Abstract
Most members of the AraC/XylS family contain a conserved carboxy-terminal DNA binding domain and a less conserved amino-terminal domain involved in binding small-molecule effectors and dimerization. However, there is no evidence that Rns, a regulator of enterotoxigenic Escherichia coli virulence genes, responds to an effector ligand, and in this study we found that the amino-terminal domain of Rns does not form homodimers in vivo. Exposure of Rns to the chemical cross-linker glutaraldehyde revealed that the full-length protein is also a monomer in vitro. Nevertheless, deletion analysis of Rns demonstrated that the first 60 amino acids of the protein are essential for the activation and repression of Rns-regulated promoters in vivo. Amino-terminal truncation of Rns abolished DNA binding in vitro, and two randomly generated mutations, I14T and N16D, that independently abolished Rns autoregulation were isolated. Further analysis of these mutations revealed that they have disparate effects at other Rns-regulated promoters and suggest that they may be involved in an interaction with the carboxy-terminal domain of Rns. Thus, evolution may have preserved the amino terminus of Rns because it is essential for the regulator's activity even though it apparently lacks the two functions, dimerization and ligand binding, usually associated with the amino-terminal domains of AraC/XylS family members.
The expression of several pilus serotypes in enterotoxigenic Escherichia coli (ETEC), an enteric pathogen that causes diarrheal disease in humans and livestock, is dependent upon the transcriptional regulator Rns (GenBank accession no. P16114). These pilus serotypes include the CS1, CS2, CS3, and CS4 pili (5, 6, 11). Rns has also been shown to repress the expression of an inner membrane lipoprotein involved in the biogenesis of outer membrane vesicles by preventing the formation of an RNA polymerase open complex at nlpAp, the lipoprotein's promoter (2, 24, 42). Rns positively autoregulates its own expression (12) and is interchangeable with several other virulence regulators, including CfaD (accession no. P25393) and CsvR (accession no. CAA42700), which are carried by some ETEC strains (6), VirF (accession no. NP_085206) from Shigella flexneri, and AggR (accession no. P43464) from enteroaggregative E. coli (EAEC) (26). In the case of VirF and Rns this is somewhat surprising because their regulons share no homologous genes. Rns may be considered the archetype for this group of conserved virulence regulators because it is the only member with a well-developed system for in vitro studies. This has facilitated characterization of its interactions with DNA and its effects upon RNA polymerase at various promoters and identification of additional genes within the Rns regulon (2, 26-28, 30).
Linker scanning mutagenesis of Rns has revealed a region within the protein that accepts insertion of 19 amino acids (M. D. Bodero and G. P. Munson, unpublished data). This region is comprised of residues 100 through 131 and probably functions as a flexible linker between two domains that are roughly the same size (see Fig. 1A). The carboxy-terminal domain (CTD) of Rns contains two putative helix-turn-helix (HTH) motifs connected by an α-helix, the signature feature of proteins belonging to the AraC/XylS superfamily of transcriptional regulators (13). Uracil interference studies suggested that Rns places a recognition helix from each HTH motif in the major groove of DNA (27, 28) in a manner similar to that seen in a DNA cocrystal of another AraC/XylS family member, MarA (PBD 1BL0) (34). Mutagenesis of either HTH motif has been shown to reduce or abolish the activity of VirF (32) and Rns in vivo (Bodero and Munson, unpublished data), presumably by disrupting DNA binding. Thus, it is likely that the CTD of Rns contains most or all of the residues that make direct contact with DNA.
FIG. 1.
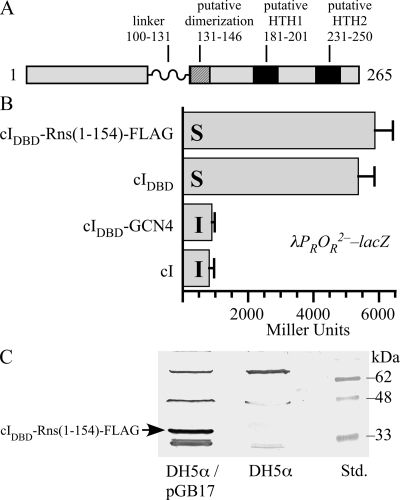
Rns(1-154) does not contain a homodimerization domain. (A) Diagram of Rns and its domain organization. The linker region between the NTD and CTD of Rns was identified by linker scanning mutagenesis (Bodero and Munson, unpublished data). Putative HTH motifs were identified by secondary and tertiary modeling. A putative dimerization helix was identified by homology to the dimerization helices of AraC, XylS, UreR, and ToxT. (B) β-Galactosidase activity (n ≥ 3) from the cI-repressed promoter λPROR2− in K-12 lysogen AG1688/λ202. cI and cIDBD-GCN4 are homodimers and were included as positive controls. cIDBD was included as a monomeric negative control. Each protein construct was also expressed in AG1688 and assayed to determine its ability to confer immunity to the lytic phage λKH54 (ΔcI). I, immune to infection and lysis by λKH54; S, sensitive to infection and lysis by λKH54. (C) Fusion protein cIDBD-Rns(1-154)-FLAG has an expected molecular mass of 34 kDa, and its expression from plasmid pGB17 was confirmed by Western blotting using a polyclonal antibody against the FLAG epitope tag. Lane Std. contained protein standards.
Unlike the CTD, the function of the amino-terminal domain (NTD) of Rns has been obscure. The NTDs of many AraC/XylS family members are known or thought to bind effector ligands. They may also contain a dimerization interface for the formation of homodimers. However, there is no evidence that the activity of Rns is modulated by exogenous ligands, and in this paper we report that the NTD of Rns lacks the ability to form homodimers. Nevertheless, the amino terminus of Rns is essential for DNA binding, and we identified two residues in the Rns NTD that may interact with the CTD of Rns in a manner that facilitates DNA binding.
MATERIALS AND METHODS
Rns and cI expression plasmids.
Plasmid pGPMRns is a derivative of pNEB193 (New England Biolabs) that expresses Rns from lacp (2). Amino-terminal truncations of Rns were constructed and epitope tagged by performing inverse PCR with pGPMRns using primer NEB-hisTag (AATGCATGCCGTGGTGGTGGTGGTGGTGCATGGTCATAGCTGTTTCCTG) and primer 057-F7 (CCAAGCATGCAGAAGCAAATTTTATCAG), 057-F8 (GGAGGCATGCTAAATGCTTGTAGAAGCATGTCAAGAA), or 057-F9 (GGAGGCATGCTAACCTTGTTGGATGAATTAAAAAAT) (underlining in the primer sequences indicates primer-template mismatches). The PCR products were then digested with SphI and circularized with T4 DNA ligase to produce plasmids pGPM1025, pGPM1036, and pGPM1037, which express His6-Rns(61-265), His6-Rns(100-265), and His6-Rns(117-265), respectively. Transposon mutagenesis of pGPMRns produced pGPMRns<Tn>2, which carries rns::kan. Plasmid pEU2030 (12) expresses Rns from a promoter within cloning vector pUC18 (accession no. L08752). Random mutagenesis of pEU2030 (see below) produced plasmid pRns100, which carries the rns-100 allele (codon 14, ATT [Ile] changed to ACT [Thr]), and plasmid pRns101, which carries rns-101 (codon 16, AAT [Asn] changed to GAT [Asp]).
Plasmids pMBPRns1, pMBPRns64, and pMBPRns58 are derivatives of the protein expression vector pMal-c2 (New England Biolabs) that express maltose binding protein (MBP)-Rns, MBP-Rns(80-265), and MBP-Rns(128-265), respectively, from the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter tacp. Construction of pMBPRns1 has been reported previously (2). Plasmids pMBPRns64 and pMBPRns58 were constructed by performing inverse PCR with pEU750 (29) using primers 057R1 (AAAGTGCATGCCCCTTCCCTCGATCCCGAGGTTGTT) and 057F1 (GGAGGCATGCTAAGGCATTTAAAGGATGCATTG) or primers 057R1 and 057F2 (ATAAGCATGCATGATAACTCAGCTTTTATATCTAGC). The PCR products were digested with SphI and then circularized with T4 DNA ligase.
The plasmids used for expression of cI included pFG157, which expresses full-length cI; pKH101, which expresses the DNA binding domain (DBD) of cI; and pJH370, which expresses a fusion between cIDBD and the leucine zipper of GCN4 (22, 23). Plasmid pGB17 expresses cIDBD-Rns(1-154)-FLAG and was constructed by amplifying a fragment of rns from pGPMRns with primers SalI-Rnsfor (AGTCAGGTCGACGATGGACTTTAAATAC) and RnsFlagBamrv3 (ACGGATCCTACTTGTCATCGTCGTCCTTGTAGTCAATTGATTCTAT), which added a FLAG epitope tag after codon 154 of rns. The PCR product was digested with SalI and BamHI and then ligated into the same sites of pJH391 (23).
Reporter plasmids, phage, and strains.
The CFA/I pilus promoter cfaAp, from position −490 to position 343 relative to cfaA, was amplified from ETEC strain H10407 (10) with primers cfaA-1for (AAAGGATCCCGAAGCCGGTACCC) and cfaA-2rev (AAAGAATTCCGCCTCAAAATATACTC). The CS2 pilus promoter cotBp, from position −303 to position 406 relative to cotB, was amplified from ETEC strain C91f (3, 40) with primers cotB-1for (CGCGGATCCATGGAGAACACTTTAT) and cotB-1rev (CCGGAATTCTCAGGGCTGGTTCTTAT). The cfaAp and cotBp PCR products were digested with EcoRI and BamHI and then ligated into the same sites immediately upstream of lacZ in pRS550 (39), resulting in pCFAILac10 and pCS2Lac1. Reporter phages λCFAILac10 and λCS2Lac1 were constructed by homologous recombination between each plasmid and λRS45 (39). Construction of the reporter phage λEU2103 (rnsp-lacZ) has been previously reported (28). Lysogen AG1688/λ202 harbors a prophage that contains the cI-repressed promoter λPROR2− fused to lacZ (23). Strain GPM1080 (2) carries the nlpAp-lacZ reporter plasmid pNLPALac1 integrated into the chromosome of MC4100 [F− araD139 Δ(argF-lac)U169 rpsL150 relA1 flhD5301 deoC1 ptsF25 rbsR] (7) at attBHK022. In pCFATet1 the promoter of tetA in pACYC184 (accession no. X06403) was replaced with cfaAp (position −490 to position 343) so that tetracycline resistance would be Rns dependent.
β-Galactosidase assays.
Reporter strains MC4100/λEU2103, MC4100/λCFAILac10, and GPM1080 were transformed with pGPMRns, pGPM1025, pGPM1036, pGPM1037, or pGPMRns<Tn>2 to determine the ability of Rns amino-terminal truncations to regulate Rns-dependent promoters. To determine the effects of mutations within Rns at Rns-dependent promoters, reporter strains MC4100/λEU2103, MC4100/λCFAILac10, MC4100/λCS2Lac1, and GPM1080 were transformed with pGPMRns, pRns100, pRns101, or vector pUC18. To determine the ability of cI and cI fusions to repress λPROR2−, reporter strain AG1688/λ202 was transformed with pGB17, pFG157, pJH370, or pKH101. All reporter strains were grown aerobically at 37°C in LB medium with 100 μg/ml ampicillin and then harvested, lysed, and assayed for β-galactosidase activity as previously described (25).
Phage immunity test.
For detection of oligomerization domains fused to cIDBD, phage immunity tests were performed by using a modification of a previously described method (23). Strain AG1688 [F′ 128(lacIq lacZ::Tn5) araD139 Δ(ara leu)7697 Δ(lac)X74 galU galK hsdR strA] was transformed with pGB17, pFG157, pJH370, or pKH101. Transformants were cultured for 3 h in tryptone broth (23) containing 100 μg/ml ampicillin and 0.3% (vol/vol) maltose with aeration at 37°C until the optical density at 600 nm was ∼0.3. They were then diluted twofold with TM buffer (23) and cultured for an additional 20 min. Aliquots of each cell culture were then combined with an equal volume of λKH54 or λimm21c that had been serially diluted in TM buffer. λKH54 lacks its own cI and is therefore a lytic phage unless the recipient cell expresses cI or a cIDBD fusion capable of forming dimers (23). λimm21c is a heteroimmune control used to test if cells are sensitive to λ (23). The mixture was incubated for 20 min at 37°C without shaking, combined with 2 ml of tryptone top agar (23), and then spread on tryptone agar plates containing 100 μg/ml ampicillin. Plaques were counted after overnight incubation at 37°C. These experiments were repeated in presence of 1 mM IPTG added to cell cultures and to plates.
Random mutagenesis of rns and selection of mutants.
The Rns expression plasmid pEU2030 was randomly mutagenized by propagation in the mutator strain XL1-Red (Stratagene) used in accordance with the manufacturer's protocol. The mutagenized plasmids were then electroporated into reporter strain MC4100/λEU2103/pCFATet1, and the transformants were plated onto Lac indicator plates (Difco antibiotic medium no. 2, 1% [vol/vol] lactose, 50 μg/ml 2,3,5-triphenyltetrazolium chloride, 100 μg/ml ampicillin, 10 μg/ml tetracycline). Lac− Tetr colonies were streak purified on the same medium to confirm that the phenotype was stable. Transformants with stable Lac− phenotypes were cultured in LB medium with 100 μg/ml ampicillin, and the plasmids were recovered and then transformed into naive reporter strains MC4100/λEU2103 and MC4100/λCFAILac10. Transformants were then plated on Lac indicator plates which did not contain tetracycline to verify that the plasmid carried a mutation that abolished expression of β-galactosidase from rnsp but not from cfaAp.
DNase I footprinting.
MBP-Rns fusion proteins were affinity purified on amylose resin (New England Biolabs) as previously described (2). The conditions used for DNase I footprinting of MBP-Rns at the CFA/I pilus promoter have been described previously (30). In brief, purified fusion proteins were equilibrated with 32P-end-labeled CFA/I pilus promoter DNA for 20 to 30 min at 37°C in footprinting buffer [10 mM Tris-HCl (pH 7.6 at room temperature), 50 mM KCl, 1 mM dithiothreitol, 0.4 mM MgCl2, 0.2 mM CaCl2, 2 ng/μl poly(dI-dC), 10 μg/ml bovine serum albumin]. After equilibration, DNase I was added to a final concentration of 100 ng/μl for 1 min at 37°C. The cleavage reaction was terminated by addition of 10 volumes of DNase I stop buffer (570 mM ammonium acetate, 50 μg/ml tRNA, 80% [vol/vol] ethanol), and then the DNA samples were precipitated and prepared as previously described (30). GA and TC sequence ladders were generated by the Maxam-Gilbert method (36).
Glutaraldehyde cross-linking assays.
MBP-Rns was initially purified by affinity chromatography as previously described (2) and then further purified by passage through a Superdex 200 (GE Healthcare) size exclusion column equilibrated with 10 mM Tris-HCl (pH 7.6), 200 mM NaCl, 1 mM EDTA, 15% (vol/vol) glycerol, 10 mM β-mercaptoethanol. His6-SpaT was bound to nickel Sepharose (GE Healthcare) in 20 mM Tris-HCl (pH 7.5), 300 mM NaCl, 20 mM imidazole and eluted from the resin by increasing the concentration of imidazole to ca. 125 mM. The hexahistidyl tag was then removed from SpaT by digestion with tobacco etch virus protease. SpaT was then bound to a Mono Q anion-exchange column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, 2 mM β-mercaptoethanol and eluted from the column by increasing the concentration of NaCl to ca. 200 mM. The eluted protein was then purified by size exclusion chromatography on a column packed with Sephacryl S200 (GE Healthcare) that had been equilibrated with 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, 2 mM β-mercaptoethanol. After purification both MBP-Rns and SpaT were equilibrated with phosphate buffer (0.1 M sodium phosphate [pH 7.6], 150 mM NaCl), and then aliquots were incubated for 30 min at room temperature with glutaraldehyde. The final concentrations of MBP-Rns and SpaT were 3 and 24 μM, respectively. The final concentration of glutaraldehyde was 0.005, 0.01, or 0.05% (vol/vol). Alternatively, MBP-Rns and SpaT at concentrations of 3, 6, and 12 μM were treated with a fixed concentration of glutaraldehyde. Cross-linking reactions were quenched by addition of 0.1 volume of stop solution (1 M Tris-HCl [pH 7.6], 1 M glycine). Denatured proteins were separated on sodium dodecyl sulfate-polyacrylamide gels and stained with Coomassie brilliant blue R-250.
Digital images.
DNase I footprinting gels were exposed to phosphor screens (Bio-Rad Laboratories) and subsequently scanned with a phosphorimager (GE Healthcare). Western blots were scanned with a flatbed optical scanner connected to a personal computer. Digital images were cropped and scaled using Canvas X (ACD Systems) running under Mac OS X (Apple Inc.). If necessary for visual clarity, brightness and contrast were uniformly adjusted across an entire image with the software mentioned above.
RESULTS
Rns is not a homodimer.
Like several other AraC family members that have been characterized, Rns contains two domains that are joined by a flexible linker (Fig. 1A). If Rns is like AraC and other dimeric family members, its NTD should contain all of the residues necessary for the formation of homodimers. However, we have also identified a potential α-helix in the CTD of Rns (Fig. 1A) that has homology, albeit limited, to the dimerization helices in the NTDs of AraC, XylS, UreR, and ToxT (8, 21, 31, 35). Therefore, in order to determine if Rns forms homodimers in vivo, we replaced the carboxy-terminal dimerization domain of the λ phage repressor, cI, with a fragment of Rns (residues 1 to 154) that contains its NTD and a portion of its CTD that includes the potential dimerization helix (Fig. 1A). We also added a FLAG epitope tag to the carboxy terminus of the fusion protein so that its expression could be confirmed by Western blotting. In the absence of its dimerization domain, cIDBD cannot prevent λ from infecting and lysing an otherwise λ-sensitive strain of E. coli, such as AG1688, because it is unable to bind DNA. However, the addition of a foreign dimerization domain to cIDBD, such as the leucine zipper from GCN4, restores cI function. When AG1688 expressing cIDBD-Rns(1-154)-FLAG was infected with λKH54, a lytic phage that lacks cI, we observed that the sensitivity of the strain was similar to that of a strain expressing the negative control protein cIDBD (Fig. 1B). In both cases, exposure to λKH54 resulted in the formation of plaques, indicating that both strains were infected and lysed by the phage. In contrast, strains expressing the dimerization-proficient positive controls cI and cIDBD-GCN4 were immune to λKH54 because plaques were not observed when they were exposed to the lytic phage. As an additional control, we infected all four strains with λimm21c, a phage whose lytic lifestyle cannot be repressed by cI, to verify that all of the strains could be infected and lysed. In each case plaques were observed, indicating that the positive control strains were immune to λKH54 solely because they expressed functional cI (data not shown).
Each of the previously described cI proteins is expressed as the result of leaky expression from lacp, which is repressed by a Lac repressor in AG1688. The low level of expression is sufficient for cI and cIDBD-GCN4 to confer immunity to λKH54 (Fig. 1B), and we were able to detect cIDBD-Rns(1-154)-FLAG expression by Western blotting in the absence of IPTG (Fig. 1C). Nevertheless, we repeated the experiment in the presence of IPTG and found that even when expression of cIDBD-Rns(1-154)-FLAG was induced, it could not confer immunity to λKH54.
In addition to immunity assays, we tested the ability of cIDBD-Rns(1-154)-FLAG to regulate the expression of β-galactosidase from the cI-repressed λPROR2− promoter in reporter strain AG1688/λ202. λ202 is a reporter phage containing lacZ driven by the λPROR2− promoter. This promoter carries a mutation in one (OR2) of the three cI operator sites that allows individual cI dimers to repress the promoter in the absence of cooperative binding (23). As expected, both cI and cIDBD-GCN4 repressed expression of β-galactosidase (Fig. 1B). In contrast, both cIDBD-Rns(1-154)-FLAG and cIDBD failed to repress expression of β-galactosidase (Fig. 1B). Because cI must dimerize to function as a repressor, the inability of Rns(1-154) to restore repressor function to cIDBD indicates that the fusion protein is not a dimer. These results are consistent with those obtained in the immunity assays and demonstrate that unlike other AraC/XylS family members that have been shown to form homodimers, the first 154 amino acids of Rns are not sufficient for dimerization.
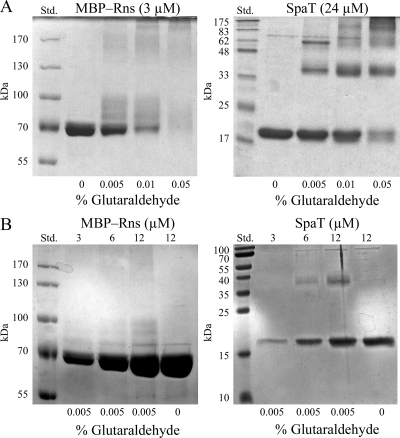
We also tested the possibility that full-length Rns maybe a dimer or other multimer by exposing MBP-Rns to the chemical cross-linker glutaraldehyde. MBP is a monomeric protein that has been previously shown to increase the solubility of Rns without interfering with its activity in vivo or in vitro (27, 28). This solubility tag is necessary for in vitro studies because Rns, for all practical purposes, is insoluble in vitro, as are most AraC/XylS family members. It has also been previously shown that MBP does not interfere with glutaraldehyde cross-linking of XylS dimers in vitro (35). Although we did observe that the mobility of MBP-Rns was decreased as a result of various amounts of glutaraldehyde bound to monomers of MBP-Rns, dimers of the 74-kDa protein were not detected (Fig. 2A). At the highest concentration of glutaraldehyde used, only large nonspecific complexes trapped near the top of the gel were observed. In contrast, dimers and trimers of SpaT (accession no. NP_902290), a multimeric chaperone from the purple-pigmented water and soil bacterium Chromobacterium violaceum, were readily detected under the same cross-linking conditions (Fig. 2A). From these initial studies with SpaT we determined that 0.005% (vol/vol) glutaraldehyde was the optimal concentration for cross-linking protein multimers. We then treated a range of protein concentrations with this optimal concentration of glutaraldehyde to determine if higher concentrations of MBP-Rns facilitated the formation of homodimers (Fig. 2B). However, we found no evidence of MBP-Rns dimers, even when the MBP-Rns concentration was raised to 12 μM. In contrast, SpaT dimers were detected with 6 μM (Fig. 2B). These results indicate that full-length Rns protein is a monomer under these conditions and are consistent with the results for cIDBD heterologous fusions, which demonstrated that the NTD of Rns does not dimerize in vivo.
FIG. 2.
Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels containing MBP-Rns and SpaT after exposure to the chemical cross-linker glutaraldehyde. (A) Static concentrations of MBP-Rns and SpaT exposed to various concentrations of glutaraldehyde. (B) Various concentrations of MBP-Rns and SpaT exposed to a static concentration of glutaraldehyde. SpaT is a multimeric protein and was included as a positive control. Monomeric MBP-Rns and SpaT have molecular masses of 73 and 19 kDa, respectively. Lane Std. contained protein standards. The glutaraldehyde concentrations are expressed as percentages (volume/volume).
Amino terminus of Rns is required for DNA binding.
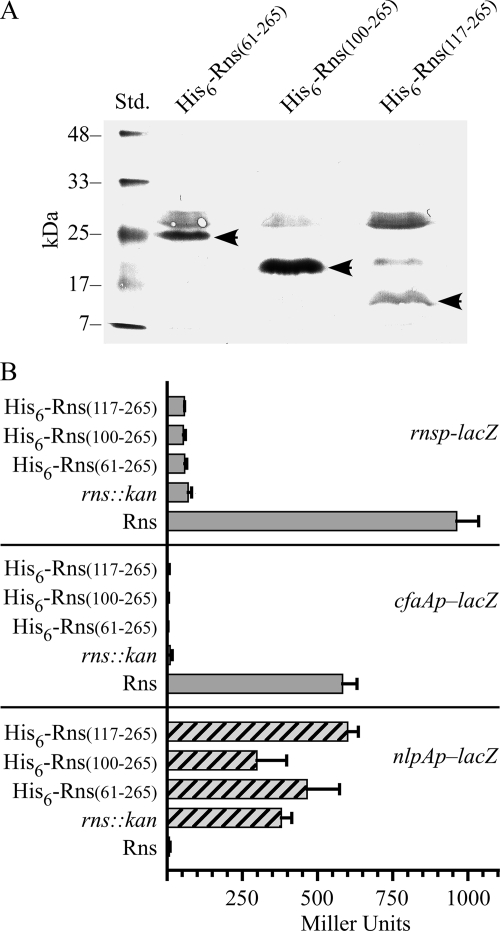
Since the NTD of Rns does not contain a homodimerization interface, we designed a series of plasmids with progressive deletions of the Rns NTD to determine if the NTD is necessary for the regulator's activity. However, only three plasmids expressed stable proteins, as determined by Western blotting using a polyclonal antibody to a His6 epitope tag (Fig. 3A). These plasmids, along with a plasmid expressing Rns or a plasmid carrying rns::kan, were then transformed into lysogens harboring rnsp-lacZ and cfaAp-lacZ reporter prophage to determine their ability to activate Rns-dependent promoters. In contrast to Rns, none of the deletion constructs were able to activate either promoter because the β-galactosidase expression was essentially the same as that of the rns::kan negative controls (Fig. 3B). We also found that none of the truncated proteins were able to efficiently repress nlpAp. With His6-Rns(100-265) we did observe that the expression of β-galactosidase from nlpAp was slightly less than that of the negative control strain carrying rns::kan; however, the difference between the two strains is within the uncertainty of the measurements and is therefore probably not significant (Fig. 3B). These results suggest that the truncated proteins cannot activate rnsp and cfaAp because they cannot bind DNA, and they eliminate the possibility that the truncated proteins bind DNA but fail to activate rnsp and cfaAp because they lack an activation domain.
FIG. 3.
Amino terminus of Rns is essential for positive autoregulation, activation of the CFA/I pilus promoter, and repression of nlpAp. (A) Western blot probed with a polyclonal antibody against the hexahistidyl epitope tag, demonstrating expression of the fusion proteins His6-Rns(61-265), His6-Rns(100-265), and His6-Rns(117-265). The arrowheads indicate the positions of the 25-, 20-, and 18-kDa fusion proteins. (B) β-Galactosidase activity (n ≥ 3) from promoter-lacZ fusions integrated into the chromosome of K-12 strain MC4100.
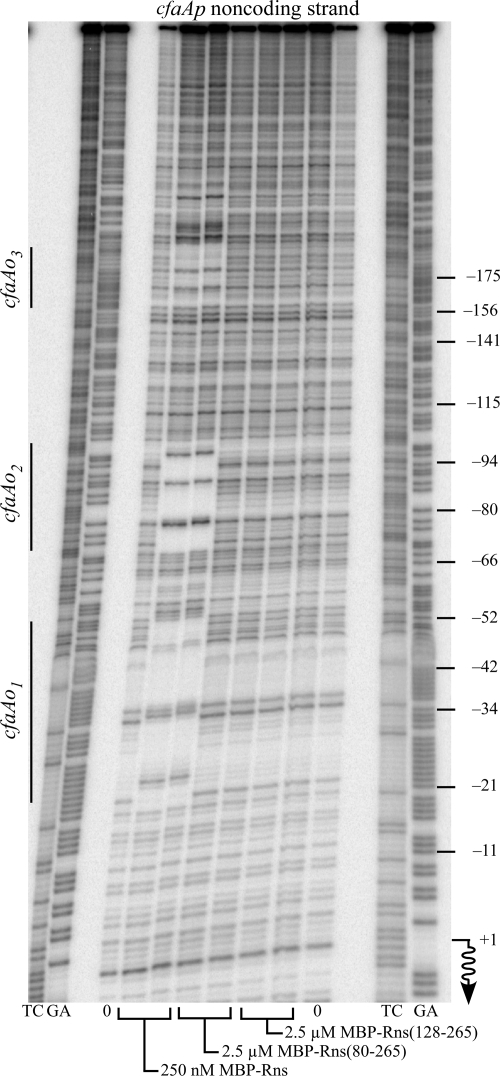
We also determined that the amino terminus of Rns is required for DNA binding in vitro by DNase I footprinting. At the CFA/I pilus promoter 250 nM MBP-Rns is sufficient to saturate both of the previously reported (30) Rns binding sites, cfaAo1 and cfaAo2, as well as an additional binding site, cfaAo3, further upstream (Fig. 4). In contrast, none of the binding sites were occupied by MBP-Rns(80-265) or MBP-Rns(128-265) even at concentrations as high as 2.5 μM (Fig. 4). As a result of different cloning strategies, the two latter MBP fusions have truncations that differ from the His6 epitope-tagged truncations reported above. Nevertheless, our conclusions from both sets of experiments are the same. The loss of its amino terminus substantially reduces the DNA binding affinity of Rns. Based upon DNase I footprinting, we estimated that the reduction is greater than 1 order of magnitude.
FIG. 4.
Amino terminus of Rns is required for DNA binding: DNase I footprinting of MBP-Rns, MBP-Rns(80-265), and MBP-Rns(128-265) bound to the noncoding strand of the CFA/I pilus promoter. Rns binding sites were designated cfaAo1, cfaAo2, and cfaAo3 based on their proximity to cfaA. The numbering on the right is relative to the transcription start site of cfaAp, which is indicated by a wavy arrow. Lanes GA and TC contained Maxam-Gilbert sequence ladders.
Isolation of mutations that abolish Rns positive autoregulation.
Since deletion analysis of Rns revealed that the first 60 amino acids of the regulator are essential for DNA binding, we next sought to identify specific residues required for Rns activity by selecting for randomly generated mutations that abolish Rns positive autoregulation. To avoid the isolation of null mutations, our selection strategy also required that these mutations did not abolish Rns-dependent expression from the CFA/I pilus promoter. We reasoned that these types of mutations could be isolated because the DNA sequences of Rns binding sites at cfaAp are not identical to those at rnsp (30). In addition, Rns binds to a site adjacent to the −35 hexamer at the pilus promoter but does not bind near the −35 hexamer of rnsp (28, 30). This suggested that Rns autoregulation may involve at least some unique residues that are not required for activation of the pilus promoter.
The Rns expression plasmid pEU2030 was randomly mutagenized by propagation in the mutator strain XL1-Red (Stratagene) and then transformed into a reporter strain that harbored an rnsp-lacZ reporter prophage and a plasmid-borne cfaAp-tetA reporter. The phenotype of this strain is Lac− Tets in the absence of Rns but Lac+ Tetr when it is transformed with pEU2030. Transformants were plated on Lac indicator media containing tetracycline, and several Lac− Tetr colonies were isolated for further analysis. False positives were eliminated by transforming naive rnsp-lacZ and cfaAp-lacZ reporter strains with plasmids recovered from Lac− Tetr isolates. Plasmids that resulted in Rns-dependent expression of β-galactosidase from the pilin promoter but not from rnsp were sequenced to determine the mutation(s) that each carried.
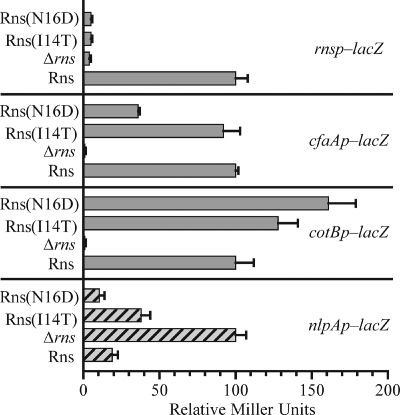
Our analysis revealed two individual mutations, I14T and N16D, near the amino terminus of Rns that abolished positive autoregulation (Fig. 5). This was expected since the mutations were selected because of their inability to activate rnsp. However, the effects of I14T and N16D at rnsp could not have been the result of poor protein expression or protein instability given that each mutation increased Rns-dependent expression from the CS2 pilus promoter, cotBp, compared to Rns. In the case of N16D, the increase was more than 150%. Rns(N16D) also repressed nlpAp as efficiently as Rns, although the repression by Rns(I14T) was less than that by the wild-type regulator (Fig. 5). Our initial selection also required that the mutations did not abolish Rns-dependent expression from the CFA/I pilus promoter, and as expected, Rns(I14T) activated cfaAp as well as Rns. However, the N16D mutation reduced expression from the CFA/I pilus promoter by as much as 70% compared to Rns (Fig. 5). Evidently, the reduced level of expression from cfaAp was sufficient for growth of Rns-dependent tetracycline-resistant colonies in our initial selection experiment.
FIG. 5.
Effects of mutations near the amino terminus of Rns vary depending on the promoter: β-galactosidase activity (n ≥ 3) from promoter-lacZ fusions integrated into the chromosome of K-12 strain MC4100. For the Rns-activated promoters rnsp, cfaAp, and cotBp, activity is expressed relative to the activity of an isogenic strain with wild-type Rns. For nlpAp, which is repressed by Rns, activity is expressed relative to the activity of an isogenic strain lacking Rns.
DISCUSSION
All members of the AraC/XylS family contain dual HTH motifs that provide binding site specificity by placing at least one recognition helix in the major groove of DNA (1, 15, 20, 34). For the majority of family members that consist of two domains, the HTH motifs are usually in the CTD of the protein. In some cases this domain also contains all of the residues necessary for DNA binding and transcriptional activation. For example, MarA and SoxS are roughly one-half the size of a typical family member and are equivalent to the CTD of Rns. Nevertheless, they are able to bind DNA and activate transcription. It has also been shown that the CTDs of XylS and RhaS are sufficient to activate transcription (19, 41). Although the CTD of MelR has also been shown to bind DNA, it is unable to activate transcription (16). In contrast, we have shown that Rns does not bind DNA after removal of residues from its amino terminus. The inability of the Rns CTD to bind DNA cannot be explained by the absence of a dimerization interface because we have also shown that the NTD of Rns does not homodimerize and that full-length Rns is not a multimer in vitro. This differs from the findings for several other family members for which it has been demonstrated that the NTD is sufficient for dimerization, such as AraC, UreR, ToxT, and XylS (4, 31, 33, 35). Because its NTD is unable to homodimerize, Rns may be analogous to PerA, another two-domain virulence regulator belonging to the AraC/XylS family that has also been shown to be a monomer (17).
The NTD of Rns may also lack the other function usually associated with the NTDs of AraC/XylS family members because there is no evidence that it contains a binding site for an exogenous ligand. In general, family members that respond to effector ligands activate the expression of genes that encode proteins that transport, catabolize, or otherwise modify the ligand. For example, in E. coli AraC positively regulates the expression of proteins for the uptake (AraE, AraF, AraG, AraH) and metabolism (AraA, AraB, AraD) of arabinose upon addition of sugar to the bacterial growth medium (38). Similarly, MelR, RhaS, and UreR activate the expression of analogous regulons in response to melibiose, rhamnose, and urea, respectively (13). In contrast, Rns is constitutively active independent of the growth medium, and the proteins in its regulon do not have homology to small-molecule transporters or metabolic enzymes. It is therefore not surprising that the amino-terminal half of Rns has little primary sequence or secondary structural homology to family members such as AraC, RhaS, RhaR, XylS, UreR, and MelR that are known to respond to effector ligands. In addition, Rns that has been purified almost to homogeneity binds to the same DNA sites in vitro as it does in vivo (2, 26, 27, 30). Although the possibility that Rns responds to an effector ligand cannot be completely eliminated, to date there is no evidence that the activity of Rns is dependent upon an exogenous ligand. The absence of an effector ligand is a feature that sets Rns and most other virulence regulators apart from the majority of AraC/XylS family members. However, several exceptions are known, including UreR, TxtR, and possibly ToxT. UreR is associated with the virulence of bacterial uropathogens and is responsive to urea (14, 31). TxtR binds cellobiose, and the virulence of the plant pathogen Streptomyces scabies is attenuated by deletion of txtR (18). ToxT is a virulence regulator of Vibrio cholerae, and some alleles may encode proteins that are responsive to bile (33).
Even though the NTD of Rns lacks the two functions usually associated with the NTDs of AraC/XylS family members, we have shown that it is nevertheless essential for the protein's activity and have identified two residues, I14 and N16, that are essential for positive autoregulation. These residues are conserved in nearly all of the regulators with which Rns is functionally interchangeable, including the ETEC virulence regulators CfaD and CsvR and AggR from EAEC. They are also conserved in the uncharacterized regulator HdaR (accession no. BAF33878) from an unusual strain of enterohemorrhagic E. coli that also exhibits some of the traits of EAEC. Surprisingly, these residues are not conserved in VirF from S. flexneri. Unlike Rns, VirF is not autoregulatory (9); nevertheless, VirF can activate rnsp in a heterologous system (26). It is not yet known whether VirF activates rnsp via a similar mechanism, albeit with alternative residues, or through a mechanism that is different than that of Rns autoregulation. However, in the case of Rns autoregulation, I14T and N16D are probably not positive control mutations because they are not transdominant over wild-type Rns (Bodero and Munson, unpublished data).
Curiously, we have also found that both I14T and N16D increase the activity of Rns at the CS2 pilus promoter but not at the CFA/I pilus promoter, even though both promoters contain an Rns binding site adjacent to the −35 hexamer. The N16D mutation also had no effect on the ability of Rns to repress nlpAp, while I14T partially relieved repression. It seems unlikely that I14T and N16D disrupt the overall organization of Rns; otherwise, their effects would be similar at each promoter or at least at promoters with similar arrangements of binding sites, such as cfaAp and cotBp. A more plausible explanation for our results is that I14 and N16 are involved in DNA binding either directly or, more likely, through an interaction between the amino terminus of Rns and its CTD. This interaction may have some similarities to the interaction between the amino-terminal arm of AraC and its CTD, which holds the protein in a conformation that binds distally spaced sites (37). In our system, we propose that an interaction between the NTD and CTD of Rns is necessary for the proper spatial orientation of the HTH motifs. This would explain the inability of the Rns CTD to bind DNA in the absence of its NTD even though Rns is not a dimer. Mutations I14T and N16D may alter the domain-domain interaction and subtly change the orientation of CTD residues that make base-specific contact. Since the sequences of the various Rns binding sites are not identical (30), these changes may decrease the occupancy of some binding sites while having minimal impact on the occupancy of other binding sites. Measuring the binding affinities of Rns, Rns(I14T), and Rns(N16D) to specific sites should allow this proposed mechanism to be evaluated in future studies. Nevertheless, the current results demonstrate that the amino terminus of Rns is essential for its activity even though the NTD of Rns lacks the ability to homodimerize and probably does not bind an effector ligand.
Acknowledgments
We thank James C. Hu for providing bacterial strains, λ phage, and cI expression plasmids.
This research was supported by NIH NIAID Public Health Service award AI057648.
Footnotes
Published ahead of print on 25 January 2008.
REFERENCES
- 1.Bhende, P. M., and S. M. Egan. 1999. Amino acid-DNA contacts by RhaS: an AraC family transcription activator. J. Bacteriol. 1815185-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodero, M. D., M. C. Pilonieta, and G. P. Munson. 2007. Repression of the inner membrane lipoprotein NlpA by Rns in enterotoxigenic Escherichia coli. J. Bacteriol. 1891627-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boylan, M., D. C. Coleman, J. R. Scott, and C. J. Smyth. 1988. Molecular cloning of the plasmid-located determinants for CS1 and CS2 fimbriae of enterotoxigenic Escherichia coli of serotype O6:K15:H16 of human origin. J. Gen. Microbiol. 1342189-2199. [DOI] [PubMed] [Google Scholar]
- 4.Bustos, S. A., and R. F. Schleif. 1993. Functional domains of the AraC protein. Proc. Natl. Acad. Sci. USA 905638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caron, J., L. M. Coffield, and J. R. Scott. 1989. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. USA 86963-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caron, J., and J. R. Scott. 1990. A rns-like regulatory gene for colonization factor antigen I (CFA/I) that controls expression of CFA/I pilin. Infect. Immun. 58874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104541-555. [DOI] [PubMed] [Google Scholar]
- 8.Childers, B. M., G. G. Weber, M. G. Prouty, M. M. Castaneda, F. Peng, and K. E. Klose. 2007. Identification of residues critical for the function of the Vibrio cholerae virulence regulator ToxT by scanning alanine mutagenesis. J. Mol. Biol. 3671413-1430. [DOI] [PubMed] [Google Scholar]
- 9.Dorman, C. J., and M. E. Porter. 1998. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 29677-684. [DOI] [PubMed] [Google Scholar]
- 10.Evans, D. G., R. P. Silver, D. J. Evans, Jr., D. G. Chase, and S. L. Gorbach. 1975. Plasmid-controlled colonization factor associated with virulence in Escherichia coli enterotoxigenic for humans. Infect. Immun. 12656-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Favre, D., S. Ludi, M. Stoffel, J. Frey, M. P. Horn, G. Dietrich, S. Spreng, and J. F. Viret. 2006. Expression of enterotoxigenic Escherichia coli colonization factors in Vibrio cholerae. Vaccine 244354-4368. [DOI] [PubMed] [Google Scholar]
- 12.Froehlich, B., L. Husmann, J. Caron, and J. R. Scott. 1994. Regulation of rns, a positive regulatory factor for pili of enterotoxigenic Escherichia coli. J. Bacteriol. 1765385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gendlina, I., D. M. Gutman, V. Thomas, and C. M. Collins. 2002. Urea-dependent signal transduction by the virulence regulator UreR. J. Biol. Chem. 27737349-37358. [DOI] [PubMed] [Google Scholar]
- 15.Hendrickson, W., and R. Schleif. 1985. A dimer of AraC protein contacts three adjacent major groove regions of the araI DNA site. Proc. Natl. Acad. Sci. USA 823129-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howard, V. J., T. A. Belyaeva, S. J. Busby, and E. I. Hyde. 2002. DNA binding of the transcription activator protein MelR from Escherichia coli and its C-terminal domain. Nucleic Acids Res. 302692-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibarra, J. A., M. I. Villalba, and J. L. Puente. 2003. Identification of the DNA binding sites of PerA, the transcriptional activator of the bfp and per operons in enteropathogenic Escherichia coli. J. Bacteriol. 1852835-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joshi, M. V., D. R. Bignell, E. G. Johnson, J. P. Sparks, D. M. Gibson, and R. Loria. 2007. The AraC/XylS regulator TxtR modulates thaxtomin biosynthesis and virulence in Streptomyces scabies. Mol. Microbiol. 66633-642. [DOI] [PubMed] [Google Scholar]
- 19.Kaldalu, N., U. Toots, V. de Lorenzo, and M. Ustav. 2000. Functional domains of the TOL plasmid transcription factor XylS. J. Bacteriol. 1821118-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon, H. J., M. H. Bennik, B. Demple, and T. Ellenberger. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 7424-430. [DOI] [PubMed] [Google Scholar]
- 21.LaRonde-LeBlanc, N., and C. Wolberger. 2000. Characterization of the oligomeric states of wild type and mutant AraC. Biochemistry 3911593-11601. [DOI] [PubMed] [Google Scholar]
- 22.Marino-Ramirez, L., L. Campbell, and J. C. Hu. 2003. Screening peptide/protein libraries fused to the lambda repressor DNA-binding domain in E. coli cells. Methods Mol. Biol. 205235-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mariño-Ramírez, L., and J. C. Hu. 2002. Using λ repressor fusions to isolate and characterize self-assembling domains, p. 375-394. In E. Golemis (ed.), Protein-protein interactions: a molecular cloning manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.McBroom, A. J., A. P. Johnson, S. Vemulapalli, and M. J. Kuehn. 2006. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 1885385-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 26.Munson, G. P., L. G. Holcomb, and J. R. Scott. 2001. Novel group of virulence activators within the AraC family that are not restricted to upstream binding sites. Infect. Immun. 69186-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munson, G. P., and J. R. Scott. 1999. Binding site recognition by Rns, a virulence regulator in the AraC family. J. Bacteriol. 1812110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munson, G. P., and J. R. Scott. 2000. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol. Microbiol. 361391-1402. [DOI] [PubMed] [Google Scholar]
- 29.Murphree, D. 1997. Regulation of CS1 expression in enterotoxigenic Escherichia coli. Ph.D. thesis. Emory University, Atlanta, GA.
- 30.Pilonieta, M. C., M. D. Bodero, and G. P. Munson. 2007. CfaD-dependent expression of a novel extracytoplasmic protein from enterotoxigenic Escherichia coli. J. Bacteriol. 1895060-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poore, C. A., C. Coker, J. D. Dattelbaum, and H. L. Mobley. 2001. Identification of the domains of UreR, an AraC-like transcriptional regulator of the urease gene cluster in Proteus mirabilis. J. Bacteriol. 1834526-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porter, M. E., and C. J. Dorman. 2002. In vivo DNA-binding and oligomerization properties of the Shigella flexneri AraC-like transcriptional regulator VirF as identified by random and site-specific mutagenesis. J. Bacteriol. 184531-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prouty, M. G., C. R. Osorio, and K. E. Klose. 2005. Characterization of functional domains of the Vibrio cholerae virulence regulator ToxT. Mol. Microbiol. 581143-1156. [DOI] [PubMed] [Google Scholar]
- 34.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA 9510413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruíz, R., S. Marques, and J. L. Ramos. 2003. Leucines 193 and 194 at the N-terminal domain of the XylS protein, the positive transcriptional regulator of the TOL meta-cleavage pathway, are involved in dimerization. J. Bacteriol. 1853036-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Saviola, B., R. Seabold, and R. F. Schleif. 1998. Arm-domain interactions in AraC. J. Mol. Biol. 278539-548. [DOI] [PubMed] [Google Scholar]
- 38.Schleif, R. 2000. Regulation of the l-arabinose operon of Escherichia coli. Trends Genet. 16559-565. [DOI] [PubMed] [Google Scholar]
- 39.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 5385-96. [DOI] [PubMed] [Google Scholar]
- 40.Smyth, C. J. 1982. Two mannose-resistant haemagglutinins on enterotoxigenic Escherichia coli of serotype O6:K15:H16 or H-isolated from travellers' and infantile diarrhoea. J. Gen. Microbiol. 1282081-2096. [DOI] [PubMed] [Google Scholar]
- 41.Wickstrum, J. R., J. M. Skredenske, A. Kolin, D. J. Jin, J. Fang, and S. M. Egan. 2007. Transcription activation by the DNA-binding domain of the AraC family protein RhaS in the absence of its effector-binding domain. J. Bacteriol. 1894984-4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, F., S. Inouye, and M. Inouye. 1986. Lipoprotein-28, a cytoplasmic membrane lipoprotein from Escherichia coli. Cloning, DNA sequence, and expression of its gene. J. Biol. Chem. 2612284-2288. [PubMed] [Google Scholar]