M. J., Y. Huang, and D. N. Wang. 2004. The structural basis of substrate translocation by the Escherichia coli glycerol-3-phosphate transporter: a member of the major facilitator superfamily. Curr. Opin. Struct. Biol. 14405-412. [DOI] [PubMed] [Google Scholar]
- 21.Mayer, S., M. Boos, A. Beyer, A. C. Fluit, and F. J. Schmitz. 2001. Distribution of the antiseptic resistance genes qacA, qacB and qacC in 497 methicillin-resistant and -susceptible European isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 47896-897. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell, B. A., I. T. Paulsen, M. H. Brown, and R. A. Skurray. 1999. Bioenergetics of the staphylococcal multidrug export protein QacA: identification of distinct binding sites for monovalent and divalent cations. J. Biol. Chem. 2743541-3548. [DOI] [PubMed] [Google Scholar]
- 23.Noguchi, N., J. Suwa, K. Narui, M. Sasatsu, T. Ito, K. Hiramatsu, and J. H. Song. 2005. Susceptibilities to antiseptic agents and distribution of antiseptic-resistance genes qacA/B and smr of methicillin-resistant Staphylococcus aureus isolated in Asia during 1998 and 1999. J. Med. Microbiol. 54557-565. [DOI] [PubMed] [Google Scholar]
- 24.Paulsen, I. T., M. H. Brown, T. G. Littlejohn, B. A. Mitchell, and R. A. Skurray. 1996. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc. Natl. Acad. Sci. USA 933630-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rouch, D. A., D. S. Cram, D. DiBerardino, T. G. Littlejohn, and R. A. Skurray. 1990. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol. Microbiol. 42051-2062. [DOI] [PubMed] [Google Scholar]
- 27.Saier, M. H., Jr. 2003. Tracing pathways of transport protein evolution. Mol. Microbiol. 481145-1156. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 29.Tamura, N., S. Konishi, S. Iwaki, T. Kimura-Someya, S. Nada, and A. Yamaguchi. 2001. Complete cysteine-scanning mutagenesis and site-directed chemical modification of the Tn10-encoded metal-tetracycline/H+ antiporter. J. Biol. Chem. 27620330-20339. [DOI] [PubMed] [Google Scholar]
- 30.Tennent, J. M., B. R. Lyon, M. T. Gillespie, J. W. May, and R. A. Skurray. 1985. Cloning and expression of Staphylococcus aureus plasmid-mediated quaternary ammonium resistance in Escherichia coli. Antimicrob. Agents Chemother. 2779-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulmschneider, M. B., and M. S. Sansom. 2001. Amino acid distributions in integral membrane protein structures. Biochim. Biophys. Acta 15121-14. [DOI] [PubMed] [Google Scholar]
- 32.Xu, Z., B. A. O'Rourke, R. A. Skurray, and M. H. Brown. 2006. Role of transmembrane segment 10 in efflux mediated by the staphylococcal multidrug transport protein QacA. J. Biol. Chem. 281792-799. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi, A., Y. Inagaki, and T. Sawai. 1995. Second-site suppressor mutations for the Asp-66→Cys mutant of the transposon Tn10-encoded metal-tetracycline/H+ antiporter of Escherichia coli. Biochemistry 3411800-11806. [DOI] [PubMed] [Google Scholar]
- 34.Yin, Y., X. He, P. Szewczyk, T. Nguyen, and G. Chang. 2006. Structure of the multidrug transporter EmrD from Escherichia coli. Science 312741-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zacharias, N., and D. A. Dougherty. 2002. Cation-π interactions in ligand recognition and catalysis. Trends Pharmacol. Sci. 23281-287. [DOI] [PubMed] [Google Scholar]