Abstract
The Wzz proteins are important for determining the length of the O-antigen side chain attached to lipopolysaccharide (LPS). Several bacteria, including Pseudomonas aeruginosa strain PAO1 (serogroup O5), produce two such proteins responsible for the preference of two different chain lengths on the surface. Our group has previously identified one wzz gene (wzz1) within the O-antigen locus of P. aeruginosa strain PA103 (serogroup O11). In this study we have identified the second wzz gene (wzz2), located in the same region of the genome and with 92% similarity to PAO1's wzz2 gene. Mutations were generated in both wzz genes by interruption with antibiotic resistance cassettes, and the effects of these mutations were characterized. Wild-type PA103 prefers two O-antigen chain lengths, referred to as long and very long. The expression of the long O-antigen chain length was reduced in the wzz1 mutant, indicating the Wzz1 protein is important for this chain length preference. The wzz2 mutant, on the other hand, was missing O-antigens of the very long chain length, indicating the Wzz2 protein is responsible for the production of very long O-antigen. The effects of the wzz mutations on virulence were also investigated. In both serum sensitivity assays and a mouse pneumonia model of infection, the wzz1 mutants exhibited greater defects in virulence compared to either wild-type PA103 or the wzz2 mutant, indicating the long chain length plays a greater role during these infectious processes.
Pseudomonas aeruginosa is a gram-negative bacterium that is ubiquitously found in the environment. It typically infects immunocompromised individuals and has been identified as the fourth leading cause of hospital-acquired infections (15). An important virulence factor for many gram-negative bacteria, including P. aeruginosa, is the lipopolysaccharide (LPS) located in the outer portion of the outer membrane. LPS is composed of lipid A, a core oligosaccharide, and O-antigen. P. aeruginosa produces two different forms of O-antigen: A-band (also known as common antigen), which is a homopolymer of d-rhamnose, and B-band, which is the heteropolymer responsible for serogroup specificity. The synthesis of each is complex and has been reviewed extensively elsewhere (25). For B-band, the Wzx translocase flips the O-antigen subunit linked to undecaprenol phosphate from the cytoplasmic to the periplasmic face of the inner membrane, where the subunits are linked together by the Wzy polymerase to form the O-antigen side chain. The entire O-antigen side chain is then added to lipid A-core by the WaaL ligase. Of particular interest for B-band synthesis are the Wzz proteins, which regulate the number of subunits added together, giving rise to a narrow strain-specific range of O-antigen chain lengths (20).
Several bacteria produce bimodal distributions of O-antigen side chains, and two wzz genes have been found within the genomes of these bacteria that are responsible for regulating the two different chain lengths (22, 29). The best-described examples are Salmonella enterica serovar Typhimurium, which has wzzst adjacent to its O-antigen locus and wzzfepE elsewhere in the genome (22), and Shigella flexneri, which has wzzsf in its O-antigen locus along with a plasmid-encoded wzzpHS-2 (29). Wild-type Salmonella predominantly expresses O-antigen chain lengths that are long (16 to 35 subunits) and very long (>100 subunits). Salmonella wzzst mutants do not express long O-antigen chain lengths and were more sensitive to serum (22). In a mouse model of infection, the wzzst mutant was also more readily cleared by the immune system, as revealed by lower splenic burdens, when compared to the wild-type organism. A Salmonella strain with a mutation in the WzzfepE protein, which regulates the very long O-antigen chain length, behaved similarly to the wild-type strain, suggesting this chain length serves another purpose than that characterized by these assays (22).
The role of different Wzz proteins and various O-antigen chain lengths has also been studied in Shigella flexneri. It was shown that the very long O-antigen chain length (>90 subunits) produced by the Wzz protein encoded on a plasmid (wzzpHS-2) was necessary for Shigella's resistance to complement (19). Previously, using O-antigen-deficient LPS mutants, Sandlin et al. (26) found that the presence of O-antigen on LPS was necessary for the cell-to-cell spread characteristic of S. flexneri infections. Strains of S. flexneri that lack O-antigen showed aberrant localization of IcsA; this protein, which is needed for actin-based motility, is normally localized to one pole of the bacterium but was found over the entire cell surface in O-antigen mutants (26). Morona et al. (19) further investigated this requirement for O-antigen in IcsA activity by showing that the short O-antigen chain length (11 to 17 subunits) regulated by Wzzsf was necessary for proper activity of IcsA. Complementation of a wzzsf mutant with Wzz proteins from other bacteria resulted in actin-based motility only when the O-antigen chain length was below a certain number as dictated by the heterologous Wzz protein. O-antigen chain lengths that were too long (>18 subunits) prevented IcsA from being able to interact with host cell proteins needed to initiate actin-based motility (19).
Based on differences in the B-band O-antigen composition, 20 international antigenic serogroups have been identified in P. aeruginosa (17). The gene sequences for the proteins involved with O-antigen biosynthesis and assembly have been identified and proposed for a representative of all 20 P. aeruginosa serogroups (24). Raymond et al. (24) identified a wzz gene within every locus using the GENEMARK gene prediction algorithm. The effects of mutations inactivating the wzz gene have been determined in the serogroup O5 strain PAO1 (3). It was noted that this Wzz protein was responsible for regulating the long O-antigen chain lengths (between 12 to 16 and 22 to 30 subunits) but not the expression of the very long chain lengths (40 to 50 subunits) in strain PAO1 (3), suggesting the presence of another wzz gene in the P. aeruginosa genome. Using the amino acid sequence of Wzz, a search of the entire PAO1 genome revealed PA0938, with 37% similarity to Wzz (5). Mutation of PA0938, renamed wzz2, resulted in a strain that was not able to regulate the very long O-antigen, which was not altered in the original wzz (renamed wzz1) mutant. Antibodies to Wzz2 were reactive with all 20 serogroups of P. aeruginosa, suggesting that a homologous protein was present across all serogroups (5).
Why P. aeruginosa maintains two Wzz proteins and two preferred O-antigen chain lengths is not known. Daniels et al. (5) suggested that Wzz2 is necessary for P. aeruginosa to maintain very long O-antigen chains that may be important for serum resistance, often a critical factor for virulence of this organism, although the effects of these PAO1 wzz mutations on virulence were not investigated. They further suggested that the long O-antigen chains regulated by Wzz1 may be important for proper localization and activity of outer membrane proteins, as has been shown for S. flexneri (19).
We have identified two wzz genes in P. aeruginosa strain PA103 (serogroup O11). To determine whether the results obtained for the wzz mutants in PAO1 (serogroup O5) apply universally to this important pathogen, we have generated strains with mutations in the wzz genes of PA103. Serogroup O11 strains are common in infections, and strain PA103 has been used extensively in animal models of infection and to define the type III secretion system of P. aeruginosa (8, 13). Additionally, our laboratory has established an attenuated Salmonella vaccine that expresses the serogroup O11 O-antigen on its surface (7). Here we investigate the effects of mutating the wzz1 and wzz2 genes in PA103 on O-antigen expression and on virulence of P. aeruginosa; these effects have not been investigated for this pathogen in the context of different O-antigen chain lengths.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains (Table 1) were grown at 37°C in LB either rotating or shaking at 200 rpm for liquid cultures or on L agar or trypticase soy agar (TSA) for plate cultures. Escherichia coli DH5α was used for all cloning experiments. Pseudomonas isolation agar (Becton Dickinson and Company, Sparks, MD) was used to recover mutants after triparental matings. Media were supplemented with tetracycline (100 μg/ml for P. aeruginosa, 10 μg/ml for E. coli), gentamicin (250 μg/ml for P. aeruginosa), kanamycin (50 μg/ml for E. coli), ampicillin (100 μg/ml for E. coli), or carbenicillin (500 μg/ml for P. aeruginosa), when necessary.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. coli DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rk− mk+) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen |
| P. aeruginosa strains | ||
| PA103 | Wild-type strain; serogroup O11 | 18 |
| Wzz1 mutant | PA103 with Gmr interruption in wzz1 | This study |
| Wzz2 mutant | PA103 with Tcr interruption in wzz2 | This study |
| Double mutant | PA103 with interruption in both wzz genes | This study |
| Plasmids | ||
| Topo2.1 | TA PCR cloning vector; Apr Kmr | Invitrogen |
| pEX18Ap | Gene replacement vector; Apr Cbr | 14 |
| pEX18-wzz1 | wzz1 interrupted with Gmr at PstI site, cloned into EcoRI site of pEX18Ap | This study |
| pEX100T | Gene replacement vector; Apr Cbr | 28 |
| pEX100-wzz2 | wzz2 interrupted with Tcr at SmaI site, cloned into SmaI site of pEX100T | This study |
| pHP45ΩTc | Source of Tcr cassette | 9 |
| pUCGM | Source of Gmr cassette | 27 |
| pRK2013 | Helper plasmid used in triparental matings; Kmr | 10 |
| pMMB66HE | Broad-host-range Ptac expression vector; Apr Cbr | 11 |
| pMMB-wzz1 | wzz1 coding region inserted into BamHI and SalI sites of pMMB66HE | This study |
| pMMB-wzz2 | wzz2 coding region inserted into HindIII and EcoRI sites of pMMB66HE | This study |
Ap, ampicillin; Km, kanamycin; Cb, carbinicillin; Gm, gentamicin; Tc, tetracycline; r, resistance.
DNA manipulations.
Chromosomal P. aeruginosa DNA was isolated using the Wizard Genome Prep kit (Promega, Madison, WI). Plasmid DNA was isolated using the Qiagen Miniprep kit (Valencia, CA). Restriction enzymes were purchased from New England Biolabs (Ipswich, MA) and used following the manufacturer's instructions. PCRs were performed using Roche HiFi polymerase (Basel, Switzerland), and amplicons were purified using Qiagen's PCR purification kit following the manufacturer's instructions.
Construction of mutants via allelic exchange.
The wzz1 coding region (∼1 kb) was amplified from genomic DNA using the primers amwwzz1 and amwwzz2 (Table 2) and inserted directly into the Topo2.1 vector (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Next, a 1-kb gentamicin resistance (Gmr) cassette from pUCGM was inserted into the PstI site within the wzz1 gene. An EcoRI digest was performed to remove the interrupted wzz1 gene from Topo2.1, which was then inserted into the EcoRI site of the gene replacement vector, pEX18Ap.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′)a | Restriction site | Purpose |
|---|---|---|---|
| amwwzz1 | CAATATATGAGCTCGGGCTTTTCCCCTGATAGAG | SacI | Creating pEX18-wzz1 |
| amwwzz2 | CAATTCAAGCATGCCAACAAGTAAGCCCTGATGCC | SphI | Creating pEX18-wzz1 |
| Wzz1 Fd | CATGTCGACGGGCTTTTCCCCTGATAGAG | SalI | Complement cloning into pMMB66HE |
| Wzz1 Rv | CATGGATCCCAACAAGTAAGCCCTGATGCC | BamHI | Complement cloning into pMMB66HE |
| Wzz2 Int Fd | GAGAACGAGCGTCAGCGAAT | Creating pEX100-wzz2 | |
| Wzz2 Int Rv | TCTCCACGCAGCTTGTCGAT | Creating pEX100-wzz2 | |
| Wzz2 start Fd | CATAAGCTTATGCCTTCCTCACAGCTTCCG | HindIII | Complement cloning into pMMB66HE |
| Wzz2 Ext Rv | CATGAATTCGAAGAACTGCGGCAGATAGG | EcoRI | Complement cloning into pMMB66HE |
| PA0937 Fd | AAGCTGGAAAGCGACGACTA | Sequencing surrounding wzz2 region | |
| PA0939 Rv | CATGCCATCTTCACACCTTG | Sequencing surrounding wzz2 region |
Underlined portions of sequences represent restriction sites.
For interruption of the wzz2 gene, a portion of the wzz2 coding region (from bp 511 to 1070) was amplified using the primers Wzz2 Int Fd and Wzz2 Int Rv. This PCR product was inserted into Topo2.1, and then the 2.1-kb tetracycline resistance (Tcr) cassette from pHP45ΩTc was digested using SmaI and inserted into the SmaI site within the wzz2 gene. The wzz2 region containing the Tcr cassette was removed from Topo2.1 with an EcoRI digest, blunt ended with mung bean nuclease (New England Biolabs) according to the manufacturer's instructions, and inserted into the SmaI site of the gene replacement vector pEX100T.
Triparental matings were performed using DH5α containing the helper plasmid pRK2013 (10). After mating, transconjugants were selected on PIA medium containing the appropriate antibiotics. Since the pEX18Ap and pEX100T vectors contain the sucrose sensitivity gene sacB and are ampicillin resistant, colonies were patched onto LB plates containing 5% sucrose and subsequently screened for carbenicillin sensitivity, indicative of loss of the plasmid backbone (28).
Insertion of the antibiotic resistance marker into the wzz1 and wzz2 genes was confirmed by PCR and Southern blot analysis. For confirmation of the wzz1 mutation by Southern blotting, an XhoI digestion was performed on the genomic DNA. Similarly, for confirmation of the wzz2 mutation, a StuI digestion was performed. Digested DNA was separated on a 0.7% agarose gel and transferred to GeneScreen nylon (Perkin-Elmer Life and Analytical Sciences, Waltham, MA). PCR amplicons were labeled and detected using the Amersham (Buckinghamshire, United Kingdom) enhanced chemiluminescence DNA labeling kit according to the manufacturer's instructions.
Complementation of wzz mutants.
Complementation was performed by amplifying the coding region of each wzz gene from genomic DNA followed by insertion into Topo2.1. wzz1 was amplified using the primers Wzz1 Fd and Wzz1 Rv. wzz2 was amplified with the primers Wzz2 start Fd and Wzz2 Ext Rv. The inserts were removed from Topo2.1 using the sites designed into the primers and inserted into the pMMB66HE vector under control of the Ptac promoter. Ligations were precipitated using isopropanol, resuspended into 1× TE (10 mM Tris-HCl pH 7.4, 1 mM EDTA pH 8.0), and electroporated directly into P. aeruginosa using an Eppendorf 2510 electroporator. P. aeruginosa was prepared for electroporation by resuspending several swabs from an overnight TSA plate into 1.0 ml of distilled H2O, washing twice with 0.5 ml H2O, and then resuspending in 200 μl H2O.
LPS preparations and visualization.
A hot aqueous phenol extraction was used to isolate LPS (31). Overnight LB cultures of P. aeruginosa strains were diluted to an optical density at 600 nm (OD600) of 0.5 followed by pelleting of 1.5 ml. Pellets were resuspended in 200 μl of 1× sodium dodecyl sulfate (SDS) buffer (0.1 M Tris-HCl pH 6.8, 2% β-mercaptoethanol, 2% SDS, 10% glycerol) and boiled for 15 min. After samples cooled, 10 μg/ml concentrations each of DNase and RNase were added and incubated at 37°C for 30 min. Proteinase K was added at 10 μg/ml, and samples were incubated for 3 h at 60°C. A 200-μl aliquot of cold Tris-saturated phenol was then added, and the samples were vortexed and incubated for 15 min at 65°C. One ml of diethyl ether was then added, and the samples were centrifuged at 14,000 rpm for 10 min. The bottom layer was transferred to a new microcentrifuge tube, and the phenol-ether extraction was performed once more. A 200-μl volume of 2× SDS buffer was added to the samples after the final extraction. For detection of the A-band, whole-cell lysates were also used: bacteria were grown as described above, followed by dilution to an OD600 of 0.5 and pelleting. Cells were resuspended in 100 μl of 2× SDS buffer and boiled for 10 min.
LPS samples were separated on either 8 or 12% SDS-polyacrylamide gels and transferred to nitrocellulose. Blots were analyzed with either A-band-specific monoclonal antibody N1F10 (16) or polyclonal serogroup O11 antiserum (Accurate Chemical & Scientific, Westbury, NY). Secondary antibodies were either anti-mouse immunoglobulin M for N1F10 antibody or goat anti-rabbit immunoglobulin G coupled to either alkaline phosphatase or horseradish peroxidase (Sigma-Aldrich, St. Louis, MO) for the polyclonal O11 antiserum.
Serum sensitivity assay.
Strains were grown at 37°C for 18 h on TSA plates and resuspended in 1% peptone-phosphate-buffered saline (PBS) to an OD600 of 0.05. A 100-μl aliquot was combined with an equal volume of normal human serum diluted in 1% peptone-PBS to final concentrations of 12.5% and 20%. A 1% peptone-PBS (0% serum) solution and heat-inactivated serum (56°C for 1 hour) served as controls. Samples were incubated for 1 hour at 37°C with shaking at 200 rpm. After incubation, samples were serially diluted, plated onto TSA plates, and grown overnight at 30°C to determine the number of CFU.
In vivo mouse model of infection.
Strains were grown on TSA plates for 12 h at 37°C, then resuspended in PBS to an OD650 of 0.5, and diluted to obtain the desired dose in 20 μl. The University of Virginia Animal Care and Use Committee approved all procedures used in this work. Six- to eight-week-old female BALB/c mice were anesthetized by intraperitoneal injection with 0.2 ml of ketamine (6.7 mg/ml) and xylazine (1.3 mg/ml) in 0.9% saline, and then 10 μl of the bacterial inoculum was placed into each nostril. To identify the 50% lethal dose (LD50) of the strains after infection, mice were monitored for up to 1 week for morbidity and mortality. The LD50s and the 95% confidence intervals (CI) were calculated by Probit analysis. The 95% CI that do not overlap differ at P level of <0.05.
For dissemination studies, bacterial strains were prepared and delivered as described above. Twenty-four hours after infection, mice were sacrificed by anesthetic overdosing followed by performing nasal washes (NW) in the volume of 0.5 ml of PBS supplemented with 1% bovine serum albumin (PBS-B). The lung, liver, and spleen were removed, weighed, and homogenized in 1 ml of PBS-B. Serial dilutions were performed in PBS-B and then plated onto TSA plates to determine CFU counts in the NW and each organ.
RESULTS AND DISCUSSION
Identification and mutagenesis of P. aeruginosa PA103 wzz genes.
Our laboratory previously identified the gene encoding the O-antigen chain length regulator, wzz, of the serogroup O11 strain P. aeruginosa PA103 in the O-antigen gene cluster. This protein has 41% identity to the PAO1 (serogroup O5) Wzz1 protein with the predicted N- and C-terminal transmembrane domains characteristic of all Wzz proteins (6). To identify the wzz2 gene from PA103, primers were designed against the PAO1 wzz2 nucleotide sequence (PA0938). These primers amplified a region of the PA103 genome of approximately the same size as that predicted for the PAO1 wzz2 fragment, about 600 bp. Sequence analysis of this fragment revealed 95% nucleotide identity to the sequence of the PAO1 wzz2 gene. Sequencing of the area surrounding the proposed wzz2 gene, using primers PA0937 Fd and PA0939 Rv (Table 2), indicated that this gene was in the same region of the genome as the wzz2 gene from PAO1, with 96% nucleotide identity.
The amino acid sequence of the PA103 Wzz2 protein was analyzed using TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) to generate a hydropathy plot and predict putative transmembrane domains. Wzz2 exhibited the amino- and carboxy-terminal transmembrane domains characteristic of all other Wzz proteins (20), including PA103 Wzz1. Morona et al. reported a correlation between the coiled-coil potential of the periplasmic loop of Wzz and the length favored by that protein and suggested this coiled-coil potential may be related to protein activity (20). The coiled-coil potential of each PA103 Wzz protein was analyzed using COILS (http://www.ch.embnet.org/software/COILS_form.html). Wzz1 is predicted to have one coiled-coil in its periplasmic region, while Wzz2 is predicted to have two to three coiled-coils, similar to the coiled-coil predictions for the PAO1 Wzz proteins. Using FASTA (http://fasta.bioch.virginia.edu), comparison of the Wzz2 sequences between PAO1 and PA103 revealed 95% nucleotide identity and 92% amino acid similarity. The PAO1 and PA103 Wzz1 proteins had 55% nucleotide identity and 75% amino acid similarity. Comparing the Wzz1 protein from either strain to either Wzz2 protein resulted in ∼50 to 75% nucleotide identity and amino acid similarity.
We constructed PA103 strains with mutations in each of the wzz genes. The wzz1 gene was interrupted by allelic exchange using a Gmr cassette; the wzz2 gene was interrupted with a Tcr cassette. A double mutant with interruptions in both wzz genes was also isolated. Interruption of these genes was confirmed by both PCR and Southern blot analyses. To confirm the insertion of the antibiotic resistance cassettes by PCR, the primers amwwzz1 and amwwzz2 were used to detect the interruption of wzz1, and primers Wzz2 Int Fd and Wzz2 Int Rv were used to detect the interruption of wzz2. Wild-type wzz1 was amplified as a 1-kb fragment, while interruption with the 1-kb Gmr cassette amplified a 2-kb fragment. Similarly, interruption of the wild-type 600-bp wzz2 fragment with the 2.1-kb Tcr cassette resulted in a 2.7-kb product. For Southern blotting, we used PCR amplicons of the wzz coding regions generated using the primers Wzz1 Fd and Wzz1 Rv for the wzz1 gene and Wzz2 Start Fd and Wzz2 Ext Rv for the wzz2 gene. Digestion of the genomic DNA with XhoI for the wzz1 gene detected a 3.7-kb band for the wild-type wzz1 gene; insertion of the Gmr cassette revealed a fragment of 4.7 kb. A StuI digestion was performed for the wzz2 gene; a 2.9-kb band for the wild-type wzz2 gene and a shift to 5 kb when the Tcr cassette was inserted were observed. None of the mutants exhibited any defect in growth compared to the wild-type PA103 strain (data not shown).
Effects of wzz mutations on LPS expression.
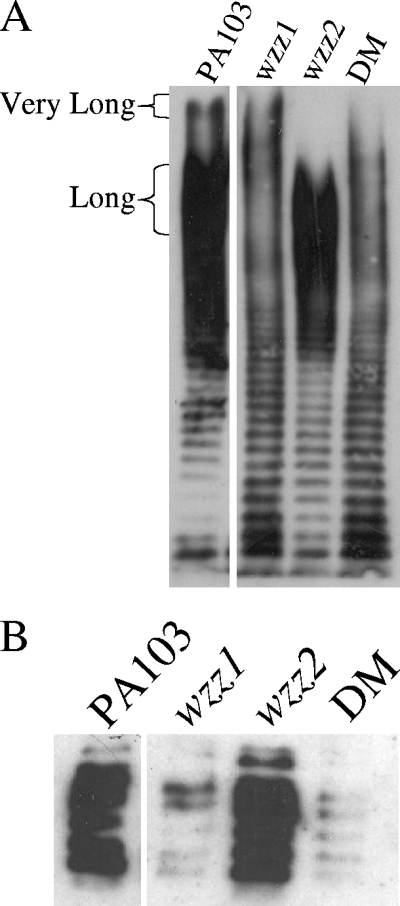
To determine the effects the mutations had on O-antigen expression, LPS was isolated from PA103 and the wzz mutant strains and analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotted with polyclonal serogroup O11 antisera. Strain PA103 has a preference for two O-antigen chain lengths, referred to as long and very long, as indicated by the intensity of the staining of these chain lengths (Fig. 1A). Each of the mutants showed a different pattern of O-antigen expression than the wild-type PA103 strain. In the wzz1 mutant, the expression of the long O-antigen chain length was reduced, suggesting that the Wzz1 protein is responsible for the preference of long O-antigen. On the other hand, the wzz2 mutant was missing very long O-antigen, suggesting that Wzz2 was responsible for the expression of the very long O-antigen chain length in strain PA103. These chain lengths correlate with the coiled-coil prediction for regulated chain length, where Wzz1 with fewer coils produces long chain lengths while Wzz2, with two to three predicted coiled-coils, produces the very long chain length in strain PA103. The double mutant had reduced expression of both chain lengths; however, antibody staining indicated it had not lost the ability to generate long O-antigen side chains.
FIG. 1.
Effects of wzz mutations. A) Effect on B-band expression. Overnight cultures of strains were diluted to an OD600 of 0.5, and a hot aqueous phenol LPS extraction was performed. Samples were run on an 8% polyacrylamide gel, and Western blotting was performed using rabbit polyclonal serogroup O11 antisera. The migration patterns of the long and very long O-antigen side chains are shown. B) Effect on A-band expression. Whole-cell extracts were prepared and run on a 12% gel. Western blotting was performed using the N1F10 common antigen monoclonal antibody. DM, double mutant.
It should be noted that the loss of Wzz1 activity only inhibits the preference for the long O-antigen side chain, while the loss of Wzz2 results in the complete absence of all very long O side chains of this length. Whether this is due to the statistical improbability of making very long O-antigen in the absence of a regulator or indicates some difference in the activity of these two proteins is not known. These results are similar to what was obtained for strain PAO1: in PAO1, Wzz1 is responsible for regulating chain lengths of 12 to 16 and 22 to 30 subunits, and Wzz2 regulates chain lengths between 40 and 50 subunits. In strain PA103, the exact range of subunits that constitute the long or very long side chains has so far not been determined; up to 30 subunits have been counted, but this number was less than the number of subunits corresponding to the long O-antigen chain lengths. Our findings that the double mutant still produces long chain lengths is in contrast to other reported double wzz mutants, which typically produce a smaller amount of a given chain length as the number of subunits added increases. For example, antibody to the O-antigen is unable to detect chain lengths much beyond the first range (12 to 16 subunits) of PAO1's Wzz1 in the PAO1 double mutant, and neither the S. enterica serovar Typhimurium nor S. flexneri double mutants produce chain lengths beyond 20 subunits (19, 22).
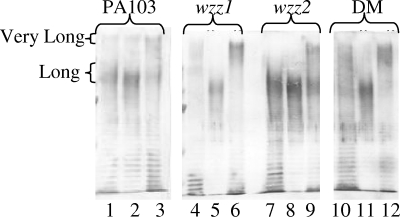
To ensure that the LPS phenotypes observed in the wzz mutants were due to the mutations in the indicated genes and not polar effects, especially since wzz1 is at the beginning of the O-antigen locus, complementation analysis was performed. The coding region of each gene was inserted into the pMMB66HE vector under control of the Ptac promoter. Isopropyl-β-d-thiogalactopyranoside (IPTG) was used to induce expression of the proteins, and LPS samples were prepared from the complemented strains (Fig. 2). The presence of vector alone in all strains did not affect the LPS banding patterns (Fig. 2, lanes 1, 4, 7, and 10). Overexpression of each gene in wild-type PA103 resulted in a slight increase in the respective chain lengths, as evidenced by darker staining; providing wild-type PA103 with wzz1 in trans led to an increased amount of long chain length on the bacterial surface, while overexpression of wzz2 led to more with very long chain length (Fig. 2, lanes 2 and 3). When the wzz1 mutant was provided with the wzz1 coding sequence in trans, darker staining similar to what is seen in the wild type was evident, indicating the preference for long chain length was reestablished (Fig. 2, lane 5). Providing the wzz2 coding sequence in trans to the wzz2 mutant allowed for the production of the very long O-antigen that was missing in the mutant (Fig. 2, lane 9). Transforming the wzz genes into the double mutant created LPS banding patterns that were similar to those of the single mutants of the gene that was still absent (Fig. 2, lanes 11 and 12). We also performed cross-complementation, which was not performed in the study by Daniels et al. (5) using the wzz mutants of PAO1. We found that providing either wzz gene in the other wzz mutant resulted in overproduction of the chain length expressed by the cloned gene (Fig. 2, lanes 6 and 8).
FIG. 2.
Complementation of wzz mutants. The coding region of either wzz1 or wzz2 was inserted under the control of the Ptac promoter into the plasmid pMMB66HE. P. aeruginosa strains containing the indicated plasmids were grown in LB overnight with appropriate antibiotic and 1 mM IPTG. LPS was extracted, and samples were run on an 8% acrylamide gel and detected using rabbit polyclonal serogroup O11 antisera. Lanes 1, 4, 7, and 10, the indicated strain plus pMMB66HE; lanes 2, 5, 8, and 11, the indicated strain plus pMMB-wzz1; lanes 3, 6, 9, and 12, the indicated strain plus pMMB-wzz2. DM, double mutant.
It is interesting that providing the wzz genes in trans in the respective wzz mutants did not simply restore the wild-type PA103 LPS banding pattern. It appeared that the overexpression of one Wzz protein inhibited the activity of the remaining chromosomally encoded Wzz protein. While this was the case for the complemented wzz2 mutant, in which there were reduced amounts of the long chain lengths when wzz2 was expressed in trans (Fig. 2, lane 9) compared to the empty vector (Fig. 2, lane 7), this was best evidenced by the complementation of the wzz1 mutant (Fig. 2, lane 5). When wzz1 was provided in trans in the wzz1 mutant, the expression of very long O-antigen was lost despite the presence of the wild-type wzz2 gene in the chromosome. This is in contrast to what was seen in the complementation of the single wzz mutants of strain PAO1; overexpression in trans of either Wzz protein had no discernible effect on the amount of the chain length regulated by the remaining chromosomally encoded wzz gene and resulted in strains with LPS that looked like the wild type (5). However, Carter et al. (4) recently described results similar to ours when they overexpressed the wzz genes of S. flexneri in a wild-type strain. They saw that overexpression of the wzz gene located within the O-antigen locus (wzzsf) inhibited the effect of the other wzz gene (wzzpHS-2), and they hypothesized that the Wzzsf protein may compete better for the proteins of the putative O-antigen assembly complex (Wzx, Wzy, and WaaL) compared to the WzzpHS-2 protein (4). Our results suggest the control of O-antigen chain length in P. aeruginosa PA103 is also mediated by the ratio of the levels of the Wzz1 and Wzz2 regulators present in the cell, as suggested by Morona et al. in their molecular chaperone theory of how the Wzz proteins regulate O-antigen chain length (21). It has been noted for Yersinia enterocolitica that overexpression of the Wzz protein helps the putative O-antigen assembly complex work more efficiently (1). Daniels et al. (5) reported cross-linking data providing evidence that the two Wzz proteins of PAO1 exist in different assembling complexes. Our results fit nicely with this model and suggest that a complex that includes one Wzz protein may interfere with the activity of complexes containing the other Wzz protein, as seen when overexpression of wzz1 inhibited production of the very long O-antigen regulated by the wzz2 gene still present in the chromosome (Fig. 2, lane 5).
The effect of the wzz mutants on A-band expression was also investigated. The A-band is assembled in a Wzy-independent manner, but both the A-band and B-band synthetic pathways share the starting glycosyltransferase, WbpL (25). Hot aqueous phenol LPS extractions immunoblotted with the A-band-specific monoclonal antibody (N1F10) demonstrated that the wzz1 mutant had a reduced amount of A-band compared to either PA103 or the wzz2 mutant (data not shown). The double mutant showed a defect similar to the wzz1 mutant. Similar results were obtained when whole-cell lysates were probed with the N1F10 antibody (Fig. 1B), indicating this result is not due to problems with isolating A-band using the hot aqueous phenol extraction method. This finding is in contrast to the wzz1 mutant of PAO1, which was not observed to affect A-band production (3), illustrating an interesting difference between the wzz1 mutants in the two strains. Effects on A-band synthesis due to mutations in proteins in the assembling complex have been described previously (2): a mutation in the wzx flippase of PAO1 also impaired A-band synthesis, although this mutation could be complemented by overexpressing the initiating glycosyltransferase, WbpL. It was hypothesized that when the B-band assembling complex is unable to flip the O-antigen subunit to the periplasmic surface, the WbpL glycosyltransferase may not be released to aid in A-band synthesis (2). Complementation of our mutants with PA103 WbpL has not been performed, but our results suggest that interfering with steps later in the B-band assembly process may also interfere with A-band synthesis, as has been seen with the wzx mutation in PAO1. Alternatively, since our PA103 wzz1 mutant behaves differently with respect to A-band synthesis than the PAO1 wzz1 mutant, the interactions between proteins involved in A-band synthesis and B-band synthesis may be different in the two different serogroups.
Serum sensitivity of wzz mutants.
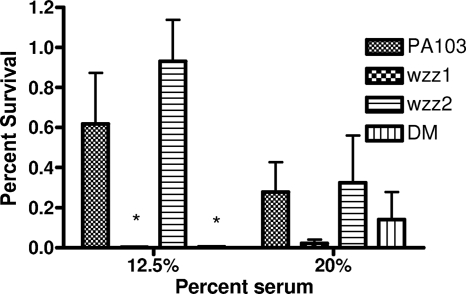
The length of the O-antigen side chain affects how gram-negative bacteria interact with the complement proteins of the innate immune system, with longer side chains preventing complement from reaching the bacterial surface to cause lysis (30). It has been shown previously in P. aeruginosa that mutants deficient in O-antigen are much more susceptible to killing by complement than isogenic wild-type strains (23). To determine whether the change in O-antigen chain length preference resulted in altered resistance to complement, PA103 and the wzz mutants were incubated in diluted normal human serum to determine sensitivity to complement. At a 12.5% serum concentration, the wzz1 mutant and double mutant exhibited reduced survival compared to wild-type PA103, but the wzz2 mutant did not differ in its survival compared to the wild-type strain (Fig. 3). The results were similar at the 20% serum concentration, where again the wzz1 and double mutant were more serum sensitive than strain PA103 and the wzz2 mutant. PA103 and the wzz2 mutant exhibited a dose-dependent decrease in survival as serum concentration increased from 12.5% to 20%, while the wzz1 and double mutant did not. This is probably due to the inherent serum sensitivity of the wild-type PA103 strain compared to other strains of P. aeruginosa (23), which makes detecting differences at this higher concentration of serum more difficult. These results indicate that the long chain length regulated by the Wzz1 protein plays a greater role in protecting P. aeruginosa from being killed by complement.
FIG. 3.
Serum sensitivities of wzz mutants. Bacteria were incubated for 1 hour at 37°C with shaking in either 12.5% or 20% human serum. Neither PBS nor heat-inactivated serum had an effect on bacterial viability. Percent survival was calculated as follows: [(number of bacteria recovered)/(number of bacteria added)] × 100. Bars represent the means of at least three experiments, and error bars represent the standard deviations. One-way analysis of variance (Kruskal-Wallis test) of the 12.5% serum concentration results showed statistically significant differences between groups (P < 0.05). *, statistically significant difference (P < 0.05) compared to wzz2 as determined with the Mann-Whitney U test. No statistical significance was determined by either one-way analysis of variance or t tests for the 20% serum data sets. DM, double mutant.
Daniels et al. (5) hypothesized that since wzz2 is present in all 20 serogroups of P. aeruginosa and is therefore more homologous among all strains compared to wzz1, it could be involved with complement resistance. Our results suggest that the long chain length, regulated by Wzz1, and not the very long chain length regulated by Wzz2, is more important for complement resistance for strain PA103. Similar results were found in S. enterica serovar Typhimurium, where the loss of the long O-antigen chain length encoded by the wzz gene (wzzst) adjacent to the O-antigen locus resulted in increased killing in serum, while the mutant in the wzz gene regulating very long O-antigen side chain (wzzfepE) behaved like the wild type (22).
Effect of O-antigen chain length on virulence in a mouse model of infection.
We next determined what role the two different chain lengths preferred by the two Wzz proteins play in the virulence of P. aeruginosa. O-antigen-deficient mutants disseminate less readily in a mouse model of infection and have higher LD50s compared to isogenic wild-type strains of P. aeruginosa (23). To determine the role of O-antigen chain length regulation on the virulence of PA103, we tested the wzz mutants in an acute pneumonia mouse model of infection. The LD50s of the wzz mutants were calculated in BALB/c mice (Table 3). The LD50 of the wzz2 mutant was about 1.5 times that of the wild-type PA103, but this was not statistically significant. The wzz1 mutant had an LD50 that was nearly 4.5 times that of wild-type strain PA103 (P < 0.05). These results suggest that the control of the long chain length plays a more critical role in this infectious process for the virulence of P. aeruginosa than does the presence of the very long chain length. The double mutant had an LD50 that was significantly higher than the wzz1 mutant.
TABLE 3.
LD50s of PA103 and wzz mutants via intranasal infection in BALB/c micea
| Strain | LD50 | Lower 95% CI | Upper 95% CI |
|---|---|---|---|
| PA103 | 1.42 × 105 | 1.06 × 105 | 1.91 × 105 |
| wzz1 | 6.4 × 105 | 6.4 × 105 | 6.4 × 105 |
| wzz2 | 2.24 × 105 | 1.28 × 105 | 3.86 × 105 |
| DM | 8.56 × 105 | 8.33 × 105 | 8.77 × 105 |
The LD50 values were calculated by Probit analysis based upon survival data of four groups of mice (n = 4) infected with a range of doses. Mice were monitored for up to 1 week after infection. DM, double mutant.
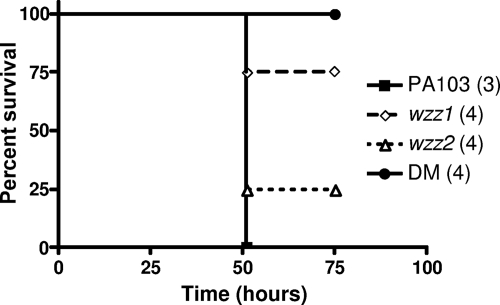
The survival curves of mice intranasally infected with wild-type PA103 and the wzz mutant strains mirror the LD50 results (Fig. 4). Wild-type PA103 was the most virulent; at the dose given (∼5.5 × 105 CFU), all infected mice eventually succumbed to the infection. At a similar dose, fewer mice infected with the wzz1 mutant succumbed to the infection compared to the wzz2 mutant, which was only slightly attenuated compared to the wild-type strain. Results were similar at higher doses (∼2.5 × 106 CFU), where wzz1-infected mice took longer to succumb to the infection than mice infected with the wzz2 mutant (data not shown). Mice infected with the double mutant survived the longest at both doses, suggesting an additive effect for the loss of virulence when both chain lengths are no longer preferred on the bacterial surface. In fact, at a dose of ∼5 × 105 CFU, the double mutant was completely avirulent. Given the attenuation of the wzz1 mutant in these assays, our results suggest that the long chain length regulated by Wzz1 plays a more important role during infection than the very long chain length regulated by Wzz2. It should be noted that the attenuation resulting from the wzz1 mutation is likely not due to the observed defect in A-band production, since the A-band is hidden by the longer B-band that is still present on the surface (12).
FIG. 4.
Survival of BALB/c mice after intranasal infection with wzz mutants. Survival of mice infected with wild-type PA103 and the wzz mutants is illustrated. Doses used for infection were as follows: PA103, 6.7 × 105; wzz1, 6.2 × 105; wzz2, 5.2 × 105; double mutant (DM), 4.9 × 105. Curves were plotted using the Kaplan-Meier method, and curve comparisons were done using the log-rank test. The PA103 and wzz2 survival curves were each significantly different from the DM curve (P < 0.05). No other statistically significant differences were detected.
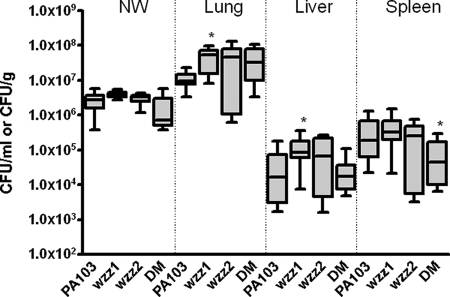
The LPS of P. aeruginosa has been implicated in the adhesion of the bacterium to host cells and therefore may affect the ability to spread from the lung in the pneumonia model of infection. To determine whether O-antigen chain length was involved in this process, we monitored nasal washes and lung tissues for the presence of bacteria 24 hours after intranasal inoculation. Dissemination of the bacterial strains to the liver and spleen was also determined at this time point (Fig. 5). There was no statistically significant difference from wild-type PA103 in the amount of bacteria present in the nasal washes for any of the wzz mutants, suggesting that these bacteria do not differ significantly in their ability to adhere to host tissues. There was also no statistical difference in the means of the samples obtained from the lungs of mice infected when comparing all strains, although the amount of wzz1 mutant was significantly higher from wild-type PA103 in this organ. Similarly, the wzz1 mutant was the only mutant significantly higher than the wild type in dissemination to the liver. Finally, the amount of bacteria reaching the spleen when mice were infected with the double mutant was significantly lower compared to wild-type PA103. Our results suggest that there may be some defect in bacterial spread from the initial site of infection when P. aeruginosa is lacking both preferred O-antigen chain lengths, but overall there appears to be little difference in dissemination between the wild type and any of our wzz mutants.
FIG. 5.
Dissemination of wzz mutants after intranasal infection of BALB/c mice. Dissemination of PA103 and the wzz mutants 24 h after infection is shown. Doses represented are as follows: PA103, 5 × 105 to 8.1 × 105 (n = 8); wzz1, 1 × 106 to 1.5 × 106 (n = 12); wzz 2, 7.7 × 105 to 8.1 × 105 (n = 7); double mutant (DM), 1.25 × 106 to 1.5 × 106 (n = 11). The number of mice for each group is indicated in parentheses. NW results are reported as CFU/ml, and organ burdens are given as CFU/gram. Data are plotted as a box-and-whiskers plot with the median and the lower and upper quartiles (25th and 75th percentile, respectively) represented by the box and the range represented by the whiskers. *, statistically significant compared to PA103 for the same sample (P < 0.05 using the Mann-Whitney U test).
Previous studies using this same mouse infection model showed that wild-type PA103 readily disseminated but PA103 mutant strains lacking O-antigen, while maintaining cytotoxicity, were unable to spread from the site of initial infection (23). Whether our wzz mutants exhibit any changes in adherence or cytotoxicity has not been tested in any in vitro assays, although our findings in the mouse model of infection suggest that the changes in the preferred length of the O-antigen side chain do not affect adherence to host tissues and are not critical for dissemination. However, our results do suggest that the O-antigen chain length is playing some other important role in the infectious process, likely related to the serum sensitivity, since our wzz1 mutants are attenuated compared to wild-type PA103.
In conclusion, we have identified the wzz genes of P. aeruginosa PA103. The wzz1 gene was previously recognized to be a part of the serogroup O11 antigen biosynthetic loci. The second wzz gene, wzz2, was identified and found to be located in the same area of the genome and 92% similar to the previously identified wzz2 gene (PA0938) from PAO1 serogroup O5. Mutants were made in PA103 wzz1 and wzz2 genes. It was determined that Wzz1 was responsible for the preference for long O-antigen chain length while Wzz2 was responsible for producing very long O-antigen chain length. It was also determined that the mutation in wzz1 affected production of the A-band common antigen. The virulence of these wzz mutants was investigated, and it was found that the loss of long chain length regulated by Wzz1 resulted in a greater ability for the bacteria to be killed by complement than the loss of very long chain length. Similarly, the loss of long O-antigen chain length had a larger effect on the LD50 of the wzz1 mutants, indicating the long chain length is more important for the virulence of P. aeruginosa strain PA103.
Acknowledgments
We thank Michael R. Davis and Jessica Davis for their technical assistance.
This work was supported by grants from the National Institutes of Health (1 R01 AI50230 and 1 R21 AI53842 to J.B.G.). E.N.K. and J.M.S. were partly supported by the National Institutes of Health through the University of Virginia Infectious Diseases training grant AI07406.
Footnotes
Published ahead of print on 7 December 2007.
REFERENCES
- 1.Bengoechea, J. A., L. Zhang, P. Toivanen, and M. Skurnik. 2002. Regulatory network of lipopolysaccharide O-antigen biosynthesis in Yersinia enterocolitica includes cell envelope-dependent signals. Mol. Microbiol. 441045-1062. [DOI] [PubMed] [Google Scholar]
- 2.Burrows, L. L., and J. S. Lam. 1999. Effect of wzx (rfbX) mutations on A-band and B-band lipopolysaccharide biosynthesis in Pseudomonas aeruginosa O5. J. Bacteriol. 181973-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrows, L. L., D. Chow, and J. S. Lam. 1997. Pseudomonas aeruginosa B-band O-antigen chain length is modulated by Wzz (Ro1). J. Bacteriol. 1791482-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter, J. A., C. J. Blondel, M. Zaldivar, S. A. Alvarez, C. L. Marolda, M. A. Valvano, and I. Contreras. 2007. O-antigen modal chain length in Shigella flexneri 2a is growth-regulated through RfaH-mediated transcriptional control of the wzy gene. Microbiology 1533499-3507. [DOI] [PubMed] [Google Scholar]
- 5.Daniels, C., C. Griffiths, B. Cowles, and J. S. Lam. 2002. Pseudomonas aeruginosa O-antigen chain length is determined before ligation to lipid A core. Environ. Microbiol. 4883-897. [DOI] [PubMed] [Google Scholar]
- 6.Dean, C. R., C. V. Franklund, J. D. Retief, M. J. Coyne Jr., K. Hatano, D. J. Evans, G. B. Pier, and J. B. Goldberg. 1999. Characterization of the serogroup O11 O-antigen locus of Pseudomonas aeruginosa PA103. J. Bacteriol. 1814275-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiGiandomenico, A., J. Rao, K. Harcher, T. S. Zaidi, J. Gardner, A. N. Neely, G. B. Pier, and J. B. Goldberg. 2007. Intranasal immunization with heterologously expressed polysaccharide protects against multiple Pseudomonas aeruginosa infections. Proc. Natl. Acad. Sci. USA 1044624-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farmer, J. J., III, R. A. Weinstein, C. H. Zierdt, and C. D. Brokopp. 1982. Hospital outbreaks caused by Pseudomonas aeruginosa: importance of serogroup O11. J. Clin. Microbiol. 16266-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52147-154. [DOI] [PubMed] [Google Scholar]
- 10.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 761648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48119-131. [DOI] [PubMed] [Google Scholar]
- 12.Hatano, K., J. B. Goldberg, and G. B. Pier. 1995. Biologic activities of antibodies to the neutral-polysaccharide component of the Pseudomonas aeruginosa lipopolysaccharide are blocked by O side chains and mucoid exopolysaccharide (alginate). Infect. Immun. 6321-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hauser, A. R., S. Fleiszig, P. J. Kang, K. Mostov, and J. N. Engel. 1998. Defects in type III secretion correlate with internalization of Pseudomonas aeruginosa by epithelial cells. Infect. Immun. 661413-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 21277-86. [DOI] [PubMed] [Google Scholar]
- 15.Jarvis, W. R., and W. J. Martone. 1992. Predominant pathogens in hospital infections. J. Antimicrob. Chemother. 29(Suppl. A)19-24. [DOI] [PubMed] [Google Scholar]
- 16.Lam, M. Y., E. J. McGroarty, A. M. Kropinski, L. A. MacDonald, S. S. Pedersen, N. Hoiby, and J. S. Lam. 1989. Occurrence of a common lipopolysaccharide antigen in standard and clinical strains of Pseudomonas aeruginosa. J. Clin. Microbiol. 27962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, P. V., M. H. Kusama, and T. Bergan. 1983. Survey of heat-stable major somatic antigens on Pseudomonas aeruginosa. Int. J. Syst. Bacteriol. 33256-264. [Google Scholar]
- 18.Liu, P. V. 1973. Exotoxins of Pseudomonas aeruginosa. I. Factors that influence the production of exotoxin A. J. Infect. Dis. 128506-513. [DOI] [PubMed] [Google Scholar]
- 19.Morona, R., C. Daniels, and L. Van Den Bosch. 2003. Genetic modulation of Shigella flexneri 2a lipopolysaccharide O antigen modal chain length reveals that it has been optimized for virulence. Microbiology 149925-939. [DOI] [PubMed] [Google Scholar]
- 20.Morona, R., L. Van Den Bosch, and C. Daniels. 2000. Evaluation of Wzz/MPA1/MPA2 proteins based on the presence of coiled-coil regions. Microbiology 1461-4. [DOI] [PubMed] [Google Scholar]
- 21.Morona, R., L. van den Bosch, and P. A. Manning. 1995. Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J. Bacteriol. 1771059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray, G. L., S. R. Attridge, and R. Morona. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 471395-1406. [DOI] [PubMed] [Google Scholar]
- 23.Priebe, G. P., C. R. Dean, T. Zaidi, G. J. Meluleni, F. T. Coleman, Y. S. Coutinho, M. J. Noto, T. A. Urban, G. B. Pier, and J. B. Goldberg. 2004. The galU gene of Pseudomonas aeruginosa is required for corneal infection and efficient systemic spread following pneumonia but not for infection confined to the lung. Infect. Immun. 724224-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raymond, C. K., E. H. Sims, A. Kas, D. H. Spencer, T. V. Kutyavin, R. G. Ivey, Y. Zhou, R. Kaul, J. B. Clendenning, and M. V. Olson. 2002. Genetic variation at the O-antigen biosynthetic locus in Pseudomonas aeruginosa. J. Bacteriol. 1843614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rocchetta, H. L., L. L. Burrows, and J. S. Lam. 1999. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 63523-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandlin, R. C., K. A. Lampel, S. P. Keasler, M. B. Goldberg, A. L. Stolzer, and A. T. Maurelli. 1995. Avirulence of rough mutants of Shigella flexneri: requirement of O antigen for correct unipolar localization of IcsA in the bacterial outer membrane. Infect. Immun. 63229-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweizer, H. P. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15831-834. [PubMed] [Google Scholar]
- 28.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 15815-22. [DOI] [PubMed] [Google Scholar]
- 29.Stevenson, G., A. Kessler, and P. R. Reeves. 1995. A plasmid-borne O-antigen chain length determinant and its relationship to other chain length determinants. FEMS Microbiol. Lett. 12523-30. [DOI] [PubMed] [Google Scholar]
- 30.Taylor, P. W. 1983. Bactericidal and bacteriolytic activity of serum against gram-negative bacteria. Microbiol. Rev. 4746-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides. Methods Carbohyr. Chem. 583-91. [Google Scholar]