Abstract
Patients suffering from cystic fibrosis (CF) commonly harbor the important pathogen Pseudomonas aeruginosa in their airways. During chronic late-stage CF, P. aeruginosa is known to grow under reduced oxygen tension and is even capable of respiring anaerobically within the thickened airway mucus, at a pH of ∼6.5. Therefore, proteins involved in anaerobic metabolism represent potentially important targets for therapeutic intervention. In this study, the clinically relevant “anaerobiome” or “proteogenome” of P. aeruginosa was assessed. First, two different proteomic approaches were used to identify proteins differentially expressed under anaerobic versus aerobic conditions. Microarray studies were also performed, and in general, the anaerobic transcriptome was in agreement with the proteomic results. However, we found that a major portion of the most upregulated genes in the presence of NO3− and NO2− are those encoding Pf1 bacteriophage. With anaerobic NO2−, the most downregulated genes are those involved postglycolytically and include many tricarboxylic acid cycle genes and those involved in the electron transport chain, especially those encoding the NADH dehydrogenase I complex. Finally, a signature-tagged mutagenesis library of P. aeruginosa was constructed to further screen genes required for both NO3− and NO2− respiration. In addition to genes anticipated to play important roles in the anaerobiome (anr, dnr, nar, nir, and nuo), the cysG and dksA genes were found to be required for both anaerobic NO3− and NO2− respiration. This study represents a major step in unraveling the molecular machinery involved in anaerobic NO3− and NO2− respiration and offers clues as to how we might disrupt such pathways in P. aeruginosa to limit the growth of this important CF pathogen when it is either limited or completely restricted in its oxygen supply.
Pseudomonas aeruginosa is a gram-negative bacterium of environmental and clinical importance that is capable of both aerobic and anaerobic respiration, the latter of which requires nitrate (NO3−), nitrite (NO2−), or nitrous oxide (N2O) as an alternative electron acceptor (24). The organism can also utilize arginine for anaerobic growth via substrate-level phosphorylation, although the final cell yield during this form of growth is abysmally low compared to that observed during anaerobic respiration (55). The most facile means to obtain anaerobic energy, however, is via respiration by NO3− reduction. The process of nitrate reduction can occur by two routes, the first of which is an assimilatory pathway where the nitrogen from NO3− is incorporated into macromolecules via formation of NH3. Assimilation can proceed under both aerobic and anaerobic conditions. In contrast, respiratory NO3− reduction (denitrification) occurs only under anaerobic conditions and involves the sequential eight-electron reduction of NO3− to nitrogen gas (N2), with intermediates including NO2−, nitric oxide (NO), and N2O. The anaerobic process generates respiratory energy for the cell.
A hallmark of the inherited fatal disease cystic fibrosis (CF) is that patients eventually succumb to lung infection by P. aeruginosa and die at an average age of 36.8 years (www.cff.org). It is now becoming increasingly evident that the oxygen tension within the thickened CF airway mucus found in patients suffering from chronic CF airway disease is either significantly reduced (<2%) (2, 35) or absent (anaerobic) (62). In fact, macrocolonies of P. aeruginosa growing in vitro as complex communities known as “biofilms” demonstrate complete oxygen depletion within the top 30 μm (11). Recently, several laboratories confirmed our findings from 2002 (62) showing that the anaerobic biofilm mode of growth can occur within pockets of the thick airway mucus of chronically infected CF patients. First, Beckmann et al. (7) showed by using phage display that narG, encoding the anaerobic respiratory nitrate reductase (NAR) α-chain, was detected in sera from CF patients within the first year of life. Palmer et al. (42) then showed that NarG was essential for growth in a synthetic CF sputum. Most recently, Son et al. (52) have shown by microarray analysis of CF sputum samples that there is abundant anaerobic gene expression, including the entire anaerobic respiratory pathway (nar, nir, nor, and arc genes), by P. aeruginosa in the CF airways. Therefore, it appears that the synthesis of denitrifying enzymes can take place even in the presence of low concentrations of oxygen as long as the levels are below the biological oxygen demand of the culture (1).
Because humans lack the enzymatic machinery for anaerobic sustenance, such enzymes and the associated processing machinery necessary for P. aeruginosa to survive under such conditions represent potentially viable targets for therapeutic intervention. Toward this end, Yoon et al. (62) revealed the following three major findings: (i) P. aeruginosa forms more robust biofilms under anaerobic than under aerobic conditions; (ii) when P. aeruginosa lacks the rhl quorum-sensing circuit, the bacterium commits a metabolic suicide by overproduction of toxic levels of NO; and (iii) organisms lacking the outer membrane protein OprF grow very poorly during anaerobic respiration. Yoon et al. (61) also found that NO2−, at what was found to be the slightly acidified pH (∼6.5) of CF airway mucus, effectively kills the antibiotic (23)- and phagocyte (14)-resistant mucoid form of P. aeruginosa. Two transcriptomic studies, although elegantly done, were somewhat limited in their breadth in the sense that they did not specifically select to rigorously assess the true “anaerobiome” of P. aeruginosa in the context of both NO3−- and NO2−-grown cells, but only with cells grown in NO3−. Specifically, Wagner et al. (57) and Filiatrault et al. (18, 19) have investigated certain aspects of anaerobic gene expression by using GeneChip microarrays. However, many of the genes that are known to be expressed under anaerobic conditions do not require supplementation with NO3− or NO2− (59). The genes that are known to be induced by strict anaerobiosis, as opposed to the simple addition of NO3− or NO2−, include narI, narK1 and -2, hemN, arcABC, and napABDFPQ, among others. However, Wagner et al. (57) also showed that anaerobic growth actually downregulated narK2, narK1, and akk as well as the napABDF genes, encoding a periplasmic NAR, which have been reported to be constitutive and are not required for anaerobic growth using NO3− (48). Because of the often-ignored differences between wild-type strains of PAO1, the conditions of growth and cell manipulation, the parameters being elucidated experimentally, the necessity for a multifaceted approach, and above all, the importance of this research, there is justification for a more detailed and thorough study, which forms the body of this work. Thus, because P. aeruginosa forms more robust biofilms during anaerobic growth and CF patients succumb to pulmonary insufficiency (60-62), again, the aforementioned anaerobic respiration pathway machinery represents plausible gene products for the development of novel therapeutic intervention. Thus, the identification of those gene products required for anaerobic growth is warranted, specifically at pH 6.5.
In this study, a collective (i) proteomic, (ii) transcriptional profiling, (iii) signature-tagged mutagenesis (STM), and (iv) targeted mutagenesis approach was initiated in order to identify P. aeruginosa gene products that are required for anaerobic NO3− and NO2− respiration at pH 6.5. Ultimately, this study represents a major step in unraveling the molecular machinery involved in P. aeruginosa anaerobic NO3− and NO2− respiration and offers clues as to how we might disrupt these pathways, which could ultimately limit the growth of this important pathogen in CF airway mucus.
MATERIALS AND METHODS
Construction of a P. aeruginosa tryptic library database.
First, each of 5,570 open reading frames carried by the P. aeruginosa genome (www.pseudomonas.com) was downloaded from http://www.pseudomonas.com/downloads/sequences/Pseudomonas_aeruginosa_PAO1_2004-Jan-14.fha in Macintosh Stuffit format. The resulting file, designated P_aeruginosa_Prot.fasta.1, was converted to a Microsoft Word document, PA.doc. The protein sequences were downloaded from PA.doc into ProFound (64) and ProteinProspector (http://prospector.ucsf.edu/), and tryptic fragment libraries were assembled.
Growth of bacteria and preparation of cell extracts.
P. aeruginosa PAO1 was grown in Luria (L) broth (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl) containing either 15 mM KNO3 or 15 mM NaNO2 at pH 6.5. Bacteria were grown either aerobically with shaking at 300 rpm (volume/flask ratio, 1:10) or anaerobically in a Coy anaerobic chamber at 37°C for 24 h (NO3−-grown cells) or 96 h (NO2−-grown cells) (see Fig. S1 in the supplemental material). Bacteria were subjected to three freeze-thaw (−80°C/37°C) cycles, followed by sonication with a Heat Systems Ultrasonics sonic disruptor (Farmingdale, NY) with the microtip at setting 5 for 20 seconds on ice. Cell extracts in 10 mM Tris-HCl, pH 7.4, were freed of membranes by centrifugation at 100,000 × g for 2 h, and samples were kept frozen at −80°C until use. Because NAR and nitric oxide reductase (NOR) are membrane bound, these proteins were not expected to be found in the membrane-free extracts.
2-D gel electrophoresis.
Two-dimensional (2-D) gel electrophoresis of P. aeruginosa cell extracts was performed according to the method of O'Farrell (40), as outlined in detail by Sauer and Camper (46). Briefly, crude protein extracts (200 μg) were solubilized in 450 μl of a solution containing urea, thiourea, dithiothreitol (DTT), 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and Pharmalyte 3-10. Samples were applied to Immobiline Dry-Strips (18 cm) (pH 3-10 NL; GE Healthcare) by in-gel rehydration. Isoelectric focusing (IEF) was performed using a Multiphor II apparatus (GE Healthcare) for a total of 48 kV-h. Upon completion of IEF, the Dry-Strips were equilibrated in a two-step process. In the first step, protein disulfide bonds were reduced by DTT for 15 min, while in the second step, cysteines were irreversibly modified by iodoacetamide treatment for 15 min. For the resolution in the second dimension, a 24- by 20-cm 2-D gel system from Genomics Solutions, Inc., was used. Crude protein extracts were separated in 11% resolving gels at 15°C. 2-D gels were stained with silver nitrate (8) and run in triplicate for each growth condition to confirm the reproducibility of the protein patterns under planktonic and biofilm growth conditions. A calibrated image scanner (GE Healthcare) was used for gel scanning to ensure even spot detection and higher accuracy for the subsequent image analysis. Computational image analysis was carried out using Image Master 2-D Platinum software (GE Healthcare). A fourfold difference in spot volume was considered significant.
MALDI-TOF MS.
Matrix-assisted laser desorption ionization-time-of-flight mass spectrometric (MALDI-TOF MS) identification of proteins was performed according to previously described strategies (50; http://proteomics.uc.edu). Briefly, protein spots of interest were excised from the 2-D gels and digested in situ with sequencing-grade, tosylsulfonyl phenylalanyl chloromethyl ketone-modified trypsin (Promega), using a ProGest workstation (Genomics Solutions Inc., MI). After digestion for 8 h at 37°C, tryptic peptides were extracted with 50% acetonitrile-0.1% trifluoroacetic acid and desalted if necessary, using ZipTips (Millipore). An aliquot of the peptide solution was spotted on a MALDI target plate, and mass spectra were recorded on an Ettan MALDI-TOF Pro mass spectrometer (GE Healthcare) operated in reflectron mode as described previously (47, 53). As little as 1 pmol of protein was sufficient for identification of candidate protein spots. Trypsin peptides were used as internal calibrants for every peptide sample to ensure high mass accuracy. The peptide mass fingerprinting spectra were processed using Ettan evaluation software (GE Healthcare). Briefly, the generated mass lists, composed of monoisotopic [M + H]+ masses, were first filtered for common contaminants (e.g., keratin) and subsequently used for database searches using the ProFound search algorithm (64). The database used in this study was composed of current, nonredundant protein sequences obtained from TIGR (comprehensive microbial resource batch download website [http://www.tigr.org/tigr-scripts/CMR2/batch_download.dbi]) and comprised the sequences of Streptococcus pneumoniae R6, S. pneumoniae TIGR4, Streptococcus pyogenes M1, Staphylococcus aureus MRSA252, Streptococcus epidermidis, Enterococcus faecalis V583, Escherichia coli K-12 MG1655, and P. aeruginosa PAO1. All proteins were identified with significant certainty (probability score of <0.03). Proteins were identified with 3 to 15 matched peptides and a minimum of 5% sequence coverage.
Protein isolation for subsequent nano-high-performance liquid chromatography-microelectrospray ionization (nHPLC-μESI) MS analyses.
P. aeruginosa strain PAO1 was grown under aerobic and anaerobic conditions in LB containing 15 mM KNO3 or NO2−, pH 6.5, at 37°C as described above. The broth-grown samples were poured over crushed ice and diluted in ice-cold buffer A (0.1 M NH4HCO3-1 mM DTT-0.05% CHAPS). The bacteria were harvested by centrifugation at 13,000 × g for 10 min at 4°C. The pellet was quick-frozen in dry ice-ethanol, thawed on ice, and resuspended in buffer A. The bacteria were then lysed twice with a French pressure cell at 12,000 lb/in2 at 4°C. The samples were treated with 20 U/ml of both DNase and RNase containing 10 mM MgCl2 for 15 min on ice. At this point, 1 mM EDTA was added. Debris was removed by centrifugation at 4°C for 15 min at 13,000 × g. An aliquot of the supernatant was removed to determine the protein concentration, and the remainder was frozen at −80°C. To enhance the recovery of membrane proteins, we added 0.3 M NaCl to the 0.1 M NH4HCO3-1 mM DTT-0.05% CHAPS.
Sample digestion for subsequent nHPLC-μESI MS.
Equal amounts of total protein (70 μg) from P. aeruginosa grown either aerobically or anaerobically as described above were diluted to a total volume of 300 μl with 100 mM ammonium bicarbonate (pH 8.5). The proteins in each solution were reduced with 200 mM DTT (5 μl) at 51°C for 1 h, carboxyamidomethylated with 450 mM iodoacetamide (5 μl) in the dark at room temperature for 1 h, and digested with modified trypsin (3.5 μg in 7 μl; Promega) at 37°C for 8 h. Proteolysis was terminated by acidifying the reaction mixture to a pH of 3 with glacial acetic acid (13 μl).
FT-ICR nHPLC-μESI MS analysis.
Aliquots of the above digests (5 μl; 1.05 μg protein) were diluted to a total volume of 100 μl with 0.1% acetic acid in water. A small amount of each solution (1 μl; 0.0105 μg total protein) was loaded separately, using a pressure bomb, onto an analytical column with an integrated ESI emitter tip (1- to 5-μm diameter) (34). Samples were analyzed in duplicate by nHPLC-μESI MS on a home-built Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer fitted with a custom-designed ESI source. The HPLC gradient (A = 100 mM acetic acid in water, B = 70% acetonitrile-100 mM acetic acid in water) was 0 to 50% B in 50 min, 50 to 100% B in 5 min, 100 to 0% B in 5 min, and 0% B for 5 min. Full-scan mass spectra (m/z 300 to 5,000) were acquired at a rate of approximately 1 scan/s. Mass resolving power ranged from 5,000 to 10,000.
Ion-trap nHPLC-μESI MS/MS analysis.
Aliquots of each of the above digests (2.5 μl; 0.525 μg total protein) were analyzed by nHPLC-μESI MS on an LCQ Deca mass spectrometer (Thermo/Finnigan, San Jose, CA). The peptides were loaded onto a custom-made C18 microcapillary precolumn by use of a pressure bomb, the precolumn was rinsed to remove salts, and the precolumn was then connected to an analytical column containing an integrated ESI emitter tip. Peptides were eluted into the mass spectrometer, using the HPLC gradient detailed above (spray voltage = 1.7 kV). The instrument was operated in data-dependent mode and cycled though a single MS (m/z 300 to 2,000) and five MS/MS experiments every 12 to 15 s. All MS/MS scans (collision energy = 35%) were performed with an isolation window of 3 atomic mass units. The dynamic exclusion option was selected, with a repeat count of 1, a repeat duration of 0.5 min, and an exclusion duration of 1 min. Peptide sequences were assigned using the SEQUEST algorithm (http://fields.scripps.edu/sequest/index.html) and/or de novo sequencing.
RNA isolation and preparation for Affymetrix GeneChip analysis.
Growth curves were generated to determine the CFU/ml and growth phase for P. aeruginosa PAO1 under anaerobic and aerobic conditions in the presence of 15 mM NO2− (see Fig. S1 in the supplemental material). Three independent cultures of P. aeruginosa PAO1 were grown in buffered (0.1 M potassium phosphate, pH 6.5) LB containing 15 mM NaNO2 for 4 days at 37°C under anaerobic conditions. Three independent cultures of P. aeruginosa PAO1 were grown aerobically for 4.5 h in the same medium. These aerobic and anaerobic growth conditions resulted in 2 × 108 CFU/ml at their respective time points and in mid-log growth phase. For growth in the presence of 15 mM KNO3, growth curves were generated to determine the CFU/ml and growth phase for P. aeruginosa PAO1 under anaerobic and aerobic conditions. Three independent cultures of P. aeruginosa PAO1 were grown in LB containing 15 mM KNO3 and 0.1 M potassium phosphate, pH 6.5, for 24 h at 37°C under anaerobic conditions. Three independent cultures of P. aeruginosa PAO1 were grown aerobically for 5.5 h in the same medium. These aerobic and anaerobic growth conditions resulted in 3 × 109 CFU/ml at their respective time points and in mid-log growth phase. After the cultures were chilled in a dry ice-ethanol bath to stop RNA synthesis, the cells were collected by centrifugation and RNA was isolated as previously described (33).
The quality of the RNA was assessed on an Agilent Bioanalyzer 2100 electrophoretic system pre- and post-DNase treatment. The RNA was treated with 2 U of DNase I (Ambion) for 15 min at 37°C to remove contaminating DNA. The reaction was stopped by the addition of 25 μl of DNase stop solution (50 mM EDTA, 1.5 M sodium acetate, 1% sodium dodecyl sulfate). The DNase I was removed by phenol-chloroform extraction followed by ethanol precipitation. Total RNA (10 μg) was used for cDNA synthesis, fragmentation, and labeling according to the Affymetrix GeneChip P. aeruginosa genome array expression analysis protocol (Affymetrix). Briefly, random hexamers (Invitrogen) were added (25 ng/μl) to the 10 μg of total RNA along with in vitro-transcribed Bacillus subtilis control spikes (as described in the Affymetrix GeneChip P. aeruginosa genome array expression analysis protocol). cDNA was synthesized using Superscript II (Invitrogen) according to the manufacturer's instructions, under the following conditions: 25°C for 10 min, 37°C for 60 min, 42°C for 60 min, and 70°C for 10 min. RNA was removed by alkaline treatment and subsequent neutralization. The cDNA was purified by a QIAquick PCR purification kit (Qiagen) and eluted in 40 μl of buffer EB (10 mM Tris-HCl, pH 8.5). The cDNA was fragmented by DNase I (0.6 U per μg cDNA; Amersham) at 37°C for 10 min and then end labeled with biotin-ddUTP, using an Enzo BioArray terminal labeling kit (Affymetrix), at 37°C for 60 min. Proper cDNA fragmentation and biotin labeling were determined by gel mobility shift assay using NeutrAvadin (Pierce) on a 5% polyacrylamide gel stained with SYBR green I (Roche).
Microarray data analysis.
Microarray data were generated using standard protocols generated by Affymetrix. Absolute transcript expression levels from data derived from three GeneChip microarrays per condition were normalized for each chip by globally scaling all probe sets to a target signal intensity of 500. Three statistical algorithms (detection, change call, and signal log ratio) were then used to identify differential gene expression in experimental and control samples. The detection metric (present, absent, or marginal) for a particular gene was determined using default parameters in MAS software (version 5.0; Affymetrix). Transcripts that were absent under both control and experimental conditions were eliminated from further consideration. The data generated in MAS were imported into Affymetrix Data Mining Tools (version 3.0) to perform batch analyses in which pairwise comparisons between individual experimental and control chips were made in order to generate a change call and a signal log ratio value for each transcript. The statistical significance of differences in signals between the control and experimental conditions (P < 0.05) for individual transcripts was determined using the t test. We defined a positive change call as one in which >50% of the transcripts had a call of increased or marginally increased for upregulated genes and decreased or marginally decreased for downregulated genes. Finally, the median value of the signal log ratios from each comparison file was calculated. Only those genes that met the above criteria and had median signal log ratios of ≥1 for upregulated transcripts and ≤1 for downregulated transcripts were kept in the final list of genes. Signal log ratio values were converted from log2 and expressed as x-fold changes.
Construction of STM library and cloning of STM tags into mini-Tn5 transposons.
Three mini-Tn5-based transposons, pUTmini-Tn5Km2, pUTmini-Tn5Tet, and pUTmini-Tn5TetGFP, were used for mutagenesis (16, 26, 36). The transposons are located on an R6K-based suicide delivery plasmid, pUT, where the Pi protein is furnished by the donor cell; the pUT plasmid provides the IS50R transposase tnp gene in cis, but external to the mobile element, and its conjugal transfer to recipients is mediated by RP4 mobilization functions in the donor (51). Plasmid DNA (0.04 pmol) was ligated with 1 pmol of double-stranded DNA tags in a final volume of 10 μl of 1× T4 DNA ligase buffer containing 40 U of T4 DNA ligase in 24 separate reaction mixtures. pUTmini-Tn5Km2 was digested with KpnI (New England Biolabs), and recombinant molecules were constructed in vitro by blunt-end fill-in with T4 DNA polymerase (Gibco BRL Products). pUTmini-Tn5Tc and -GFP were digested with NotI (New England Biolabs), and recombinant molecules were constructed in vitro by blunt-end fill-in with Klenow fragment (New England Biolabs). Ligated products were purified using Microcon PCR (Millipore) and resuspended in 5 μl of water. The entire 5 μl of ligated products was transformed into E. coli S17-λpir by electroporation using a Bio-Rad apparatus operated at 2.5 kV, 200 Ω, and 25 μF with a 2-mm electroporation gap cuvette. Transformed bacteria containing tagged plasmids were selected on tryptic soy agar supplemented with 50 μg/ml of ampicillin and 50 μg/ml of kanamycin. Single colonies were selected, purified, and screened using 10 pmol of one of the oligonucleotide tags (29) used to construct the DNA tags as the 5′ primer and 10 pmol of pUTKanaR1 (5′-GCGGCCTCGAGCAAGACGTTT-3′) as the 3′ primer in the kanamycin resistance gene. Thermal cycling conditions were set for touchdown PCR in a DNA thermal cycler (Perkin-Elmer Cetus) by using a hot start for 7 min at 95°C; 2 cycles at 95°C for 1 min, a temperature ranging from 70°C to 60°C for 1 min, and 72°C for 1 min; and 10 cycles at 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min. Ten microliters of amplified products was analyzed by electrophoresis in a 1% agarose gel with 1× Tris-borate-EDTA buffer and stained for 10 min in 0.5-μg/ml ethidium bromide solution (45). The amplified product had a size of 500 base pairs.
Conjugation and insertion of mini-Tn5 into P. aeruginosa and arraying of mutant libraries.
E. coli S17-λpir containing the pUTminiTn5 plasmids with tags was used as a donor for conjugal transfer into the recipient P. aeruginosa strain PAO1, using a ratio of 1 donor cell of E. coli to 10 recipient cells of P. aeruginosa. Bacteria were mixed, and 50 μl was spotted on a sterile nylon membrane that was then placed on a nonselective brain heart infusion (BHI) agar plate. Plates were incubated at 30°C for 8 h. Filters were washed with 10 ml of sterile phosphate-buffered saline to recover bacteria. Five 100-μl aliquots of the phosphate-buffered saline solution containing the transconjugants were plated on five BHI agar plates supplemented with the appropriate antibiotics. Kanamycin was used to select transconjugants with mini-Tn5Km2, and tetracycline was used to select those with mini-Tn5Tc and mini-Tn5GFP. Plates were incubated overnight at 37°C. The selected colonies were picked on BHI agar supplemented with ampicillin to exclude bacterial colonies carrying the suicide donor pUT plasmid inserted into the chromosome by homologous recombination. Exconjugants were selected on BHI agar supplemented with chloramphenicol (5 μg ml−1) (Sigma) and kanamycin (500 μg ml−1) for mini-Tn5Km2 or with tetracycline (15 μg ml−1) for mini-Tn5Tc and mini-Tn5GFP. Kanamycin-resistant and ampicillin-sensitive exconjugants were arrayed as libraries of 96 clones in 96-well microtiter plates, using 1.5 ml of BHI supplemented with kanamycin and appropriate antibiotics. To assemble the mutant library, one mutant from each library was picked to form 96 pools of 72 unique tagged mutants in each 2-ml well, labeled, and arrayed. In a defined library, each mutant had the same tag, but it was theoretically inserted at a different location in the bacterial chromosome.
Cloning and analysis of disrupted STM gene mutants.
Chromosomal DNAs from the STM mutants were prepared as described by the manufacturer (Qiagen). Chromosomal DNA (1 μg) was digested with PstI, giving DNA fragments ranging in size from 1 to 6 kb. Digested chromosomal DNA was cloned into pTZ18R (Amersham Pharmacia Biotech). Ligations were performed using 1 μg of digested chromosomal DNA mixed with 50 ng of digested pTZ18R in 20 μl of T4 DNA ligase buffer with 40 units of T4 DNA ligase. Ligated products were incubated overnight at 16°C and purified using Microcon PCR (Millipore) as described by the manufacturer. The recombinant plasmid was electroporated into E. coli DH5α. Bacterial clones were purified and analyzed for plasmid content with a Qiagen Mini preparation kit as described by the manufacturer (Qiagen). Plasmids were sequenced using the complementary primer of the corresponding tagged mutant or the 3′-conserved transposon primers encoding antibiotic resistance. Automated sequencing was performed as suggested by the manufacturer. The DNA sequences obtained were assembled and subjected to database searches using BLAST, included in the GCG Wisconsin package (version 11.0). Similarity searches with complete genomes were performed at the NCBI website, using microbial genome sequences (http://www.ncbi.nlm.nih.gov), or in this specific case, the P. aeruginosa sequence (http://www.pseudomonas.com).
Screening of STM mutants under aerobic versus anaerobic conditions.
Approximately 140 microtiter dishes containing the entire P. aeruginosa PAO1 STM library were screened for the ability to grow under aerobic conditions, with each well containing either LB-100 mM NO3− or LB-15 mM NO2− at pH 6.5 for 24 h at 37°C. Anaerobic NO3−- and NO2−-grown cultures were grown in a Coy anaerobic chamber for 48 h (NO3−) and 96 h (NO2−). The criterion for a lack of growth indicated as a negative sign in Table 8 is based upon little or no change in optical density of the control anaerobic mutant, the Δanr mutant, which is known to be incapable of anaerobic growth via denitrification or arginine substrate-level phosphorylation (65).
TABLE 8.
Growth of P. aeruginosa STM mutants and wild-type PAO1 in NO3- and NO2-containing medium with or without O2
| STM mutanta | PA no.b | Gene namec | Growthd
|
|||
|---|---|---|---|---|---|---|
| KNO3 at 24 h
|
NaNO2 at 4 days
|
|||||
| +O2 | −O2 | +O2 | −O2 | |||
| STM525 | PA0525 | norD | + | − | + | − |
| STM527 | PA0527 | dnr | + | − | + | − |
| STM1544 | PA1544 | anr | + | − | + | − |
| STM2611 | PA2611* | Siroheme synthase gene cysG | + | − | + | − |
| STM2611 | PA2611* | Siroheme synthase gene cysG | + | − | + | − |
| STM2611 | PA2611* | Siroheme synthase gene cysG | + | − | + | − |
| STM2639 | PA2639* | nuoD | + | − | + | − |
| STM2639 | PA2639* | nuoD | + | − | + | − |
| STM2642 | PA2642 | nuoG | + | − | + | − |
| STM2648 | PA2648 | nuoM | + | − | + | − |
| STM3872 | PA3872 | narI | + | − | + | + |
| STM3876 | PA3876 | narK2 | + | − | + | + |
| STM4723 | PA4723 | dksA | + | − | + | − |
| STM5407 | PA5407 | Hypothetical | + | − | + | − |
| STM525 | PA0525 | norD | + | + | + | + |
| STM3617 and -18* | PA3617 and -3618 | Intergenic upstream gene recA | + | + | + | + |
| STM3810* | PA3810 | hscA | + | + | + | + |
P. aeruginosa STM mutant strains found to be defective in anaerobic growth in L broth containing KNO3 or NaNO2.
Asterisks represent genes into which the transposon was inserted at different locations.
Underlined genes are consistent with those found by Wagner et al. (56) to be essential for anaerobic growth, using KNO3 as the terminal electron acceptor.
+, growth; −, no growth.
RESULTS
Anaerobic versus aerobic expression of P. aeruginosa proteins, determined using 2-D gel and MALDI-TOF identification.
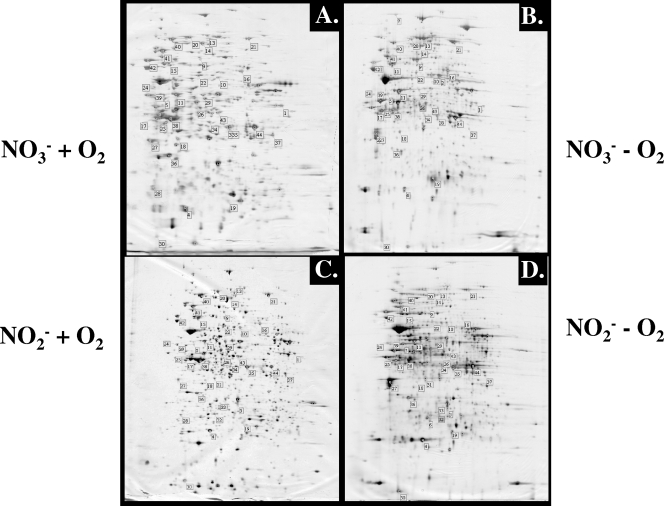
We first assessed aerobic versus anaerobic protein expression in P. aeruginosa via 2-D gel electrophoresis followed by MALDI-TOF MS identification of candidate protein spots. We used media strategically buffered at pH 6.5, a value representing the slightly acidic pH of the mucus lining the CF airways (15, 61). Images of 2-D gels derived from cell extracts obtained from all aerobically and anaerobically grown organisms in L broth with NO3− and NO2− were compared and analyzed. On average, >1,200 protein spots were detected per 2-D image. When 2-D images of P. aeruginosa grown in the presence of NO3− and oxygen (Fig. 1A and C) were compared to those obtained in the absence of oxygen (Fig. 1B and D), 68 proteins were significantly more expressed with nitrate in the absence of oxygen and 71 were less expressed. Furthermore, >100 de novo synthesized protein spots were detected anaerobically with NO3− that were absent in images obtained from organisms grown in the presence of NO3− and oxygen. When cell extracts from NO2−-grown P. aeruginosa with and without oxygen were compared, >140 proteins were more abundant with oxygen and >50 proteins were more abundant without oxygen (Fig. 1B and D). In addition, several de novo synthesized proteins were detected. Comparison of 2-D images obtained from P. aeruginosa grown anaerobically in the presence of NO3− or NO2− also revealed differential protein production. Overall, >170 proteins were induced at least fourfold in the presence of NO2− compared to those in the presence of NO3− under anaerobic conditions. In contrast, only 30 proteins appeared to be more abundant in the presence of NO3−, primarily in the class of metabolic enzymes (e.g., pyruvate dehydrogenase [PDH], aconitate hydratase 2, l-cysteine desulfurase, N-succinylglutamase 5-semialdehyde dehydrogenase, trigger factor, glutamate-asparaginase, hypothetical protein, and elongation factors G, P, Ts, and Tu) (Table 1). In contrast, a total of nine proteins whose production was less or undetectable under aerobic conditions were identified. These proteins included ATP synthase β-chain, ribonucleoside reductase, flagellin type B, a probable transcriptional regulator (PA4902), a probable peroxidase, an organic solvent tolerance protein (OstA), two hypothetical proteins, and NarH, a component respiratory nitrate reductase that donates electrons to NarG. Interestingly, two isoforms of NarH were detected that differed with respect to their isoelectric point and production profile. One isoform was detected only in 2-D images of bacteria that were grown under anaerobic growth conditions in the presence of NO3−, while the other NarH isoform was expressed at ∼16-fold higher levels under anaerobic conditions in the presence of NO3− than in the presence of NO2−. Furthermore, five proteins were identified that were absent in the presence of NO3−, including a probable transcriptional regulator (PA4902), leucine dehydrogenase, and three hypothetical proteins (Table 1).
FIG. 1.
2-D gels of aerobically (A and C) and anaerobically (B and D) expressed P. aeruginosa proteins. Cell extracts (30 μg) derived from the same growth phase (see Fig. S1 in the supplemental material) grown with NO3− (A and B)- or NO2− (C and D)-containing medium were separated by 2-D gel electrophoresis and stained with silver nitrate. The numbered spots correspond to proteins identified by MALDI-TOF analysis (Table 1).
TABLE 1.
MALDI-TOF MS analysis of aerobic versus anaerobic proteins identified from 2-D gels, in order of PA number from www.pseudomonas.com
| Protein no. | Protein name | Gene | PA no. | PseudoCAP functional class | Expectation value | Coverage (%) | Mass (kDa)/pI | Aerobic vs anaerobic growtha | Growth in nitrate vs nitrite in absence of oxygena |
|---|---|---|---|---|---|---|---|---|---|
| 29 | Succinate-semialdehyde dehydrogenase | gabD | PA0265 | Central intermediary metabolism; amino acid biosynthesis and metabolism; carbon compound catabolism | 0.042 | 6.4 | 51.9/5.6 | 3.6 | 0.4 |
| 15 | Transketolase | tktA | PA0548 | Energy metabolism | 0.05 | 7.7 | 72.6/5.2 | 2.5 | 3.0 |
| 7 | Organic solvent tolerance protein OstA | ostA | PA0595 | Membrane proteins; transport of small molecules | 0.07 | 5.7 | 104.6/5.4 | ND | Not detectable with nitrite |
| 30 | Probable DNA-binding stress protein | PA0862 | Hypothetical, unclassified, unknown | 0.011 | 25.0 | 17.5/5.0 | 0.6 | 0.28 | |
| 34 | N-Succinylglutamate 5-semialdehyde dehydrogenase | aruC | PA0895 | Amino acid biosynthesis and metabolism | 0.022 | 8.1 | 43.8/5.6 | 5.3 | 0.15 |
| 5 | Flagellin type B | fliC | PA1092 | 0.004 | 13.3 | 49.2/5.4 | 0.14 | 0.14 | |
| 13 | Ribonucleoside reductase | nrdA | PA1156 | Nucleotide biosynthesis and metabolism | 0.028 | 7.9 | 107.6/5.6 | 0.33 | 9.7 |
| 33 | Hypothetical protein | PA1191 | Hypothetical, unclassified, unknown | 0.017 | 17.8 | 23.0/6.3 | 1.4 | Not detectable with nitrate | |
| 37 | Glutamate-asparaginase | ansB | PA1337 | Amino acid biosynthesis and metabolism | 0.0 | 13.0 | 38.6/6.7 | 11.7 | 0.14 |
| 16 | Succinate dehydrogenase | sdhA | PA1583 | Energy metabolism | 0.05 | 9.3 | 64.1/6.0 | 5.6 | 1.9 |
| 21 | 2-Oxoglutarate dehydrogenase | sucA | PA1585 | Amino acid biosynthesis and metabolism; energy metabolism | 0.001 | 7.1 | 106.4/6.1 | 2.2 | 2.5 |
| 35 | Succinyl-CoA synthetase β chain | sucC | PA1588 | Energy metabolism | 0.19 | 10.6 | 41.8/5.8 | 3.0 | 0.7 |
| 27 | Outer membrane protein OprF | oprF | PA1777 | Membrane proteins; transport of small molecules | 0.001 | 22.9 | 37.9/5.0 | 1.1 | 2.4 |
| 40 | Aconitate hydratase 2 | acnB | PA1787 | Energy metabolism | 0.005 | 7.9 | 94.2/5.2 | 5.3 | 1.1 |
| 24 | Trigger factor | tig | PA1800 | Cell division; chaperones and heat shock proteins | 0.0 | 24.8 | 48.6/4.8 | 10.8 | 0.16 |
| 1 | Isocitrate dehydrogenase | icd | PA2623 | Carbon compound catabolism; amino acid biosynthesis and metabolism; energy metabolism | 0.169 | 9.8 | 45.7/5.1 | 3.1 | Not detectable with nitrite |
| 28 | Translation elongation factor P | efp | PA2851 | Translation, posttranslational modification, degradation | 0.008 | 21.3 | 21.1/4.8 | NDA | Not present under anaerobic conditions |
| 32 | Leucine dehydrogenase | ldh | PA3418 | 0.008 | 7.6 | 35.9/5.6 | 1.5 | Not detectable with nitrate | |
| 25 | Hypothetical protein | PA3515 | Hypothetical, unclassified, unknown | 0.024 | 14.3 | 40.1/5.1 | 14.7 | 0.05 | |
| 4 | Probable peroxidase | PA3529 | Adaptation, protection; putative enzymes | 0.0 | 31.5 | 21.9/5.4 | 0.5 | 0.3 | |
| 36 | Elongation factor Ts | tsf | PA3655 | Translation, posttranslational modification, degradation | 0.5 | 14.2 | 30.7/5.2 | 4.8 | 0.14 |
| 26 | l-Cysteine desulfurase | iscS | PA3814 | Amino acid biosynthesis and metabolism; biosynthesis of cofactors, prosthetic groups, and carriers | 0.004 | 11.6 | 44.8/5.7 | 7.6 | 0.5 |
| 2 | Respiratory nitrate reductase β-subunit (isoform 1) | narH | PA3874 | Energy metabolism | 0 | 20.3 | 59.11/5.8 | ND | Not detectable with nitrite |
| 10 | Respiratory nitrate reductase β-subunit (isoform 2) | narH | PA3874 | Energy metabolism | 0.0 | 15.8 | 59.1/5.7 | ND | 2.2 |
| 3 | Conserved hypothetical protein | PA3944 | Hypothetical, unclassified, unknown | 0.029 | 9.9 | 21.9/5.4 | 0.8 | Not detectable with nitrate | |
| 17 | DNA-directed RNA polymerase α chain | rpoA | PA4238 | Transcription, RNA processing and degradation | 0.0 | 23.1 | 36.8/4.9 | 0.6 | 1.1 |
| 38 | Elongation factor Tu | tufA | PA4265 | Translation, posttranslational modification, degradation | 0.0 | 25.4 | 43.7/5.2 | 8.6 | 0.14 |
| 41 | Elongation factor G | fusA1 | PA4266 | Translation, posttranslational modification, degradation | 0.033 | 10.2 | 78.1/5.1 | 4.3 | 0.6 |
| 20 | Secretion protein SecA | secA | PA4403 | Protein secretion/export apparatus | 0.004 | 7.1 | 104.1/5.4 | 2.0 | 3.6 |
| 18 | Rod shape-determining protein MrqqqqeB | mreB | PA4481 | Cell wall/lipopolysaccharide/ capsule; cell division | 0.001 | 20.3 | 37.2/5.3 | 0.9 | 4.1 |
| 6 | Hypothetical protein | PA4495 | Hypothetical, unclassified, unknown | 0.07 | 16.5 | 24.9/5.8 | ND | Not detectable with nitrate | |
| 43 | Serine hydroxymethyltransferase | glyA3 | PA4602 | Amino acid biosynthesis and metabolism | 0.068 | 9.8 | 45.4/5.7 | 2.6 | 0.5 |
| 42 | DnaK protein | dnaK | PA4761 | DNA replication, recombination, modification, and repair; adaptation and protection; chaperones and heat shock proteins | 0.016 | 10.8 | 68.5/4.8 | 0.7 | 0.4 |
| 31 | Probable transcriptional regulator | PA4902 | Transcriptional regulators | 0.029 | 12.4 | 33.6/6.1 | 0.54 | Not detectable with nitrate | |
| 14 | Pyruvate dehydrogenase | aceE | PA5015 | Amino acid biosynthesis and metabolism; energy metabolism | 0.0 | 18.5 | 99.9/5.6 | 4.1 | 0.4 |
| 44 | Ornithine carbamoyltransferase | arcB | PA5172 | Amino acid biosynthesis and metabolism | 0.054 | 13.4 | 38.6/6.1 | 0.7 | 0.6 |
| 22 | Probable transcarboxylase subunit | PA5435 | Central intermediary metabolism | 0.032 | 8.2 | 66.3/5.6 | 3.5 | 0.48 | |
| 19 | Hypothetical protein | PA5496 | Hypothetical, unclassified, unknown | 0.0 | 18.3 | 25.3/6.0 | 1.8 | 22.0 | |
| 9 | Hypothetical protein | PA5497 | Hypothetical, unclassified, unknown | 0.0 | 18.7 | 83.3/5.8 | 0.15 | 0.12 | |
| 39 | ATP synthase β chain | atpD | PA5554 | Energy metabolism | 0.02 | 15.1 | 49.5/5.0 | 0.4 | 0.4 |
| 11 | ATP synthase α chain | atpA | PA5556 | Energy metabolism | 0.003 | 17.5 | 55.5/5.3 | 0.7 | 0.6 |
ND, not detectable under aerobic conditions. NDA, not detectable under anaerobic conditions. Note that the last two columns represent x-fold changes expressed as ratios. In the second-to-last column, we compared the overall difference between 2-D images obtained under aerobic and anaerobic conditions (independent of whether nitrate or nitrite was used). Thus, the expression ratio data show averages between aerobic nitrate and nitrite cultures versus anaerobic nitrate and nitrite cultures.
Under anaerobic conditions, several proteins were not detected in the presence of NO3− (a probable transcriptional regulator, leucine dehydrogenase, and three hypothetical proteins), while some others were not produced in the presence of NO2−. The latter included the organic solvent tolerance protein OstA and the nitrate reductase subunit NarH. However, the isoform of NarH was detectable under both growth conditions, but protein expression was greater in the presence of NO3− than in the presence of NO2−. A similar protein production profile was observed for the rod shape-determining protein MreB, ribonucleoside reductase, and a hypothetical protein. Proteins that were produced in greater abundance under anaerobic growth conditions in the presence of NO2− included flagellin type B, elongation factors Ts and Tu, N-succinylglutamate 5-semialdehyde dehydrogenase, a probable DNA-binding stress protein, trigger factor, a probable peroxidase, glutamate-asparaginase, and two hypothetical proteins. In contrast, those proteins that were demonstrably lower in abundance in the presence of NO2− were MreB (rod shape-determining protein), ribonucleotide reductase (NrdA), the outer membrane protein OprF, succinate dehydrogenase (SdhA), and another putative ribonucleotide reductase (NrdJb), encoded by PA5496. Finally, proteins that were detected in NO3−-grown bacteria but not in NO2−-grown organisms were NarH (respiratory NAR), OstA (organic solvent tolerance protein), and isocitrate dehydrogenase (Icd).
Identification of differentially expressed peptides/proteins using nHPLC-μESI MS and MS/MS.
A second, more sensitive MS approach coupling nHPLC-μESI MS and MS/MS was employed to improve on our initial 2-D gel/MALDI-TOF analyses. Because of the time and effort required for these experiments, we focused on protein expression that was most dramatically upregulated anaerobically only in the presence of NO3−. Cell extracts from aerobically and anaerobically grown P. aeruginosa cultures were reduced, alkylated, and treated with trypsin, in solution, to generate peptide fragments. These samples were then subjected to nHPLC-ESI MS analysis on an FT-ICR mass spectrometer to determine the peptide m/z ratios. Figure 2A and B show the chromatograms from the two replicate analyses of the anaerobic samples and demonstrate the complexity of the samples as well as the reproducibility of the FT-ICR analysis. A comparison of the peak areas for a tryptic peptide (AQAAEIVEQAK; m/z 579.31) from the constitutively expressed ATP synthase β-subunit revealed that the level of this peptide in every sample did not differ by more than a factor of 4 (Fig. 3). Using this information, a “differentially expressed peptide” was defined as a peptide whose peak area increased by at least a factor of 10 in the anaerobic sample relative to each of the aerobic samples. One peptide peak (m/z 488.77) meeting the criteria for being considered differentially expressed is shown in Fig. 3. Using in-house software, the peptide masses from all of the FT-ICR analyses were deconvoluted to their +1 monoisotopic masses and compared to one another. As expected, the comparisons between similar samples (both anaerobic or both aerobic samples) revealed only 370 differences, on average, while the comparisons between dissimilar samples revealed over 1,360 differentially expressed peptides, on average. Further analysis of these comparisons yielded a list of 489 differentially expressed peptide masses that appeared consistently in each aerobic-anaerobic analysis. Utilizing 120 of these highly accurate mass measurements as target values in an nHPLC-μESI MS/MS analysis on an ion-trap instrument allowed the sequence for each of these peptide m/z ratios to be determined. The MS/MS spectrum of the differentially expressed peptide peak shown in Fig. 3 (m/z 488.77) is shown in Fig. 4. As depicted, these data were used to assign the sequence VVQPEYNK to this m/z ratio, revealing that this differentially expressed peptide peak was derived from NirS (also known as cytochrome cd1 or nitrite reductase [NIR]). A list of all differentially expressed proteins obtained by nHPLC-μESI MS/MS is shown in Table 2. We found that the most commonly identified peptides were those derived from NirS. In fact, there were 13 “hits” of NirS alone, with scores ranging from 2 (++) to 4 (++++) (from 10- to 1,000-fold induction). The next most abundant peptides were those derived from ArcA, ArcB, NosZ, NrdB, and cobalamin. Recall that ArcA and ArcB are required for anaerobic arginine substrate-level phosphorylation (20), NosZ is a regulator involved in the disposal of anaerobically produced N2O (5), NrdB is part of the class II ribonucleotide reductase complex that is required for anaerobic growth (27, 56), and cobalamin, too, has been purported to be required for this process (44). Some paradoxical results included enhanced anaerobic production of the major catalase KatA (PA4236) (32) and an alkyl hydroperoxide reductase, AhpCF (PA0139-140) (39), under anaerobic conditions. Finally, the magnitude of upregulation of the anaerobically induced proteins was consistent, in most cases, with the transcriptional profiling experiments that are discussed below. Finally, using both MALDI-TOF and nHPLC-μESI MS/MS techniques, a synopsis of proteins was assembled in Table 3 for comparative purposes. Note that far more peptides were identified using the sensitive nHPLC-μESI MS/MS system. However, some were identified only using MALDI-TOF MS. It should be noted here that MALDI-TOF data are more quantitative, while LC-MS data are far more sensitive. Therefore, for comparative purposes, it was important to compare both methodologies to expose the most consistent findings using both techniques.
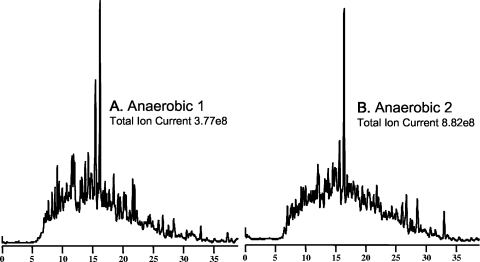
FIG. 2.
Chromatograms from FT-ICR analysis of anaerobic P. aeruginosa digests. Total ion chromatograms 1 (A) and 2 (B) from anaerobic FT-ICR analysis are shown. Note the complexity as well as the reproducibility of the chromatography samples between replicate analyses.
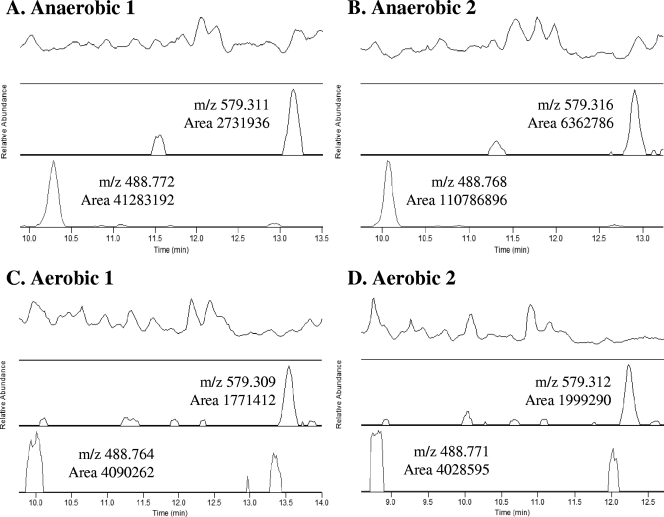
FIG. 3.
Determination of differentially expressed peptide peak areas. The peak at m/z 579.31, corresponding to the constitutively expressed tryptic peptide AQAAEIVEQAK from the P. aeruginosa ATP synthase β-subunit, was observed at approximately the same level in each of the four analyses (anaerobic [A and B] and aerobic [C and D]). Since the peak areas varied maximally by a factor of 4, a 10-fold change in area was used in the subsequent determination of differentially expressed m/z values. The peak shown at m/z 488.77 represents one such differentially expressed peptide m/z ratio. Note the consistency of the chromatographic elution profiles that easily permits alignment of the proper peaks.
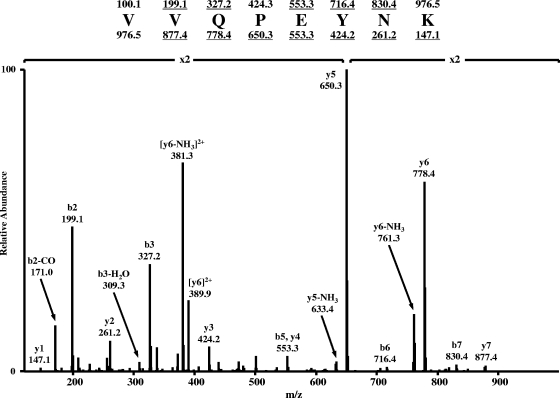
FIG. 4.
MS/MS spectrum of the differentially expressed peptide VVQPEYNK at m/z 488.77. Predicted masses for the ions of type b and y are shown above and below the sequence, respectively. Ions observed in the spectrum are underlined.
TABLE 2.
Identification of peptides by nHPLC-μESI MS, using both ion-trap and FT-ICR MS, in anaerobically grown P. aeruginosa in the presence of NO3−
| Exptl FT mass (Da) | FT retention time (min) | Charge state | Exptl LC mass (Da) | LCQ retention time (min) | Tryptic peptide | Calculated monoisotopic mass (Da) | Calculated avg mass (Da) | Protein | Relative intensity changea | Gene nameb |
|---|---|---|---|---|---|---|---|---|---|---|
| 365.70 | 6.48 | 2 | 366.28 | 10.37 | R.AGQIEGR.I | 365.70 | 365.90 | PA5427 | ++ | adhA |
| 449.25 | 7.98 | 2 | 449.61 | 11.07 | K.LGVHSEAGK.L | 449.24 | 449.51 | PA5171 | +++ | arcA |
| 431.22 | 8.27 | 3 | 431.62 | 11.26 | K.SKDGLSEDERR.I | 431.21 | 431.46 | PA1155 | ++ | nrdB |
| 431.56 | 8.27 | 3 | 431.69 | 12.06 | K.HPGQLSGGQQQR.V | 431.55 | 431.80 | PA1339 | ++ | |
| 385.17 | 8.38 | 3 | 385.57 | 16.05 | K.IEKPAGISNPK.D | 385.22 | 385.46 | PA0588 | ++ | |
| 360.21 | 9.04 | 2 | 360.28 | 14.82 | K.APGFGDR.R | 360.18 | 360.39 | PA4385 | ++ | groEL |
| 485.72 | 9.07 | 2 | 486.12 | 12.32 | K.TEETDGYR.A | 485.71 | 485.99 | PA4263 | ++ | rplC |
| 448.55 | 9.17 | 3 | 449.02 | 14.35 | R.IHEVASCENER.E | 448.54 | 448.82 | PA3874 | ++ | narH |
| 394.25 | 9.71 | 2 | 394.80 | 13.77 | K.VLVKPEDRPK.K | 394.24 | 394.48 | PA0519 | +++ | nirS |
| 366.23 | 10.05 | 2 | 366.71 | 14.58 | K.VAVIDSK.D | 366.22 | 366.44 | PA0519 | +++ | nirS |
| 488.77 | 10.28 | 2 | 489.00 | 14.53 | R.VVQPEYNK.R | 488.76 | 489.06 | PA0519 | ++ | nirS |
| 588.79 | 10.38 | 2 | 589.17 | 14.75 | R.AVSADDSDAVAR.A | 588.78 | 589.11 | PA1155 | +++ | nrdB |
| 468.25 | 10.65 | 3 | 468.65 | 20.26 | R.EVTEDAVVGRDGR.R | 468.24 | 468.51 | PA2097 | − | |
| 686.39 | 11.67 | 2 | 686.50 | 17.18 | K.VVNDTAVAVNQGGK.R | 686.37 | 686.77 | PA1156 | ++ | nrdA |
| 639.29 | 12.07 | 2 | 639.69 | 18.05 | R.FAASTCYNSEK.A | 639.28 | 639.70 | PA3392 | +++ | nosZ |
| 565.31 | 12.11 | 2 | 565.61 | 18.13 | K.NMQAPEAVAAK.R | 565.29 | 565.66 | PA4246 | ++ | rpsE |
| 598.28 | 12.18 | 2 | 598.53 | 18.58 | K.LNPAGSPSVEPK.S | 598.32 | 598.68 | PA1800 | ++ | tig |
| 598.28 | 12.18 | 2 | 598.68 | 19.00 | K.VCSDTWGGSAR.A | 598.26 | 598.65 | PA4265 | ++ | tufA |
| 527.78 | 12.95 | 2 | 528.18 | 24.27 | R.ATGCDFDLR.K | 527.74 | 528.08 | PA2639 | ++ | nuoD |
| 527.78 | 12.95 | 2 | 528.18 | 24.27 | K.CEFVEGADK.L | 527.73 | 528.08 | PA3482 | ++ | metG |
| 527.78 | 12.99 | 2 | 528.18 | 24.27 | R.ISDTTTFGGR.K | 527.76 | 528.07 | PA1092 | ++ | fliC |
| 668.85 | 13.18 | 2 | 668.87 | 19.93 | R.VVETGGNSFAAER.E | 668.83 | 669.22 | PA5171 | ++ | arcA |
| 458.76 | 13.28 | 2 | 458.92 | 20.55 | K.LTLTEDPK.E | 458.75 | 459.03 | PA5172 | +++ | arcB |
| 389.72 | 13.33 | 2 | 390.04 | 20.56 | R.GTFANIR.I | 389.71 | 389.95 | PA1562 | ++ | acnA |
| 389.72 | 13.33 | 2 | 390.04 | 20.56 | K.IFSQVGK.K | 389.73 | 389.97 | PA4236 | ++ | katA |
| 630.81 | 13.45 | 2 | 631.06 | 20.77 | R.YFMTAANNSNK.V | 630.79 | 631.21 | PA0519 | +++ | nirS |
| 491.26 | 13.56 | 2 | 491.40 | 20.24 | R.FSAADVDEK.G | 491.23 | 491.52 | PA4266 | ++ | fusA1 |
| 433.75 | 14.04 | 2 | 434.11 | 22.38 | R.FVTLGDSK.V | 433.74 | 434.00 | PA3392 | ++ | nosZ |
| 708.90 | 14.35 | 2 | 709.16 | 22.62 | R.DAGQIALVDGDSKK.I | 708.87 | 709.29 | PA0519 | ++ | nirS |
| 536.77 | 14.52 | 2 | 537.15 | 22.80 | K.QYSFEEAAK.L | 536.75 | 537.08 | PA4273 | ++ | rplA |
| 587.33 | 14.62 | 2 | 587.75 | 22.21 | K.VLDEQVSEVR.V | 587.31 | 587.65 | PA1596 | ++ | htpG |
| 463.28 | 14.68 | 3 | 463.48 | 22.80 | R.VQLPPVSAGQHVR.R | 463.27 | 463.54 | PA3914 | ++ | moeA1 |
| 648.33 | 14.68 | 2 | 648.61 | 22.95 | R.FDYPAAQQEAR.I | 648.30 | 648.70 | PA0520 | ++ | nirQ |
| 584.84 | 15.10 | 2 | 585.24 | 23.60 | K.PLTPDITQQR.G | 584.82 | 585.17 | PA0519 | +++ | nirS |
| 584.84 | 15.10 | 2 | 585.24 | 26.49 | R.AEIAGELPAAVK.I | 584.83 | 585.19 | PA2945 | +++ | cobW |
| 495.26 | 15.14 | 2 | 495.45 | 24.64 | R.EDGLDAELK.S | 495.24 | 495.53 | PA1927 | + | metE |
| 467.27 | 15.40 | 2 | 467.51 | 24.28 | K.TALSGDELK.E | 467.25 | 467.52 | PA0316 | ++ | serA |
| 467.27 | 15.40 | 2 | 467.51 | 24.28 | K.EQITLMAK.Y | 467.26 | 467.58 | PA0519 | ++ | nirS |
| 401.20 | 15.52 | 2 | 401.81 | 24.45 | K.GTNLLGVK.A | 401.25 | 401.49 | PA1562 | + | acnA |
| 631.33 | 15.65 | 2 | 632.12 | 25.35 | K.NDSSALVVVDDK.T | 631.32 | 631.69 | PA0519 | ++++ | nirS |
| 512.28 | 15.96 | 2 | 512.43 | 26.28 | K.TDDIYQIR.A | 512.26 | 512.56 | PA3915 | ++ | moaB1 |
| 644.85 | 16.38 | 2 | 645.05 | 26.43 | R.DAGQIALVDGDSK.K | 644.82 | 645.20 | PA0519 | +++ | nirS |
| 539.60 | 17.00 | 3 | 540.00 | 26.77 | R.GAQMGTFDVSHPDVR.E | 539.59 | 539.94 | PA5497 | ++ | nrdJa |
| 652.67 | 17.48 | 3 | 653.06 | 27.22 | K.AGGPRPTEVDWVDDGAEGK.L | 652.64 | 653.03 | PA3875 | ++ | thiD |
| 458.78 | 17.92 | 2 | 458.98 | 28.52 | R.FLPAILSR.F | 458.78 | 459.08 | PA2846 | ++ | |
| 458.78 | 17.92 | 2 | 459.12 | 33.04 | R.VPVILFTK.G | 458.80 | 459.09 | PA5034 | ++ | hemE |
| 420.28 | 19.60 | 2 | 420.68 | 30.43 | R.LPVELLR.Q | 420.27 | 420.53 | PA2445 | ++ | gcvP2 |
| 557.35 | 20.49 | 2 | 557.77 | 32.86 | K.TLAPSLDLVGK.I | 557.33 | 557.67 | PA0139 | ++ | ahpC |
| 603.85 | 20.70 | 2 | 603.98 | 34.51 | R.ISQSVAVFDLK.N | 603.84 | 604.21 | PA0519 | +++ | nirS |
| 732.05 | 20.91 | 3 | 732.36 | 33.97 | K.YIQHTPPQPPEWGMPEMR.E | 732.01 | 732.52 | PA0519 | ++ | nirS |
| 601.38 | 21.99 | 3 | 601.78 | 33.82 | R.IEEVKVPLPGPGQVLVK.I | 601.36 | 601.74 | PA5427 | ++ | |
| 684.35 | 22.47 | 2 | 684.32 | 35.93 | K.AGLPVGVPAMTLNK.V | 684.39 | 684.85 | PA2001 | + | atoB |
| 684.35 | 22.47 | 2 | 684.32 | 35.93 | K.GGDCTFLVEELK.A | 684.33 | 684.78 | PA5496 | ++ | |
| 657.85 | 22.98 | 2 | 658.25 | 38.63 | K.QILYLLGPVGGGK.S | 657.89 | 658.31 | PA0588 | ++ | |
| 527.27 | 24.27 | 3 | 527.73 | 41.02 | K.RGDEVWFSVWNGK.N | 527.26 | 527.59 | PA0519 | + | nirS |
| 643.35 | 26.30 | 3 | 643.75 | 49.79 | K.NPPAGEEEFLLDLITNR.V | 643.33 | 643.72 | PA1787 | ++ | |
| 712.36 | 26.59 | 2 | 712.66 | 43.49 | R.GDEVWFSVWNGK.N | 712.34 | 712.78 | PA0519 | ++ | nirS |
| 649.70 | 26.82 | 3 | 650.06 | 42.87 | K.AGIGQAPALPAHLADLFER.E | 649.69 | 650.08 | PA3735 | + | thrC |
| 658.39 | 29.60 | 2 | 658.79 | 48.30 | R.LEGLDSSALASPR.E | 658.35 | 658.73 | PA2463 | + | |
| 620.02 | 31.02 | 3 | 620.47 | 48.92 | K.QLNDLDLPNLFSVTLR.D | 620.01 | 620.38 | PA0519 | ++ | nirS |
| 605.65 | 31.98 | 3 | 605.81 | 51.58 | K.VFDAAGFADYLAGLTQR.Y | 605.64 | 606.01 | PA3635 | + | eno |
| 557.99 | 32.25 | 3 | 558.32 | 51.73 | K.DAVIGSLADIVAITEGK.A | 557.98 | 558.31 | PA5173 | + | arcC |
Relative frequencies of isolated peptides. PA2097 (shown in bold) is a probable flavin-binding monooxygenase. Thus, a lower expression level was expected. The reason that there are no other downregulated peptides shown is that we focused on those that were upregulated in the anaerobic sample. −, <10-fold decrease in that particular peptide (anaerobic relative to aerobic); +, <10-fold increase; ++, >10-fold but <100-fold increase; +++, >100-fold but <1,000-fold increase; ++++, >1,000-fold increase.
Unfilled cells represent genes encoding hypothetical proteins.
TABLE 3.
Summary of MALDI-TOF and LC-MS analyses of differentially expressed proteins
| PA no. (gene name) | Protein | Anaerobic vs aerobic expression in NO3−-containing mediuma
|
|
|---|---|---|---|
| MALDI-TOF | LC-MS | ||
| PA0040 | Hypothetical, similar to hemolysin activator protein | — | >10 |
| PA0139 (ahpC) | Alkyl hydroperoxide reductase subunit C | — | >10 |
| PA0265 (gabD) | Succinate-semialdehyde dehydrogenase | 0.3 | — |
| PA0316 (serA) | d-3-Phosphoglycerate dehydrogenase | — | >10 |
| PA0519 (nirS) | Nitrate reductase precursor | — | >10 |
| PA0520 (nirQ) | Denitrification regulatory protein (NirQ) | — | >10 |
| PA0548 (tktA) | Transketolase | 0.4 | — |
| PA0588 | Conserved hypothetical protein | — | >10 |
| PA0595 (ostA) | Organic solvent tolerance protein OstA | ND | — |
| PA0895 (aruC) | N-Succinylglutamate 5-semialdehyde dehydrogenase | 0.2 | — |
| PA0962 | Probable DNA-binding stress protein | 1.7 | — |
| PA1092 (fliC) | Flagellin type B | 7.1 | >10 |
| PA1155 (nrdB) | Ribonucleoside reductase, small chain | — | >10 |
| PA1156 (nrdA) | Ribonucleoside reductase, large chain | 3.0 | — |
| PA1191 | Hypothetical protein | 0.7 | — |
| PA1337 (ansB) | Glutamate-asparaginase | 0.2 | — |
| PA1339 | Probable ATP-binding component of ABC transporter | — | >10 |
| PA1562 (acnA) | Aconitate hydratase 1 | — | >10 |
| PA1583 (sdhA) | Succinate dehydrogenase | 0.9 | — |
| PA1585 (sucA) | 2-Oxoglutarate dehydrogenase | 0.7 | — |
| PA1588 (sucC) | Succinyl-CoA synthetase β chain | 0.3 | — |
| PA1596 (htpG) | Heat shock protein HtpG | — | >10 |
| PA1777 (oprF) | Outer membrane protein OprF | 0.9 | — |
| PA1787 (acnB) | Aconitate hydratase 2 | 0.2 | >10 |
| PA1800 (tig) | Trigger factor | 0.2 | >10 |
| PA1927 (metE) | Cobalamin-independent methionine synthase | — | >10 |
| PA2001 (atoB) | Acetyl-CoA acetyltransferase | — | >10 |
| PA2445 (gcvP2) | Glycine cleavage system protein P2 | — | >10 |
| PA2623 (icd) | Isocitrate dehydrogenase | 0.3 | — |
| PA2639 (nuoD) | NADH dehydrogenase I chains C and D | — | >10 |
| PA2846 | Probable transcriptional regulator | — | >10 |
| PA2851 (efp) | Translation elongation factor P | NDA | — |
| PA2945 | Cobalamin biosynthetic protein | — | >10 |
| PA3392 (nosZ) | Nitrous oxide reductase | — | >10 |
| PA3418 (ldh) | Leucine dehydrogenase | 0.7 | ND |
| PA3482 (metG) | Methionyl-tRNA synthetase | — | >10 |
| PA3515 | Hypothetical protein | 0.1 | — |
| PA3529 | Probable peroxidase | 2.0 | — |
| PA3635 (eno) | Enolase | — | >10 |
| PA3655 (tsf) | Elongation factor Ts | 0.2 | — |
| PA3735 (thrC) | Threonine synthase | — | >10 |
| PA3814 (iscS) | l-Cysteine desulfurase | 0.1 | — |
| PA3874 (narH) | Respiratory nitrate reductase beta subunit | ND | >10 |
| PA3875 (narG) | Respiratory nitrate reductase alpha chain | — | >10 |
| PA3914 (moeA1) | Molybdenum cofactor biosynthetic protein A1 | — | >10 |
| PA3915 (moaB1) | Molybdopterin biosynthetic protein | — | >10 |
| PA3944 | Conserved hypothetical protein | 1.3 | — |
| PA4236 (katA) | Catalase A | — | >10 |
| PA4238 (rpoA) | DNA-directed RNA polymerase α chain | 1.7 | — |
| PA4246 (rpsE) | 30S ribosomal protein | — | >10 |
| PA4263 (rplC) | 50S ribosomal protein L3 | — | >10 |
| PA4265 (tufA) | Elongation factor Tu | 0.3 | >10 |
| PA4266 (fusA1) | Elongation factor G | 0.2 | >10 |
| PA4273 (rplA) | 50S ribosomal protein L1 | — | >10 |
| PA4385 (groEL) | GroEL protein | — | >10 |
| PA4403 (secA) | Secretion protein SecA | 0.2 | — |
| PA4481 (mreB) | Rod shape-determining protein MreB | 1.1 | — |
| PA4495 | Hypothetical protein | ND | — |
| PA4602 (glyA3) | Serine hydroxymethyltransferase | 0.4 | — |
| PA4761 (dnaK) | DnaK protein | 1.0 | — |
| PA4902 | Probable transcriptional regulator | 1.9 | — |
| PA5015 (aceE) | Pyruvate dehydrogenase | 0.2 | — |
| PA5034 (hemE) | Uroporphyrinogen decarboxylase | — | >10 |
| PA5171 (arcA) | Arginine deaminase | — | >10 |
| PA5172 (arcB) | Ornithine carbamoyltransferase | 1.4 | >10 |
| PA5173 (arcC) | Carbamate kinase | — | >10 |
| PA5427 (adhA) | Alcohol dehydrogenase | — | >10 |
| PA5435 | Probable transcarboxylase subunit | 0.3 | — |
| PA5496 | Hypothetical protein | 3.6 | >10 |
| PA5497 | Hypothetical protein | 6.7 | >10 |
| PA5554 (atpD) | ATP synthase β chain | 2.5 | — |
| PA5556 (atpA) | ATP synthase α chain | 1.0 | — |
Proteins identified by both MALDI-TOF and LC-MS are shown in bold. ND, not detected aerobically; NDA, not detected anaerobically; —, not tested.
Microarray analysis of P. aeruginosa anaerobic gene expression: effect of NO3− versus NO2−.
Transcriptional profiling experiments were initiated using P. aeruginosa PAO1 grown to the same phase in LB-NO3− or LB-NO2− under aerobic versus anaerobic conditions. Organisms were harvested at the same optical density for isolation of RNA as described in Materials and Methods. Because of the shear mass of data collected in these experiments, all genes, separated by (i) an arbitrary level of induction or repression, (ii) their putative gene products, and (iii) whether they were induced, as assessed by nHPLC-μESI MS/MS (given as “LC-MS” is tables), are provided in Tables S1 to S4 in the supplemental material. For this paper, we elected to simplify the data by presenting it in tabular form and by curated PseudoCyc metabolic pathways for ease of interpretation.
(i) Genes regulated by NO3− (anaerobically induced).
As shown in Table 4 and Table S1 in the supplemental material, the genes most activated by anaerobic relative to aerobic growth are those that would be predicted to be involved in classical anaerobic NO3− reduction. According to an analysis of curated PseudoCyc metabolic pathways, disproportionately more of the activated genes are involved in the processes of denitrification (P = 0.0013) and heme d1 biosynthesis (P = 0.014) than would be expected. Of the top 100 upregulated genes, the most induced class of anaerobic respiratory genes are norCBD, encoding subunits of the protective NOR. NOR functions to detoxify potentially harmful NO during anaerobic respiration in P. aeruginosa (63). Not surprisingly, these genes are conveniently localized in a predicted operon on the P. aeruginosa genome (Fig. 5). The next most extensively represented group of genes include an operon involved in biosynthesis of the respiratory NO3− reductase complex, narK1-narK2-narGHJI (PA3871). The narK1 and narK2 genes encode extrusion pumps to rid the cell of potentially toxic levels of NO2− (49). PA3871 encodes a putative probable peptidyl-prolyl cis-trans isomerase, and moaA1 encodes a molybdenum cofactor biosynthesis protein. Molybdenum cofactor biosynthesis proteins are required for NO3− reduction in P. aeruginosa (38). Another predicted operon that was highly transcribed and involved in the reduction of NO2− was the nirSMCFDLGHJEN operon. Activation of this operon required, among others, the transcriptional activator NirQ (PA0520), whose gene expression was also upregulated but was below the 30-fold induction cutoff used for Table S1 in the supplemental material. The final gene class involved in the denitrification pathway that was in the top 100 most activated genes was nosRZDFYL, encoding members of the nitrous oxide reductase enzyme and regulators. Interestingly, the class of genes that we did not expect to be upregulated dramatically were bacteriophage-related genes. In fact, within the top 100 most activated genes, 44 were related to bacteriophage production. Most of these genes, encompassing PA0613 through PA0648, are localized between trpG and trpE on the chromosome. However, transcription of another set of phage-related genes, PA0717 to PA0729, was also dramatically induced.
TABLE 4.
Genes activated in NO3−-containing PseudoCyc metabolic pathwaysa
| Functional category or metabolic pathway | Genes activated in NO3−-containing medium
|
Genes in P. aeruginosa genome
|
P valueb | ||
|---|---|---|---|---|---|
| No. of genes identified | % of genes identified | No. of genes | % of genes | ||
| COG functional categories | |||||
| Unclassified | 20 | 37.74 | 1,054 | 16.99 | 0.001739* |
| Energy production and conversion | 8 | 15.09 | 320 | 5.16 | 0.03622* |
| General function prediction only | 8 | 15.09 | 604 | 9.74 | 0.3502 |
| Function unknown | 6 | 11.32 | 498 | 8.03 | 0.5835 |
| Inorganic ion transport and metabolism | 4 | 7.55 | 303 | 4.88 | 0.5679 |
| Transcription | 3 | 5.66 | 482 | 7.77 | 0.7828 |
| Coenzyme metabolism | 1 | 1.89 | 204 | 3.29 | 1 |
| DNA replication, recombination, and repair | 1 | 1.89 | 132 | 2.13 | 1 |
| Nucleotide transport and metabolism | 1 | 1.89 | 108 | 1.74 | 1 |
| Posttranslational modification, protein turnover, chaperones | 1 | 1.89 | 191 | 3.08 | 1 |
| Amino acid transport and metabolism | 0 | 0 | 486 | 7.83 | 0.006755* |
| Signal transduction mechanisms | 0 | 0 | 337 | 5.43 | 0.05938 |
| PseudoCyc metabolic pathways | |||||
| Unclassified | 41 | 71.93 | 4,701 | 77.64 | 0.4433 |
| Denitrification | 7 | 12.28 | 17 | 0.28 | 0.001346* |
| Biosynthesis of heme d1 | 4 | 7.02 | 9 | 0.15 | 0.01402* |
| Nitrogen metabolism | 4 | 7.02 | 35 | 0.58 | 0.06486 |
| Molybdopterin biosynthesis | 1 | 1.75 | 11 | 0.18 | 0.4975 |
A significantly greater proportion of genes were involved in denitrification than that for the whole genome (P = 0.0013); 7 of 17 genes in the genome were identified in this study. Also, a significantly greater proportion of genes were involved in biosynthesis of heme d1 than that for the genome (P = 0.014); four of nine genes in the genome were identified in this study. A significantly greater proportion of genes were classified as being involved in energy production and conversion than that for the genome (P = 0.036).
Pearson's chi-square test with Yates's continuity correction. For those categories with small values (<5), Fisher's exact test was performed instead. Asterisks indicate statistical significance (P < 0.05).
FIG. 5.
Schematic depiction of genes whose transcription is either activated anaerobically (x-fold activation [in green]) or repressed/not activated (in red) in the presence of NO3− or NO2−. The boxes that are empty represent those whose cutoff values were below the designated detection limit of Tables S1 to S4 in the supplemental material. The figure includes genes involved in the transcription of nar genes (A), nir/nor genes (B), NADH dehydrogenase genes (nuo) (C), and two different predicted operons encoding PF1 bacteriophages (D and E) and TCA cycle genes that were repressed when organisms were grown in the presence of NO2− (in red) (F).
(ii) Genes regulated by NO2− (anaerobically induced).
We next examined gene expression by P. aeruginosa grown aerobically versus anaerobically in 15 mM NO2− at pH 6.5. By the same analysis using curated PseudoCyc metabolic pathways, disproportionately more of these activated genes are involved in denitrification (P = 0.000060) and biosynthesis of heme d1′ (P = 0.00070) (Table 5; see Table S2 in the supplemental material), similar to the case for NO3−-grown cells. Of particular note, all nine genes in the genome classified as being involved in biosynthesis of heme d1′ were identified in this study. Interestingly, similar to the case for anaerobic NO3−-grown organisms, 32 of the top 100 most activated genes were genes involved in bacteriophage production. However, unlike the two bacteriophage classes observed with the NO3−-grown bacteria discussed above, only the genes from the PA0612-to-PA0648 operon were activated. Similar to the case for NO3−-grown bacteria, the nirSMCFDLGHJEN, norCBD, and nosRZDFYL operons were found to be activated. The arcDABC operon was significantly more activated in NO2−-grown bacteria than in NO3−-grown organisms. One noticeable difference between NO3−- and NO2−-induced gene expression during anaerobic growth was overexpression of the adhA gene in NO2−-grown cells but not in NO3−-grown organisms.
TABLE 5.
Genes activated in NO2−-containing mediuma
| Functional category or metabolic pathway | Genes activated in NO2−-containing medium
|
Genes in P. aeruginosa genome
|
P valueb | ||
|---|---|---|---|---|---|
| No. of genes identified | % of genes identified | No. of genes | % of genes | ||
| COG functional category | |||||
| Unclassified | 18 | 31.58 | 1,054 | 16.99 | 0.02506* |
| General function prediction only | 16 | 28.07 | 604 | 9.74 | 0.001746* |
| Function unknown | 7 | 12.28 | 498 | 8.03 | 0.4464 |
| Energy production and conversion | 5 | 8.77 | 320 | 5.16 | 0.4679 |
| Transcription | 4 | 7.02 | 482 | 7.77 | 1 |
| Inorganic ion transport and metabolism | 2 | 3.51 | 303 | 4.88 | 1 |
| Nucleotide transport and metabolism | 2 | 3.51 | 108 | 1.74 | 0.6827 |
| Amino acid transport and metabolism | 1 | 1.75 | 486 | 7.83 | 0.1005 |
| Coenzyme metabolism | 1 | 1.75 | 204 | 3.29 | 1 |
| DNA replication, recombination, and repair | 1 | 1.75 | 132 | 2.13 | 1 |
| Signal transduction mechanisms | 0 | 0 | 337 | 5.43 | 0.05938 |
| PseudoCyc metabolic pathways | |||||
| Unclassified | 42 | 60.87 | 4,701 | 77.64 | 0.01568* |
| Denitrification | 12 | 17.39 | 17 | 0.28 | 0.00005975* |
| Biosynthesis of heme d1 | 9 | 13.04 | 9 | 0.15 | 0.0007025* |
| Arginine and proline metabolism | 1 | 1.45 | 27 | 0.45 | 1 |
| Bile acid biosynthesis | 1 | 1.45 | 2 | 0.03 | 1 |
| Fatty acid metabolism | 1 | 1.45 | 7 | 0.12 | 1 |
| Glycerolipid metabolism | 1 | 1.45 | 17 | 0.28 | 1 |
| Glycolysis/ gluconeogenesis | 1 | 1.45 | 21 | 0.35 | 1 |
| Tyrosine metabolism | 1 | 1.45 | 4 | 0.07 | 1 |
Significantly greater proportions of genes were involved in denitrification (P = 0.000060) and biosynthesis of heme d1 (P = 0.00070) than those for the genome. Of particular note, all nine genes in the genome classified as being involved in biosynthesis of heme d1 were identified in this study.
Pearson's chi-square test with Yates's continuity correction. For those categories with small values (<5), Fisher's exact test was performed instead. Asterisks indicate statistical significance (P < 0.05).
(iii) Genes regulated by NO3− (anaerobically repressed).
We next examined genes that were repressed by anaerobic growth in NO3−-containing medium. According to a COG-based analysis, significantly more of these repressed genes are classified as being involved in amino acid transport and metabolism than would be expected (P = 0.0069) (Table 6; see Table S3 in the supplemental material). Of note, the identified genes were classified in many PseudoCyc amino acid metabolic pathways, supporting these COG findings. For example, the most downregulated genes were those involved in branched-chain amino acid transport, i.e., PA1070 to PA1074. However, none of these specific PseudoCyc pathway categories reached statistical significance. Among the most logically repressed genes were those encoding two dioxygenases, homogentisate-1,2-dioxygenase (PA2009) and 4-hydroxyphenylpyruvate dioxygenase (PA0865), both of which are members of the tyrosine degradation pathway and dependent upon the presence of oxygen (63). Two other gene members of the tyrosine degradation pathway, maleylacetoacetate isomerase (PA2007) and fumarylacetoacetase (PA2008), were also downregulated. The oxidative stress gene katB, which is only responsive to aerobic H2O2 in an OxyR-dependent fashion (39), was also repressed 7.5-fold.
TABLE 6.
Genes repressed in NO3−-containing mediuma
| Functional category or metabolic pathway | Genes activated in NO3−-containing medium
|
Genes in P. aeruginosa genome
|
P valueb | ||
|---|---|---|---|---|---|
| No. of genes identified | % of genes identified | No. of genes | % of genes | ||
| COG functional category | |||||
| Amino acid transport and metabolism | 24 | 23.53 | 495 | 7.98 | 0.0069* |
| Unclassified | 16 | 15.69 | 1,054 | 16.99 | 0.9534 |
| Energy production and conversion | 8 | 7.84 | 320 | 5.16 | 0.629 |
| General function prediction only | 8 | 7.84 | 604 | 9.74 | 0.8233 |
| Inorganic ion transport and metabolism | 8 | 7.84 | 303 | 4.88 | 0.5705 |
| Function unknown | 6 | 5.88 | 498 | 8.03 | 0.75 |
| Transcription | 5 | 4.9 | 482 | 7.77 | 0.5876 |
| Carbohydrate transport and metabolism | 4 | 3.92 | 222 | 3.58 | 1 |
| Lipid metabolism | 4 | 3.92 | 190 | 3.06 | 1 |
| DNA replication, recombination, and repair | 3 | 2.94 | 132 | 2.13 | 1 |
| Posttranslational modification, protein turnover, chaperones | 3 | 2.94 | 191 | 3.08 | 1 |
| Secondary metabolite biosynthesis, transport, and catabolism | 3 | 2.94 | 158 | 2.55 | 1 |
| Signal transduction mechanisms | 3 | 2.94 | 337 | 5.43 | 0.721 |
| Cell envelope biogenesis, outer membrane | 2 | 1.96 | 254 | 4.09 | 0.6827 |
| Coenzyme metabolism | 2 | 1.96 | 204 | 3.29 | 1 |
| Nucleotide transport and metabolism | 2 | 1.96 | 108 | 1.74 | 1 |
| Intracellular trafficking and secretion | 1 | 0.98 | 171 | 2.76 | 0.6212 |
| PseudoCyc metabolic pathway | |||||
| Unclassified | 80 | 74.07 | 4,701 | 77.64 | 0.6718 |
| Aromatic compound catabolism | 5 | 4.63 | 58 | 0.96 | 0.2516 |
| Butanoate metabolism | 3 | 2.78 | 11 | 0.18 | 0.2462 |
| Valine, leucine, and isoleucine degradation | 3 | 2.78 | 11 | 0.18 | 0.2462 |
| Glycine, serine, and threonine metabolism | 2 | 1.85 | 40 | 0.66 | 1 |
| Lysine degradation | 2 | 1.85 | 10 | 0.17 | 0.4975 |
| One carbon pool by folate | 2 | 1.85 | 16 | 0.26 | 0.4975 |
| Arginine and proline metabolism | 1 | 0.93 | 27 | 0.45 | 1 |
| Cyanoamino acid metabolism | 1 | 0.93 | 6 | 0.1 | 1 |
| Fatty acid biosynthesis (path 2) | 1 | 0.93 | 3 | 0.05 | 1 |
| Fatty acid metabolism | 1 | 0.93 | 7 | 0.12 | 1 |
| Methane metabolism | 1 | 0.93 | 9 | 0.15 | 1 |
| Nitrogen metabolism | 1 | 0.93 | 35 | 0.58 | 1 |
| Peptidoglycan biosynthesis | 1 | 0.93 | 18 | 0.3 | 1 |
| Propanoate metabolism | 1 | 0.93 | 9 | 0.15 | 1 |
| Pyruvate metabolism | 1 | 0.93 | 33 | 0.55 | 1 |
| Tryptophan metabolism | 1 | 0.93 | 8 | 0.13 | 1 |
| Anthranilate | 1 | 0.93 | 1 | 0.02 | 1 |
A significantly greater proportion of genes were involved in amino acid transport and metabolism (P = 0.0069) than that for the genome. For the PseudoCyc metabolic pathway analysis, identified genes were classified in many amino acid metabolism pathways, supporting the COG findings. However, none of the categories reached statistical significance, most likely because the genes are spread out among multiple categories.
Pearson's chi-square test with Yates's continuity correction. For those categories with small values (<5), Fisher's exact test was performed instead. Asterisks indicate statistical significance (P < 0.05).
(iv) Genes regulated by NO2− (anaerobically repressed).
Table 7 and Table S4 in the supplemental material indicate that a disproportionately high level of repressed genes are classified by COG as being involved in energy production and conversion (P = 3.18 × 10−8) and in translation, ribosomal structure, and biogenesis (P = 0.025). According to PseudoCyc analysis, a disproportionately higher level of repressed genes are involved in the trichloroacetic acid (TCA) cycle (P = 0.0019) than would be expected, supporting the above COG data that significantly more of these repressed genes are involved in energy production and conversion. Interestingly, the entire postglycolytic metabolic machinery, including members of the PDH complex, the TCA cycle, and the electron transport (oxidative phosphorylation) cascade, appears to be repressed significantly. For example, the aceEF genes (PA5015 and PA5016) encode the PDH and dihydrolipoamide dehydrogenase components of the PDH complex. Genes encoding TCA cycle enzymes, including sucCD (PA1588 and PA1589; succinyl coenzyme A [CoA] synthase), icd (PA2623; isocitrate dehydrogenase), gltA (PA1580; citrate synthase), acnB (PA1787; aconitase), and sdhAC (PA1583; succinate dehydrogenase), were all downregulated. Finally, the majority of the nuo class of genes, encoding subunits of the NADH dehydrogenase (complex I) of the electron transport chain, were also substantially repressed. Two putative cytochome oxidase components (PA1856 and PA1553) were downregulated 14- and 11-fold, respectively. Interestingly, the anr gene (PA1544), encoding the FNR/CRP-like transactivator ANR that is absolutely required for anaerobic growth of P. aeruginosa (65), was also repressed in NO2−-containing medium. This essentially indicates that the entire TCA cycle and electron transport chain are repressed. Potential reasons for decreased expression of these genes are offered in Discussion. Also, note that many genes involved in translation, ribosomal structure, and biogenesis were disproportionately repressed but, similar to the case in Table 5, were distributed relatively evenly among multiple categories, and no specific PseudoCyc annotated pathway reached statistical significance.
TABLE 7.
Genes repressed in NO2−-containing medium
| Functional category or metabolic pathway | Genes activated in NO2−-containing medium
|
Genes in P. aeruginosa genome
|
P value | ||
|---|---|---|---|---|---|
| No. of genes identified | % of genes identified | No. of genes | % of genes | ||
| COG functional category | |||||
| Energy production and conversion | 15 | 38.46 | 320 | 5.16 | 3.18E-08* |
| Translation, ribosomal structure, and biogenesis | 5 | 12.82 | 198 | 3.19 | 0.02457* |
| Unclassified | 3 | 7.69 | 1,054 | 16.99 | 0.08562 |
| Lipid metabolism | 3 | 7.69 | 190 | 3.06 | 0.2134 |
| Carbohydrate transport and metabolism | 2 | 5.13 | 222 | 3.58 | 1 |
| Posttranslational modification, protein turnover, chaperones | 2 | 5.13 | 191 | 3.08 | 0.721 |
| Amino acid transport and metabolism | 1 | 2.56 | 486 | 7.83 | 0.2134 |
| Cell envelope biogenesis, outer membrane | 1 | 2.56 | 254 | 4.09 | 1 |
| Coenzyme metabolism | 1 | 2.56 | 204 | 3.29 | 1 |
| DNA replication, recombination, and repair | 1 | 2.56 | 132 | 2.13 | 1 |
| Defense mechanisms | 1 | 2.56 | 77 | 1.24 | 0.6212 |
| General function prediction only | 1 | 2.56 | 604 | 9.74 | 0.0818 |
| Secondary metabolite biosynthesis, transport, and catabolism | 1 | 2.56 | 158 | 2.55 | 1 |
| Signal transduction mechanisms | 1 | 2.56 | 337 | 5.43 | 0.721 |
| Transcription | 1 | 2.56 | 482 | 7.77 | 0.2134 |
| Function unknown | 0 | 0 | 498 | 8.03 | 0.006755* |
| PseudoCyc metabolic pathways | |||||
| Unclassified | 15 | 25.42 | 4,701 | 77.64 | 4.28E−13* |
| Citrate cycle (TCA cycle) | 7 | 11.86 | 22 | 0.36 | 0.00194* |
| Pyruvate metabolism | 4 | 6.78 | 33 | 0.55 | 0.06486 |
| Glyoxylate and dicarboxylate metabolism | 3 | 5.08 | 18 | 0.3 | 0.05938 |
| Oxidative phosphorylation | 3 | 5.08 | 24 | 0.4 | 0.05938 |
| Aminoacyl-tRNA biosynthesis | 2 | 3.39 | 20 | 0.33 | 0.2462 |
| C5 branched dibasic acid metabolism | 2 | 3.39 | 4 | 0.07 | 0.2462 |
| Fatty acid biosynthesis (path 1) | 2 | 3.39 | 10 | 0.17 | 0.2462 |
| Glycolysis/gluconeogenesis | 2 | 3.39 | 21 | 0.35 | 0.2462 |
| Lysine degradation | 2 | 3.39 | 10 | 0.17 | 0.2462 |
| Reductive carboxylate cycle (CO2 fixation) | 2 | 3.39 | 8 | 0.13 | 0.2462 |
| Valine, leucine, and isoleucine biosynthesis | 2 | 3.39 | 14 | 0.23 | 0.2462 |
| Aminophosphonate metabolism | 1 | 1.69 | 1 | 0.02 | 0.4975 |
| Amino sugar metabolism | 1 | 1.69 | 5 | 0.08 | 0.4975 |
| Butanoate metabolism | 1 | 1.69 | 11 | 0.18 | 0.4975 |
| Glutathione metabolism | 1 | 1.69 | 8 | 0.13 | 0.4975 |
| Glycerolipid metabolism | 1 | 1.69 | 17 | 0.28 | 0.4975 |
| Lysine biosynthesis | 1 | 1.69 | 10 | 0.17 | 0.4975 |
| Methionine metabolism | 1 | 1.69 | 11 | 0.18 | 0.4975 |
| Propanoate metabolism | 1 | 1.69 | 9 | 0.15 | 0.4975 |
| Purine metabolism | 1 | 1.69 | 37 | 0.61 | 1 |
| Pyrimidine metabolism | 1 | 1.69 | 27 | 0.45 | 0.4975 |
| Selenoamino acid metabolism | 1 | 1.69 | 10 | 0.17 | 0.4975 |
| Tryptophan metabolism | 1 | 1.69 | 8 | 0.13 | 0.4975 |
| Urea cycle and metabolism of amino groups | 1 | 1.69 | 16 | 0.26 | 0.4975 |
aSignificantly greater proportions of genes were classified as being involved in energy production and conversion (P = 3.18E−08) and translation, ribosomal structure, and biogenesis (P = 0.025). A significantly greater proportion of genes were involved in the TCA cycle (P = 0.0019) than that for the genome, supporting the above COG data showing that significantly more genes are involved in energy production and conversion. Also, note that many genes are involved in pathways related to translation, ribosomal structure, and biogenesis, but similar to the case in Table 5, they are spread out among multiple categories and did not reach statistical significance.
bPearson's chi-square test with Yates's continuity correction. For those categories with small values (<5), Fisher's exact test was performed instead. Asterisks indicate statistical significance (P < 0.05).
Simplified synopsis of transcriptional profiling results from an operonic perspective.
Given the shear mass of data collected in our transcriptional profiling experiments, we next elected to select predicted operons in the P. aeruginosa genome that collectively were either up- or downregulated for the purpose of consistency. These operons were downloaded from the P. aeruginosa genome website (www.pseudomonas.com), and genes that were activated (green numbers) or repressed (red numbers) are boxed in Fig. 5. Figure 5A indicates that the narK1-narK2-narGHJI (PA3871)-moaA1 operon was upregulated 37- to 72-fold in anaerobic NO3− but not NO2− culture. Surprisingly, the narX-narL genes, encoding a two-component regulatory system that was recently shown to be required for activation of narK1-narK2-narGHJI (PA3871)-moaA1 in cooperation with the second-tier regulator DNR (48), were not upregulated during anaerobic NO3− growth. Unlike the genes encoding the NAR machinery, several, but not all, genes involved in production of NIR and NOR activity were significantly (between 32- and 244-fold) induced by both anaerobic NO3− and NO2− growth (Fig. 5B). NO2−, but not NO3−, was found to significantly repress the nuo genes encoding the NADH dehydrogenase complex (complex I) of the respiratory chain (Fig. 5C). One of two major gene regions on the P. aeruginosa chromosome encoding bacteriophage, from PA0713 to PA0729, was upregulated only by NO2− (Fig. 5D). This was in contrast to the very large chromosomal region encompassing PA0613 to PA0648, which was activated by both NO3− and NO2− (Fig. 5E). Finally, anaerobic NO2− growth significantly downregulated two PDH genes, aceE and aceF, and eight genes encoding components of five enzymes within the TCA cycle (Fig. 5F).
Transposon library screen for genes required for anaerobic growth using NO3− or NO2− as a terminal electron acceptor.
An STM library comprised of 7,968 mutants (43) was next screened for mutants that could not grow anaerobically using NO3− or NO2− as a terminal electron acceptor. As shown in Table 8, the [4Fe-4S]2+ transactivator ANR and its downstream regulator, DNR, were required for anaerobic growth using both NO3− and NO2−. Similar mutants harboring transposons within the cysG (encoding siroheme synthase), nuoD, nuoG, nuoM (encoding components of the NADH dehydrogenase complex), and dskA (encoding a C4 TraR zinc finger protein) genes were isolated. The narI (encoding the membrane-binding γ component of NAR) and narK2 (encoding NO2− extrusion pump 2) genes were required for anaerobic growth using NO3− but not NO2−. No mutants were identified that could grow anaerobically with NO3− but not with NO2−.
DISCUSSION
Although many processes in humans can occur under anaerobic conditions (e.g., glycolysis during vigorous aerobic exercise), humans require oxygen for life, and oxygen limitation or excess can damage human and even bacterial cells (25). In contrast, P. aeruginosa can easily survive and even thrive without oxygen if it is provided with NO3−, NO2−, and to a lesser extent, arginine. Arguably, proteins and, most importantly, enzymes that are essential for survival or growth under such conditions could represent targets for novel therapeutic intervention strategies. Importantly, the treatment of highly refractory P. aeruginosa biofilms in CF airway disease has been a source of immense frustration for both basic and clinical scientists since one initial feature of the disease is manifested, in part, as pancreatic insufficiency (17). In 2002, Worlitszch et al. (60) and our research group (62) discovered that P. aeruginosa growing in the thickened CF airway mucus is capable of growth under reduced oxygen tension and even anaerobic conditions, especially during chronic, end-stage CF airway disease. Given that humans cannot undergo anaerobic respiration, this study was initiated with the goal of identifying P. aeruginosa genes and gene products that are modulated during anaerobic growth by NO3− or NO2− respiration specifically at the slightly acidic pH (∼6.5) of the CF airway mucus. Given that highly advanced techniques and diverse expertise were available to our collaborative research groups, we elected to use a combined and exhaustive proteomic, microarray, and STM mutagenesis approach to identify potential therapeutic targets for intervention of the anaerobic biofilm mode of P. aeruginosa.
Differences in anaerobic growth patterns between NO3− and NO2− growth. (i) NO3− or NO2− reduction machinery and arginine substrate-level phosphorylation.
For anaerobic respiration using NO3− or NO2−, P. aeruginosa has dissimilatory NAR, NIR, NOR, and nitric oxide synthase activities for the purpose of ATP generation. During both our proteomic and microarray studies, we found clear evidence of gene and protein expression patterns that are consistent with some but not all members of the denitrification pathway being activated during anaerobic but not aerobic growth with NO3− as well as with NO2−. Predictably, the norCBD genes, encoding the protective enzyme NOR, were induced under both conditions. We have recently shown that a P. aeruginosa mutant lacking NOR (e.g., norCB) grows abysmally slowly during anaerobic growth but does not die (63). This is due to an elegant two-pronged mechanism. It involves (i) an NO-mediated inactivation of the master anaerobic regulator ANR coupled with (ii) overexpression of two paradoxically oxygen-dependent gene products that are typically the most repressed proteins, namely, homogentisate-1,2-dioxygenase (HmgA) and 4-hydroxyphenylpyruvate dioxygenase (Hpd), both of which possess inherent NO-scavenging activity (63). Despite the fact that only NO3− and NO2− were used as alternative electron acceptors in these studies, genes/proteins involved in the arginine deiminase pathway, including arcA/arcB, were also activated, especially transcriptionally, by NO2− (see Table S2 in the supplemental material), and proteomically using LC-MS (Table 2), confirming previous results indicating that some of the master anaerobic regulators, including ANR, DNR, and ArgR, are fully operative as regulators of the arginine deiminase pathway (20, 31). Furthermore, our LC-MS proteomic data indicated that NirS and Arc(ABC) represent some of the most frequently encountered proteins during anaerobic NO3− respiration (Table 2). The anaerobic transcriptional profiling results of Filiatrault et al. (19) also confirm the activation of arcA and arcB (and arcD) by NO3− alone. The transcription of the various nar genes was elegantly shown to be under the control of DNR and NarX/L (4, 48), both of which require activation by the global transcriptional activator ANR by oxygen limitation (48). In contrast, DNR, NarX/L, and NirQ each participate in the regulation of nir, nor, and nos genes (3, 48).
(ii) Somewhat unexpected findings.
Why are genes and proteins involved in postglycolytic energy metabolism repressed during anaerobic NO2− respiration? The mucus lining the CF airways was recently found to be slightly acidic (pH ∼6.5) (61), and such conditions allow a product of anaerobic NO3− respiratory metabolism, namely, NO2−, to be reduced nonenzymatically to NO. The elegance of this finding is that the formidable mucA mutant mucoid strains of P. aeruginosa, which are resistant to antibiotics and phagocytes, are exquisitely susceptible to anaerobic NO (61). This was recently shown by Yoon et al. (61) and offers some hope for eradication of the mucoid form of P. aeruginosa from the CF airways. One of the major frontline antibiotics used to treat P. aeruginosa biofilm infections is the aminoglycoside tobramycin. The efficacy of tobramycin is either markedly reduced or absent in the absence of oxygen (11, 13, 37, 54), yet a recent study indicates that biofilm P. aeruginosa is more susceptible to tobramycin when cultures are amended with either nitrate or arginine (10). Our study reveals that the entire postglycolytic metabolism of P. aeruginosa is shut down under these conditions. For example, in the presence of NO2−, PDH, five of eight genes encoding TCA cycle enzymes, and various members of the NADH dehydrogenase complex are downregulated (Fig. 5F). The potential rationale behind these metabolic events is as follows. Two enzymes, in particular (aconitase and fumarase), are strategically positioned immediately prior to the production of TCA cycle enzymes (isocitrate dehydrogenase and α-ketoglutarate dehydrogenase) that generate reducing power in the form of NADH. Both aconitase and fumarase have been shown to be susceptible to inactivation not only by aerobic free radicals, such as the superoxide anion (O2−) (22, 30), but also by NO (21). The inherent cellular logic for these events is that the cell uses a “circuit-breaker” mechanism to compromise the activity of enzymes immediately preceding those that generate NADH. The “circuit-breaker” term was coined in the metabolic sense by Irwin Fridovich and colleagues at Duke University 16 years ago, after they revealed the O2− and NO sensitivity of these enzymes (21, 22). This allows cells to survive while growing slowly in the presence of 15 mM NO2− (see Fig. S1 in the supplemental material), an amount that was shown nearly 30 years ago to be the limit of toxicity for P. aeruginosa (59). This feature of NO-mediated inactivation of cellular proteins is not unique to aconitase and fumarase. In fact, the ribonucleotide reductase NrdA is an iron-sulfur protein that is known to be poisoned by NO (D. J. Hassett, unpublished results).
Anaerobic bacteriophage gene activation.
One of the most intriguing results was the discovery of a dramatic increase in bacteriophage gene activation during anaerobic growth using both NO3− and NO2− as terminal electron acceptors (see Tables S4 and S5 in the supplemental material). Specifically, of the top 62 most upregulated genes during anaerobic growth in the presence of NO3−, 32 (NO3−) and 33 (NO2−) were the phage genes PA613 to PA648, while 11 genes were of the PA713-to-PA729 cluster when cells were grown in the presence of NO3−. Interestingly, PA0713, the first gene of the PA713-to-PA729 cluster, was the most upregulated gene in the presence of NO2− (121-fold). These phage genes encode Pf1-type (pilus/flagellum) proteins that are known to be upregulated in biofilms (58). Because slightly acidified NO2− generates NO, the latter gas could be a cellular signal indicating that the prophage incorporated into the bacterial genome is under attack and that it would behoove the phage to find a suitable host that is not under such stress. Thus, it is not surprising that bacteriophages have been shown to cause death in P. aeruginosa biofilms (58). Relatedly, NO has also been shown to cause dispersal of P. aeruginosa from biofilms (6). We also have shown that an absence of the rhl quorum-sensing circuit causes metabolic suicide by overproduction of NO via anaerobic respiration in biofilms and planktonic culture (62). In addition, we also showed that slightly acidified NO2− actually kills the formidable mucoid, alginate-overproducing form that is highly resistant to conventional antibiotics and phagocytic killing (61). Thus, both the quorum-sensing mutants and the mucoid strains may perish by an as yet unappreciated mechanism involving activation of bacteriophage(s), a hypothesis that certainly merits experimental testing.
Clearly, organisms exposed to NO2− during anaerobic growth exhibited the most revealing metabolic pattern with respect to the therapeutic potential of acidified NO2−. Remarkably, the organism appeared to be shutting down the production of reducing power in the form of NADH and FADH from a dramatically reduced PDH complex (aceEF genes), a reduced TCA cycle (gltA, acnB, icd, sucABCD, and sdhA), and also reduced ATP generation by a reduction in NADH dehydrogenase activity (nuoBDEFHIJKM genes). In fact, even two genes encoding putative cytochrome oxidases (PA1553 and PA1856) were the most repressed genes during anaerobic growth in the presence of NO2− at pH 6.5. In reducing the proton motive force by reduction of NADH dehydrogenase I activity, there would be reduced transport of NO2− into P. aeruginosa. With reduced NO2− transport coupled with a reduction in cellular NADH because of the likely poisoning of P. aeruginosa aconitase by NO (21), the organism grows poorly. As such, we have shown that acidified NO2− at pH 6.5 can act almost in a bacteriostatic fashion (61). These results are consistent with those of Wagner et al. (57) showing that members of the NADH dehydrogenase complex (nuo genes) were absolutely required for anaerobic growth in the presence of NO3−. However, two new genes that were found to be required for anaerobic growth in the presence of both NO3− and NO2− were completely unexpected. One of these genes, dksA, controls the intercellular signaling process of quorum sensing in P. aeruginosa (12, 28). It is for this reason that a dksA mutant is not able to perform quorum sensing, and perhaps its inability to grow in NO3− or NO2− medium may be a function of a defect similar to the metabolic overproduction of NO akin to anaerobic rhlR mutant bacteria (62). Finally, the multifunction product of the cysG gene, siroheme synthase, was shown by Zumft and colleagues (56) to be essential for incorporation of protoporphyrin IX into heme (9). Since heme is required for many enzymes involved in the anaerobic respiratory pathway, these results are consistent with their cellular function.
Limitations of this study. (i) Differences in growth rate and cell yield between NO3−- and NO2−-grown bacteria.
Recall that the growth characteristics of P. aeruginosa in NO3− versus NO2− medium are very different. The anaerobic growth rate of P. aeruginosa in NO2− medium is significantly lower than that in NO3− medium, and the organisms do not achieve the same maximal cell density (see Fig. S1 in the supplemental material). NO2− at concentrations approaching 20 mM are not well tolerated by P. aeruginosa, and the cells cannot generate the ATP of NO3−-respiring cells because they have no substrate (NO3−) for the NAR (proton-pumping) coupling step. These differences in growth rate could provide an alternative explanation for some of the differences in gene expression. Bacteria that are growing more rapidly may increase the expression of genes in the central pathways (e.g., PDH complex and TCA cycle), which is essential for synthesis of many cellular metabolites and structures relating to anaplerotic functions. This may also be the reason for differences in expression of genes for energy production and conversion as well as for translation, ribosomal structure, and biogenesis.
(ii) Results derived from rich medium versus those derived from the host during infection.
Despite the exhaustive and comprehensive nature of this study, we must stress caution in attempting to relate the significance and potential correlates of our results to the behavior of P. aeruginosa in the CF airway and other disease settings. First, the proteome and transcriptome were determined in vitro and in complex growth media. Recently, a synthetic medium containing many of the factors in CF sputum was used to monitor the behavior of P. aeruginosa (41), and this medium may be useful for further studies involving a similar research theme. The argument that the “proteogenome” of P. aeruginosa in situ is identical or even similar to the results of these work conditions would be considered tenuous at best. However, we would also argue that the basic challenge of a slightly decreased pH (∼6.5) (61) and anaerobic or oxygen-limiting conditions (2, 60) would in all probability require similar regulation of cellular pathways irrespective of additional environmental stress that would ensue in an actual infection. Finally, we must also note that the P. aeruginosa research community almost always uses strain PAO1. This strain can behave very differently from laboratory to laboratory, and while the vast majority of results using this strain are highly reproducible, some differences may occur.
In conclusion, we have accomplished comprehensive (i) MALDI-TOF/LC-MS proteomic analysis, (ii) transcriptional profiling using GeneChip microarrays, and finally, (iii) STM analysis of genes that are required for anaerobic growth of P. aeruginosa at pH 6.5. We hope that these studies provide a partial framework for future investigations to unravel new clues as to mechanisms by which we might help to eradicate this notorious pathogen during the course of various disease states.
Supplementary Material
Acknowledgments
This work was supported in part by Public Health Service grants GM-69845 (D.J.H.) and GM-37537 (D.F.H.), Cystic Fibrosis Foundation grants HASSETT07G0 and R457-CR01 (D.J.H.), a Cure Finders grant (D.J.H.), the National Science Foundation 0311307 (K.S.), National Institutes of Health grant HL-073835 (K.S.), The Canadian Institute for Health Research (R.C.L.), and Cystic Fibrosis Foundation pilot funds.
Footnotes
Published ahead of print on 18 January 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aida, T., S. Hata, and H. Kusunoki. 1986. Temporary low oxygen conditions for the formation of nitrate reductase and nitrous oxide reductase by denitrifying Pseudomonas sp. G59. Can. J. Microbiol. 32543-547. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez-Ortega, C., and C. S. Harwood. 2007. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65153-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arai, H., Y. Igarashi, and T. Kodama. 1995. Expression of the nir and nor genes for denitrification of Pseudomonas aeruginosa requires a novel CRP/FNR-related transcriptional regulator, DNR, in addition to ANR. FEBS Lett. 37173-76. [DOI] [PubMed] [Google Scholar]
- 4.Arai, H., T. Kodama, and Y. Igarashi. 1997. Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol. Microbiol. 251141-1148. [DOI] [PubMed] [Google Scholar]
- 5.Arai, H., M. Mizutani, and Y. Igarashi. 2003. Transcriptional regulation of the nos genes for nitrous oxide reductase in Pseudomonas aeruginosa. Microbiology 14929-36. [DOI] [PubMed] [Google Scholar]
- 6.Barraud, N., D. J. Hassett, S. H. Hwang, S. A. Rice, S. Kjelleberg, and J. S. Webb. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 1887344-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beckmann, C., M. Brittnacher, R. Ernst, N. Mayer-Hamblett, S. I. Miller, and J. L. Burns. 2005. Use of phage display to identify potential Pseudomonas aeruginosa gene products relevant to early cystic fibrosis airway infections. Infect. Immun. 73444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blum, H., H. Bier, and H. J. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 893-99. [Google Scholar]
- 9.Bollivar, D. W., T. Elliott, and S. I. Beale. 1995. Anaerobic protoporphyrin biosynthesis does not require incorporation of methyl groups from methionine. J. Bacteriol. 1775778-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borriello, G., L. Richards, G. D. Ehrlich, and P. S. Stewart. 2006. Arginine or nitrate enhances antibiotic susceptibility of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 50382-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borriello, G., E. Werner, F. Roe, A. M. Kim, G. D. Ehrlich, and P. S. Stewart. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 482659-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Branny, P., J. P. Pearson, E. C. Pesci, T. Kohler, B. H. Iglewski, and C. Van Delden. 2001. Inhibition of quorum sensing by a Pseudomonas aeruginosa dksA homologue. J. Bacteriol. 1831531-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryan, L. E., S. K. Kowand, and H. M. Van Den Elzen. 1979. Mechanism of aminoglycoside antibiotic resistance in anaerobic bacteria: Clostridium perfringens and Bacteroides fragilis. Antimicrob. Agents Chemother. 157-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabral, D. A., B. A. Loh, and D. P. Speert. 1987. Mucoid Pseudomonas aeruginosa resists nonopsonic phagocytosis by human neutrophils and macrophages. Pediatr. Res. 22429-431. [DOI] [PubMed] [Google Scholar]
- 15.Coakley, R. D., B. R. Grubb, A. M. Paradiso, J. T. Gatzy, L. G. Johnson, S. M. Kreda, W. K. O'Neal, and R. C. Boucher. 2003. Abnormal surface liquid pH regulation by cultured cystic fibrosis bronchial epithelium. Proc. Natl. Acad. Sci. USA 10016083-16088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 1726568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.diSant'Agnese, P. A., R. C. Darling, G. A. Perera, and E. Shea. 1953. Abnormal electrolyte composition of sweat in cystic fibrosis of the pancreas: clinical significance and relationship to the disease. Pediatrics 12549-563. [PubMed] [Google Scholar]
- 18.Filiatrault, M. J., K. F. Picardo, H. Ngai, L. Passador, and B. H. Iglewski. 2006. Identification of Pseudomonas aeruginosa genes involved in virulence and anaerobic growth. Infect. Immun. 744237-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filiatrault, M. J., V. E. Wagner, D. Bushnell, C. G. Haidaris, B. H. Iglewski, and L. Passador. 2005. Effect of anaerobiosis and nitrate on gene expression in Pseudomonas aeruginosa. Infect. Immun. 733764-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gamper, M., A. Zimmermann, and D. Haas. 1991. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J. Bacteriol. 1734742-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner, P. R., G. Costantino, C. Szabo, and A. L. Salzman. 1997. Nitric oxide sensitivity of the aconitases. J. Biol. Chem. 27225071-25076. [DOI] [PubMed] [Google Scholar]
- 22.Gardner, P. R., and I. Fridovich. 1991. Superoxide sensitivity of the Escherichia coli aconitase. J. Biol. Chem. 26619328-19333. [PubMed] [Google Scholar]
- 23.Govan, J. R. W., and J. A. M. Fyfe. 1978. Mucoid Pseudomonas aeruginosa and cystic fibrosis: resistance of the mucoid form to carbenicillin, flucloxacillin and tobramycin and the isolation of mucoid variants in vitro. J. Antimicrob. Chemother. 4233-240. [DOI] [PubMed] [Google Scholar]
- 24.Hassett, D. J., J. Cuppoletti, B. Trapnell, S. V. Lymar, J. J. Rowe, S. Sun Yoon, G. M. Hilliard, K. Parvatiyar, M. C. Kamani, D. J. Wozniak, S. H. Hwang, T. R. McDermott, and U. A. Ochsner. 2002. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv. Drug Deliv. Rev. 541425-1443. [DOI] [PubMed] [Google Scholar]
- 25.Hassett, D. J., and J. A. Imlay. 2006. Oxidative stress systems in bacteria: four model systems, p. 544-573. In C. Nickerson and M. J. Schurr (ed.), Kluwer Academic-Plenum Publishers, New York, NY.
- 26.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1726557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan, A., E. Torrents, I. Sala, U. Hellman, I. Gibert, and P. Reichard. 1999. Ribonucleotide reduction in Pseudomonas species: simultaneous presence of active enzymes from different classes. J. Bacteriol. 1813974-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jude, F., T. Kohler, P. Branny, K. Perron, M. P. Mayer, R. Comte, and C. van Delden. 2003. Posttranscriptional control of quorum-sensing-dependent virulence genes by DksA in Pseudomonas aeruginosa. J. Bacteriol. 1853558-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehoux, D. E., F. Sanschagrin, and R. C. Levesque. 2001. Discovering essential and infection-related genes. Curr. Opin. Microbiol. 4515-519. [DOI] [PubMed] [Google Scholar]
- 30.Liochev, S. I., and I. Fridovich. 1993. Modulation of the fumarases of Escherichia coli in response to oxidative stress. Arch. Biochem. Biophys. 301379-384. [DOI] [PubMed] [Google Scholar]
- 31.Lu, C. D., H. Winteler, A. Abdelal, and D. Haas. 1999. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 1812459-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, J.-F., U. A. Ochsner, M. G. Klotz, V. K. Nanayakkara, M. L. Howell, Z. Johnson, J. Posey, M. L. Vasil, J. J. Monaco, and D. J. Hassett. 1999. Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J. Bacteriol. 1813730-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin, D. W., M. J. Schurr, H. Yu, and V. Deretic. 1994. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to sE and stress response. J. Bacteriol. 1766688-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin, S. E., J. Shabanowitz, D. F. Hunt, and J. A. Marto. 2000. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 724266-4274. [DOI] [PubMed] [Google Scholar]
- 35.Matsui, H., V. E. Wagner, D. B. Hill, U. E. Schwab, T. D. Rogers, B. Button, R. M. Taylor II, R. Superfine, M. Rubinstein, B. H. Iglewski, and R. C. Boucher. 2006. A physical linkage between cystic fibrosis airway surface dehydration and Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 10318131-18136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthysse, A. G., S. Stretton, C. Dandie, N. C. McClure, and A. E. Goodman. 1996. Construction of GFP vectors for use in gram-negative bacteria other than Escherichia coli. FEMS Microbiol. Lett. 14587-94. [DOI] [PubMed] [Google Scholar]
- 37.Mingeot-Leclercq, M. P., Y. Glupczynski, and P. M. Tulkens. 1999. Aminoglycosides: activity and resistance. Antimicrob. Agents Chemother. 43727-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noriega, C., D. J. Hassett, and J. J. Rowe. 2005. The mobA gene is required for assimilatory and respiratory nitrate reduction but not xanthine dehydrogenase activity in Pseudomonas aeruginosa. Curr. Microbiol. 51419-424. [DOI] [PubMed] [Google Scholar]
- 39.Ochsner, U. A., M. L. Vasil, E. Alsabbagh, K. Parvatiyar, and D. J. Hassett. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB, ahpB, and ahpCF. J. Bacteriol. 1824533-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 2504007-4021. [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer, K. L., L. M. Aye, and M. Whiteley. 2007. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 1898079-8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer, K. L., S. A. Brown, and M. Whiteley. 2007. Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J. Bacteriol. 1894449-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potvin, E., D. E. Lehoux, I. Kukavica-Ibrulj, K. L. Richard, F. Sanschagrin, G. W. Lau, and R. C. Levesque. 2003. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ. Microbiol. 51294-1308. [DOI] [PubMed] [Google Scholar]
- 44.Roth, J. R., J. G. Lawrence, M. Rubenfield, S. Kieffer-Higgins, and G. M. Church. 1993. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J. Bacteriol. 1753303-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 1836579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sauer, K., M. C. Cullen, A. H. Rickard, L. A. Zeef, D. G. Davies, and P. Gilbert. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 1867312-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schreiber, K., R. Krieger, B. Benkert, M. Eschbach, H. Arai, M. Schobert, and D. Jahn. 2007. The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J. Bacteriol. 1894310-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma, V., C. E. Noriega, and J. J. Rowe. 2006. Involvement of NarK1 and NarK2 proteins in transport of nitrate and nitrite in the denitrifying bacterium Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 72695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68850-858. [DOI] [PubMed] [Google Scholar]
- 51.Simon, R., U. Priefer, and A. Puehler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1784-791. [Google Scholar]
- 52.Son, M. S., W. J. Matthews, Jr., Y. Kang, D. T. Nguyen, and T. T. Hoang. 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 755313-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Southey-Pillig, C. J., D. G. Davies, and K. Sauer. 2005. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 1878114-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tresse, O., T. Jouenne, and G. A. Junter. 1995. The role of oxygen limitation in the resistance of agar-entrapped, sessile-like Escherichia coli to aminoglycoside and beta-lactam antibiotics. J. Antimicrob. Chemother. 36521-526. [DOI] [PubMed] [Google Scholar]
- 55.Vander Wauven, C., A. Pierard, M. Kley-Raymann, and D. Haas. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vollack, K. U., E. Hartig, H. Korner, and W. G. Zumft. 1999. Multiple transcription factors of the FNR family in denitrifying Pseudomonas stutzeri: characterization of four fnr-like genes, regulatory responses and cognate metabolic processes. Mol. Microbiol. 311681-1694. [DOI] [PubMed] [Google Scholar]
- 57.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 1852080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 1854585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams, D. R., J. J. Rowe, P. Romero, and R. G. Eagon. 1978. Denitrifying Pseudomonas aeruginosa: some parameters of growth and active transport. Appl. Environ. Microbiol. 36257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Wei, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Doring. 2002. Reduced oxygen concentrations in airway mucus contribute to the early and late pathogenesis of Pseudomonas aeruginosa cystic fibrosis airway infection. J. Clin. Investig. 109317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoon, S. S., R. Coakley, G. W. Lau, S. V. Lymar, B. Gaston, A. C. Karabulut, R. F. Hennigan, S. H. Hwang, G. Buettner, M. J. Schurr, J. E. Mortensen, J. L. Burns, D. Speert, R. C. Boucher, and D. J. Hassett. 2006. Anaerobic killing of mucoid Pseudomonas aeruginosa by acidified nitrite derivatives under cystic fibrosis airway conditions. J. Clin. Investig. 116436-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoon, S. S., R. F. Hennigan, G. M. Hilliard, U. A. Ochsner, K. Parvatiyar, M. C. Kamani, H. L. Allen, T. R. DeKievit, P. R. Gardner, U. Schwab, J. J. Rowe, B. H. Iglewski, T. R. McDermott, R. P. Mason, D. J. Wozniak, R. E. Hancock, M. R. Parsek, T. L. Noah, R. C. Boucher, and D. J. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3593-603. [DOI] [PubMed] [Google Scholar]
- 63.Yoon, S. S., A. C. Karabulut, J. D. Lipscomb, R. F. Hennigan, S. V. Lymar, S. L. Groce, A. B. Herr, M. L. Howell, P. J. Kiley, M. J. Schurr, B. Gaston, K. H. Choi, H. P. Schweizer, and D. J. Hassett. 2007. Two-pronged survival strategy for the major cystic fibrosis pathogen, Pseudomonas aeruginosa, lacking the capacity to degrade nitric oxide during anaerobic respiration. EMBO J. 263662-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, S., and B. T. Chait. 2000. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 722482-2489. [DOI] [PubMed] [Google Scholar]
- 65.Zimmermann, A., C. Reimmann, M. Galimand, and D. Haas. 1991. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol. Microbiol. 51483-1490. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.