Abstract
CodY is a global regulatory protein that was first discovered in Bacillus subtilis, where it couples gene expression to changes in the pools of critical metabolites through its activation by GTP and branched-chain amino acids. Homologs of CodY can be found encoded in the genomes of nearly all low-G+C gram-positive bacteria, including Staphylococcus aureus. The introduction of a codY-null mutation into two S. aureus clinical isolates, SA564 and UAMS-1, through allelic replacement, resulted in the overexpression of several virulence genes. The mutant strains had higher levels of hemolytic activity toward rabbit erythrocytes in their culture fluid, produced more polysaccharide intercellular adhesin (PIA), and formed more robust biofilms than did their isogenic parent strains. These phenotypes were associated with derepressed levels of RNA for the hemolytic alpha-toxin (hla), the accessory gene regulator (agr) (RNAII and RNAIII/hld), and the operon responsible for the production of PIA (icaADBC). These data suggest that CodY represses, either directly or indirectly, the synthesis of a number of virulence factors of S. aureus.
A global transcriptional regulator, CodY, controls the expression of Bacillus subtilis genes whose products allow the cell to adapt to nutrient limitation and metabolic stress (53). CodY represses the synthesis of catabolic enzymes (15, 59), genetic competence factors (46), biosynthetic enzymes for certain amino acids (35, 49), antibiotics (22), sporulation proteins (35), and transport systems for amino acids, peptides and sugars (35, 47, 49, 50). CodY is also a positive regulator of some carbon overflow pathways (48). B. subtilis CodY senses nutrient availability through interaction with two types of effector molecules, branched-chain amino acids (BCAAs; isoleucine, valine, and leucine) and GTP (44, 49). The effectors act independently and additively to increase the affinity of CodY for DNA (49). Thus, when the bacterium encounters nutrient-depleted conditions, the intracellular BCAA and GTP pools decrease, CodY loses its activity as a DNA-binding protein, and CodY-repressed genes are induced. In Lactococcus lactis, CodY regulates genes for amino acid biosynthesis, nitrogen uptake, peptide degradation, transporters, intermediary carbon metabolism, and some regulatory proteins (21). The L. lactis CodY is activated by BCAAs in vivo and in vitro but not by GTP (10, 43).
Homologs of CodY are encoded in the genomes of nearly all of the low-G+C gram-positive bacteria, including Staphylococcus aureus (28, 44). In several of these species, a codY mutation affects virulence. In Listeria monocytogenes, a ΔrelA mutant is highly attenuated for virulence, likely due to an inability to induce the stringent response (3). As a result, the GTP pool is thought to remain high, despite amino acid limitation (3). Introduction of a ΔcodY mutation into a ΔrelA mutant strain restores virulence to the ΔrelA mutant, suggesting that the reduced virulence of the ΔrelA mutant is due, at least in part, to failure to control the activity of CodY (3). In Streptococcus pyogenes, CodY has a broad impact on virulence gene expression during growth in laboratory culture or in human blood (33, 34). Although the direct CodY targets in S. pyogenes are not known, analysis of 51 selected transcriptional units showed that 23 regulatory genes, virulence genes, transporters, and metabolic enzymes are differentially regulated in the codY mutant (33). In Clostridium difficile, CodY directly regulates transcription of the gene for the alternative sigma factor, TcdR, which is essential for the activation of toxin synthesis (11). A codY mutation in C. difficile leads to significant derepression of tcdR and toxin-encoding genes (11).
S. aureus causes an array of diseases, ranging from localized skin lesions and tissue damage to systemic infections, such as pneumonia, endocarditis, and exotoxin syndromes (14). A contributing factor to the diversity of S. aureus-associated disease is its ability to adapt efficiently to varying environmental conditions and to produce a plethora of secreted and cell wall-associated virulence factors. S. aureus regulates expression of virulence genes in response to signals reflecting both cell population density and nutritional availability. The well-characterized, quorum-sensing, two-component regulatory system (TCS) agr (for accessory gene regulator) plays a critical role in the growth-phase-dependent regulation of S. aureus virulence factor synthesis (5, 13, 60). The agr locus is transcribed from bidirectional promoters P2 and P3 (23, 42). The transcript originating from P2, known as RNAII, encodes four proteins, AgrB, AgrD, AgrC, and ArgA, which are the functional components of the quorum-sensing TCS (5, 37). High cell population density causes the activation of AgrA, the response regulator of the TCS, which induces transcription of the P3 promoter (37). P3 drives transcription of RNAIII, a regulatory RNA that is both a positive and a negative regulator of virulence factor production (38). RNAIII also serves as an mRNA for the synthesis of delta-toxin, a hemolysin (23). Activation of RNAIII transcription in response to an increase in cell population density marks a transition in gene expression correlated with a need for metabolic and stress adaptations (5). Although several TCS and other regulatory proteins, such as the SarA family of proteins, have been shown to regulate virulence genes (5), there is little known about the mechanisms by which S. aureus senses and responds to the nutritional status of the environment.
The regulation of the virulence factors of S. aureus in response to growth phase and nutrient availability is reminiscent of regulation by CodY in other gram-positive bacteria. The CodY homologue of S. aureus shares 64% overall amino acid similarity with the B. subtilis protein (16); the regions predicted to be important for dimer association, isoleucine interaction, and DNA binding share 100, 70, and 97% amino acid similarity, respectively (25, 28). Taken together, these observations suggest conservation of function between the homologues of the CodY proteins in B. subtilis and S. aureus. Although S. aureus CodY has not been studied extensively, a transposon mutagenesis screen in the clinical isolate S30, selecting for deficits in biofilm formation, yielded an insertion in the codY gene (55). By constructing codY-null mutants of two additional clinical isolates of S. aureus, UAMS-1 and SA564, we show here that CodY regulates virulence gene expression. Strains UAMS-1 and SA564 were chosen because they represent two genetically and phenotypically distinct lineages (51, 61). In both clinical isolates, a codY-null mutation resulted in an increase in hemolytic activity of stationary-phase culture supernatants. Transcript analysis revealed that the gene encoding alpha-toxin, hla, was overexpressed in both the exponential and the stationary phases of growth in the codY mutants. In addition, the levels of the agr locus transcripts, RNAII and RNAIII, were derepressed during the exponential phase of growth. In contrast to what was seen for the clinical isolate S30 (55), the codY mutants of UAMS-1 and SA564 displayed a greatly increased ability to form biofilms, apparently resulting from high-level expression of the icaADBC operon and accumulation of polysaccharide intercellular adhesin (PIA).
MATERIALS AND METHODS
Strains and growth conditions.
Strains and plasmids used in this study are listed in Table 1. S. aureus strains UAMS-1 and SA564 are clinical isolates. S. epidermidis 1457 is a biofilm-producing strain, and S. carnosus TM300 is a control strain that does not produce biofilm. The Staphylococcus strains were grown in tryptic soy broth (TSB; Becton-Dickinson Co.) with shaking (250 rpm) at 37°C, maintaining a flask-to-medium volume ratio of 10:1. Antibiotics needed for plasmid maintenance were used at the following concentrations: erythromycin, 10 μg/ml; and chloramphenicol, 10 μg/ml. When indicated, IPTG (isopropyl-β-d-thiogalactopyranoside; Diagnostic Chemicals, Ltd.) was added to the growth medium to a final concentration of 1 mM.
TABLE 1.
Strains and plasmids used in this study
| Plasmid or strain | Relevant genotype and/or characteristic(s) | Source or reference |
|---|---|---|
| Plasmids | ||
| pTS1 | Shuttle vector, pE194orits, ColE1 bla cat | 20 |
| pCD7-d | Derivative of pTS1, with ΔcodY::ermC fragment, and deletion of plasmid-encoded 3′ region of ermC | This study |
| pCL15 | Expression shuttle vector, derivative of pSI-1, PSPAC, cat | 29 |
| pTL6936 | pCL15, PSPAC-codY | This study |
| pGEM-T Easy | Cloning vector | Promega |
| Strains | ||
| RN4220 | S. aureus restriction-deficient derivative of RN450 | 26, 39 |
| UAMS-1 | S. aureus clinical isolate | 18 |
| MS1 | UAMS-1 ΔcodY::ermC | This study |
| SA564 | S. aureus clinical isolate | 52 |
| CDM7 | SA564 ΔcodY::ermC | This study |
| MS2 | UAMS-1, pTL6936 | This study |
| MS3 | MS1, pTL6936 | This study |
| 1457 | S. epidermidis biofilm + | 31 |
| TM300 | S. carnosus biofilm − | 19 |
Allelic replacement of codY.
A codY mutation was generated by using the following strategy. A 4.2-kb amplification product, consisting of a 2.2-kb sequence encoding the ermC gene, flanked by 1 kb of sequence normally found upstream of the codY gene and 1 kb of sequence normally downstream of codY, was generated using overlap PCR. The ermC gene was amplified from pCN51 (7) by using the primers OCD23 and OCD24 (Table 2), which also incorporated sequences homologous to the regions immediately flanking the codY gene in S. aureus. Genomic DNA (gDNA) from strain SA564 was used as the template for amplification of the gDNA regions flanking codY using the primers P−codY and P+codY and the ermC fragment generated as described above. The primers specific for codY were generated based on the sequence of S. aureus strain 8325 (GenBank ID ABD30331.1). The engineered fragment, which also contained XmaI sites on the 5′ and 3′ ends, was ligated to the XmaI site of pTS1 (20) and introduced by electroporation into E. coli DH5α. To eliminate intraplasmid recombination with the plasmid-encoded 3′ region of the ermC gene, a derivative of the plasmid was made by PCR and self-ligation by using the primers pTS1dErm-f and pTS1dErm-r. The resulting plasmid, pCD7-d, was introduced by electroporation into the restriction-deficient strain RN4220 according to methods previously described (45). After isolation of pCD7-d from RN4220, the plasmid was used to transform the clinical isolates by the same method. An erythromycin resistant transformant was grown in TSB medium supplemented with erythromycin and chloramphenicol for 12 h at 30°C. A 1:100 dilution of the culture was grown at 42°C for 12 h in TSB with no added antibiotics. After six 12-h passages at 42°C, each representing a 1:100 dilution of the previous culture, the cultures were shifted to 30°C, and the bacteria were passaged six additional times without antibiotics. Bacteria were then plated on tryptic soy agar (TSA) with erythromycin, and erythromycin-resistant, chloramphenicol-sensitive clones were selected.
TABLE 2.
Primers used in this study
| Primer | Nucleotide sequence (5′→3′) |
|---|---|
| P-codY | GATCGACCCGGGTCTCAAAGCAATAATCC |
| P+codY | TCGTCACCCGGGCATCTGCCATTTTAGCA |
| pTS1dErm-r | AGAGTGTCTTGTCAGACCTAAATTC |
| pTS1dErm-f | CGACACGGATATATAGTGGATGTG |
| codY22 | GGATCCACAAAAGGAGAAAAATTCATGAGC |
| codY27 | CCGCGGACATCGTCACGACTAGGACAT |
| P-hla | GCATTTCAATTTCGAGGG |
| OCD14 | GAACCGCATGGTTCAAAA |
| OCD15 | CATTTCACCGCTACACATGG |
| OCD23 | ACCCCACAATATGTTGATGATAAAAATTGAATGAGACATGCTAC |
| OCD24 | TGTCCCAGACTCATCGACAAAACTGGTTTAAGCCGAC |
| OCM43 | TGAAATTGTTCACTGTGTCGAT |
| OCM49 | CATTGAACAAGAAGCCTGACA |
| OCM50 | GAGTGCGTTGGCTTTACCTC |
| OCM51 | CAGAAAATTCCTCAGGCGTA |
| OCM52 | AAGGGGTATGACGGTACAACA |
| OCM57 | CAGAGATGTGATGGAAAATAGTTGA |
| OCM58 | TGACCAGTTTGCCACGTATC |
| OCM59 | CCAAAAGGAAGAAGGTGCAT |
Construction of the codY complementation strains.
The codY gene and its ribosome-binding site were amplified by using the primers codY22 and codY27. The PCR fragment was introduced into pGEMT-easy (Promega) and then recloned in pCL15 (29), using the restriction sites SstI and BamHI. In the resulting plasmid, pTL6936, the codY gene is under the control of the PSPAC promoter. pTL6936 was introduced into the strain RN4220 as described above. After plasmid isolation from RN4220, pTL6936 was introduced into UAMS-1 and SA564 by electroporation, as described above. Chloramphenicol-resistant colonies were selected and characterized.
Hemolytic assays.
S. aureus colonies from TSA plates were used to inoculate a 3-ml culture in TSB. In addition, four successive 100-fold dilutions of the initial inoculum were incubated overnight. The next day, flasks were selected that contained cultures with an optical density at 600 nm (OD600) of 3 to 7 and used to inoculate fresh TSB to a starting OD600 of 0.05. After the bacteria reached an OD600 between 0.5 and 2, they were diluted again in fresh TSB to an OD600 of 0.05 to be sure that the experimental cultures were well adapted to steady-state exponential growth phase. During subsequent growth, culture supernatant fluids were harvested from bacterial cells by centrifugation at 6,000 rpm for 10 min at 4°C. Supernatant samples were frozen immediately and stored at −20°C. The hemolytic activity of culture supernatants was determined by using a 2% suspension of rabbit red blood cells (rRBCs) prepared from rabbit defibrinated blood (HemoStat, Dixon, CA) in phosphate-buffered saline (PBS). The rRBCs were washed three times prior to resuspension in PBS. Serial dilutions (1.5-fold) of bacterial supernatant fluids were made in PBS plus 0.5% bovine serum albumin (MP Biomedicals, Inc.) in 96-well V-bottom plates (Corning, Inc.). PBS plus 0.5% BSA was used as a negative control, and 1% saponin (Sigma-Aldrich) was used as a positive control. An equal volume of 2% rRBCs was added to each sample and allowed to incubate for 10 min at 37°C. rRBCs were pelleted at 1,000 rpm for 5 min at 4°C, and the OD540 of the supernatant fluid was determined. For each sample, the dilution values were plotted (μl of culture fluid versus OD540), and the slope was calculated for the linear range of the lysis reaction. Using the linear equation, the hemoglobin release (OD540) for 50 μl of the lysis reaction fluid was determined for each sample. This value was then used to determine the hemolytic units (HU) per sample by using the equation HU = test OD540/50% total lysis OD540. This equation defines one HU as 50% of the total RBC lysis.
Western blot analysis.
S. aureus strains were grown, and supernatant samples were prepared in the same manner as for the hemolytic assay. Samples (4 μl) of supernatant fluid were loaded on a 12% sodium dodecyl sulfate-polyacrylamide gel after boiling in Laemmli sample buffer (27). The Western blot protocol was performed as described by Ausubel et al. (1) and the product guide for Amersham ECL Western blotting detection reagents (GE Healthcare). Alpha-toxin was detected with rabbit anti-staphylococcal alpha-toxin (Sigma-Aldrich) according to the manufacturer's recommendations. Human immunoglobulin G (Sigma-Aldrich) at 25 μg per 10 ml total volume was added during all antibody incubations and washes to block nonspecific binding by protein A (4).
Primer extension and reverse transcription-PCR (RT-PCR) analysis.
S. aureus strains were grown in the same way as for the hemolytic assay. For RNA isolation, 109 bacteria from exponential (OD600 = 0.6 to 0.8, 100 min)- or stationary (OD600 = 8 to 11, 330 min)-phase cultures were mixed with an equal volume of a 1:1 mixture of ice-cold acetone and ethanol and frozen at −80°C for 24 h. The suspensions were thawed on ice and centrifuged at 6,000 rpm for 10 min. The samples were washed twice with TE (10 mM Tris [pH 8.0], 1 mM EDTA) and resuspended in 1 ml of RLT (plus 1% β-mercaptoethanol) from the RNeasy kit (Qiagen). Cells were transferred to assembled 2-ml screw-cap tubes (Sarstedt) filled to 1/4 volume with 0.1-mm silica beads (BioSpec Products, Inc.) and broken by agitation in a Mini-Beadbeater (BioSpec Products, Inc.) for two 60-s intervals at the maximum disruption frequency (setting of 48) separated by a 5-min incubation on wet ice. The suspensions were then centrifuged at 13,000 × g for 15 min at 4°C, and the supernatant fluid was used for RNA isolation with an RNeasy mini-column (Qiagen), according to the manufacturer's recommendations, without the on-column DNase treatment. RNA concentrations were determined from the OD260, and the RNA was run on an RNase-free 2% agarose gel to test for generalized degradation.
For primer extension analysis, 10 to 20 μg of total RNA was used per reaction. Radioactive [γ-32P]ATP (Perkin-Elmer) was used to label the primers P-hla for the hla gene and OCM43 for the RNAIII/hld gene with T4 polynucleotide kinase (New England Biolabs, Inc.) according to the manufacturer's recommendations for primer extension analysis. cDNA was synthesized with Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's recommendations and then collected by precipitation with ethanol. Samples were run on a denaturing 6% urea-polyacrylamide gel according to the procedure suggested by the SequaGel sequencing system kit (National Diagnostics). The dried gel was exposed to a Phosphor Screen (Kodak) and analyzed with a Storm 820 Scanner and software (Amersham Biosciences). Using ImageQuant TL (Amersham Biosciences), the appropriate bands were quantified by using the rolling-ball method and normalized to the total amount of RNA in the sample.
For RT-PCR analysis, gDNA contamination was removed from the RNA samples by treatment with Turbo DNase (Ambion) according to the manufacturer's recommendations. Removal of DNA was confirmed by the absence of a product in a PCR using the treated RNA as a template. A 0.5-μg portion of gDNA-free RNA was then subjected to cDNA synthesis using Superscript II reverse transcriptase (see above). Subsequent PCRs used Taq DNA polymerase (Invitrogen) and incubation in the PTC-100 programmable thermal controller (MJ Research, Inc.). A total of 1 μl of the cDNA reaction was used for each sample when testing for icaA and icaR transcripts; 1 μl of a 1:10 dilution of the cDNA reaction was used for each sample when testing for 16S rRNA, RNAII, and RNAIII. To determine the linear range of the PCR for each cDNA sample, samples were removed after cycles 15, 20, 25, 30, and 35. Then, 10 μl of the PCR product was run on a 0.8% agarose gel containing ethidium bromide for detection. After determination of the linear range of the PCR for each transcript, samples from each cDNA synthesis reaction were chosen for comparison within the same cycle number. The following primers were used for cDNA sysnthesis: OCD15 (16S rRNA), OCM50 (icaA), OCM51 (icaR), OCM43 (RNAIII), and OCM59 (RNAII). The following additional primers were used for PCR amplification: OCD14 (16S rRNA), OCM49 (icaA), OCM52 (icaR), OCM57 (RNAIII), and OCM58 (RNAII).
PIA immunoblot assay.
PIA accumulation was determined as previously described (58). Briefly, TSB medium supplemented with 0.25% dextrose was inoculated with equal numbers of S. aureus bacteria from a 13-h preculture. The starting inoculum was determined by using OD (i.e., OD600) to measure the amount of wild-type bacteria in a 1:200 dilution of the culture. The starting inoculum for non-wild-type strains was then normalized to the wild-type inoculum. Every 2 h, equal numbers of cells (equivalent to an absorbance of 0.5 OD600 units) were harvested by centrifugation, and PIA was extracted in 0.5 M EDTA (pH 8.0) by boiling for 5 min. Samples of PIA-containing extracts were applied to a polyvinylidene difluoride membrane (Immobilon-P, Millipore) and blocked with 5% skim milk overnight. The polyvinylidene difluoride membrane was incubated for 2 h with PIA-specific antiserum (kindly provided by Michael Otto) and then for 2 h with an anti-rabbit immunoglobulin G-peroxidase conjugate. The presence of PIA-antibody complexes was detected by using SuperSignal West Pico chemiluminescent substrate (Pierce). The integrated density values of bands on autoradiograms were determined with TotalLab software (Nonlinear Dynamics, Ltd.). Error bars reflect replicates from two separate experiments. The amount of PIA produced by a wild-type sample at 2 h in one experiment was used as a standard (100%) for relative PIA accumulation.
Static in vitro biofilm assay.
The static biofilm assay was performed as previously described (2). Briefly, overnight cultures of Staphylococcus strains were grown in TSB. Samples (100 μl) of 1:100 dilutions of the overnight cultures in fresh TSB supplemented with 0.5% glucose and 3% sodium chloride were used to inoculate wells of a sterile, polystrene 96-well, flat-bottom microtiter plate (Corning, Inc.) and then incubated at 37°C for 24 h. Microtiter plates were not coated with human plasma. In each experiment, each strain was represented at least four times in the microtiter plate. S. epidermidis strain 1457 was included as a biofilm-producing positive control, and S. carnosus strain TM300 was included as a non-biofilm-producing control. After incubation, the medium and nonadherent bacteria were decanted, and the dishes were washed two times with PBS. Adherent bacteria were fixed to the microtiter plate with 100% ethanol for 20 min. The fluid was then removed by aspiration, and the plate was exposed to 100% crystal violet (CV) for 8 min. After the CV was decanted, the plate was washed two times with PBS and then incubated with 95% ethanol for 10 min. The OD600 of the CV released from adherent bacteria was determined. All samples were normalized to the negative control, S. carnosus TM300, and 100% biofilm formation was determined based on the values determined for S. epidermidis 1457.
RESULTS
Construction of the codY deletion mutants.
codY-null mutations were created in two clinical isolates, UAMS-1 and SA564, by allelic replacement of the codY gene with the ermC gene. The ermC gene was cloned in a temperature-sensitive plasmid and was flanked by 1-kb regions upstream and downstream of the codY gene. The replacement of codY with ermC and the loss of the plasmid backbone were verified by PCR and Southern blot analysis (data not shown). Western blot analysis showed that in the parental strains, UAMS-1 and SA564, CodY is produced at similar levels during exponential and stationary phases and confirmed the absence of CodY in the mutant strains (data not shown). In TSB medium, strains MS1 (ΔcodY::ermC derivative of UAMS-1) and CDM7 (ΔcodY::ermC derivative of SA564) showed a slight growth defect compared to UAMS-1 and SA564, respectively (Fig. 1A and data not shown). The codY mutants also had similar, yet smaller, colony morphology compared to the wild-type isolates (data not shown).
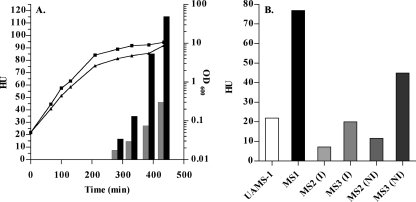
FIG. 1.
Effect of a ΔcodY mutation on the hemolytic activity of the culture fluid of S. aureus. The hemolytic activity of the culture fluid of each strain was tested on rabbit erythrocytes as described in Materials and Methods. One HU is defined as 50% total erythrocyte lysis per 50 μl of culture fluid. (A) The culture fluids of CDM7 (SA564 ΔcodY::ermC) (▪) and SA564 (wild type) (░⃞) were tested throughout growth for hemolytic activity. The growth of strains SA564 (▪) and CDM7 (▴) were measured as the OD600. (B) The culture fluids of MS1 (UAMS-1 ΔcodY::ermC), UAMS-1 (wild type), MS2 (UAMS-1 containing pTL6936), and MS3 (MS1 containing pTL6936) were tested in the late stationary phase for hemolytic activity. Strains carrying the codY+ complementation plasmid, pTL6936, were grown with (I) or without (NI) IPTG to induce codY expression.
CodY represses the production of hemolytic activity in culture supernatants.
To characterize the codY mutants, the culture fluids of UAMS-1 (codY +), MS1 (ΔcodY::ermC), SA564 (codY +), and CDM7 (ΔcodY::ermC) were assayed for hemolytic activity toward rabbit erythrocytes. Hemolytic activity in the culture fluid of SA564 was undetectable during exponential phase but increased to 46 HU in stationary-phase cultures (Fig. 1A). CDM7 showed increased levels of hemolytic activity compared to the isogenic wild-type strain, SA564. No hemolytic activity was detected in exponential-phase cultures of CDM7 but reached 115 HU in stationary phase (Fig. 1A). The culture fluids of UAMS-1 and its codY mutant, MS1, showed no hemolytic activity in exponential phase but increased to 22 and 77 HU in the stationary phase, respectively (Fig. 1B and data not shown). In summary, postexponential-phase and stationary-phase culture supernatants of CDM7 and MS1 had twofold-higher hemolytic activities than did their parent strains.
The codY mutation was complemented in the UAMS-1 background with pTL6936, a plasmid containing the codY gene under the control of PSPAC, an IPTG-inducible promoter. Upon induction, strain MS3 (MS1 carrying pTL6936) produced CodY protein as determined by Western blot analysis (data not shown). The hemolytic activities of MS2 (UAMS-1 carrying pTL6936) and MS3 were tested under inducing (I) and noninducing (NI) conditions (Fig. 1B). The hemolytic activity of the stationary-phase culture fluid of MS3 (complemented codY mutant treated with IPTG) was fourfold less than that of the culture fluid of MS1 (uncomplemented codY mutant). Even without IPTG, the presence of the wild-type codY gene on a plasmid caused a reduction in the hemolytic activity of the codY mutant (compare MS1 and MS3 [NI]). Thus, when CodY is supplied in trans, the phenotype of increased hemolytic activity in strain MS1 is suppressed. Similarly, when MS2 (codY + plus pTL6936) was treated with IPTG, the hemolytic activity of this strain decreased compared to UAMS-1 and MS2 (NI) (Fig. 1B), suggesting that increased levels of CodY protein result in repressed hemolytic activity even in a wild-type background.
CodY represses alpha-toxin levels in the culture fluid.
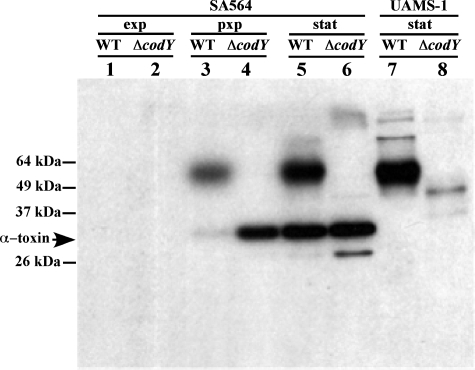
To determine whether the increased hemolytic activity of the culture supernatant of the codY mutants was due to increased production of alpha-toxin, the culture fluid samples were subjected to Western blot analysis (Fig. 2). It has previously been reported that UAMS-1 has a nonsense mutation in the hla gene (6). Anti-alpha-toxin Western blot analysis of culture fluid from strains UAMS-1 and its codY mutant derivative MS1 in stationary phase confirmed that no alpha-toxin was made in these strains (Fig. 2, lanes 7 and 8), suggesting that the hemolytic activity of these strains is due to a different hemolysin. In contrast, Western blot analysis showed that SA564 and CDM7 produced alpha-toxin. This result correlates well with the observation that SA564 and CDM7 had higher HU values than did UAMS-1 and MS1. During exponential phase, there was no detectable alpha-toxin antigen in the culture fluids of SA564 or CDM7. In postexponential phase, CDM7 had much more alpha-toxin in the culture fluid than did SA564. In stationary-phase culture fluids, alpha-toxin was present at similarly high levels in SA564 and CDM7.
FIG. 2.
Effect of growth phase on alpha-toxin production in the culture supernatants of SA564, CDM7, UAMS-1, and MS1. Proteins in equal volumes of culture fluids of wild-type and codY mutant strains were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to an Immobilon-P membrane, and probed with rabbit antibody to alpha-toxin. Lanes 1 to 6 contained culture fluids from SA564 (wild type [WT]) and CDM7 (SA564 ΔcodY::ermC); lanes 7 to 8 contained culture fluids from UAMS-1(wild type) and MS1 (UAMS-1 ΔcodY::ermC). Culture fluids in lanes 1 and 2 were from exponentially growing cells (exp), culture fluids in lanes 3 and 4 were from postexponentially growing cells (pxp), and culture fluids in lanes 5 through 8 were from stationary-phase cells (stat). The bands appearing above 50 kDa are of unknown origin but appear to be unrelated to alpha-toxin.
codY represses hla transcription.
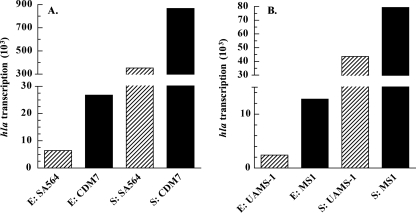
To determine whether the increased hemolytic activity of the codY mutants was due to an increase in hla transcription, primer extension analysis was used to quantitate hla mRNA in the various strains (Fig. 3). In exponential-phase cells, CDM7 had 4.2-fold more hla transcript than did SA564. Similarly, MS1 was derepressed for hla transcription in the exponential phase, having 5.2-fold more transcript than did UAMS-1. In stationary-phase cells, the codY mutants continued to have more hla transcript than did the WT samples. CDM7 had 2.5-fold more hla transcript than did SA564, and MS1 had 1.8-fold more hla transcript than did UAMS-1. We note that strains UAMS-1 and MS1 expressed an hla transcript even though the hla gene contains a nonsense mutation (6) and produces no alpha-toxin protein.
FIG. 3.
Effect of a ΔcodY mutation on hla transcription. hla transcription values represent quantified reverse transcripts normalized to total 16S rRNA from primer extension analysis of the hla promoter. RNA was extracted from cells harvested in the exponential (E) or stationary (S) phases of growth. (A) SA564 (wild type) and CDM7 (SA564 ΔcodY::ermC); (B) UAMS-1 (wild type) and MS1 (UAMS-1 ΔcodY::ermC).
Although the codY mutants showed significant derepression of hla transcription in the exponential phase, hla transcription in the codY mutants still showed growth-phase-dependent regulation. The parental strains UAMS-1 and SA564 showed 18- and 55-fold stationary-phase inductions of hla transcription, respectively, whereas MS1 and CDM7 showed 6.2- and 32-fold stationary-phase inductions of hla transcription, respectively. These data suggest that, even in the codY mutants, the hla promoter is subject to CodY-independent growth-phase-dependent regulation.
CodY represses synthesis of RNAII and RNAIII.
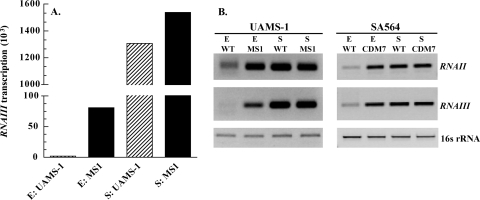
RNAIII positively regulates hla transcription (13) and translation (36, 38) and also encodes a hemolysin, delta-toxin (5, 23). RNAIII transcription peaks in postexponential and early stationary phase (57). Because RNAIII is expressed in a growth-phase-dependent manner, contributes to alpha-toxin activity, and also encodes delta-toxin, we speculated that synthesis of RNAIII might be CodY-regulated. Transcript analysis using primer extension revealed that the level of RNAIII in exponential-phase cells was 40-fold higher in MS1 than in its parent UAMS-1 (Fig. 4A). In stationary-phase cells, RNAIII transcript was greatly increased in both MS1 and UAMS-1, showing a <2-fold difference in expression (Fig. 4A).
FIG. 4.
Effect of a ΔcodY mutation on transcription of the agr locus. Transcript analysis was done on RNA extracted from exponential-phase (E) or stationary-phase (S) cells. (A) RNAIII transcription values represent quantified reverse transcripts normalized to total 16S rRNA from primer extension analysis of the RNAIII promoter of the strains UAMS-1 (wild type [WT]) and MS1 (UAMS-1 ΔcodY::ermC). (B) RT-PCR analysis of RNAII, RNAIII, and 16S rRNA from strains UAMS-1, MS1, SA564 (wild type), and CDM7 (SA564 ΔcodY::ermC) harvested during the exponential phase (E) or the stationary phase (S).
RT-PCR analysis was performed on the wild-type and codY mutant strains of SA564 and UAMS-1 to determine RNAII transcript levels and to further characterize RNAIII transcription. RT-PCR analysis showed that RNAIII transcript levels were derepressed in exponential-phase cells of CDM7 compared to its parent SA564 (Fig. 4B). Similarly, RT-PCR confirmed the primer extension analysis results, which showed that RNAIII was derepressed in exponential-phase cells of MS1 compared to UAMS-1 (Fig. 4B). In addition, in exponential-phase cells, RNAII transcript levels were derepressed in the codY mutants, MS1 and CDM7 compared to the parental strains, UAMS-1 and SA564, respectively (Fig. 4B). In summary, transcript analysis of the agr locus showed that in exponential phase, P2 (RNAII) and P3 (RNAIII) were both derepressed in the codY mutants of the clinical isolates UAMS-1 and SA564.
CodY represses biofilm formation in a static biofilm assay.
Biofilm formation on implanted medical devices and mammalian tissues is a significant contributor to the pathogenicity of S. aureus (12). Although there are many environmental and bacterial factors that influence biofilm attachment and accumulation, the PIA, also known as polymeric N-acetyl-glucosamine, is the best understood contributor to S. aureus biofilm formation (30, 32, 40). The enzymes responsible for PIA production are encoded by the genes of the ica operon; an ica mutant of S. aureus is deficient in PIA production and biofilm formation (9). Divergent from the promoter of the icaADBC operon is a promoter driving transcription of the icaR gene. IcaR is a potent repressor of icaADBC transcription, limiting the production of PIA (24). Other factors, such as SarA (2), TcaR (24), Spx (41), and general stress responses (40) also regulate biofilm formation.
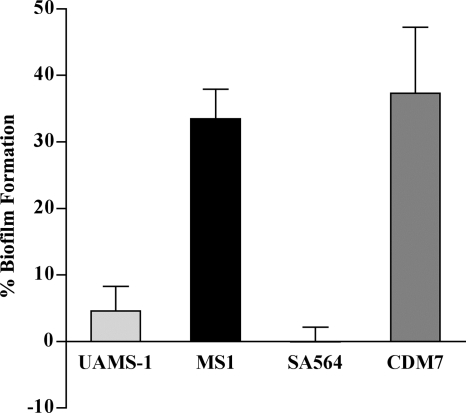
Having observed that the codY mutants were much more adherent than their parents when cultured in glass tubes and flasks, we tested the codY mutants and wild-type strains in a static assay that is used as a surrogate for tests of biofilm formation (2). Cultures in TSB supplemented with 0.5% glucose and 3% NaCl were used to inoculate wells of polystyrene plates, which were then incubated for 24 h at 37°C. After several washing steps, adherent bacteria were detected by using CV. By this assay, the codY mutant MS1 produced 7.5-fold more biofilm than did its parent UAMS-1 (Fig. 5). SA564 produced no detectable biofilm, but the codY mutant of this isolate, CDM7, showed high biofilm formation (Fig. 5). The data in Fig. 5 represent four independent experiments for UAMS-1 and MS1 and three independent experiments for SA564 and CDM7.
FIG. 5.
In vitro static biofilm formation. All strains were grown in TSB medium supplemented with glucose and sodium chloride. The percent biofilm formation was determined as described in Materials and Methods after 24 h of growth of the strains UAMS-1 (wild type), MS1 (UAMS-1 ΔcodY::ermC), SA564 (wild type), and CDM7 (SA564 ΔcodY::ermC). The non-biofilm-producing strain S. carnosus TM300 was used as a negative control. One-hundred percent biofilm production was defined as that produced by S. epidermidis strain 1457.
CodY represses PIA production during exponential growth.
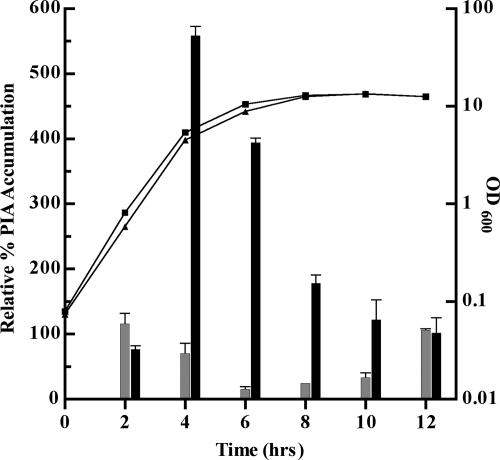
PIA is a major component of S. aureus biofilm formation. Cultures of UAMS-1 and MS1 were tested for PIA production by using a PIA immunoblot assay. MS1 showed derepressed PIA synthesis that peaked at 560% relative PIA accumulation in late exponential phase (time [t] = 4 h) (Fig. 6). At this time, the level of PIA accumulation for MS1 was eightfold higher than for UAMS-1. In postexponential phase (t = 6 h), PIA accumulation for MS1 was ∼20-fold higher than for UAMS-1. After t = 6 h, the extent of derepression in MS1 was reduced; at t = 12 h, PIA production in both the MS1 and the UAMS-1 strains had decreased to ca. 100% relative PIA accumulation (Fig. 6). In summary, PIA production was derepressed in late exponential to early stationary phase in the codY mutant compared to the isogenic wild-type strain.
FIG. 6.
Relative PIA accumulation. Samples were taken throughout growth and prepared for PIA release. A PIA immunoblot assay was performed on samples prepared from equal cell numbers, and the results were quantified. PIA accumulation of a wild-type sample at t = 2 h was set as a 100% standard for relative PIA accumulation. UAMS-1 (wild type) (░⃞) and MS1 (UAMS-1 ΔcodY::ermC) (▪).
codY represses icaA transcription.
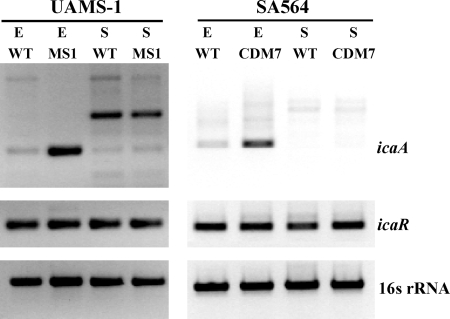
To know whether CodY regulates PIA production at the transcriptional level, we analyzed the transcript levels of icaA, the first gene in the icaADBC operon. RT-PCR analysis was performed on RNA isolated from samples grown to exponential or stationary phases. In both strain backgrounds, a codY mutation resulted in strong derepression of icaA transcription in exponential-phase cells (Fig. 7). In stationary-phase cells, little to no icaA transcript could be detected in the wild-type or codY mutant strains. The codY mutant strains showed no difference in icaR transcript levels compared to the wild-type isolates in either the exponential or the stationary phases (Fig. 7). These data indicate that the exponential-phase derepression of icaA in the codY mutants is independent of the levels of icaR transcript and also that a CodY-independent regulator is responsible for the stationary-phase repression of icaA transcription and PIA production.
FIG. 7.
Effect of a ΔcodY mutation on transcription of the ica locus. Transcripts corresponding to icaA, icaR, and 16S rRNA were analyzed by RT-PCR in samples extracted from exponential-phase (E) or stationary-phase (S) cells of strains UAMS-1 (wild type), MS1 (UAMS-1 ΔcodY::ermC), SA564 (wild type), and CDM7 (SA564 ΔcodY::ermC). The upper band that occurs in the RT-PCR analysis of icaA is likely to reflect nonspecific RT products. WT, wild type.
DISCUSSION
The present study shows that introduction of a codY mutation in two clinical isolates of S. aureus, UAMS-1 and SA564, resulted in derepressed synthesis of hla mRNA and its product, alpha-toxin, derepressed transcription of the accessory gene regulator (agr) locus, and derepressed PIA-dependent biofilm formation. Although transcript analysis of the genes encoding alpha-toxin (hla) and delta-toxin (RNAIII/hld) showed that their transcription was most significantly derepressed during exponential-phase growth, increased hemolytic activity and alpha-toxin production could only be detected in the culture fluids of postexponential-phase cells. Because both RNAIII and hla transcripts are derepressed in the codY mutant strains and RNAIII is an activator of hla translation (36, 38), it is unclear why the appearance of hemolytic activity and alpha-toxin production is delayed. It is possible that the hemolytic assay and Western blot analysis are not sensitive enough to detect differences in low levels of hemolysin(s) present in culture fluids from low numbers of bacteria. Since UAMS-1 does not produce alpha-toxin, in keeping with a nonsense mutation in the UAMS-1 hla gene (6), the hemolytic activity of the culture fluid of MS1 is likely due to the derepression of another hemolysin, such as delta-toxin. The culture fluids of strains SA564 and CDM7 did contain alpha-toxin. Thus, the high HU values seen in the culture fluid of CDM7 are likely due to derepression of both alpha-toxin and delta-toxin. In addition, although the codY mutants showed significant exponential-phase derepression of hla and RNAIII/hld transcription, both genes were still subject to growth-phase-dependent regulation. This is not surprising since these loci are known to be regulated by a number of other factors, such as numerous TCS (5, 17), SarA (8, 13), SarS (54), and other members of the SarA family (5).
Since our studies show that RNAII transcription is also altered in the codY mutants, the CodY-mediated regulation of RNAIII/hld transcription may be an indirect effect of regulation of RNAII transcription. In addition, our present results are consistent with either direct or indirect repression by CodY of hla transcription. Ongoing studies aim to understand how CodY participates in the regulatory network of hla and RNAIII/hld transcription, specifically by identifying the direct CodY targets. Since CodY has been shown to control many aspects of metabolism important for adaptation to nutritional and metabolic changes in other bacteria, such as B. subtilis (35) and L. lactis (21), we assume that CodY will prove to play a central role in these processes in S. aureus as well. Thus, the effects of codY mutations on virulence gene expression may reflect direct interaction of CodY with virulence gene promoters or indirect effects of alterations of metabolic activities.
CodY acts as a repressor of PIA-dependent biofilm formation. In looking at quantitative measures of biofilm formation, such as PIA production and icaA transcription, it is clear that the CodY-mediated control of biofilm formation in UAMS-1 and SA564 is specific to the growth phase and is likely to occur through transcriptional regulation of the icaADBC operon. In the postexponential phase of in vitro growth, PIA production is derepressed in the codY mutant of UAMS-1. Similarly, in both clinical isolates, a codY mutation results in derepression of icaA transcription during exponential phase. Interestingly, in stationary phase, the CodY-mediated derepression is abolished, as icaA transcript levels and PIA production in the codY mutant drop to the very low levels seen for wild-type cells. In the codY mutants, this transient, yet potent, derepression of the icaADBC operon during exponential phase likely leads to a substantial increase in biofilm formation after 24 h of growth. Thus, CodY is certainly not the sole regulator of this locus. IcaR, TcaR (24), SarA (2, 56), and likely numerous other factors (40) contribute to the complex regulation of the icaADBC operon.
IcaR is a transcriptional repressor that binds to the promoter region of icaADBC (24) and represses this operon in both the exponential phase (41) and the stationary phase (24). Transcriptional analysis of icaR in the codY mutants of SA564 and UAMS-1 showed that even though icaA was highly derepressed in the exponential phase, there was no difference in icaR transcript levels between the codY mutants and the wild-type strains. This suggests that CodY-mediated regulation of the icaADBC locus is independent of IcaR and that CodY-mediated exponential-phase repression of icaA transcription supersedes IcaR-mediated repression.
Tu Quoc et al. recently reported that a codY mutation in the clinical isolate S30 results in reduced biofilm formation and PIA production (55). These data are in contrast to the effect of a codY mutation in UAMS-1 and SA564. Other than citing strain-to-strain variation among S. aureus isolates, it is difficult to explain the different results obtained by the two groups. One possibility is that CodY's regulatory role in biofilm formation is multifactorial. CodY may act, directly or indirectly, to repress icaADBC transcription in isolates of S. aureus such as SA564 and UAMS-1. However, in high-biofilm-producing isolates, such as S30 (55), the regulatory network of PIA-dependent biofilm formation may be different than in SA564 and UAMS-1. If this is the case, the effect of CodY on biofilm formation in a high-biofilm-producing strain may be an indirect reflection of its role in regulating various aspects of intermediary metabolism. As we learn more about the mechanism of CodY-mediated regulation of the icaADBC locus, we hope be able to explain the different phenotypes resulting from codY mutations in S30, UAMS-1, and SA564.
In other gram-positive bacteria, CodY is a regulatory protein that responds to the nutrient status of the cell by sensing the intracellular pools of GTP and/or the BCAAs. Interaction with these effector molecules activates CodY as a DNA-binding protein. In this manner, CodY is an important regulator of stationary phase adaptation that is activated and inactivated by environmental cues. Amino acid sequence similarity with the B. subtilis protein and in vitro DNA-binding studies indicate that the affinity of S. aureus CodY for it binding sites is enhanced by the BCAAs and GTP (C. Majerczyk and A. L. Sonenshein, unpublished results), raising the possibility that CodY serves in S. aureus to link environmental nutrient availability with regulation of pathogenesis. Moreover, since transcription of RNAIII is derepressed in the codY mutants, S. aureus appears to regulate this characterized virulence gene regulator through both quorum-sensing and nutrient-sensing mechanisms.
Acknowledgments
We thank Michael Otto for the gift of antibody to PIA, Timothy J. Foster for the pTS1 plasmid, Paul Fey for the strains S. epidermidis 1457 and S. carnosus TM300, and Luke Handke for many helpful discussions and assistance with the biofilm assay.
This study was supported by research grants from the U.S. Public Health Service (GM042219 to A.L.S., GM076585 to G.A.S., and AI037027 to C.L.) and from the American Heart Association (0760005Z to G.A.S.).
Footnotes
Published ahead of print on 21 December 2007.
REFERENCES
- 1.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1997. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 2.Beenken, K., J. Blevins, and M. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, H., D. Pearce, S. Glenn, C. Taylor, M. Kuhn, A. Sonenshein, P. Andrew, and I. Roberts. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 63:1453-1467. [DOI] [PubMed] [Google Scholar]
- 4.Blevins, J., A. Gillaspy, T. Rechtin, B. Hurlburt, and M. Smeltzer. 1999. The staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner. Mol. Microbiol. 33:317-326. [DOI] [PubMed] [Google Scholar]
- 5.Bronner, S., H. Monteil, and G. Prevost. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183-200. [DOI] [PubMed] [Google Scholar]
- 6.Cassat, J., P. Dunman, E. Murphy, S. Projan, K. Beenken, K. Palm, S. Yang, K. Rice, K. Bayles, and M. Smeltzer. 2006. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology 152:3075-3090. [DOI] [PubMed] [Google Scholar]
- 7.Charpentier, E., A. Anton, P. Barry, B. Alfonso, Y. Fang, and R. Novick. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 70:6078-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A., J. Koomey, C. Butler, S. Projan, and V. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cramton, S., C. Gerke, N. Schnell, W. Nichols, and F. Gotz. 1999. The intercellular adhesin (ica) locus is present in Staphylococcus aurues and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.den Hengst, C. D., P. Curley, R. Larsen, G. Buist, A. Nauta, D. van Sinderen, O. P. Kuipers, and J. Kok. 2005. Probing direct interactions between CodY and the oppD promoter of Lactococcus lactis. J. Bacteriol. 187:512-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dineen, S., A. Villapakkam, J. Nordman, and A. Sonenshein. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66:206-219. [DOI] [PubMed] [Google Scholar]
- 12.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunman, P. M., E. Murphy, S. Haney, D. Palacois, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischetti, V. 2000. Gram-positive pathogens. American Society for Microbiology, Washington, DC.
- 15.Fisher, S., K. Rohrer, and A. Ferson. 1996. Role of CodY in regulation of the Bacillus subtilis hut operon. J. Bacteriol. 178:3779-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frees, D., L. Thomsen, and H. Ingmer. 2005. Staphylococcus aureus ClpYQ plays a minor role in stress survival. Arch. Microbiol. 183:286-291. [DOI] [PubMed] [Google Scholar]
- 17.Georke, C., U. Fluckiger, A. Steinhuber, W. Zimmerll, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA, and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40:1439-1447. [DOI] [PubMed] [Google Scholar]
- 18.Gillaspy, A., S. Hickmon, R. Skinner, J. Thomas, C. Nelson, and M. Smeltzer. 1995. Role of the accessory gene regulatory (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gotz, F. 1990. Staphylococcus carnosus: a new host organism for gene cloning and protein production. Soc. Appl. Bacteriol. Symp. Ser. 19:49S-53. [DOI] [PubMed] [Google Scholar]
- 20.Greene, C., D. McDevitt, P. Francois, P. E. Vaudauz, D. P. Lew, T. J. Foster, D. McDevitt, P. Francois, P. E. Vaudauz, D. P. Lew, and T. J. Foster. 1995. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17:1143-1152. [DOI] [PubMed] [Google Scholar]
- 21.Guedon, E., B. Sperandio, N. Pons, S. Dusko-Ehrlich, and P. Renault. 2005. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models of CodY regulation in Firmicutes. Microbiology 151:3895-3909. [DOI] [PubMed] [Google Scholar]
- 22.Inaoka, T., K. Takahashi, M. Ohnishi-Kameyama, M. Yoshica, and K. Ochi. 2003. Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 278:2169-2176. [DOI] [PubMed] [Google Scholar]
- 23.Janzon, L., S. Lofdahl, and S. Arvidson. 1989. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol. Gen. Gent. 219:480-485. [DOI] [PubMed] [Google Scholar]
- 24.Jefferson, K., D. Pier, D. Goldman, and G. Pier. 2004. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joseph, P., M. Ratnayake-Lecamwasam, and A. L. Sonenshein. 2005. A region of Bacillus subtilis CodY protein required for interaction with DNA. J. Bacteriol. 187:4127-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreiswirth, B., S. Lofdahl, M. Betley, M. O'Reilly, P. Schlievert, M. Bergdoll, and R. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli, U. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Levdikov, V. M., E. Blagova, P. Joseph, A. L. Sonenshein, and A. J. Wilkinson. 2006. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria. J. Biol. Chem. 281:11366-11373. [DOI] [PubMed] [Google Scholar]
- 29.Luong, T., and C. Lee. 2006. The arl locus positively regulates Staphylococcus aureus type 5 capsule via an mgrA-dependent pathway. Microbiology 152:3123-3131. [DOI] [PubMed] [Google Scholar]
- 30.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mack, D., N. Siemssen, and R. Laufs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maira-Litran, T., A. Kropec, C. Abeygunawardana, J. Joyce, G. Mark III, D. Goldmann, and G. Pier. 2002. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 70:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malke, H., and J. Ferretti. 2007. CodY-affected transcriptional gene expression of Streptococcus pyogenes during growth in human blood. J. Med. Microbiol. 56:707-714. [DOI] [PubMed] [Google Scholar]
- 34.Malke, H., K. Steiner, W. McShan, and J. Ferretti. 2006. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int. J. Med. Microbiol. 296:259-275. [DOI] [PubMed] [Google Scholar]
- 35.Molle, V., Y. Nakaura, R. Shivers, H. Yamaguchi, R. Losik, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morfeldt, E., D. Taylor, A. von Gabain, and S. Arvidson. 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 14:4569-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novick, R., S. Projan, J. Kornblum, H. Ross, G. Ji, B. Kreiswirth, F. Vandenesch, and S. Moghazeh. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Gent. 248:446-458. [DOI] [PubMed] [Google Scholar]
- 38.Novick, R., H. Ross, S. Projan, J. Kornblum, B. Kreiswirth, and S. Moghaseh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novick, R. P. 1990. The Staphylococcus as a molecular genetic system, p. 587-636. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, NY.
- 40.O'Gara, J. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270:179-188. [DOI] [PubMed] [Google Scholar]
- 41.Pamp, S., D. Frees, S. Englemann, M. Hecker, and H. Ingmer. 2006. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 188:4861-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng, H., R. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petranovic, D., E. Guedon, B. Sperandio, C. Delorme, D. Ehrlich, and P. Renault. 2004. Intracellular effectors regulating the activity of the Lactococcus lactis CodY pleiotropic transcription regulator. Mol. Microbiol. 53:613-621. [DOI] [PubMed] [Google Scholar]
- 44.Ratnayake-Lecamwasam, M., P. Serror, K. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schenk, S., and R. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 94:133-138. [DOI] [PubMed] [Google Scholar]
- 46.Serror, P., and A. L. Sonenshein. 1996. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 178:5910-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serror, P., and A. L. Sonenshein. 1996. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol. Microbiol. 20:843-852. [DOI] [PubMed] [Google Scholar]
- 48.Shivers, R. P., S. S. Dineen, and A. L. Sonenshein. 2006. Positive regulation of Bacillus subtilis ackA by CodY and CcpA: establishing a potential hierarchy in carbon flow. Mol. Microbiol. 62:811-822. [DOI] [PubMed] [Google Scholar]
- 49.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53:599-611. [DOI] [PubMed] [Google Scholar]
- 50.Slack, F., P. Serror, E. Joyce, and A. L. Sonenshein. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15:689-702. [DOI] [PubMed] [Google Scholar]
- 51.Somerville, G., S. B. Beres, J. R. Fitzgerald, F. R. DeLeo, R. L. Cole, J. S. Hoff, and J. M. Musser. 2002. In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence. J. Bacteriol. 184:1430-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Somerville, G., M. Chaussee, C. Morgan, R. Fitzgerald, D. Dorward, L. Reitzer, and J. Musser. 2002. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70:6373-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonenshein, A. L. 2005. CodY, a global regulator of stationary phase and virulence in gram-positive bacteria. Curr. Opin. Microbiol. 8:203-207. [DOI] [PubMed] [Google Scholar]
- 54.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 55.Tu Quoc, P., P. Genevaux, M. Pajunen, H. Savilahti, C. Georgopoulos, J. Schrenzel, and W. Kelley. 2007. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect. Immun. 74:1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valle, J., A. Toledo-Arana, C. Berasain, J. Ghigo, B. Amorena, J. Penades, and I. Lasa. 2003. SarA and not Sigma B is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 57.Vandenesch, F., J. Kornblum, and R. P. Novick. 1991. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J. Bacteriol. 173:6313-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vuong, C., C. Gerke, G. Somerville, E. Fischer, and M. Otto. 2003. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188:706-718. [DOI] [PubMed] [Google Scholar]
- 59.Wray, L., A. Ferson, and S. Fisher. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and SpoOH. J. Bacteriol. 179:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright, J., III, R. Jin, and R. Novick. 2005. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. USA 102:1691-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu, Y., E. Weiss, M. Otto, P. D. Fey, M. Smeltzer, and G. A. Somerville. 2007. Staphylococcus aureus biofilm metabolism and the influence of arginine on polysaccharide intercellular adhesin synthesis, biofilm formation, and pathogenesis. Infect. Immun. 75:4219-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]