Abstract
We constructed a Bacillus subtilis Marburg strain that harbors deletion mutations in all seven extracytoplasmic function (ECF) sigma genes. The strain shows wild-type growth at 37°C both in a complex and in a synthetic medium and exhibits wild-type sporulation. ECF sigma genes of B. subtilis are dispensable as long as no stress is imposed, although they seem to be required for quick response to stresses.
Since the assignment of sigma E of Streptomyces coelicolor to a subfamily of eubacterial RNA polymerase sigma factors (14), many sigma factors belonging to this family have been described (3, 4, 9, 18, 22). These sigma factors were called extracytoplasmic function (ECF) sigma factors, as they were found to direct the expression of proteins functioning in the outer membrane or in periplasmic space (14). Further analysis, however, revealed that these sigma factors direct expression of proteins functioning in cytoplasmic space, as well (2, 5, 9, 21, 28).
The numbers of ECF sigma genes vary among bacterial species. Some bacteria have lost ECF sigma genes, and some bacteria possess a lot of them (9). These ECF sigma genes are believed to be nonessential and auxiliary in nature, although inactivation of rpoE in Escherichia coli and sigG in Synechocystis sp. strain PCC6803 was proven to be lethal (8, 11). In Bacillus subtilis W23, simultaneous inactivation of sigX and sigM causes a growth defect (17). However, the exact roles of those sigma genes in the maintenance of growth have still not been elucidated, and there is no study in which all stress sigma genes have been systematically inactivated in an organism. For this study, we successively interrupted all ECF sigma genes of B. subtilis to construct an ECF sigmaless strain in order to examine whether the genes are indispensable or merely auxiliary.
B. subtilis has seven ECF sigma genes (sigM, sigV, sigW, sigX, sigY, sigZ, and ylaC), which are negatively regulated by anti-sigma proteins, except for sigZ (2, 6, 7, 9, 13, 16, 25, 26, 27). Inactivation of these sigma genes one at a time does not affect viability because the sigma factors seem to overlap functionally (6, 9, 10, 16, 26). A quadruple mutant (ΔsigV ΔsigY ΔsigZ ΔylaC) of B. subtilis was constructed and was found to be viable (15).
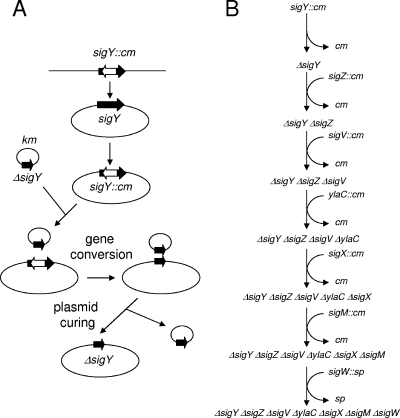
In order to examine whether there is a minimal set of ECF sigma genes required for B. subtilis growth under nonstress conditions, we constructed deletion mutations in all seven ECF sigma genes on the genome of B. subtilis Marburg 168 (trpC2) as outlined in Fig. 1. Only one construction pathway is shown. There are many other possible pathways and many combinations of multiple disruptions, and we constructed a large number of different multiple disruptants (data not shown). During successive disruptions of ECF sigma genes, sufficient quantities of wild-type size transformants or segregants were obtained to suggest that there were no suppressor mutations, concerning colony size on LB agar, associated with the disruption of the ECF sigma genes. The sigW gene was disrupted at the end of the procedure because inactivation of sigW reduced competence (see below). All ECF sigma genes disrupted with the cm cassette (cat gene) were obtained from K. Kobayashi. The first step, ΔsigY strain construction, is shown in Fig. 1A. The kanamycin-resistant (Kmr) plasmid pCHE11(ΔsigY) bearing a deletion in sigY was constructed by PCR and introduced into the sigY::cm strain by transformation. Kmr transformants were selected at 30°C. Among Kmr transformants, a few colonies [pCHE11 (ΔsigY)/ΔsigY] were chloramphenicol sensitive (Cms) due to gene conversion between ΔsigY on pCHE11 and chromosomal sigY::cm (12). Kmr Cms transformants thus obtained were grown in drug-free LB broth at 37°C to cure the plasmid and to obtain a plasmid-free ΔsigY strain, as pCHE11 (obtained from F. Kawamura) has a mutation leading to temperature-sensitive replication (19). The deletion mutation ΔsigY was confirmed by PCR. Next, the sigZ::cm ΔsigY strain was constructed by transformation of the ΔsigY strain with DNA from the sigZ::cm strain (Fig. 1B). Plasmid pCHE11 (ΔsigZ) was introduced into the sigZ::cm ΔsigY strain. A plasmid-free ΔsigZ ΔsigY strain was derived from pCHE11 (ΔsigZ)/ΔsigY sigZ::cm cells through gene conversion and curing, as shown in Fig. 1A. These procedures were repeated successively, as depicted in Fig. 1B. The resulting strain was named BSU2007 (ΔsigY ΔsigZ ΔsigV ΔylaC ΔsigX ΔsigM ΔsigW trpC2).
FIG. 1.
One path to an ECF sigma-free strain. (A) The first step, ΔsigY strain construction. Detailed procedures are described in the text. (B) Successive procedures used to construct the B. subtilis mutant strain BSU2007 carrying deletions in all ECF genes. The initial strain, carrying sigY::cm, was B. subtilis Marburg 168 trpC2.
For construction of plasmid pCHE11 carrying a deletion mutation in an ECF sigma gene, 5′ and 3′ regions of each ECF sigma gene were PCR synthesized with primer pairs of each primer set (Table 1) and template wild-type chromosomal DNA. The resulting DNA fragments bearing 5′ and 3′ regions of each ECF sigma gene were annealed and subjected to PCR amplification with distal 5′ and 3′ primers. The synthesized DNA fragment carrying a deletion followed by a stop codon in each ECF sigma gene was cloned into plasmid pCHE11 in E. coli cells.
TABLE 1.
Primers used for construction of deletion mutations in ECF sigma genes
| Primer set | Sequencea
|
|
|---|---|---|
| F | R | |
| sigM5′ | 5′ GGAGGATCCCTTCAATTGGCACAAGCG | 5′ CAGCCGCTGTTAATCGATCGTCACTGCCTT |
| sigM3′ | 5′ ACGATCGATTAACAGCGGCTGAAAGCACTT | 5′ GGAGGATCCGCTTTTGCGGTCGTTTGT |
| sigV5′ | 5′ GAAGAATTCAGGAGATAGAAGCAGAGC | 5′ CGTCAACTGTTATTTCTTCATTGCAATAAAGGG |
| sigV3′ | 5′ ATGAAGAAATAACAGTTGACGAAGGAGGAT | 5′ GGAGGATCCGACTTTGATATAGCGCTC |
| sigW5′ | 5′ GAAGAATTCTAACACGGGTTCGAATCC | 5′ AAGATCCCTTTAAATCATCATTTCCATATTTATCTA |
| sigW3′ | 5′ ATGATGATTTAAAGGGATCTTTAAGTGGGG | 5′ GGAGGATCCTCCCCGCAGACTTCATCT |
| sigX5′ | 5′ GAAGAATTCAAGTTTTCCGGTGTTGCG | 5′ CCTCAAAAGTTAGGTTTCTTCCATTTGAAACCC |
| sigX3′ | 5′ GAAGAAACCTAACTTTTGAGGGAGGAGCTA | 5′ GGAGGATCCCCGAACCCCAGTCTCATT |
| sigY5′ | 5′ GAAGAATTCGATCCCGGGATGGGAACA | 5′ CTTTCTGATTTATTGTGTATCCAATGACCG |
| sigY3′ | 5′ GATACACAATAAATCAGAAAGGAGTGGGAT | 5′ GGAGGATCCCGTTCGTCCGCTCTTCTT |
| sigZ5′ | 5′ GAAGAATTCAGTCTACTTTGCCGGAGC | 5′ ATCCGCTTCTTATTGATCCCATAGATCCTC |
| sigZ3′ | 5′ TGGGATCAATAAGAAGCGGATCGATATGGA | 5′ GGAGGATCCCGTATCCGTCATACGTAT |
| ylaC5′ | 5′ GAAGAATTCGCAGTCAGGGTGACATGG | 5′ CTCTCTACTTTAGGAATCCCTATGCTTCAT |
| ylaC3′ | 5′ AGGGATTCCTAAAGTAGAGAGGAGGAGCGG | 5′ GGAGGATCCGCAGTTGAACTTGGCTTC |
EcoRI and BamHI sites are underlined. Boldface letters indicate TAA stop codons.
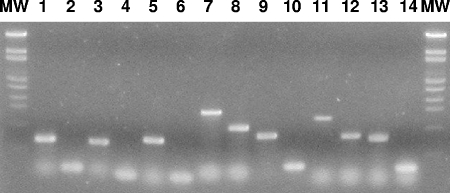
Confirmation of the deletion mutations in all seven ECF sigma genes was carried out by PCR amplification of the deleted or wild-type genes with primer pairs described in the figure legend and template DNA from strain BSU2007 or wild-type strain 168 (Fig. 2). BSU2007 carried a deletion mutation in each of the seven ECF sigma genes.
FIG. 2.
PCR confirmation of deletion mutations in ECF sigma genes of B. subtilis BSU2007. Lanes 1 and 2, sigM; lanes 3 and 4, sigV; lanes 5 and 6, sigW; lanes 7 and 8, sigX; lanes 9 and 10, sigY; lanes 11 and 12, sigZ; lanes 13 and 14, ylaC. Odd numbers indicate PCR products with 168 DNA as a template. Even numbers indicate PCR products with BSU2007 DNA as a template. The primer pairs were as follows: 5′ GTCGTCGACGTGTATAACATAGAGGGG/5′ GCAGCATGCAGTCATTTCCTGGTCG for sigM, 5′ GTCGTCGACGTATATTCAAAAAGGAGCCC/5′ GCAGCATGCTGTAATCTCTTATCCATTAAG for sigV, 5′ GTCGTCGACCGGTGAAGGCAGAGG/5′ GCAGCATGCATAAGCTGCACAATTTG for sigW, 5′ GAAGAATTCCGACAAAACGGTAAAATCGAG/5′ GGAGGATCCTTTTCAGGAACGACAAAC for sigX, 5′ GAAGAATTCAATGCATCGTCCTCC/5′ GCAGCATGCTTATTCATCATCCCACTCC for sigY, 5′ GTCGTCGACGCAAAATTATAGGAGG/5′ GCAGCATGCAAACATCAAAGGAAAAATGC for sigZ, and 5′ GTCGTCGACATTTGGATGACGGTGTGG/5′ GCAGCATGCTTTCCAATTCAAATGGC for ylaC. The lengths of the PCR products were 594 bases (159 bases), 558 bases (93 bases), 647 bases (110 bases), 1424 bases (938 bases), 759 bases (261 bases), 1,204 bases (751 bases), and 709 bases (229 bases) for sigM (ΔsigM), sigV (ΔsigV), sigW (ΔsigW), sigX (ΔsigX), sigY (ΔsigY), sigZ (ΔsigZ), and ylaC (ΔylaC), respectively.
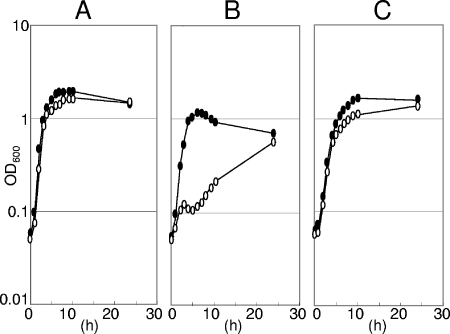
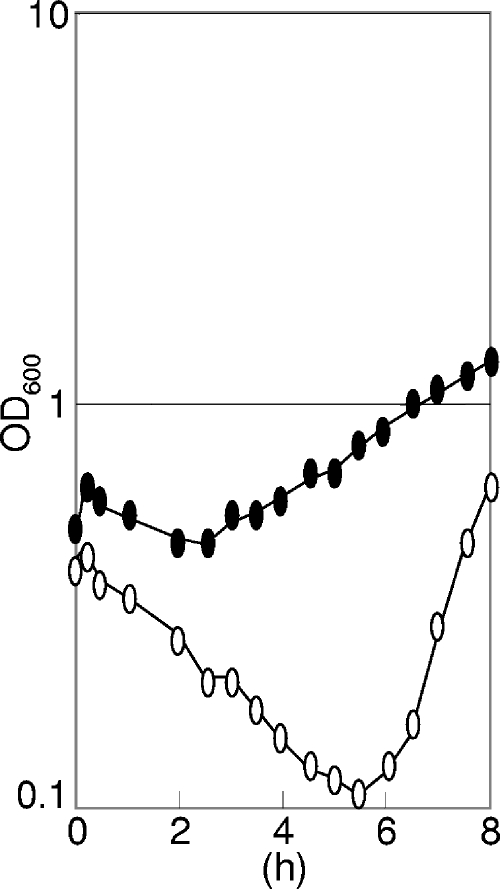
The BSU2007 strain showed no obvious phenotype in LB broth (23) containing yeast extract, tryptone, and NaCl; the colony-forming ability of the ECF sigma-free strain BSU2007 on LB broth agar was similar to that of the wild-type strain at all temperatures tested (20°C, 37°C, and 50°C). We studied several characteristic features of the mutant BSU2007 under some other conditions. As shown in Fig. 3, the ECF sigma-free strain BSU2007 showed wild-type growth in a complex LB broth or synthetic minimal glucose medium (1) but was slow to start growing in a complex nutrient broth (Difco) containing beef extract and peptone. However, addition of MgSO4 (1 mM) to Difco nutrient broth restored the wild-type growth of BSU2007 (data not shown). It grew like the wild-type strain in a complex sporulation medium (24) containing complex nutrient broth (Difco) supplemented with metals and salts [1 mM MgSO4, 1 mM FeSO4, 10 mM MnCl2, 13.4 mM KCl, 1 mM Ca(NO3)2]. The capacity to sporulate was not lost in BSU2007 (Table 2. On the other hand, BSU2007 showed defective competence development (Table 3) in the above-mentioned synthetic minimal glucose medium, due to the null function of sigW leading to low competence of strain BSU2000. BSU2006 carrying deletion mutations in all six ECF sigma genes except sigW showed wild-type competence. Even in LB broth, BSU2007 showed slow growth recovery after a temperature shift from 37°C to 52°C, as shown in Fig. 4. The slow recovery did not seem to be due to suppressor mutations, as recovered culture again showed slow growth recovery at 52°C. The involvement of ECF sigma factors in heat adaptation has not yet been studied. We also examined the stress sensitivity of BSU2007. The stresses imposed were acetic acid, NaCl, HCl, NaOH, EDTA, H2O2, sodium dodecyl sulfate, toluene, and ethanol. Paper discs soaked with samples were placed on LB agar inoculated with a culture of either the wild-type strain or the ECF sigmaless BSU2007 strain. We examined the growth inhibition of the bacterial lawn after an overnight incubation at 37°C. There were no differences in the sensitivities of both strains to the above-mentioned stresses, except for acetate and toluene. BSU2007 was more sensitive than the wild-type strain only to acetate and toluene, as shown in Table 4, although the sensitivity difference was small with toluene.
FIG. 3.
Slow growth of the ECF sigmaless strain BSU2007 of B. subtilis in Difco nutrient broth medium. A portion of an overnight culture in LB medium was transferred to fresh medium and incubated at 37°C. Filled ovals, wild-type strain 168; open ovals, BSU2007. (A) LB broth. (B) Bacto nutrient broth (Difco Laboratories, Detroit, MI). (C) Minimal glucose medium (1).
TABLE 2.
Sporulation frequencies of the ECF sigmaless mutant BSU2007 of B. subtilisa
| Conditions | CFU (108/ml) for strain:
|
|
|---|---|---|
| 168 | BSU2007 | |
| −Heat | 4.95 | 5.50 |
| +Heat | 2.30 | 3.50 |
| +/− | 0.46 | 0.69 |
Cells were grown in Difco sporulation medium at 37°C overnight. The culture sample was exposed to heat (75°C for 20 min) or not, and the numbers of CFU were determined on LB agar that had been incubated at 37°C overnight.
TABLE 3.
Defective competence development in the ECF sigmaless mutant BSU2007 of B. subtilisa
| Parameter for donor | Value for strain:
|
|||
|---|---|---|---|---|
| 168 | BSU2000 | BSU2006 | BSU2007 | |
| Plasmid TF (103/ml) | 2.89 | 0.53 | 7.69 | 0.02 |
| (pDG148) CFU (108/ml) | 7.50 | 1.06 | 8.30 | 1.98 |
| Frequency (10−6) | 3.85 | 0.50 | 9.27 | 0.09 |
| Ratio | 1.00 | 0.13 | 2.40 | 0.02 |
| Chromosome TF (103/ml) | 4.15 | 0.59 | 3.36 | 0.05 |
| (TrpC2) CFU (108/ml) | 9.55 | 11.60 | 6.40 | 1.58 |
| Frequency (10−6) | 4.35 | 0.51 | 5.25 | 0.32 |
| Ratio | 1.00 | 0.12 | 1.21 | 0.07 |
Donor DNA was 1 μg/ml. Competent cells were prepared as described previously (20). BSU2000 (ΔsigW trpC2) and BSU2006 (ΔsigY ΔsigZ ΔsigV ΔylaC ΔsigX ΔsigM trpC2) carry one and six deletion mutations.
FIG. 4.
Slow growth recovery from temperature shift of ECF sigmaless strain BSU2007. Rapidly growing cells of wild-type strain 168 (filled ovals) or BSU2007 (open ovals) in LB broth were transferred from 37°C to 52°C at time zero. OD600, optical density at 600 nm.
TABLE 4.
Acetate and toluene sensitivities of BSU2007a
| Strain | Inhibition zone (mm)
|
|
|---|---|---|
| Acetate | Toluene | |
| 168 | 10.0 | 2.0 |
| BSU2007 | 12.0 | 3.5 |
Fresh culture was poured on LB agar, and 30 μl of acetate (diluted 1/4 with distilled water) or toluene was placed on a paper disc (diameter, 8 mm). The inhibition zone was measured after overnight incubation at 37°C.
The B. subtilis strain BSU2007, which lacks seven functional ECF sigma genes, is viable and shows no obvious defects as long as no stress is imposed, although it exhibits slow growth in Difco broth. However, it showed wild-type growth and sporulation in a complex sporulation medium containing metals and salts in Difco broth. The strain BSU2007 may not be able to compete for growth in liquid medium with the wild-type strain; however, prolonged mixed culture of both strains in LB broth at 37°C for 3 days contained similar numbers of CFU of both strains. BSU2007 showed slow recovery from high-temperature stress, although its ability to form colonies was not affected. Some combination of ECF sigma genes must be required for a quick response to heat stress.
We are currently analyzing the phenotypes of various combinations of ECF sigma gene deletions to see which ECF sigma gene is responsible for which phenotype. Beyond that, since target gene candidates of the seven ECF sigma genes share only one target gene with those of the general stress sigma B (2), we are constructing multiple-stress-responsive sigma mutants based on BSU2007 in order to elucidate the nature of the stress response of B. subtilis.
Footnotes
Published ahead of print on 25 January 2008.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1969. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai, K., H. Yamaguchi, C.-M. Kang, K. Yoshida, Y. Fujita, and Y. Sadaie. 2003. DNA microarray analysis of Bacillus subtilis sigma factors of extracytoplasmic function family. FEMS Microbiol. Lett. 220155-160. [DOI] [PubMed] [Google Scholar]
- 3.Bashvam, M. D., and S. F. Hasnain. 2004. The extracytoplasmic function sigma factors: role in bacterial pathogenesis. Infect. Genet. Evol. 4301-308. [DOI] [PubMed] [Google Scholar]
- 4.Brooks, B. E., and S. K. Buchanan. 15 June 2007. Signaling mechanisms for activation of extracytoplasmic function (ECF) sigma factors. 0Biochim. Biophys. Acta. doi: 10.1016/j.bbamem2007.06.005. [DOI] [PMC free article] [PubMed]
- 5.Butcher, B. G., and J. D. Helmann. 2006. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol. Microbiol. 44206-216. [DOI] [PubMed] [Google Scholar]
- 6.Cao, M., L. Salzberg, C. S. Tsai, T. Mascher, C. Bonilla, T. Wang, R. W. Ye, L. Marquez-Magana, and J. D. Helmann. 2003. Regulation of the Bacillus subtilis extracytoplasmic function protein σY and its target promoters. J. Bacteriol. 1854883-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, M., T. Wang, R. Ye, and J. D. Helmann. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol. Microbiol. 451267-1276. [DOI] [PubMed] [Google Scholar]
- 8.DeLa Penas, A., L. Connolly, and C. A. Gross. 1997. Sigma E is an essential sigma factor in Escherichia coli. J. Bacteriol. 1796862-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 4647-110. [DOI] [PubMed] [Google Scholar]
- 10.Horsburgh, M. J., and A. Moir. 1999. Sigma M, an ECF RNA polymerase sigma factor of Bacillus subtilis 168, is essential for growth and survival in high concentrations of salt. Mol. Microbiol. 3241-50. [DOI] [PubMed] [Google Scholar]
- 11.Huckauf, J., C. Nomura, K. Forchhammer, and M. Hagemann. 2000. Stress responses of Synechocystis sp. strain PCC 6803 mutants impaired in genes encoding putative alternative sigma factors. Microbiology 1462877-2889. [DOI] [PubMed] [Google Scholar]
- 12.Kawamura, F., L. F. Wang, and R. H. Doi. 1985. Catabolite-resistant sporulation (crsA) mutations in the Bacillus subtilis RNA polymerase sigma 43 gene (rpoD) can suppress and be suppressed by mutations in spo0 genes. Proc. Natl. Acad. Sci. USA 828124-8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunst, F., N. Ogasawara, and I. Moszer. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390249-256. [DOI] [PubMed] [Google Scholar]
- 14.Lonetto, M. A., K. L. Brown, K. E. Rudd, and M. J. Buttner. 1994. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial s factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. USA 917573-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mascher, T., A.-B. Hachmann, and J. D. Helmann. 2007. Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function σ factors. J. Bacteriol. 1896919-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto, T., K. Nakanishi, K. Asai, and Y. Sadaie. 2005. Transcriptional analysis of the ylaABCD operon of Bacillus subtilis encoding a sigma factor of extracytoplasmic function family. Genes Genet. Syst. 80385-393. [DOI] [PubMed] [Google Scholar]
- 17.Minning, K., J. L. Barblan, S. Kehl, S. B. Moller, and C. Mauel. 2003. In Bacillus subtilis W23, the duet sigma X sigma M, two sigma factors of the extracytoplasmic function subfamily, are required for septum and wall synthesis under batch culture conditions. Mol. Microbiol. 491435-1447. [DOI] [PubMed] [Google Scholar]
- 18.Missiakas, D., and S. Raina. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 281059-1066. [DOI] [PubMed] [Google Scholar]
- 19.Nanamiya, H., K. Kasai, A. Nozawa, C.-S. Yun, T. Narisawa, K. Murakami, Y. Natori, F. Kawamura, and Y. Tozawa. 2008. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 67291-304. [DOI] [PubMed] [Google Scholar]
- 20.Ohshima, H., S. Matsuoka, K. Asai, and Y. Sadaie. 2002. Molecular organization of intrinsic restriction and modification genes BsuM of Bacillus subtilis Marburg. J. Bacteriol. 184381-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paget, M. S., J. G. Kang, J. H. Roe, and M. J. Buttner. 1998. Sigma R, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2). EMBO J. 175776-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55591-624. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schobel, S., S. Zelleier, W. Schumann, and T. Wiegert. 2004. The Bacillus subtilis sigma W anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol. Microbiol. 521091-1105. [DOI] [PubMed] [Google Scholar]
- 26.Tojo, S., M. Matsunaga, T. Matsumoto, C. M. Kang, H. Yamaguchi, K. Asai, Y. Sadaie, K. Yoshida, and Y. Fujita. 2003. Organization and expression of the Bacillus subtilis sigY operon. J. Biochem. 134935-946. [DOI] [PubMed] [Google Scholar]
- 27.Yoshimura, M., K. Asai, Y. Sadaie, and H. Yoshikawa. 2004. Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology 150591-599. [DOI] [PubMed] [Google Scholar]
- 28.Zellmeier, S., C. Hofmann, S. Thomas, T. Wiegert, and W. Schumann. 2005. Identification of σV-dependent genes of Bacillus subtilis. FEMS Microbiol. Lett. 253221-229. [DOI] [PubMed] [Google Scholar]