Abstract
The riboflavin kinase in Methanocaldococcus jannaschii has been identified as the product of the MJ0056 gene. Recombinant expression of the MJ0056 gene in Escherichia coli led to a large increase in the amount of flavin mononucleotide (FMN) in the E. coli cell extract. The unexpected features of the purified recombinant enzyme were its use of CTP as the phosphoryl donor and the absence of a requirement for added metal ion to catalyze the formation of FMN. Identification of this riboflavin kinase fills another gap in the archaeal flavin biosynthetic pathway. Some divalent metals were found to be potent inhibitors of the reaction. The enzyme represents a unique CTP-dependent family of kinases.
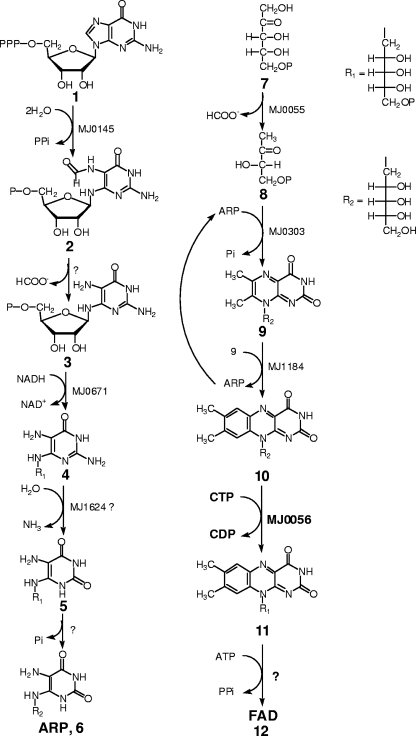
Work on the biosynthesis of riboflavin, flavin mononucleotide (FMN), and flavin adenine dinucleotide (FAD) in the archaea has revealed a number of surprises in terms of both the genes encoding the pathway enzymes and the pathway itself. Analysis of archaeal genomes has generally shown the absence of ribA, the gene encoding GTP cyclohydrolase II, the first enzyme in the presently established pathways to riboflavin in bacteria. In the archaea, this reaction may be catalyzed by at least two separate enzymes. The first enzyme, GTP cyclohydrolase III, produces 2-amino-5-formylamino-6-ribofuranosylamino-4(3H)-pyrimidinone 5′-phosphate (compound 2 in Fig. 1) and inorganic pyrophosphate by hydrolysis of GTP (11). This intermediate is subsequently hydrolyzed to 2,5-diamino-6-ribofuranosylamino-4(3H)-pyrimidinone 5′-phosphate (compound 3) by a currently unknown enzyme. Compound 3 is then converted into 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione (ARP, compound 6 in Fig. 1) by a dehydrogenase (MJ0671) (5), a deaminase, and a phosphatase following the eukaryotic pathway (13). The involvement of compound 2 does not occur in either the bacterial or the eukaryotic pathway, and the dehydrogenase and deaminase steps are in the same order as in the eukaryotic pathway, which is the reverse of that found in the bacterial pathway. The product of this series of reactions, ARP, is the precursor of F420 and riboflavin (10). In the conversion of ARP to riboflavin in the archaea, there is a fundamental difference in the stereochemistry of the pentacyclic intermediate involved in the dismutation of 6,7-dimethyl-8-ribityllumazine into riboflavin and 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione in the last step of riboflavin biosynthesis (17).
FIG. 1.
Riboflavin biosynthetic pathway in archaea.
Two different groups of enzymes are known to be involved in the conversion of riboflavin first to FMN and then to FAD. In the first group, the reaction is catalyzed by FAD synthetase, a bifunctional enzyme (RibF or RibC) that first acts as a kinase converting riboflavin to FMN in the presence of ATP and then acts as a nucleotidyltransferase by using a second ATP to convert the FMN to FAD and PPi (8, 21). In the other group, these reactions are catalyzed by two separate enzymes, riboflavin kinase (RibR, flavokinase, or FMN1) and FAD synthetase (FAD1 in yeast) (32). Enzymes homologous to the yeast flavokinase (FMN1) (27) are widely distributed (29). Aside from the differences in the enzymes used to generate FAD, other differences in the pathways and/or enzymes have been noted. It has been reported that the ribF gene in Bacillus subtilis uses reduced riboflavin as the substrate (19, 28), Corynebacterium ammoniagenes RibF can use meta-phosphate as the phosphoryl donor (23), and the macrolide resistance gene mreA of Streptococcus agalactiae has also been shown to function as a riboflavin kinase (6). The absence of identifiable genes encoding any of these enzymes in the archaea has prompted a search for the genes and reactions required to carry out these last two steps of FAD biosynthesis in the methanogens. Here we report that the MJ0056-derived protein is an archaeon-specific riboflavin kinase that catalyzes the reaction shown in Fig. 1. We designate it RibK to indicate the unique character of this archaeal riboflavin kinase. To date, orthologs of MJ0056 have been found only in archaea.
It was noticed that the crystal structure of the MJ0056 gene product recently deposited in the Protein Data Bank (code 2oyn) has a structural fold similar to that found in the human and yeast riboflavin kinases (4, 18), as well as the riboflavin kinase domain of the flavin-binding protein from Thermotoga maritima, annotated as an FAD synthetase (30). This crystal structure also contains a bound CDP in the same position as the ATP bound in the human enzyme. In addition, in the archaeal genomes of Archaeoglobus fulgidus DSM 4304, Haloarcula marismortui ATCC 43049, Halobacterium sp. strain NRC-1, Methanothermobacter thermautotrophicus strain Delta H, Methanococcus maripaludis S2, Methanococcoides burtonii DSM 6242, Methanosarcina acetivorans C2A, Methanosarcina mazei Go1, and Methanopyrus kandleri AV19, the MJ0056 gene (ribK) is next to the MJ0055 gene (ribB), which encodes 3,4-dihydroxy-2-butanone-4-phosphate synthase, which is involved in the biosynthesis of riboflavin. These findings indicated that this gene product could be the archaeal kinase despite the fact that it has no detectable sequence similarity to any known kinase.
The M. jannaschii gene at locus MJ0056 (Swiss-Prot accession number Q60365) was amplified by PCR from genomic DNA with oligonucleotide primers pMJ0056Fwd (5′-GGTCATATGATTATTGAGGGAGAAG-3′) and pMJ0056Rev (5′-GATCGGATCCTTATTCATCTTTATCTCCC-3′). The PCR was performed as described previously, with a 55°C annealing temperature (12). The primers introduced an NdeI and a BamHI site at the 5′ and 3′ ends, respectively. The amplified PCR product was purified with a QIAquick spin column (Qiagen), digested with restriction enzymes NdeI and BamHI, and then ligated into the compatible sites in plasmid pT7-7 to make recombinant plasmid pMJ0056. DNA sequences were verified by dye terminator sequencing at the University of Iowa DNA facility. The resulting plasmid, pMJ0056, was transformed into Escherichia coli BL21-CodonPlus(DE3)-RIL cells (Stratagene). The transformed cells were grown in Luria-Bertani medium at 37°C until an absorbance at 600 nm of 1.0 was reached. The protein was induced by adding 2% (wt/vol) lactose and culturing the cells for an additional 4 h. The protein was then extracted from the cells, heated at 70°C, and purified on a MonoQ column as previously described (14). Elution of the protein was monitored by UV absorbance at 280 nm. One major UV-absorbing peak eluting at 0.37 M NaCl contained the desired protein, based on activity measurement and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. The protein purified by this method was >95% pure by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with Coomassie staining and showed a molecular mass of 15 kDa. Electrospray mass spectrometry (MS) measured a mass of 15,218.1 ± 10 Da, which agrees with the mass of 15,213.8 Da calculated from the gene sequence for a protein of 132 amino acids. All of the kinetic analyses were done with protein purified in this manner.
The FMN eluting from MonoQ-separated cell extractions was used to measure the amount of FMN present in the extracts. The MonoQ fractions containing FMN were identified by fluorescence, and the total amount was quantified by high-performance liquid chromatography (HPLC). Extracts derived from cells expressing the MJ0056 gene were found to have 8.7 μM FMN. This value was sixfold higher than that observed in extracts from cells expressing other enzymes not related to FMN production.
The standard assay for RibK was performed by incubation of 47 mM TES buffer (pH 7.2), 1.2 μg of RibK, 1.7 mM CTP, and 0.37 mM riboflavin in a total volume of 60 μl at 70°C for 10 min. Under this assay condition, 42% of the riboflavin was consumed and converted into FMN. Analysis of the enzyme-catalyzed reaction was conducted by HPLC with detection of riboflavin and FMN with a Shimadzu HPLC system with a C18 reverse-phase column (Varian PursuitXRs; 250 by 4.6 mm, 5-μm particle size) and detection by fluorescence with an excitation λmax of 450 nm and an emission λmax of 520 nm. The elution profile consisted of 5 min in 95% sodium acetate buffer (25 mM [pH 6.0], 0.2% NaN3)-5% methanol, followed by a linear gradient to 20% sodium acetate buffer-80% methanol over 40 min at 0.5 ml/min. Specifically, the ratio of riboflavin to FMN was measured from the areas of the HPLC fluorescence signals generated by the separated compounds. Under these conditions, riboflavin was eluted at 31.1 min, FMN at 29 min, and FAD at 26.5 min. The FMN product gave the same electrospray MS spectrum as authentic FMN with an MH+ ion at 457.0 m/z and an MNa+ ion at 479.0 m/z in the positive mode and an M− ion at 454.9 m/z in the negative mode. The MS-MS of the 457.0 m/z ion for both the known and enzymatically generated FMN gave the same ions at 439.0 m/z for [M-H2O]+, 377.1 m/z for [riboflavin]+, 359.1 m/z for [riboflavin-H2O]+, and 243.1 m/z for [flavin ring system]+. The MS-MS of 454.9 m/z ion for both the known and enzymatically generated FMN gave the same ions at 241.0 m/z for [flavin ring system]−, 212.9 m/z for [M-flavin ring system]−, and 96.9 m/z for [phosphate]−.
Size exclusion chromatography was performed on a Superose 12HR column (1 by 30 cm; Amersham Biosciences) with the same conditions and protein standards as previously described (20). The data indicated that the enzyme was a monomer (19.7 kDa). When 1.2 mg of the enzyme was heated in 40 μl of extraction buffer at different temperatures of up to 100°C for 10 min, no significant change in enzyme activity was observed. Although dithiothreitol was used in these thermal experiments, we have no indication that dithiothreitol is required to maintain the enzyme's activity. The enzyme retained 40% (0.13 nmol min−1) of its activity compared to the control (0.32 nmol min−1) after treatment with 0.1 M HCl (pH ∼1.05) for 10 min at room temperature, followed by dilution in acetate buffer (pH 6.0). In these experiments, the detection limit was about 2% of the control activity.
The enzyme was most active with CTP as the substrate. The activities with ATP and GTP at the same concentration were 30% and 11% of that seen with CTP, respectively.
The kinetics of RibK were determined under steady-state conditions in the presence of a saturating concentration of either CTP or riboflavin. KeleidaGraph 4.0 was used to calculate the kinetic parameters from the Michaelis-Menten plot. The measured kinetic parameters for RibK with CTP as the phosphate donor are as follows for riboflavin: Kmapp = 159 μM, kcat/Km = 2.1 × 103 M−1 s−1. They are as follows for CTP: Kmapp = 1.8 mM, kcat/Km = 190 M−1 s−1, and Vmax = 1.3 μmol min−1 mg−1. There is a wide range of kinetic values reported for riboflavin kinases from different organisms. The lowest Km values reported for riboflavin and ATP are 120 nM and 210 nM, respectively, for the flavokinase from Neurospora crassa (24), and the highest Km values for riboflavin and ATP are reported as 420 μM and 4.55 mM, respectively, for the rat liver enzyme (3). Also, the range of specific activities for flavokinases is enormous at 0.00023 μmol min−1 mg−1 for the flavokinase in rat liver (3) to 2.95 μmol min−1 mg−1 for the flavokinase from N. crassa (24). Using the standard assay method supplemented with ether no metal ion or 1.6 mM Mg2+, Mn2+, Zn2+, Ni2+, Cu2+, or Co2+, the specific activity observed was 0.7, 0.6, 0.35, 0.12, 0.4, 0.02, or 0.27 μmol min−1 mg−1, respectively. Rat liver riboflavin kinase was active with these cations and had the highest activity with Zn2+ (22).
The MonoQ-purified protein was analyzed for magnesium and zinc at the Virginia Tech Soil Testing Laboratory by inductively coupled plasma emission spectrophotometry. The result showed 4 to 5 mol of Mg and 0.0013 mol of Zn per mol of protein.
The reasons why this enzyme preferentially uses CTP as the phosphate donor instead of ATP, as is found in almost all known kinases, is not clear. 1H nuclear magnetic resonance analysis of M. jannaschii cell extracts indicates that the concentration of CTP is comparable to that of ATP (R. H. White, unpublished data). Thus, the enzyme could simply be taking advantage of the high CTP concentration. The only known example of a CTP-dependent kinase is dolichol kinase (1). There are a few GTP-dependent kinases, e.g., phosphoglycerate kinase (25) and polyribonucleotide 5′-hydroxyl-kinase (31), and some kinases are probably promiscuous and can use either GTP or CTP. There are also some other non-ATP-dependent kinases such as a few ADP-dependent and pyrophosphate kinases (16, 26).
The evolutionary lineage of this archaeal riboflavin kinase is not clear, as it is so different from all other kinases. This alternation in kinases is common in the archaeal biosynthetic pathways where we find a new shikimate kinase in the shikimate pathway (7), a new kinase in a new pathway to isoprenoids (15), and a new predicted pantothenate kinase in coenzyme A biosynthesis (9).
An important biological implication of the presence of RibK in those archaeal genomes that have no genes for de novo riboflavin synthesis, such as “Pyrococcus abyssi” GE5, Pyrococcus horikoshii OT3, and Thermoplasma acidophilum DSM 1728, provides a clear interpretation of FMN and FAD biogenesis via a salvage pathway from exogenous riboflavin.
ADDENDUM
While this work was under review, Ammelburg et al. published data showing that the MJ0056 gene product is a riboflavin kinase and proposed an evolutionary lineage of MJ0056 to other riboflavin kinases (2).
Acknowledgments
We thank Kim Harich for assistance with obtaining the mass spectral data and Walter Niehaus and Laura Grochowski for assistance in editing the manuscript.
National Science Foundation grant MCB-0722787 supported this work.
Footnotes
Published ahead of print on 1 February 2008.
REFERENCES
- 1.Allen, C. M., Jr., J. R. Kalin, J. Sack, and D. Verizzo. 1978. CTP-dependent dolichol phosphorylation by mammalian cell homogenates. Biochemistry 175020-5026. [DOI] [PubMed] [Google Scholar]
- 2.Ammelburg, M., M. D. Hartmann, S. Djuranovic, V. Alva, K. K. Koretke, J. Martin, G. Sauer, V. Truffault, K. Zeth, A. N. Lupas, and M. Coles. 2007. A CTP-dependent archaeal riboflavin kinase forms a bridge in the evolution of cradle-loop barrels. Structure 151577-1590. [DOI] [PubMed] [Google Scholar]
- 3.Bandyopadhyay, D., A. K. Chatterjee, and A. G. Datta. 1997. Effect of cadmium on purified hepatic flavokinase: involvement of reactive-SH group(s) in the inactivation of flavokinase by cadmium. Life Sci. 601891-1903. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, S., K. Kemter, A. Bacher, R. Huber, M. Fischer, and S. Steinbacher. 2003. Crystal structure of Schizosaccharomyces pombe riboflavin kinase reveals a novel ATP and riboflavin-binding fold. J. Mol. Biol. 3261463-1473. [DOI] [PubMed] [Google Scholar]
- 5.Chatwell, L., T. Krojer, A. Fidler, W. Romisch, W. Eisenreich, A. Bacher, R. Huber, and M. Fischer. 2006. Biosynthesis of riboflavin: structure and properties of 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate reductase of Methanocaldococcus jannaschii. J. Mol. Biol. 3591334-1351. [DOI] [PubMed] [Google Scholar]
- 6.Clarebout, G., C. Villers, and R. Leclercq. 2001. Macrolide resistance gene mreA of Streptococcus agalactiae encodes a flavokinase. Antimicrob. Agents Chemother. 452280-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daugherty, M., V. Vonstein, R. Overbeek, and A. Osterman. 2001. Archaeal shikimate kinase, a new member of the GHMP-kinase family. J. Bacteriol. 183292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Efimov, I., V. Kuusk, X. Zhang, and W. S. McIntire. 1998. Proposed steady-state kinetic mechanism for Corynebacterium ammoniagenes FAD synthetase produced by Escherichia coli. Biochemistry 379716-9723. [DOI] [PubMed] [Google Scholar]
- 9.Genschel, U. 2004. Coenzyme A biosynthesis: reconstruction of the pathway in archaea and an evolutionary scenario based on comparative genomics. Mol. Biol. Evol. 211242-1251. [DOI] [PubMed] [Google Scholar]
- 10.Graham, D. E., and R. H. White. 2002. Elucidation of methanogenic coenzyme biosynthesis: from spectroscopy to genomics. Nat. Prod. Rep. 19133-147. [DOI] [PubMed] [Google Scholar]
- 11.Graham, D. E., H. Xu, and R. H. White. 2002. A member of a new class of GTP cyclohydrolases produces formylaminopyrimidine nucleotide monophosphates. Biochemistry 4115074-15084. [DOI] [PubMed] [Google Scholar]
- 12.Graupner, M., H. Xu, and R. H. White. 2000. Identification of an archaeal 2-hydroxy acid dehydrogenase catalyzing reactions involved in coenzyme biosynthesis in methanoarchaea. J. Bacteriol. 1823688-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graupner, M., H. Xu, and R. H. White. 2002. The pyrimidine nucleotide reductase step in riboflavin and F420 biosynthesis in archaea proceeds by the eukaryotic route to riboflavin. J. Bacteriol. 1841952-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grochowski, L. L., H. Xu, K. Leung, and R. H. White. 2007. Characterization of an Fe2+-dependent archaeal specific GTP cyclohydrolase, MptA, from Methanocaldococcus jannaschii. Biochemistry 466658-6667. [DOI] [PubMed] [Google Scholar]
- 15.Grochowski, L. L., H. Xu, and R. White. 2006. Methanocaldococcus jannaschii uses a modified mevalonate pathway for the biosynthesis of isopentenyl diphosphate. J. Bacteriol. 1883192-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen, T., and P. Schonheit. 2004. ADP-dependent 6-phosphofructokinase, an extremely thermophilic, non-allosteric enzyme from the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus strain 7324. Extremophiles 829-35. [DOI] [PubMed] [Google Scholar]
- 17.Illarionov, B., W. Eisenreich, N. Schramek, A. Bacher, and M. Fischer. 2005. Biosynthesis of vitamin B2: diastereomeric reaction intermediates of archaeal and non-archaeal riboflavin synthases. J. Biol. Chem. 28028541-28546. [DOI] [PubMed] [Google Scholar]
- 18.Karthikeyan, S., Q. Zhou, F. Mseeh, N. V. Grishin, A. L. Osterman, and H. Zhang. 2003. Crystal structure of human riboflavin kinase reveals a beta barrel fold and a novel active site arch. Structure 11265-273. [DOI] [PubMed] [Google Scholar]
- 19.Kearney, E. B., J. Goldenberg, J. Lipsick, and M. Perl. 1979. Flavokinase and FAD synthetase from Bacillus subtilis specific for reduced flavins. J. Biol. Chem. 2549551-9557. [PubMed] [Google Scholar]
- 20.Li, H., M. Graupner, H. Xu, and R. H. White. 2003. CofE catalyzes the addition of two glutamates to F420-0 in F420 coenzyme biosynthesis in Methanococcus jannaschii. Biochemistry 429771-9778. [DOI] [PubMed] [Google Scholar]
- 21.Manstein, D. J., and E. F. Pai. 1986. Purification and characterization of FAD synthetase from Brevibacterium ammoniagenes. J. Biol. Chem. 26116169-16173. [PubMed] [Google Scholar]
- 22.Merrill, A. H., Jr., and D. B. McCormick. 1980. Affinity chromatographic purification and properties of flavokinase (ATP:riboflavin 5′-phosphotransferase) from rat liver. J. Biol. Chem. 2551335-1338. [PubMed] [Google Scholar]
- 23.Nakagawa, S., T. Hagihara, T. Fujio, and K. Aisaka. 1995. Metaphosphate-dependent phosphorylation of riboflavin to FMN by Corynebacterium ammoniagenes. Appl. Microbiol. Biotechnol. 43325-329. doi: 10.1007/BF00172833. [DOI] [PubMed] [Google Scholar]
- 24.Rajeswari, S. R., V. S. Jonnalagadda, and S. Jonnalagadda. 1999. Purification and characterization of flavokinase from Neurospora crassa. Indian J. Biochem. Biophys. 36137-142. [PubMed] [Google Scholar]
- 25.Reeves, R. E., and D. J. South. 1974. Phosphoglycerate kinase (GTP). An enzyme from Entamoeba histolytica selective for guanine nucleotides. Biochem. Biophys. Res. Commun. 581053-1057. [DOI] [PubMed] [Google Scholar]
- 26.Ronimus, R. S., and H. W. Morgan. 2001. The biochemical properties and phylogenies of phosphofructokinases from extremophiles. Extremophiles 5357-373. [DOI] [PubMed] [Google Scholar]
- 27.Santos, M. A., A. Jimenez, and J. L. Revuelta. 2000. Molecular characterization of FMN1, the structural gene for the monofunctional flavokinase of Saccharomyces cerevisiae. J. Biol. Chem. 27528618-28624. [DOI] [PubMed] [Google Scholar]
- 28.Solovieva, I. M., R. A. Kreneva, D. J. Leak, and D. A. Perumov. 1999. The ribR gene encodes a monofunctional riboflavin kinase which is involved in regulation of the Bacillus subtilis riboflavin operon. Microbiology 145(Pt. 1)67-73. [DOI] [PubMed] [Google Scholar]
- 29.von Mering, C., L. J. Jensen, M. Kuhn, S. Chaffron, T. Doerks, B. Kruger, B. Snel, and P. Bork. 2007. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 35D358-D362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, W., R. Kim, H. Yokota, and S. H. Kim. 2005. Crystal structure of flavin binding to FAD synthetase of Thermotoga maritima. Proteins 58246-248. [DOI] [PubMed] [Google Scholar]
- 31.Westaway, S. K., H. G. Belford, B. L. Apostol, J. Abelson, and C. L. Greer. 1993. Novel activity of a yeast ligase deletion polypeptide. Evidence for GTP-dependent tRNA splicing. J. Biol. Chem. 2682435-2443. [PubMed] [Google Scholar]
- 32.Wu, M., B. Repetto, D. M. Glerum, and A. Tzagoloff. 1995. Cloning and characterization of FAD1, the structural gene for flavin adenine dinucleotide synthetase of Saccharomyces cerevisiae. Mol. Cell. Biol. 15264-271. [DOI] [PMC free article] [PubMed] [Google Scholar]