Abstract
A number of gram-negative bacteria regulate gene expression in a cell density-dependent manner by quorum sensing via N-acylhomoserine lactones (AHLs). Gluconacetobacter intermedius NCI1051, a gram-negative acetic acid bacterium, produces three different AHLs, N-decanoyl-l-homoserine lactone, N-dodecanoyl-l-homoserine lactone, and an N-dodecanoyl-l-homoserine lactone with a single unsaturated bond in its acyl chain, as determined by liquid chromatography-tandem mass spectrometry. Two genes encoding an AHL synthase and a cognate regulator were cloned from strain NCI1051 and designated ginI and ginR, respectively. Disruption of ginI or ginR abolished AHL production, indicating that NCI1051 contains a single set of quorum-sensing genes. Transcriptional analysis showed that ginI is activated by GinR, which is consistent with the finding that there is an inverted repeat whose nucleotide sequence is similar to the sequence bound by members of the LuxR family at position −45 with respect to the transcriptional start site of ginI. A single gene, designated ginA, located just downstream of ginI is transcribed by read-through from the GinR-inducible ginI promoter. A ginA mutant, as well as the ginI and ginR mutants, grew more rapidly in medium containing 2% (vol/vol) ethanol and accumulated acetic acid at a higher rate with a greater final yield than parental strain NCI1051. In addition, these mutants produced larger amounts of gluconic acid than the parental strain. These data demonstrate that the GinI/GinR quorum-sensing system in G. intermedius controls the expression of ginA, which in turn represses oxidative fermentation, including acetic acid and gluconic acid fermentation.
Diverse bacteria communicate intercellularly to regulate the transcription of specific target genes in a cell density-dependent manner. This cell-cell communication is termed quorum sensing. Bacteria monitor their cell density by measuring the concentration of self-produced diffusible signal molecules termed autoinducers or pheromones. Quorum sensing is used to regulate diverse physiological functions, including secondary metabolite production, swimming and swarming motility, conjugal plasmid transfer, biofilm formation, and virulence (25, 27).
Many gram-negative bacteria use N-acylhomoserine lactones (AHLs) as quorum sensors to regulate gene expression in concert with cell density (7, 8). Two important proteins are involved in AHL-dependent quorum sensing: a LuxR-type protein, which is an AHL-dependent transcriptional regulator, and a LuxI-type protein, which synthesizes AHLs. In general, LuxR-type proteins bind their cognate AHLs once the concentration of the AHL reaches a critical level, and the resulting complex activates the transcription of specific target genes.
Acetic acid bacteria are gram-negative, obligately aerobic bacteria with the ability to oxidize ethanol and sugars into their corresponding organic acids. Acetobacter and Gluconacetobacter are used to produce vinegar because of their ability to oxidize ethanol into acetic acid and their strong resistance to acetic acid and ethanol. Two membrane-bound enzymes, alcohol dehydrogenase and aldehyde dehydrogenase, catalyze the oxidation of ethanol. The discovery by a research group at Mizkan Group Co., Ltd., of luxI and luxR homologues in Gluconacetobacter polyoxogenes, using genome sequencing, indicates that an AHL-mediated quorum-sensing system is present in acetic acid bacteria (K. Kondo, personal communication). Here, we report cloning of the luxI and luxR homologues ginI and ginR and identification of several AHLs produced by Gluconacetobacter intermedius NCI1051. The ginA gene, located just downstream of ginI, was found to be a target of GinR and to control two important characteristics of acetic acid bacteria: the growth of the strain in ethanol-containing medium and acetic acid production. This is the first report of a relationship between quorum sensing and oxidative fermentation in acetic acid bacteria.
MATERIALS AND METHODS
Bacterial strains and plasmids.
G. intermedius NCI1051 and G. polyoxogenes NCI1028 were obtained from the Central Research Institute, Mizkan Group Co., Ltd. The Acetobacter/Gluconacetobacter-Escherichia coli shuttle vector pMV24 was described previously (6). E. coli strain JM109 and pUC19, which were used for cloning, were purchased from Takara Bio. Agrobacterium tumefaciens NTL4(pZLR4), obtained from S. K. Farrand, and Chromobacterium violaceum CV026, obtained from P. Williams, were used to assay AHL production.
Media and culture conditions.
YPG medium (pH 6.5) consisted of 5 g of yeast extract (Wako Pure Chemicals), 3 g of polypeptone (Wako Pure Chemicals), and 30 g of glucose in 1 liter of water. The Gluconacetobacter strains were grown at 30°C in YPG medium with 1% (vol/vol) cellulase (Celluclast 1.5 L; Novozymes) with or without 2% (vol/vol) ethanol.
For the acetic acid fermentation tests, the Gluconacetobacter strains were first cultured in 5 ml of YPG medium containing 1% (vol/vol) cellulase in a test tube with shaking at 30°C for 24 h. A portion (5 ml) of each culture was then inoculated into 1.5 liters of YPG medium supplemented with 3% (vol/vol) ethanol, 1% (vol/vol) cellulase, and 0.001% (vol/vol) silicone (KM72; Shin-Etsu Chemical Co., Ltd.) in a 3-liter mini-jar fermentor (Bioneer300 3L; B. E. Marubishi Co., Ltd.) at 30°C with agitation at 500 rpm and aeration at a rate of 1.0 liter/min. The ethanol concentration was automatically maintained at 2% (vol/vol) by addition of ethanol during cultivation. The acetic acid and gluconic acid concentrations in the culture broth were determined by high-performance liquid chromatography. Bacterial growth was monitored by measuring the turbidity of the cultures at 660 nm with a photometer.
For polysaccharide production, the Gluconacetobacter strains were first cultured in 5 ml of FPY medium (20 g/liter fructose, 10 g/liter polypeptone, 5 g/liter yeast extract, 2.5 g/liter K2HPO4; pH 5.0) containing 1% (vol/vol) cellulase in a test tube with shaking at 30°C for 24 h. A portion (0.5 ml) of each culture was then inoculated into 50 ml of FPY medium containing 0.15% (vol/vol) lactate in a 300-ml flask and further cultured statically at 30°C for 3 to 7 days.
A. tumefaciens NTL4(pZLR4) was cultured at 30°C in AB medium containing 0.2% (wt/vol) glucose, 0.1% (wt/vol) yeast extract, and 5 μg/ml gentamicin (12). C. violaceum CV026 was cultured at 30°C in LB medium (14). E. coli was cultured at 37°C in LB medium. Ampicillin and kanamycin were used at a final concentration of 100 μg/ml when necessary to maintain the plasmids.
DNA manipulation.
Restriction enzymes, T4 DNA ligase, and other DNA-modifying enzymes were purchased from Takara Bio. All DNA manipulations in E. coli were performed as described previously (2, 13). The Gluconacetobacter strains were transformed by electroporation (28). Chromosomal DNA from the Gluconacetobacter strains was isolated using a GenomicPrep cell and tissue DNA isolation kit (Amersham Bioscience). Chromosomal DNA from G. intermedius NCI1051 and G. polyoxogenes was used as the template for PCR. All nucleotide sequences were determined with a CEQ dye terminator cycle sequencer using a Quick Start kit (Beckman Coulter).
Cloning of ginI and ginR.
A 0.2-kb fragment containing a portion of the luxI homologue GP1166 from G. polyoxogenes NCI1028 was amplified by PCR using primers GP-IF and GP-IR (Table 1). Similarly, a 0.4-kb fragment containing a portion of the luxR homologue GP1165 from G. polyoxogenes NCI1028 was amplified by PCR using primers GP-RF and GP-RR. The resulting fragments were made into 32P-labeled probes for Southern hybridization with PstI-digested chromosomal DNA from G. intermedius NCI1051. By using standard methods, including colony hybridization using the 0.2-kb fragment as a probe, a 2.8-kb PstI fragment containing ginI and ginR was cloned into the PstI site of pUC19.
TABLE 1.
Primers used
| Primer | Sequence (5′ to 3′)a |
|---|---|
| GP-IF | TGTGGCCAATGAGCAGTGGG |
| GP-IR | ACCGGGTGATTTCCCAGACC |
| GP-RF | GGATATCCAACGGAATGGGTC |
| GP-RR | GTCAAAGTTAACGGTTCGCTC |
| A-F | CCGGAATTCACATGATGTCGCTTCTGCCG |
| A-R | CCCAAGCTTCTGCGATGGAGAGCGAGTTC |
| S1-IF | CCGGAATTCGGATATGTCGCTCCCCATTC |
| S1-IR | GGGGACTTCCCACTGCTCATTAG |
| S1-RF | TCCGCAATAGCCTGCCTCTC |
| S1-RR | GTGGCTGCGTTCTCAAGGTC |
| UF | TCCGCAATAGCCTGCCTCTC |
| RF | GACCTTGAGAACGCAGCCAC |
| IF | CCGATATGATGCCCGATCCG |
| AF | CGGAGAACTCGCTCTCCATC |
| RR | ATCAGCCCTGCGCAGATAGC |
| IR | TGCATGCTGCTGCTCAATGC |
| AR | GGTGCCAGCCTGATAATTGC |
| DR | ACCAGGTGCGTGAGGGCATG |
Restriction sites are underlined.
Plasmid construction.
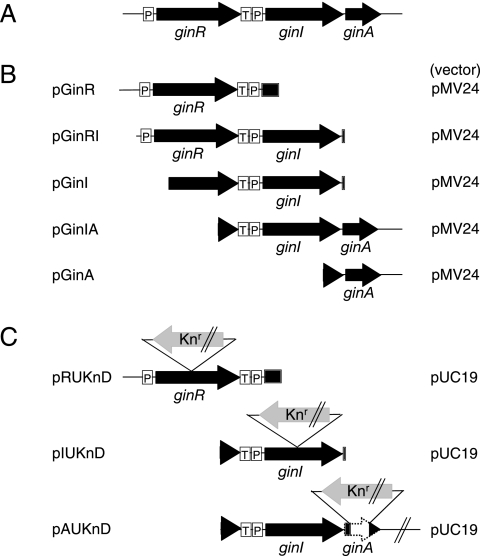
The expression plasmids (pGinR, pGinI, pGinIA, pGinA, pGinRI, pMGinR, and pMGinA) and the plasmids used for gene disruption (pRUKnD, pIUKnD, and pAUKnD) were constructed using standard methods. Details are described in the supplemental material.
Disruption of ginI, ginR, and ginA.
Gene disruption was accomplished by insertion of a neomycin/kanamycin resistance gene into the target gene by homologous recombination (double crossover). For disruption of ginI, pIUKnD was introduced into G. intermedius NCI1051 by electroporation, and kanamycin-resistant colonies were selected as candidate ginI mutants. The desired mutants were identified by Southern hybridization using a 0.2-kb fragment amplified using primers GP-IF and GP-IR as a 32P-labeled probe (data not shown). Similarly, the ginR and ginA mutants were constructed using pRUKnD and pAUKnD, respectively. The desired ginR and ginA mutants were identified by Southern hybridization with a 0.4-kb fragment amplified using primers GP-RF and GP-RR and a 0.2-kb fragment amplified using primers A-F and A-R, respectively, as 32P-labeled probes (data not shown).
AHL assay.
The Gluconacetobacter strains were first grown at 30°C for 24 h in YPG medium containing 1% cellulase and 2% ethanol. The cells were then removed by centrifugation, and the AHLs were extracted from the supernatants using acidified ethyl acetate, as described previously (19). The AHLs were detected in well diffusion assays (19) using A. tumefaciens NTL4(pZLR4) (12) or C. violaceum CV026 (14) as an AHL indicator strain. A portion of the ethyl acetate extract was placed into the well of an agar plate inoculated with the AHL indicator strain. The time course of AHL production by strain NCI1051 was determined by measuring the β-galactosidase activity of A. tumefaciens NTL4(pZLR4), as described by Luo et al. (12).
AHL production by E. coli JM109 harboring pGinRI was detected on agar medium using the A. tumefaciens NTL4(pZLR4) reporter system. E. coli JM109 harboring pGinRI was streaked on LB agar containing 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) perpendicular to A. tumefaciens NTL4(pZLR4). AHL production was detected by production of a blue pigment by A. tumefaciens NTL4(pZLR4). As a negative control, E. coli JM109 harboring the empty vector pMV24d, which was pMV24 containing a lacZ-disrupted gene, was streaked on the same medium.
The chemical structures of the AHLs produced by strain NCI1051 were determined by liquid chromatography-mass spectrometry (LC-MS) and liquid chromatography-tandem mass spectrometry (LC-MS/MS) using the concentrated ethyl acetate extract, as described previously (15). The standards N-decanoyl-l-homoserine lactone (C10-AHL) and N-dodecanoyl-l-homoserine lactone (C12-AHL) were purchased from Sigma.
S1 nuclease mapping.
Total RNA was isolated by the hot phenol method (18) from cells grown at 30°C for 8 h in YPG medium containing 2% ethanol and 1% cellulase in a shaking flask. S1 nuclease mapping was conducted as described previously (10). The hybridization probes were prepared by PCR using pairs of 32P-labeled and unlabeled primers. The primers used were S1-IF and S1-IR for ginI and S1-RF and S1-RR for ginR (Table 1). Primers S1-IR and S1-RR were labeled with 32P at their 5′ ends using T4 polynucleotide kinase before PCR. All sequencing reactions were performed using a BcaBEST dideoxy sequencing kit (Takara Bio).
RT-PCR.
Total RNA was isolated as described above. Reverse transcription (RT) was performed using the total RNA with SuperScript III reverse transcriptase (Invitrogen). The cDNA was then amplified by PCR using the RT reaction mixture as the template and specific primers (Table 1). The RNA samples were tested with and without RT to verify the absence of genomic DNA. PCR using G. intermedius NCI1051 genomic DNA as the template was performed to ensure the fidelity of each primer pair.
Enzyme assay.
Alcohol dehydrogenase (EC 1.1.99.8) activity was measured as described previously using the cytoplasmic membrane (1, 20). Briefly, the oxidation of potassium ferricyanide, resulting in Prussian blue coloring, was measured by determining the absorbance at 660 nm.
Production of cellulose and water-soluble polysaccharides.
The production of cellulose and water-soluble polysaccharides was measured as described previously (9, 23). To purify the cellulose, a precipitate of culture broth obtained by centrifugation was washed with deionized water and treated with 0.1 M NaOH at 80°C for 20 min. The cellulose was then purified by three washes with deionized water. The purified cellulose was dried at 80°C and weighed. To purify the water-soluble polysaccharides, the supernatant of a culture broth obtained by centrifugation was treated with 2 volumes of ethanol. The water-soluble polysaccharides that precipitated were air dried, and the residue was dissolved in 0.1 M NaOH. An equal volume of chloroform was added to the water-soluble polysaccharide solution, and the liquids were gently mixed. After the tube was centrifuged, the aqueous phase was precipitated with ethanol and air dried; the remaining material was weighed.
Nucleotide sequence accession numbers.
The nucleotide sequences of ginRIA, GP1165, and GP1166 have been deposited in the DDBJ, EMBL, and GenBank databases under accession numbers AB330995, AB330993, and AB330994, respectively.
RESULTS
Identification of quorum-sensing systems in acetic acid bacteria.
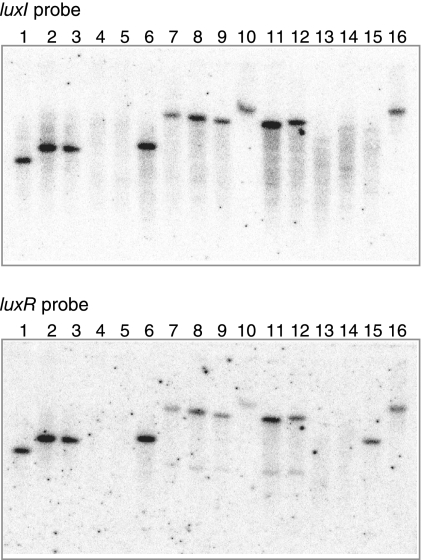
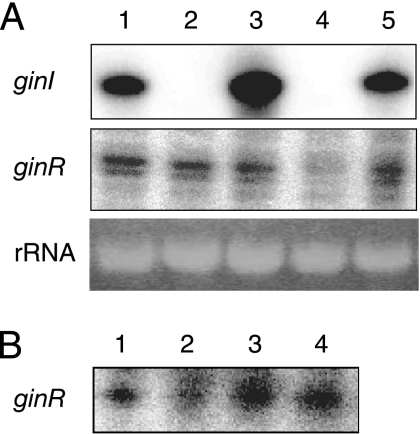
To establish the presence of AHL-based quorum-sensing systems in acetic acid bacteria, we prepared a 32P-labeled probe based on the nucleotide sequence of GP1166, which is a luxI homologue in G. polyoxogenes NCI1028 (Kondo, personal communication). Southern hybridization of this probe with PstI-digested chromosomal DNA from various acetic acid bacteria, including G. intermedius and Gluconacetobacter xylinus, indicated that nucleotide sequences homologous to the probe were present in this group of bacteria (Fig. 1). Each positive strain produced a single signal, indicating the presence of a single luxI homologue highly similar to the luxI probe used. A separate experiment using a 32P-labeled probe for GP1165, which is a luxR homologue, produced similar results (Fig. 1). These observations indicate that most acetic acid bacteria have a system homologous to GinI-GinR.
FIG. 1.
Southern blots to determine the distribution of luxI and luxR homologues in acetic acid bacteria. A luxI probe (a 0.2-kb fragment amplified by PCR using primers GP-IF and GP-IR) and a luxR probe (a 0.4-kb fragment amplified by PCR using primers GP-RF and GP-RR) were used for hybridization with PstI-digested total DNA from each Gluconacetobacter strain. Lane 1, G. intermedius NCI1051; lane 2, G. xylinus NCI1061; lane 3, G. xylinus NCI1062; lane 4, G. xylinus NCI1381; lane 5, G. xylinus NCI1382; lane 6, G. xylinus NCI1447; lane 7, G. xylinus ATCC 53263; lane 8, G. xylinus ATCC 53264; lane 9, G. xylinus ATCC 53524; lane 10, G. xylinus ATCC 53582; lane 11, G. xylinus ATCC 53749; lane 12, G. xylinus ATCC 53750; lane 13, G. xylinus ATCC 12528; lane 14, G. xylinus IFO3288; lane 15, G. xylinus BPR2001; lane 16, G. polyoxogenes NCI1028.
Cloning of ginI-ginR, a luxI-luxR homologue.
We used the probe described above to clone the 2.8-kb luxI-positive fragment from G. intermedius NCI1051 by colony hybridization. The isolated fragment was subsequently cloned into the PstI site of pUC19.
Sequencing of the 2.8-kb fragment revealed the presence of three complete open reading frames (ORFs) (Fig. 2A). The luxI homologue encodes a protein consisting of 209 amino acids, whereas the luxR homologue, which is located upstream of and in the same direction as the luxI homologue, encodes a protein consisting of 236 amino acids. We designated the luxI and luxR homologues ginI and ginR, respectively, because of their functions in quorum sensing (see below). The primary sequence of GinI shares about 30% identity with diverse members of the LuxI family, including LuxI from Vibrio fischeri (3), AlpI from Azospirillum lipoferum (24), CepI from Burkholderia cepacia (11), and AfeI from Acidithiobacillus ferrooxidans (5). Similarly, GinR shares about 40% identity with several LuxR family members, including LuxR from V. fischeri (3), BpsR from Burkholderia pseudomallei (21), LuxR1 from Vibrio salmonicida (17), and AhyR from Aeromonus hydrophila (22). Sequence alignments of the GinI and GinR homologues are shown in Fig. S1 in the supplemental material.
FIG. 2.
Schematic representation of the genes at the gin locus (A) and the fragments inserted into the plasmids used (B and C). Arrows represent the direction and length of each gene. Knr indicates the neomycin/kanamycin resistance gene. P and T indicate the promoter and terminator, respectively. The pMV24-derived plasmids (B) were used for expression of the gin genes in G. intermedius NCI1051. The pUC19-derived plasmids (C) were used for gene disruption.
An additional ORF, designated ginA, that was oriented in the same direction as the ginI and ginR ORFs, was present just downstream of ginI (Fig. 2A). The ginA gene encodes a small protein consisting of 89 amino acids with a predicted pI of 10.1 and no significant similarity to any known protein. As described below, ginA is cotranscribed with ginI.
ginI and ginR genes mediate AHL production.
To detect the production of AHLs by G. intermedius, we extracted candidate AHLs from the supernatant of a stationary-phase culture of strain NCI1051 using ethyl acetate and subjected them to well diffusion assays with A. tumefaciens NTL4(pZLR4) (12) and C. violaceum CV026 (14) as indicators. A. tumefaciens was used to detect AHLs containing acyl chains longer than C6 by a lacZ reporter system (12), whereas C. violaceum was used to detect AHLs with acyl chains shorter than C8 by induction of violacein synthesis (14). The ethyl acetate extract of strain NCI1051 caused A. tumefaciens to express the reporter gene, resulting in a blue color around the well. In contrast, no induction of violacein synthesis was observed for the C. violaceum system. These results indicate that NCI1051 produces AHLs with acyl chains longer than C8.
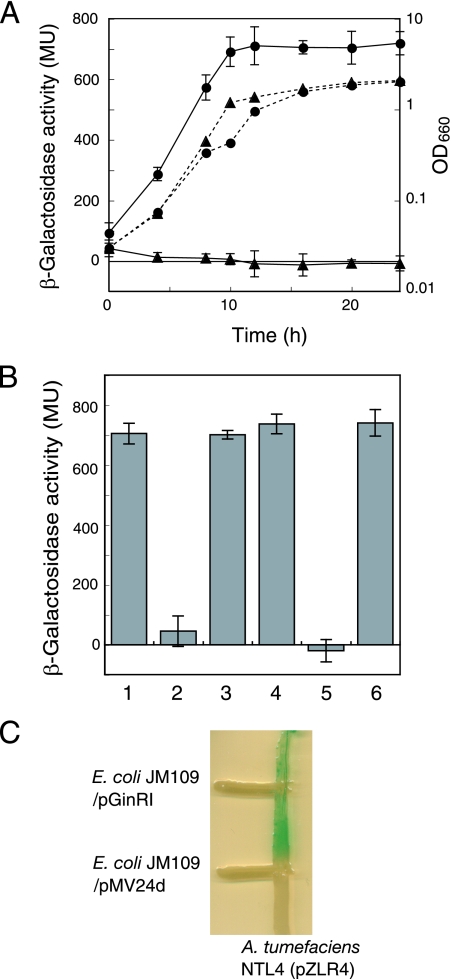
The identification of AHL production by strain NCI1051 was followed by preparation of ethyl acetate extracts during growth and measurement of the induction of lacZ expression as described above. NCI1051 accumulated AHLs in a growth- and cell density-dependent manner, whereas a ginI mutant produced no AHLs (Fig. 3A). The introduction of intact ginI-ginA from pGinIA (Fig. 2B) into the ginI mutant restored its ability to produce AHLs (Fig. 3B). However, ginA is not involved in AHL production because the introduction of ginI alone from pGinI (Fig. 2B) into the ginI mutant restored AHL production (Fig. 3B). A ginR mutant was also unable to produce AHLs; however, introduction of intact ginR from pGinR (Fig. 2B) restored AHL production (Fig. 3B). Thus, ginI and ginR constitute a quorum-sensing system based on AHLs with acyl chains longer than C8.
FIG. 3.
Accumulation of AHLs in the culture broth of G. intermedius NCI1051 and AHL production by E. coli JM109 harboring pGinRI. (A) Parental strain NCI1051 (•) and the ginI mutant (▴) were grown at 30°C for 24 h in YPG medium containing 2% (vol/vol) ethanol and 1% (vol/vol) cellulase. These two strains had similar growth curves (dotted lines), which were determined by measuring the optical density at 660 nm (OD660) over time. At each time point, the cultures were sampled, and extracts were prepared with ethyl acetate for use in assays to determine the production of AHLs (solid line) by measuring the β-galactosidase activity in A. tumefaciens NTL4(pZLR4). The values are means ± standard deviations (n = 3). MU, Miller units. (B) AHL activities in the culture broth of the strains grown for 24 h, as assayed by measuring the β-galactosidase activity in the AHL indicator strain. Bar 1, wild-type strain NCI1051 harboring pMV24; bar 2, ginI mutant harboring pMV24; bar 3, ginI mutant harboring pGinI; bar 4, ginI mutant harboring pGinIA; bar 5, ginR mutant harboring pMV24; bar 6, ginR mutant harboring pGinR. The values are means ± standard deviations (n = 3). (C) AHL production by E. coli JM109 harboring pGinRI as detected by the agar plate assay using A. tumefaciens NTL4(pZLR4) as the AHL indicator strain. E. coli JM109 harboring pGinRI was streaked on LB agar containing X-Gal and IPTG perpendicular to A. tumefaciens NTL4(pZLR4). AHL production was detected by blue pigmentation in A. tumefaciens NTL4(pZLR4). E. coli JM109 harboring the empty vector pMV24d was used as a negative control.
E. coli JM109 harboring pGinIR containing ginR and ginI was streaked on LB agar containing X-Gal and IPTG perpendicular to the AHL indicator strain A. tumefaciens NTL4(pZLR4). E. coli JM109 harboring pGinIR caused A. tumefaciens to express the reporter gene, resulting in blue pigmentation (Fig. 3C). In contrast, A. tumefaciens did not respond to E. coli JM109 harboring the empty vector pMV24d, which was a pMV24 derivative containing a lacZ-disrupted gene. These results showed that ginI encoded an enzyme for biosynthesis of the AHLs.
Identification of AHLs produced by G. intermedius.
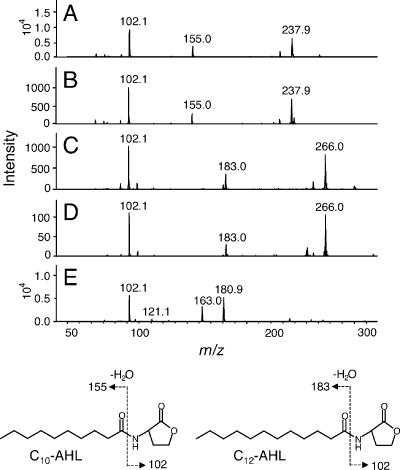
To investigate the structure of the AHLs produced by strain NCI1051, we used LC-MS and LC-MS/MS. LC-MS resulted in three ion peaks in the extract at m/z 256, 284, and 282 [M+H]+ (data not shown). Fragmentation of the parental ion at m/z 256 by LC-MS/MS generated product ions at m/z 102, representing a homoserine lactone moiety; m/z 155, representing a decanoyl side chain; and m/z 238, representing an [M+H − H2O]+ ion (Fig. 4A). This fragmentation pattern was identical to that of synthetic C10-AHL (Fig. 4B). Fragmentation of the parental ion at m/z 284 generated product ions at m/z 102, representing a homoserine lactone moiety; m/z 183, representing a dodecanoyl side chain; and m/z 266, representing an [M+H − H2O]+ ion (Fig. 4C). This fragmentation pattern was identical to that of synthetic C12-AHL (Fig. 4D). Fragmentation of the parental ion at m/z 282 generated product ions at m/z 102, representing a homoserine lactone moiety; and m/z 181, representing a dodecanoyl side chain with a single unsaturated carbon bond (Fig. 4E). Thus, we concluded that G. intermedius NCI1051 produces three different AHLs, N-decanoyl-l-homoserine lactone, N-dodecanoyl-l-homoserine lactone, and N-dodecanoyl-l-homoserine lactone with a single unsaturated bond in its acyl chain.
FIG. 4.
AHL analysis by tandem mass spectrometry: tandem mass spectra of the AHLs produced by NCI1051. (A) Parental ion at m/z 256; (C) m/z 284; (E) m/z 282; (B) synthetic C10-AHL; (D) synthetic C12-AHL. The fragmentation patterns of C10-AHL and C12-AHL are shown schematically below the graphs.
Dependence of ginI transcription on ginR.
In other luxI/luxR systems, the transcription of luxI depends on LuxR (25). We examined ginI transcription in wild-type NCI1051 and in the ginI and ginR mutants, all of which harbored pMV24, by low-resolution S1 nuclease mapping (Fig. 5A). RNA was isolated by the hot phenol method from cells grown at 30°C for 8 h in YPG medium containing 1% (vol/vol) cellulase and 2% (vol/vol) ethanol. rRNA was used to check the amount of RNA used. The ginI gene was actively transcribed in wild-type strain NCI1051 (Fig. 5A, lane 1) but was largely undetected in the ginR mutant (Fig. 5A, lane 4). Introduction of pGinR into the ginR mutant restored ginI transcription (Fig. 5A, lane 5), showing that GinR is essential for ginI transcription.
FIG. 5.
Transcriptional analysis of ginI and ginR. (A) S1 nuclease mapping of ginI and ginR in wild-type strain NCI1051 harboring pMV24 (lane 1), the ginI mutant harboring pMV24 (lane 2), the ginI mutant harboring pGinIA (lane 3), the ginR mutant harboring pMV24 (lane 4), and the ginR mutant harboring pGinR (lane 5). (B) S1 nuclease mapping of ginR in wild-type strain NCI1051 harboring pMV24 (lane 1), the ginR mutant harboring pMV24 (lane 2), the ginR mutant harboring pGinR (lane 3), and the ginR mutant harboring pMGinR (lane 4).
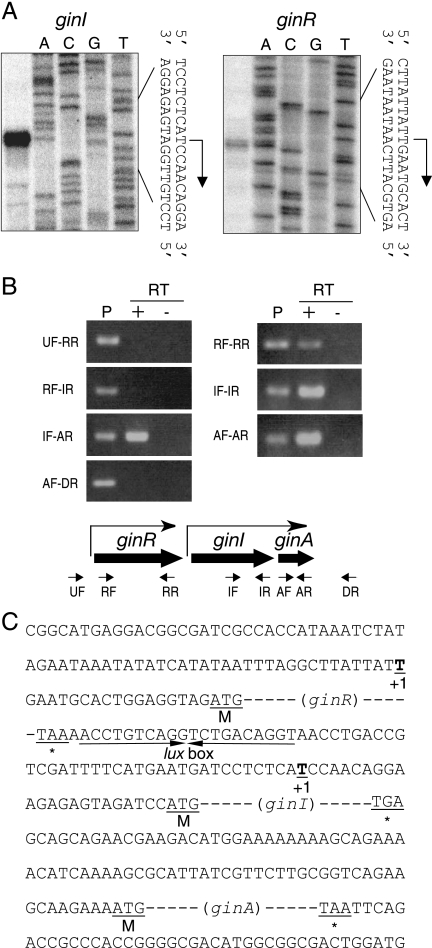
We used high-resolution S1 nuclease mapping to identify the transcription start site of ginI. A single start site was identified 23 nucleotides upstream of the translational start site of ginI (Fig. 6A). A 20-bp lux box-like sequence was identified in the putative ginI promoter region (positions −54 to −35 relative to the transcriptional start site at position +1) (Fig. 6C). Because LuxR-inducible promoters contain a lux box at this position, GinR presumably binds the ginI promoter to activate its transcription, as almost all LuxR family proteins do. In contrast, the ginI promoter was inactive in the ginI mutant (Fig. 5A, lane 2); however, transcription from the ginI promoter was detected in a ginI mutant harboring pGinIA (Fig. 5A, lane 3) at a level greater than that observed in the wild-type strain (Fig. 5A, lane 1). This enhanced transcription was probably caused by the increased copy number of the ginI promoter because we failed to distinguish between the signals from the endogenous ginI promoter and those from the exogenous ginI promoter on pGinIA. These results suggest that GinI is also required for ginI transcription. This is probably because GinR activates the ginI promoter in the presence of AHLs, as has been suggested for most LuxR homologues in quorum-sensing systems. Together, these observations suggest that ginI is positively regulated by GinR in conjunction with AHLs, similar to other luxI-type genes (26).
FIG. 6.
Transcriptional start sites of ginI and ginR and transcriptional units within the gin locus. (A) The transcriptional start sites of ginI and ginR were determined by high-resolution S1 nuclease mapping. The resulting DNA fragment (left panel) was analyzed by polyacrylamide gel electrophoresis with the sequencing ladder (A, C, G, and T) of the probe. (B) Agarose gel electrophoresis of the RT-PCR products. RT-PCR was performed using total RNA from G. intermedius NCI1051. The RNA samples were analyzed with (+) and without (−) reverse transcriptase (RT) to verify the absence of genomic DNA. Amplification was confirmed using G. intermedius NCI1051 genomic DNA as the template (lane P). The positions and directions of the primers are shown schematically below the gels. (C) Sequence of the region upstream of ginR, ginI, and ginA. The lux box-like sequence, which forms an imperfect inverted repeat, is indicated by converging arrows. The transcriptional start site (+1), start codon (M), and stop codon (asterisk) are also indicated.
We also examined ginR transcription in wild-type strain NCI1051 and in the ginI and ginR mutants, all of which harbored pMV24, by low-resolution S1 nuclease mapping (Fig. 5A). Weak ginR transcription was detected in wild-type strain NCI1051 (Fig. 5A, lane 1). Of the two signals detected, the fast-moving signal was a degradation product of the major transcript because this signal was not clearly detected by high-resolution S1 nuclease mapping (Fig. 6A). In accordance with this, only one signal was detected by additional low-resolution S1 nuclease mapping (Fig. 5B). Thus, a single transcription start site for ginR is located 18 nucleotides upstream from the ginR translation start site (Fig. 6C). In the ginI mutant, ginR transcription was detected (Fig. 5A, lane 2) at a level similar to that in the wild-type strain (Fig. 5A, lane 1), suggesting that GinI is not needed for ginR transcription. In contrast, in the ginR mutant, ginR transcription was substantially reduced (Fig. 5A, lane 4, and Fig. 5B, lane 2). To clarify whether GinR activates its own transcription, we introduced a frameshift mutation into ginR on pGinR, creating pMGinR. In the mutant construct, the Ile27 codon (ATC) of GinR was changed to ACTC. Introduction of pMGinR (Fig. 5B, lane 4), as well as introduction of pGinR (Fig. 5A, lane 5, and Fig. 5B, lane 3), into the ginR mutant drove ginR transcription at a level similar to or greater than that in the parental strain. Thus, GinR is not required for its own transcription. The endogenous ginR promoter in the ginR mutant may have been partially inactivated by a polar effect from the kanamycin/neomycin resistance gene inserted at the ginR locus in the opposite direction. We assumed that ginR was constitutively transcribed at a low level in wild-type strain NCI1051.
ginA gene is cotranscribed with ginI.
A small ORF, designated ginA, is present just downstream of ginI. Because there are only 78 nucleotides between the termination codon of ginI and the start codon of ginA and because the phenotypes of the ginI mutant (growth in medium containing ethanol and antifoam activity) could be partially complemented by pGinIA but not by pGinI (see below), we assumed that ginA was cotranscribed with ginI, and we confirmed this by RT-PCR (Fig. 6B). Using cDNA synthesized by RT from the 3′ regions of ginR, ginI, and ginA, each gene was successfully amplified by PCR (primers RF and RR for ginR, IF and IR for ginI, and AF and AR for ginA) (Fig. 6B, right panel), confirming that there was a sufficient amount of mRNA for each gene in the original sample. Using the same RNA sample, ginI-ginA was amplified by PCR using the primers IF and AR (Fig. 6B, IF-AR) from cDNA synthesized from the 3′ region of ginA. Our results indicate that ginI and ginA are cotranscribed as an operon.
In contrast, ginR-ginI was not amplified by PCR using the primers RF and IR (Fig. 6B, RF-IR) using cDNA synthesized from the 3′ region of ginI. Thus, we concluded that there is no read-through between ginR and ginI and that these two genes are individually transcribed from their own promoters. A transcriptional termination sequence must therefore be present in the 56-bp intervening region between the termination codon of ginR and the transcriptional start site of ginI.
Phenotypes of ginI and ginR mutants.
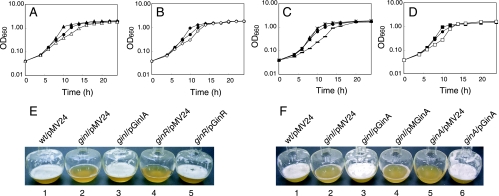
Acetic acid bacteria are characterized by their ability to ferment large amounts of acetic acid from ethanol and by their ability to produce exopolysaccharides, such as cellulose. The growth of the ginI and ginR mutants in medium lacking ethanol was the same as that of the parental strain (data not shown); however, the growth rates and cell masses of the mutants during the exponential growth phase in YPG medium containing 2% (vol/vol) ethanol were greater than those of the parental strain (Fig. 7A and B). The high growth rates of the mutants were caused by the mutation in either ginI or ginR because the introduction of ginI-ginA or ginR into the corresponding mutant reduced the growth rate to a value below that of the parental strain (Fig. 7A and B). The reduced growth rates of the mutants were probably caused by gene dosage effects of GinI-GinA and GinR; Southern blotting performed using the method of Zhang et al. (29) indicated that the copy number of pMV24 was roughly 10 (data not shown).
FIG. 7.
Growth of G. intermedius in YPG medium containing ethanol. (A to D) Wild-type strain NCI1051 harboring pMV24 (•), the ginI mutant harboring pMV24 (▴), the ginI mutant harboring pGinIA (▵), the ginR mutant harboring pMV24 (⧫), the ginR mutant harboring pGinR (⋄), the ginI mutant harboring pGinA (−), the ginI mutant harboring pMGinA (×), the ginA mutant harboring pMV24 (▪), and the ginA mutant harboring pGinA (□) were cultured as described in Materials and Methods. OD660, optical density at 660 nm. (E) Foam production following growth in YPG medium containing 2% (vol/vol) ethanol in a shaking flask for 10 h. Flask 1, wild-type strain NCI1051 harboring pMV24; flask 2, ginI mutant harboring pMV24; flask 3, ginI mutant harboring pGinIA; flask 4, ginR mutant harboring pMV24; flask 5, ginR mutant harboring pGinR. (F) Foam production following growth in YPG medium containing 2% (vol/vol) ethanol in a shaking flask for 10 h. Flask 1, wild-type strain NCI1051 harboring pMV24; flask 2, ginI mutant harboring pMV24; flask 3, ginI mutant harboring pGinA; flask 4, ginI mutant harboring pMGinA; flask 5, ginA mutant harboring pMV24; flask 6, ginA mutant harboring pGinA. wt, wild type.
To determine whether a GinI-GinR quorum-sensing system is involved in the production of bacterial cellulose and water-soluble polysaccharides, the amounts of polysaccharides produced by wild-type strain NCI1051 and by the ginI and ginR mutants were compared. When cultured statically on FPY medium containing lactate, the three strains produced nearly identical amounts of cellulose and water-soluble polysaccharides (data not shown), suggesting that a loss of either ginI or ginR has no effect on extracellular polysaccharide production.
During side-by-side culture of the parental strain and the ginI and ginR mutants, we noticed that the mutant cultures always contained less foam than the parental strain cultures in the late exponential phase (Fig. 7E). The reduced foam production in the mutants was restored to the level of the parental strain when intact ginI-ginA or ginR was introduced into the respective mutants. Interestingly, when the cells were removed by centrifugation and filtration and the supernatant was vigorously shaken in a flask, almost the same amount of foam was produced by all strains. This indicates that the quorum-sensing-defective mutant cells had higher antifoam activity than the wild-type strain cells in the late exponential phase. We assumed that some cell surface characteristic affected the antifoam activity. From these results, we concluded that quorum sensing in G. intermedius NCI1051 represses the antifoam activity of the cells in the late exponential phase.
GinI/GinR controls acetic acid fermentation.
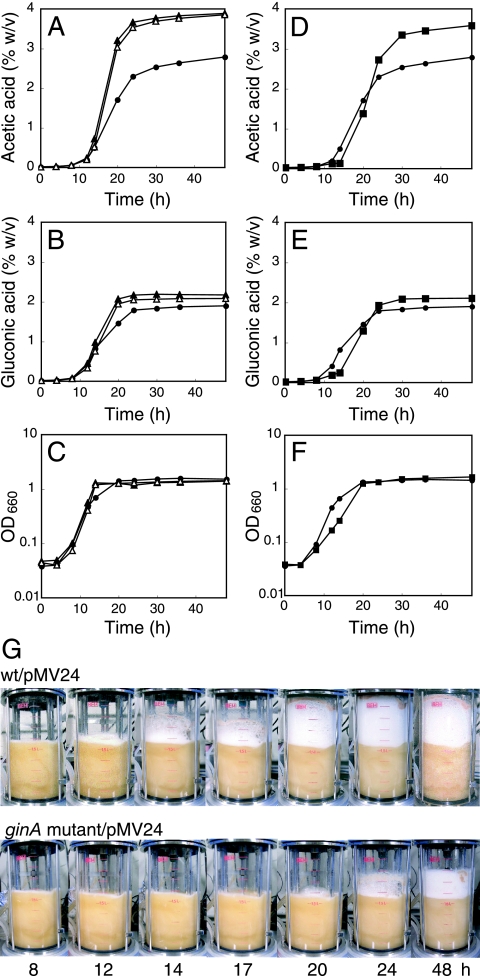
Because the ginI/ginR system affected cell growth in medium containing ethanol, we measured acetic acid production from ethanol in wild-type strain NCI1051 and the ginI and ginR mutants, all of which harbored pMV24. The strains were cultured at 30°C in YPG medium containing 3% ethanol in a 3-liter mini-jar fermentor. The ginI and ginR mutants accumulated more acetic acid than parental strain NCI1051 (Fig. 8A). The total amounts of acetic acid produced by the ginI mutant (3.94% ± 0.07%, wt/vol) and the ginR mutant (3.98% ± 0.11%) were larger than the amount produced by parental strain NCI1051 (2.78% ± 0.11%). The observed increases in the rate of acetic acid production and in the total amount of acetic acid produced by the ginI and ginR mutants are in agreement with the increased growth rates of the mutants compared to that of the parental strain during the late exponential growth phase (i.e., from approximately 12 to 20 h after inoculation) (Fig. 8C).
FIG. 8.
Acetic acid and gluconic acid fermentation by G. intermedius. (A to C) Wild-type strain NCI1051 harboring pMV24 (•), the ginI mutant harboring pMV24 (▴), and the ginR mutant harboring pMV24 (▵) were cultured as described in Materials and Methods. (D to F) Wild-type strain NCI1051 harboring pMV24 (•) and the ginA mutant harboring pMV24 (▪) were cultured as described in Materials and Methods. OD660, optical density at 660 nm. (G) Foam production following growth in YPG medium in a jar fermentor. The medium contained 3% (vol/vol) ethanol initially, and the ethanol concentration of the medium was automatically maintained at 2% (vol/vol) by addition of ethanol during cultivation. wt, wild type.
Alcohol dehydrogenase is involved in the oxidation of ethanol into acetic acid. We therefore measured the levels of alcohol dehydrogenase activity in the ginI and ginR mutants. However, no significant difference was observed between the mutants and the parental strain (data not shown).
GinI/GinR also controls gluconic acid fermentation.
Acetic acid fermentation is called oxidative fermentation; however, gluconic acid is also produced by oxidative fermentation in acetic acid bacteria (1). We therefore measured gluconic acid production by the ginI and ginR mutants to see whether the repressive effect of ginI/ginR on acetic acid fermentation was also observed for gluconic acid fermentation. The ginI and ginR mutants accumulated more gluconic acid than parental strain NCI1051 (Fig. 8B). Similarly, the amounts of gluconic acid produced by the ginI mutant (2.20% ± 0.04% wt/vol) and the ginR mutant (2.15% ± 0.06%) were larger than the amount produced by the parental strain (1.89% ± 0.08%). Thus, we concluded that the quorum-sensing system in G. intermedius NCI1051 represses oxidative fermentation, including acetic acid and gluconic acid fermentation.
Involvement of ginA in growth and antifoam activity.
As described above, the increased growth rate and reduced foam production of the ginI mutant were restored to the wild-type levels when the ginI and ginA genes from pGinIA were introduced into the mutant. However, the wild-type levels were not restored by introduction of ginI into the mutant (data not shown), whereas AHL production by the ginI mutant was rescued by both pGinI and pGinIA. Therefore, we assumed that ginA, which is induced by GinR, might contribute to the phenotypes of the quorum-sensing-defective mutants of G. intermedius NCI1051. To examine this hypothesis, we introduced ginA from pGinA (Fig. 2A) into the ginI mutant. The ginA gene on pGinA is transcribed from a heterologous promoter (the lac promoter of pMV24). The introduction of ginA into the ginI mutant reduced the growth rate of the mutant to a level below that of the parental strain (Fig. 7C) and restored foam production (Fig. 7F, flask 3). We then introduced a frameshift mutation into the ginA gene of pGinA, creating pMGinA. In pMGinA, the sequence ATGCGG (underlining indicates the start codon of ginA) was changed to ATCCCGG (underlining indicates the altered nucleotides). The introduction of pMGinA into the ginI mutant had no effect on the phenotype of the mutant (Fig. 7C and F, flask 4), indicating that a functional GinA protein, instead of ginA RNA, is essential for restoring the wild-type phenotype in the ginI mutant.
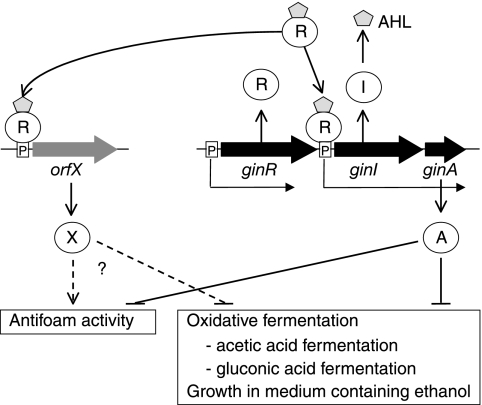
To clarify the function of ginA, we disrupted the ginA gene in G. intermedius NCI1051, resulting in the ginA mutant. As we expected, the ginA mutant produced almost the same amount of AHLs as the wild-type strain (data not shown). The growth rate and cell mass of the ginA mutant during the exponential growth phase in YPG medium containing 2% (vol/vol) ethanol were greater than those of the parental strain (Fig. 7D). The high growth rate of the mutant was caused by the mutation in ginA because introduction of pGinA into the mutant decreased the growth rate to a level lower than that of the parental strain (Fig. 7D). The ginA mutant also produced a smaller amount of foam during culture (Fig. 7F, flask 5) than the parent strain, just like the ginI and ginR mutants; however, this phenotype was rescued by introduction of pGinA into the ginA mutant (Fig. 7F, flask 6). Thus, we concluded that a lack of functional GinA in G. intermedius grown in YPG medium containing 2% (vol/vol) ethanol causes an increase in the growth rate and antifoam activity of the cells in the late exponential phase (Fig. 9).
FIG. 9.
Model for quorum sensing in G. intermedius. The GinI/GinR quorum-sensing system in G. intermedius activates ginA expression, which in turn represses oxidative fermentation, including acetic acid and gluconic acid fermentation, and growth in the presence of ethanol. The GinA protein also represses the antifoam activity of the cells. Other target genes of GinR may be present that also affect these phenotypes. These target genes are expected to have a negative effect on oxidative fermentation and growth in the presence of ethanol and a positive effect on antifoam activity.
Oxidative fermentation by the ginA mutant.
We next measured acetic acid and gluconic acid production in the ginA mutant and wild-type strain NCI1051, both of which harbored pMV24. The strains were cultured at 30°C in YPG medium containing 3% ethanol in a 3-liter mini-jar fermentor. The ginA mutant accumulated acetic acid and gluconic acid with higher yields than parental strain NCI1051(Fig. 8D and E). The amounts of acetic acid (3.67% ± 0.09%, wt/vol) and gluconic acid (2.16% ± 0.05%) produced by the ginA mutant also were larger the amounts produced by parental strain NCI1051 (acetic acid, 2.78% ± 0.11%; gluconic acid, 1.89% ± 0.08%). However, in contrast to the ginI and ginR mutants (Fig. 8A and B), the ginA mutant began to produce both acetic acid and gluconic acid after a delay of about 2 h compared to the parental strain. Because of this delay, the growth rate of the ginA mutant during the exponential growth phase was lower than that of the parent strain, and the cell mass was reduced (Fig. 8F). In addition, the ginA mutant had much greater antifoam activity (Fig. 8G) than the ginI and ginR mutants. In a 3-liter mini-jar fermentor, the foam production by the ginI and ginR mutants was slightly less than that by the parental strain (data not shown). Thus, the phenotypes of the quorum-sensing-defective mutants (the ginI and ginR mutants) were not the same as those of the ginA mutant. This raises the possibility that some other target gene(s) of GinR that affects the phenotype of the cells is present (Fig. 9).
DISCUSSION
We demonstrated that G. intermedius contains an AHL-based quorum-sensing system, like that found in many gram-negative bacteria. Southern hybridization using the luxI-luxR homologue from G. polyoxogenes NCI1028 and chromosomal DNA from various acetic acid bacteria suggested that a similar quorum-sensing system is present in several bacteria in this group. Because industrial vinegar fermentation has traditionally been performed using a mixed culture containing multiple acetic acid bacteria (4), it is possible that the quorum-sensing system in each individual cell plays a role in cell-cell communication between members of the same species and among different species in the fermentor.
In most quorum-sensing systems, LuxR activates multiple genes for cell-cell communication in addition to luxI, which mediates the biosynthesis of the cognate AHLs. Although GinR in G. intermedius activates the transcription of ginI-ginA, it is unclear whether GinR has an additional target gene(s). However, because the phenotypes of the ginI-ginR mutant were not identical to those of the ginA mutant, we assumed that another target gene(s) of GinR might be present and that such genes also influence the phenotype of the cells (Fig. 9). Taking the phenotypic difference between the ginI mutant (or the ginR mutatnt) and the ginA mutant into consideration, the target gene(s) likely has a negative effect on oxidative fermentation and growth in the presence of ethanol and a positive effect on the antifoam activity of the cells. In contrast, it is unclear whether the characteristics that are affected by the GinI/GinR system, such as growth in the presence of ethanol, the rate of acetic acid production, the amount of acetic acid produced, and the antifoam activity of the cells, are directly controlled by ginA. GinA is a small protein consisting of 89 amino acids with no significant homology to any known protein. Nevertheless, its importance in the GinI/GinR system is apparent because complementation of the phenotype of the ginI mutant, such as growth in the presence of ethanol and antifoam activity, required not only intact ginI but also intact ginA. The molecular function of GinA remains to be elucidated.
This is the first study to demonstrate a relationship between quorum sensing and oxidative fermentation in acetic acid bacteria. The GinI/GinR quorum-sensing system in G. intermedius represses oxidative fermentation, including acetic acid and gluconic acid fermentation. What is the biological significance of this phenomenon? Oxidative fermentation is necessary to produce energy for growth. G. intermedius synthesizes ATP via an electron transport system coupled with oxidative fermentation. Thus, the repression of oxidative fermentation leads to a decrease in the growth rate of this microorganism. When the cell density increases, a decrease in growth rate may provide some advantage to the microbial community, especially during acetic acid fermentation, because acetic acid is toxic to the microorganism.
We successfully increased both the rate of acetic acid production and the final yield of acetic acid by inactivating the quorum-sensing system or by disrupting ginA during acetic acid fermentation in G. intermedius. It is very difficult to enhance the production of acetic acid by fermentation, because acetic acid, which is a toxic agent, inhibits the growth of acetic acid bacteria. Only aco encoding an aconitase from Acetobacter aceti (16) and aldh encoding an aldehyde dehydrogenase from G. polyoxogenes (6) have been reported to be genes that enhance the production of acetic acid. The discovery of ginRIA is therefore important and useful for practical purposes in the vinegar fermentation industry. Overexpression of aco increased the yield of acetic acid 1.25-fold (8.4 to 10.5%, wt/vol) and overexpression of aldh increased the yield 1.41-fold (6.84 to 9.66%, wt/vol). In the present study, we observed that disruption of ginI led to a 1.42-fold increase in the yield of acetic acid (2.78 to 3.94, wt/vol) (Fig. 8). In addition to enhancement of acetic acid fermentation, ginA appears to enhance oxidative fermentation, including gluconic acid fermentation (Fig. 8). Because acetic acid bacteria contain various oxidative enzymes (1), manipulation of the quorum-sensing system is expected to be applicable to the industrial production of not only acetic acid and gluconic acid but also various other materials. In addition, the increased antifoam activity of the cells (or the reduced amount of foam in the culture), which also results from inactivation of the quorum-sensing system or disruption of ginA, is advantageous for the industrial production of acetic acid; when foam production is reduced, the volume of medium in a jar fermentor can be increased. Thus, our findings should lead to strain improvement in industrial acetic acid fermentation.
Supplementary Material
Acknowledgments
We thank Stephen K. Farrand (University of Illinois), Paul Williams (University of Nottingham), and Tsukasa Ikeda and Tomohiro Morohoshi (Utsunomiya University) for providing the AHL reporter strains. We thank Nobutaka Funa (The University of Tokyo) for the LC-MS analysis.
Footnotes
Published ahead of print on 1 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ameyama, M. 1982. Enzymic microdetermination of d-glucose, d-fructose, d-gluconate, 2-keto-d-gluconate, aldehyde, and alcohol with membrane-bound dehydrogenase. Methods Enzymol. 8920-29. [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingstone, D. O. Moore, J. S. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 3.Engebrecht, J., and M. Silverman. 1984. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA 814154-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Entani, E., S. Ohmori, H. Masai, and K. Suzuki. 1985. Acetobacter polyoxogenes sp. nov., a new species of an acetic acid bacterium useful for producing vinegar with high acidity. J. Gen. Appl. Microbiol. 31475-490. [Google Scholar]
- 5.Farah, C., M. Vera, D. Morin, D. Haras, C. A. Jerez, and N. Guiliani. 2005. Evidence for a functional quorum-sensing type AI-1 system in the extremophilic bacterium Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 717033-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukaya, M., K. Tayama, T. Tamaki, H. Tagami, H. Okumura, Y. Kawamura, and T. Beppu. 1989. Cloning of the membrane-bound aldehyde dehydrogenase gene of Acetobacter polyoxogenes and improvement of acetic acid production by use of the cloned gene. Appl. Environ. Microbiol. 55171-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50727-751. [DOI] [PubMed] [Google Scholar]
- 8.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishida, T., Y. Sugano, T. Nakai, and M. Shoda. 2002. Effects of acetan on production of bacterial cellulose by Acetobacter xylinum. Biosci. Biotechnol. Biochem. 661677-1681. [DOI] [PubMed] [Google Scholar]
- 10.Kelemen, G. H., P. Brian, K. Flärdh, L. Chamberlin, K. F. Chater, and M. J. Buttner. 1998. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 1802515-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo, Z. Q., S. Su, and S. K. Farrand. 2003. In situ activation of the quorum-sensing transcription factor TraR by cognate and noncognate acyl-homoserine lactone ligands: kinetics and consequences. J. Bacteriol. 1855665-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 14.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camera, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1433703-3711. [DOI] [PubMed] [Google Scholar]
- 15.Morin, D., B. Grasland, K. Vallée-Réhel, C. Dufau, and D. Haras. 2003. On-line high-performance liquid chromatography-mass spectrometric detection and quantification of N-acylhomoserine lactones, quorum sensing signal molecules, in the presence of biological matrices. J. Chromatogr. A 100279-92. [DOI] [PubMed] [Google Scholar]
- 16.Nakano, S., M. Fukaya, and S. Horinouchi. 2004. Enhanced expression of aconitase raises acetic acid resistance in Acetobacter aceti. FEMS Microbiol. Lett. 235315-322. [DOI] [PubMed] [Google Scholar]
- 17.Nelson, E. J., H. S. Tunsjo, P. M. Fidopiastis, H. Sørum, and E. G. Ruby. 2007. A novel lux operon in the cryptically bioluminescent fish pathogen Vibrio salmonicida is associated with virulence. Appl. Environ. Microbiol. 731825-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamoto-Kainuma, A., W. Yan, S. Kadono, K. Tayama, Y. Koizumi, and F. Yanagida. 2002. Cloning and characterization of groESL operon in Acetobacter aceti. J. Biosci. Bioeng. 94140-147. [DOI] [PubMed] [Google Scholar]
- 19.Ravn, L., A. B. Christensen, S. Molin, M. Givskov, and L. Gram. 2001. Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods 44239-251. [DOI] [PubMed] [Google Scholar]
- 20.Saeki, A., K. Matsushita, S. Takeno, M. Taniguchi, H. Toyama, G. Theeragool, N. Lotong, and O. Adachi. 1999. Enzymes responsible for acetate oxidation by acetic acid bacteria. Biosci. Biotechnol. Biochem. 632102-2109. [DOI] [PubMed] [Google Scholar]
- 21.Song, Y., C. Xie, Y. M. Ong, Y. H. Gan, and K. L. Chua. 2005. The BpsIR quorum-sensing system of Burkholderia pseudomallei. J. Bacteriol. 187785-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swift, S., A. V. Karlyshev, L. Fish, E. L. Durant, M. K. Winson, S. R. Chhabra, P. Williams, S. Macintyre, and G. S. Stewart. 1997. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 1795271-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyosaki, H., T. Naritomi, A. Seto, M. Matsuoka, T. Tsuchida, and F. Yoshinaga. 1995. Screening of bacterial cellulose-producing Acetobacter strains suitable for agitated culture. Biosci. Biotechnol. Biochem. 591498-1502. [Google Scholar]
- 24.Vial, L., C. Cuny, K. Gluchoff-Fiasson, G. Comte, P. M. Oger, D. Faure, Y. Dessaux, R. Bally, and F. Wisniewski-Dyé. 2006. N-Acyl-homoserine lactone-mediated quorum-sensing in Azospirillum: an exception rather than a rule. FEMS Microbiol. Ecol. 58155-168. [DOI] [PubMed] [Google Scholar]
- 25.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21319-346. [DOI] [PubMed] [Google Scholar]
- 26.Weingart, C. L., C. E. White, S. Liu, Y. Chai, H. Cho, C. S. Tsai, Y. Wei, N. R. Delay, M. R. Gronquist, A. Eberhard, and S. C. Winans. 2005. Direct binding of the quorum sensing regulator CepR of Burkholderia cenocepacia to two target promoters in vitro. Mol. Microbiol. 57452-467. [DOI] [PubMed] [Google Scholar]
- 27.Williams, P. 2006. Quorum sensing. Int. J. Med. Microbiol. 29657-59. [DOI] [PubMed] [Google Scholar]
- 28.Wong, H. C., A. L. Fear, R. D. Calhoon, G. H. Eichinger, R. Mayer, D. Amikam, M. Benziman, D. H. Gelfand, J. H. Meade, A. W. Emerick, R. Bruner, A. Ben-Bassat, and R. Tal. 1990. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc. Natl. Acad. Sci. USA 878130-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, Y., J. Praszkier, A. Hodgson, and A. J. Pittard. 1994. Molecular analysis and characterization of a broad-host-range plasmid, pEP2. J. Bacteriol. 1765718-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.