Abstract
Archaea and eukaryotes share a dolichol phosphate-dependent system for protein N-glycosylation. In both domains, the acetamido sugar N-acetylglucosamine (GlcNAc) forms part of the core oligosaccharide. However, the archaeal Methanococcales produce GlcNAc using the bacterial biosynthetic pathway. Key enzymes in this pathway belong to large families of proteins with diverse functions; therefore, the archaeal enzymes could not be identified solely using comparative sequence analysis. Genes encoding acetamido sugar-biosynthetic proteins were identified in Methanococcus maripaludis using phylogenetic and gene cluster analyses. Proteins expressed in Escherichia coli were purified and assayed for the predicted activities. The MMP1680 protein encodes a universally conserved glucosamine-6-phosphate synthase. The MMP1077 phosphomutase converted α-d-glucosamine-6-phosphate to α-d-glucosamine-1-phosphate, although this protein is more closely related to archaeal pentose and glucose phosphomutases than to bacterial glucosamine phosphomutases. The thermostable MJ1101 protein catalyzed both the acetylation of glucosamine-1-phosphate and the uridylyltransferase reaction with UTP to produce UDP-GlcNAc. The MMP0705 protein catalyzed the C-2 epimerization of UDP-GlcNAc, and the MMP0706 protein used NAD+ to oxidize UDP-N-acetylmannosamine, forming UDP-N-acetylmannosaminuronate (ManNAcA). These two proteins are similar to enzymes used for proteobacterial lipopolysaccharide biosynthesis and gram-positive bacterial capsule production, suggesting a common evolutionary origin and a widespread distribution of ManNAcA. UDP-GlcNAc and UDP-ManNAcA biosynthesis evolved early in the euryarchaeal lineage, because most of their genomes contain orthologs of the five genes characterized here. These UDP-acetamido sugars are predicted to be precursors for flagellin and S-layer protein modifications and for the biosynthesis of methanogenic coenzyme B.
Most cells use acetamido sugars in their cell walls, glycolipids, or protein posttranslational modifications. Bacterial peptidoglycan contains cross-linked polysaccharides of N-acetylglucosamine (GlcNAc) and its derivative, N-acetylmuramic acid (11). Lipopolysaccharides of many gram-negative proteobacteria and the capsules of both gram-positive and gram-negative bacteria include N-acetylmannosaminuronate (ManNAcA) and other oxidized acetamido sugars (15). In eukaryotes, the core oligosaccharide of protein N-linked glycosylation contains several GlcNAc subunits (39). O-GlcNAc modifications to soluble eukaryotic proteins provide another level of posttranslational regulation (43).
Archaea also use GlcNAc and its derivatives. While archaea do not have true peptidoglycan or produce lipopolysaccharides, genome sequences suggest they have a dolichol phosphate-dependent pathway for oligosaccharide synthesis (5, 9), and dolichol phosphate-linked sugars were detected in the euryarchaeon Haloferax volcanii (17). Compared to eukaryotes, these archaea are predicted to use a simplified glycosylation pathway to modify extracellular proteins (1). 2-Acetamido sugars have been identified in Methanococcus voltae as archaetidyl lipids (10) and the N-linked glycosylation of flagellin and S-layer proteins (7). Gene disruption studies suggest that glycosylation stabilizes S-layer assembly in H. volcanii (1) and flagellar assembly in Methanococcus spp. (7). The alg7 GlcNAc glycosyltransferase gene appears to be essential in the methanococci (35). While no specific function has been identified for these glycosylations, they could promote cellular recognition, increase solubility, or alter the cell surface hydrophobicity. Archaea also use these sugars to modify cytoplasmic factors: folates from Pyrococcus furiosus have GlcNAc appendages (40), and methanogenic coenzyme B was reported to contain a ManNAcA (β-1→4) UDP-GlcNAc disaccharide head group (33). Remarkably, P. furiosus accumulates up to 14 mM UDP-GlcNAc and UDP-N-acetylgalactosamine during the late log phase of growth (29).
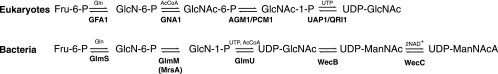
Based on its universal distribution, GlcNAc biosynthesis probably evolved early. Both eukaryotes and bacteria produce 2-acetamido sugars as UDP-activated compounds, but they differ in their biosynthetic pathways and enzymologies (Fig. 1). Both lineages use the universally conserved enzyme glucosamine synthase to convert fructose-6-phosphate (Fru-6-P) to α-d-glucosamine-6-phosphate (GlcN-6-P). Eukaryotes acetylate this product using acetyl-coenzyme A (Ac-CoA), and then they transfer the phosphate group to the anomeric position of the sugar. A nucleotidyltransferase enzyme uses UTP to activate the sugar, forming UDP-GlcNAc. Alternatively, bacteria convert GlcN-6-P to glucosamine-1-phosphate (GlcN-1-P) and then use a bifunctional enzyme to acetylate the sugar and transfer it to UTP. Some bacteria also use an epimerase to convert UDP-GlcNAc to UDP-ManNAc and a four-electron transferring oxidoreductase to produce UDP-ManNAcA. These bacteria present the ManNAcA sugar acid in characteristic surface antigens (15). The analogous hexosamine pathway in the archaea has not been demonstrated.
FIG. 1.
Eukaryotes use the Leloir pathway to produce UDP-GlcNAc (22). The yeast Gfa1p protein is a d-Fru-6-P amidotransferase that produces GlcN-6-P. The Gna1p acetyltransferase produces N-acetylglucosamine 6-phosphate (GlcNAc-6-P), which can be converted to GlcNAc-1-P by the Agm1p (Pcm1p) phosphomutase. Finally, the Uap1p (Qri1p) uridylyltransferase produces UDP-GlcNAc. Yeast cells can also incorporate exogenous glucosamine, using separate acetyltransferase and kinase enzymes. Bacteria use the GlmS amidotransferase, the GlmM phosphomutase, and the bifunctional GlmU acetyltransferase/uridylyltransferase enzymes to produce UDP-GlcNAc (21). The WecB epimerase and the WecC dehydrogenase make UDP-ManNAcA for enterobacterial common-antigen biosynthesis (in some gram-negative proteobacteria) and capsule formation (in some gram-positive bacteria).
The proteins in this pathway have homologs that use different substrates; therefore, current genome sequence annotations do not accurately describe the genes’ functions. For example, most archaeal genomes include homologs of the proteobacterial glucosamine-6-phosphate phosphomutase (GlmM) (see Fig. S1 in the supplemental material) (14). However, members of this phosphohexomutase family catalyze the isomerization of different sugar phosphates, making it difficult to predict substrate specificity (34). The previously characterized archaeal homologs from Pyrococcus horikoshii (2), Sulfolobus solfataricus (32), and Thermococcus kodakaraensis (31) have both phosphoglucomutase and phosphomannomutase activities but lack significant phosphoglucosamine phosphomutase activity. Another member of this family, from T. kodakaraensis, has phosphopentomutase activity (30). Therefore, no archaeal GlmM protein had been identified.
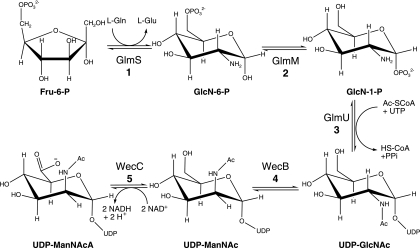
To identify archaeal proteins involved in hexosamine biosynthesis, we compared the sequences of known eukaryotic and bacterial acetamido sugar-biosynthetic enzymes to the set of predicted proteins from Methanococcus maripaludis and Methanocaldococcus jannaschii. No homologs of the eukaryotic GNA1, AGM1, or UAP1 proteins were conserved in the methanogens. In contrast, we identified proteins that could catalyze each reaction in the bacterial pathway. Four of the five targeted genes have paralogs in the Methanococcales, so phylogenetic distribution and gene cluster analyses were key to distinguishing homologs involved specifically in this pathway. Candidate genes were cloned from M. maripaludis or M. jannaschii and heterologously expressed in Escherichia coli, and the proteins were purified to homogeneity. The results from biochemical assays and spectroscopy confirmed the predicted reactions catalyzed by the following proteins. The MMP1680 protein catalyzed the isomerization of Fru-6-P and its transamination from the l-glutamine amide, producing GlcN-6-P (Fig. 2). The MMP1077 protein had phosphoglucosamine phosphomutase activity. The MJ1101 enzyme catalyzed the acetylation of the 2-amino group of glucosamine 1-phosphate using Ac-CoA and the transfer of N-acetylglucosamine-1-phosphate (GlcNAc-1-P) to UTP, releasing pyrophosphate and UDP-GlcNAc. Subsequently, the MMP0705 protein catalyzed the isomerization of UDP-GlcNAc to produce UDP-N-acetyl-α-d-mannosamine (UDP-ManNAc). Finally, the MMP0706 protein catalyzed the four-electron oxidation of UDP-ManNAc, reducing NAD+ and releasing UDP-ManNAcA. These results show that methanogens and most euryarchaea use the bacterial-type pathway to produce UDP-GlcNAc and UDP- ManNAcA as substrates for glycosyltransferases.
FIG. 2.
Biosynthesis of UDP-ManNAcA in the Methanococcales. The isomerizing glutamine-Fru-6-P transaminase (GlmS; EC 2.6.1.16; MJ1420 or MMP1680) catalyzes the isomerization of Fru-6-P and its transamination from the l-glutamine amide, producing GlcN-6-P. Phosphoglucosamine mutase (GlmM; EC 5.4.2.10; MJ1100 or MMP1077) catalyzes the transfer of phosphate from GlcN-6-P to the anomeric position, forming α-d-glucosamine-1-phosphate (GlcN-1-P). The bifunctional GlcN-1-P uridylyltransferase/acetyltransferase (GlmU; EC 2.7.7.23/2.3.1.57; MJ1101 or MMP1076) catalyzes the acetylation of the 2-amino group using acetyl-CoA and the transfer of GlcNAc-1-P to UTP, releasing pyrophosphate and UDP-GlcNAc. The UDP-GlcNAc 2-epimerase (WecB; EC 5.1.3.14; MJ1504 or MMP0705) catalyzes the isomerization of UDP-GlcNAc to produce UDP-ManNAc. Finally, the UDP-ManNAc 6-dehydrogenase (WecC; EC 1.1.1.-; MJ0468 or MMP0706) catalyzes the four-electron oxidation of UDP-ManNAc, reducing NAD+ and releasing UDP-ManNAcA.
MATERIALS AND METHODS
Chemicals and reagents.
UDP sugars and hexose phosphates were purchased from Sigma-Aldrich. Oligonucleotides were purchased from Operon, and PrimeStar DNA polymerase was from Takeda. Restriction enzymes, Vent DNA polymerase, and thermostable pyrophosphatase were from New England Biolabs. Rabbit muscle lactate dehydrogenase and glucose-6-phosphate dehydrogenase enzymes were from USB. Other reagent grade chemicals were purchased from various distributors and used without further purification.
Strains.
M. maripaludis 900 was a gift from John Leigh (University of Washington) (24), and M. jannaschii JAL-1 DSM 2661 was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen. Cultures were grown anaerobically using a modified autolithotrophic growth medium (26).
Cloning and molecular biology.
The MMP1680 gene was amplified by PCR using the oligonucleotide primers 5MMP1680BN and 3MMP1680B (see Table S1 in the supplemental material) and M. maripaludis chromosomal DNA. The product was cloned in the NdeI and BamHI sites of vector pET-19b to produce vector pDG348 and into the same sites of vector pET-11a to produce pDG430 (see Table S2 in the supplemental material). The MMP1077 gene was similarly amplified by PCR using primers 5MMP1077BN and 3MMP1077B. The product was cloned into pET-19b to produce pDG432 and into pET-11a to produce pDG435. The MJ1100 ortholog of MMP1077 was amplified from M. jannaschii chromosomal DNA using primers 5MJ1100BN and 3MJ1100B. The product was cloned into pET-19b to produce pDG301, and the PCR product was cloned into pET-11a to produce pDG431. The MJ1101 gene was amplified from M. jannaschii DNA using primers 5MJ1101BN and 3MJ1101B, and the product was cloned into pET-19b to produce pDG300. The genes at loci MMP0706 and MMP0705 are adjacent on the chromosome of M. maripaludis, oriented in the same direction, and separated by only 5 base pairs (13). Oligonucleotide primers 5MMP0706KNc and 3MMP0705X were used to amplify a DNA segment containing both genes from M. maripaludis. The PCR product was cloned into KpnI and XhoI sites in plasmid pET-30a to create pDG295. The MMP0705 gene was separately amplified using primers 5MMP0705N and 3MMP0705X. The PCR product was cloned into NdeI and XhoI sites of pET-43.1b to produce pDG327. The MMP0706 gene was separately amplified using primers 5MMP0706KNc and 3MMP0706H; the PCR product was cloned into NcoI and HindIII sites of pET-30a to create pDG308.
Plasmids were propagated in E. coli DH5α cells. Recombinant DNA was sequenced at the Institute for Cellular and Molecular Biology Core Laboratories DNA Sequencing Facility (University of Texas at Austin). Except for the MJ1100 and MMP0706 genes, the cloned DNA sequences were identical to those reported by the genome-sequencing projects (4, 13). In plasmid pDG308, a single base change of T945G in MMP0706 is predicted to be a silent mutation. In pDG301, a transversion mutation, C625A, is predicted to cause a Leu209Ile substitution in the MJ1100 protein. Because the C625A variant was found in clones from two independent PCR products (obtained using different DNA polymerases), the variation appears to be a natural polymorphism.
Protein expression and purification.
The polyhistidine-tagged proteins His10-MJ1100, His10-MMP1077, His10-MJ1101, MMP0705-His6, and His6-MMP0706 were heterologously expressed in E. coli BL21(DE3) strains transformed with the respective expression vectors (see Table S2 in the supplemental material). The cells were grown with continuous shaking at 37°C (250 rpm) in Luria-Bertani broth with the antibiotic ampicillin (100 μg ml−1) or kanamycin (50 μg ml−1) to an optical density at 600 nm of 0.6 to 0.8. Protein expression was induced by adding 1% (wt/vol) α-d-lactose, and then the cells were grown for an additional 3 h. The cells were harvested by centrifugation at 4°C; cell lysis and Ni2+ affinity chromatography were performed using standard methods (8). Cell lysate containing the His10-MJ1101 protein was heated at 70°C for 10 min, followed by centrifugation at 14,000 × g for 10 min at 4°C before affinity purification.
The untagged MMP1680 protein was expressed in E. coli BL21(DE3)(pDG430), and cells were lysed in 50 mM TES [N-[tris(hydroxymethyl)methyl]-2-aminoethanesulfonate]-NaOH (pH 7.2). Streptomycin sulfate (2.5% wt/vol) was added to the crude extract with stirring at 4°C for 35 min, followed by centrifugation at 23,000 × g for 20 min at 4°C. The MMP1680 protein was purified by loading the supernatant onto a HiPrep 16/10 Q Sepharose Fast Flow (20 ml) column (GE Healthcare), which was equilibrated with binding buffer consisting of 20 mM Tris-HCl (pH 8.0) using a flow rate of 5 ml min−1. Protein elution was achieved using a linear gradient from 0 to 100% elution buffer (binding buffer with 1 M NaCl) over 40 min.
Protein concentrations were measured using the Bradford protein assay with bovine serum albumin (fraction V) as a standard. After the addition of 1 mM dithiothreitol (DTT) or tris-(2-carboxyethyl)phosphine hydrochloride (TCEP), most of the purified proteins were stored in 20% glycerol at −80°C. The His10-MJ1101 protein was stored at 4°C. The MMP1680 protein was stored in 20% glycerol at −80°C without reductant; this protein lost significant activity after 5 days. For all enzymes, 1 unit of enzymatic activity catalyzed the conversion of 1 μmol substrate to product per min.
Protein mass measurements.
Apparent molecular masses of purified, denatured proteins were measured by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 12% acrylamide and 2.7% cross-linker (18). Proteins in the gels were stained with Coomassie brilliant blue R-250 dye. Native protein sizes were measured using analytical gel filtration chromatography (12).
Glutamine-Fru-6-P transamidase activity measurements.
Amidotransferase reaction mixtures (100 μl) contained 50 mM potassium phosphate (pH 7.8), 200 mM KCl, 2.5 mM EDTA, 2 mM TCEP, 10 mM l-glutamine, 10 mM Fru-6-P, and 5 μg purified MMP1680 protein. The reaction mixtures were incubated at 37°C; after 20 min, samples of the reaction mixture were removed and derivatized with naphthalene-2,3-dicarboxaldehyde and cyanide. N-substituted 1-cyanobenz[f]-isoindole derivatives were separated by high-performance liquid chromatography (HPLC) and detected using a fluorescence detector as described previously (12). Product concentrations were determined by standard addition.
Phosphohexomutase activity measurements.
Activity in the reverse direction (converting a hexose 1-phosphate to hexose 6-phosphate) was measured in continuous coupled assays using glucose 6-phosphate dehydrogenase (14). Activity in the forward direction (converting GlcN-6-P to GlcN-1-P) was measured in discontinuous coupled assays using the MJ1101 acetyltransferase/uridylyltransferase enzyme (see the supplemental material).
Acetyltransferase and nucleotidyltransferase activity measurements.
A discontinuous HPLC assay was used to detect UDP sugar production. Reaction mixtures (30 μl) contained 0.5 mM UTP, 0.5 mM Ac-CoA, 1 mM DTT, 5 mM MgCl2, 50 mM Tris-HCl (pH 7.5), 2 units of inorganic pyrophosphatase, and 50 ng of purified His10-MJ1101 protein. The mixtures were preincubated at 50°C for 2 min, followed by addition of 0.5 mM GlcN-1-P substrate. Complete reaction mixtures were incubated for 5 min and then terminated by the addition of EDTA (10 mM at pH 8.0). Nucleotides were detected by analytical HPLC (44). Assays for alternative nucleotide utilization included GlcN-1-P and 0.5 mM dTTP, dUTP, CTP, ATP, or GTP; assays for sugar phosphate utilization included UTP and 0.5 mM Glc-1-P, mannose-1-phosphate (Man-1-P), galactose-1-phosphate, or galactosamine-1-phosphate. The reverse reaction was determined in assays containing 0.5 mM UDP-GlcNAc, 1 mM pyrophosphate, 1 mM DTT, 5 mM MgCl2, 50 mM Tris-HCl (pH 7.5), and 50 ng His10-MJ1101 in a 30-μl reaction mixture. UTP production was estimated from the integrated peak area of the chromatogram. Electrospray ionization-liquid chromatography-mass spectrometry (ESI-LC-MS) was performed by the Mass Spectrometry Facility in the Department of Chemistry and Biochemistry at the University of Texas at Austin. An ion pair gradient with triethylamine and 5% acetonitrile to 100% acetonitrile was used to elute compounds from a reversed-phase column. ESI-MS was performed in the negative ion mode.
UDP-GlcNAc epimerase and dehydrogenase activity measurements.
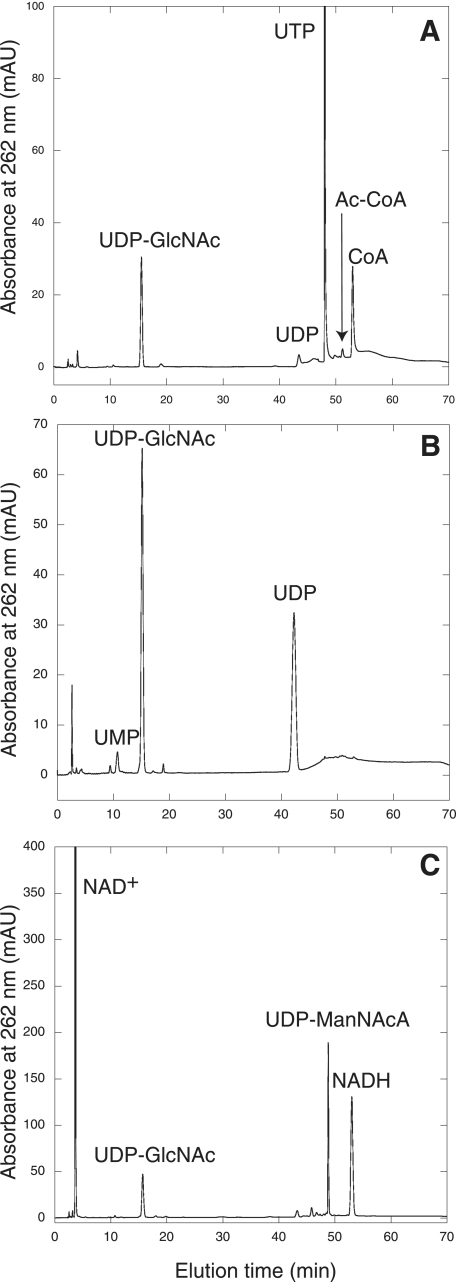
Reaction mixtures (200 μl) containing 1 mM UDP-GlcNAc, 1 mM TCEP, 200 mM KCl, 1 mM NAD+, 100 mM TES-NaOH (pH 8.5), and 80 μg of coexpressed, purified MMP0705-His6 and His6-MMP0706 proteins were incubated at 37°C for 5 h. The nucleotide products were separated using a strong anion-exchange column (5 μm; 150 by 4.6 mm; Phenosphere; Phenomenex) with a guard column (5 μm; 4 by 3 mm). The column was equilibrated with 100 mM ammonium phosphate buffer (pH 5.0). Isocratic elution at 0.7 ml min−1 separated the nucleotides, which were detected by their absorbance at 262 nm and collected in 1-ml fractions. Reaction products separated by HPLC using a CarboPac PA1 column (44) eluted with the following retention times: 3.6 min (NAD+), 43.0 min (UDP), 48.8 min (UDP-ManNAcA), and 53.0 min (NADH). Rates of UDP-GlcNAc epimerization catalyzed by MMP0705 were determined in a continuous coupled assay with the MMP0706 dehydrogenase (see the supplemental material) (16).
Identification of reaction products.
The reaction product of the MMP0705 and MMP0706 enzymes was purified (see the supplemental material) and analyzed by ESI-LC-MS operated in negative ion mode and by high-resolution ESI-MS. For 1H nuclear magnetic resonance (NMR), samples were dried under vacuum and dissolved in D2O with sodium 3-trimethylsilylpropionate-2,2,3,3-d4. Samples for 31P-NMR were treated with alkaline EDTA in D2O and referenced to an external phosphoric acid standard.
UDP-ManNAc dehydrogenase activity assay.
Rates of UDP-ManNAc oxidation to UDP-ManNAcA were measured in the coupled assay described above, using limiting amounts of MMP0706. Reaction mixtures (300 μl) containing 1 mM UDP-GlcNAc, 1 mM TCEP, 200 mM KCl, 100 mM TES-NaOH (pH 8.5), 30 μg MMP0705-His6, and 2 μg His6-MMP0706 were preincubated at 37°C for 4 min before they were initiated with 3 mM NAD+. Reactions initiated with 0.5 to 3 mM NAD+ provided the initial rate data, which were obtained from the linear part of the reaction progress curve. Apparent steady-state kinetic constants were measured by fitting the initial rate data to the Michaelis-Menten-Henri equation.
Stoichiometry of the dehydrogenase reaction.
A reaction mixture containing 1 mM UDP-GlcNAc, 1 mM TCEP, 3 mM NAD+, 200 mM KCl, 100 mM TES-NaOH (pH 8.5), 50 μg MMP0705-His6, and 100 μg His6-MMP0706 was incubated at 37°C for 4.5 h. The analytes were separated by analytical anion-exchange chromatography using a gradient elution scheme from 5 mM ammonium phosphate (pH 2.8) to 750 mM ammonium phosphate (pH 3.7) over 20 min, maintaining the final buffer conditions for 5 min at a flow rate of 0.7 ml min−1. NAD+ and UDP-ManNAcA concentrations were determined by the method of standard addition.
Inhibitor screening.
For UDP-GlcNAc epimerase inhibition assays, epimerase reactions were initiated with a mixture containing 0.4 mM UDP-GlcNAc and potential inhibitors. For UDP-ManNAc dehydrogenase inhibition assays, a 1-ml reaction mixture containing 5 mM UDP-GlcNAc, 5 mM TCEP, 200 mM KCl, and 150 μg MMP0705-His6 in 100 mM TES-NaOH (pH 7.5) was incubated at 37°C overnight. The reaction mixture was then centrifuged at 7,500 × g for 50 min at 4°C using a 10-kDa molecular mass cutoff ultrafiltration device (Microcon). Reaction mixtures containing 1 mM of a UDP-GlcNAc and UDP-ManNAc mixture, potential inhibitor (0.3 mM to 3 mM), 1 mM TCEP, and 2 μg His6-MMP0706 in 100 mM TES-NaOH (pH 8.5) were initiated with 3 mM NAD+.
Accession numbers.
The C625A variant sequence was submitted to GenBank with accession number EU430076. The RefSeq accession numbers for the proteins described here are MMP1680 (NP_988800.1), MMP1077 (NP_988197.1), MJ1101 (NP_248094.1), MJ1100 (NP_248093.1), MMP0705 (NP_987825.1), and MMP0706 (NP_987826.1).
RESULTS
Glucosamine-6-phosphate synthase activity of MMP1680.
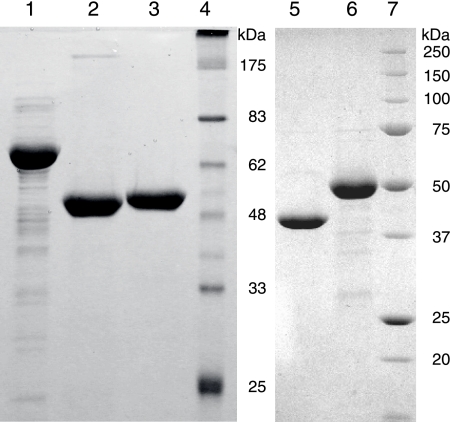
The M. maripaludis protein at locus MMP1680 is 39% identical to the previously characterized E. coli glucosamine-6-phosphate synthase, GlmS (38). Proteins in this isomerizing amidotransferase family are highly conserved throughout the bacterial, eukaryotic, and archaeal domains, although no archaeal homolog has been characterized. E. coli BL21(DE3) cells express the untagged MMP1680 protein from plasmid pDG430 at high levels of soluble protein. Protein purified by ion-exchange chromatography had an apparent molecular mass of 61 kDa measured by SDS-PAGE (Fig. 3), close to its predicted mass of 67 kDa. This protein eluted from a size exclusion column with an apparent molecular mass of 171 kDa and a Stokes radius of 45 Å, consistent with a dimeric structure.
FIG. 3.
Purified proteins analyzed by SDS-PAGE on 12% acrylamide gels and stained with Coomassie blue. Lane 1, 10 μg of MMP1680; lane 2, 8.5 μg of His10-MMP1077; lane 3, 8 μg of His10-MJ1101; lane 4, protein markers corresponding to the indicated molecular masses; lane 5, 8 μg of MMP0705-His6; lane 6, 10 μg of His6-MMP0706; lane 7, protein markers.
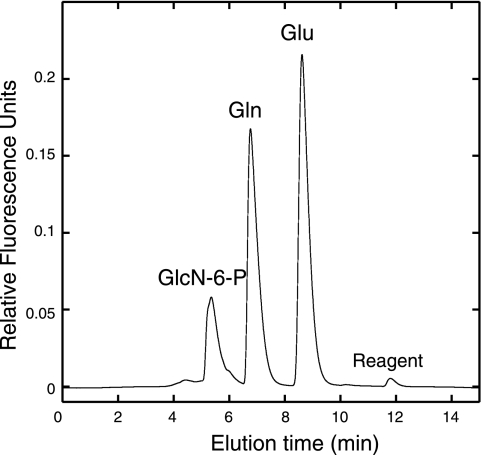
In reactions containing Fru-6-P and l-glutamine, purified MMP1680 protein catalyzed transamidation and epimerization reactions that produced GlcN-6-P and glutamate (Fig. 4). 1H-NMR analysis of the reaction product (400 MHz, D2O) showed a characteristic resonance of the anomeric proton of GlcN-6-P at 5.24 ppm (d, J = 3.3 Hz). Proton-decoupled 31P-NMR analysis (162 MHz, D2O) of base-treated reaction mixtures showed peaks corresponding to d-glucosamine-6-phosphate (5.75 ppm, s) and d-Fru-6-P (5.12 ppm, s). The enzyme was active over a pH range of 6.5 to 7.8, and activity was enhanced by potassium chloride and TCEP reductant. At pH 7.8 and saturating substrate concentrations, the enzyme had a specific activity of 0.59 ± 0.08 U mg−1, corresponding to a rate of 0.66 s−1. An amino-terminal polyhistidine-tagged protein had no catalytic activity, presumably because the tag interfered with the catalytic Cys2 residue. Since the archaeal GlmS protein sequences are remarkably similar to well-studied bacterial sequences, no further characterization experiments were performed.
FIG. 4.
The MMP1680 protein catalyzes the isomerization and transamidation of Fru-6-P to form GlcN-6-P. Amines from reactions containing purified protein, l-glutamine (Gln), and Fru-6-P were derivatized to form fluorescent 1-cyanobenz[f]-isoindole derivatives of GlcN-6-P, Gln, and glutamate (Glu) products, which were separated by reversed-phase HPLC. Reactions without MMP1680 protein showed only a Gln peak, but no GlcN-6-P or Glu products. The experimental conditions are described in the text.
GlmM activity of MMP1077.
The genomes of M. maripaludis and M. jannaschii encode homologs of putative phosphohexomutases in genes adjacent to bifunctional nucleotidyltransferase genes (described below). This MJ1100 protein formed large aggregates and did not catalyze the phosphohexomutase reaction in standard reactions with α-d-glucose-6-phosphate (Glc-6-P), mannose-6-phosphate, or GlcN-6-P, consistent with previous results (41). To circumvent this aggregation problem, we expressed the homologous protein from M. maripaludis (MMP1077). The His10-MMP1077 protein migrated with an apparent molecular mass of 50 kDa by SDS-PAGE, close to its predicted mass of 52 kDa (Fig. 3). Nickel affinity purification separated this protein from the highly expressed E. coli GlmM protein (14). Analytical size exclusion chromatography showed that most of the His10-MMP1077 protein formed a 340-kDa aggregate, while 11% of the protein eluted in monomeric form with an apparent molecular mass of 51 kDa and a Stokes radius of 31 Å.
Phosphohexomutase activity was measured in coupled reactions containing His10-MMP1077 and Glc-6-P dehydrogenase (Table 1). The initial rate of conversion of GlcN-1-P to GlcN-6-P (0.52 s−1) was fivefold higher than the rate of conversion of Glc-1-P to Glc-6-P (0.10 s−1), assuming all of the protein was catalytically active. Fructose-1,6-bisphosphate was required to activate the enzyme. Activity was 90% lower when glucose-1,6-bisphosphate was used instead, and no activity was observed with ATP as an activator. Activity was 95% lower when MgCl2 was used instead of MnCl2, and 1 mM DTT was required in reactions for maximal activity.
TABLE 1.
Specific activities of phosphohexomutase homologs
| Enzyme | Sp act on substratea:
|
Reference | ||
|---|---|---|---|---|
| GlcN-6-P | GlcN-1-P | Glc-1-P | ||
| His10-MMP1077b | 0.037 ± 0.006 | 0.60 ± 0.09 | 0.12 ± 0.02 | |
| S. solfataricus PMM/PGM-His6 | - | - | 0.0026 | 32 |
| P. aeruginosa His6-GlmM | 2.5 | ND | 0.06 | 37 |
| E. coli His6-GlmM | 3 | 10 | 0.007 | 14 |
Specific activities are shown in units of μmol substrate min−1 mg−1. -, not detected; ND, not determined.
Data from this work.
In the forward, physiologically relevant direction, GlcN-6-P phosphomutase activity was measured in coupled assays with His10-MMP1077, the bifunctional His10-MJ1101 protein described below, and inorganic pyrophosphatase. Together, these enzymes catalyzed the conversion of GlcN-6-P to UDP- GlcNAc at a rate of 1.9 min−1 (Fig. 5A and Table 1). In contrast to results from the reverse reaction, activation of MMP1077 was 12-fold lower when fructose-1,6-bisphosphate was used instead of glucose-1,6-bisphosphate, and MgCl2 was required for activity; it could not be replaced by MnCl2. A control reaction without any MMP1077 protein did not indicate any UDP-GlcNAc formation.
FIG. 5.
Chromatogram showing the separation of nucleotides by HPLC on a CarboPac PA1 column. (A) Reactions containing His10-MMP1077, His10-MJ1101, UTP, Ac-CoA, GlcN-6-P, and pyrophosphatase produced UDP-GlcNAc and CoA. No UDP-GlcNAc or CoA was observed in reactions without His10-MMP1077. (B) The MMP0705-His6 protein catalyzed the epimerization of UDP-GlcNAc to form UDP-ManNAc (not resolved) and UDP, an intermediate in the epimerase reaction formed by the enzymatic cleavage of UDP sugars. No UDP or early-eluting product was detected in control reactions without MMP0705-His6 protein. (C) Reactions containing MMP0705-His6, His6-MMP0706, UDP-GlcNAc, and NAD+ produced UDP-ManNAcA and two equivalents of NADH. Reactions without the His6-MMP0706 protein did not produce UDP-ManNAcA or NADH. The minor unlabeled peaks in the chromatograms were detected in both enzymatic and control reactions. The full experimental conditions are described in the text.
Bifunctional acetyltransferase and nucleotidyltransferase activities of MJ1101.
In both M. maripaludis and M. jannaschii, the phosphoglucosamine phosphomutase gene is adjacent to a putative nucleotidyltransferase gene on the chromosome. Thermostable nucleotidyltransferases are desirable for the enzymatic synthesis of UDP sugars (23); therefore, we cloned and expressed the MJ1101 nucleotidyltransferase homolog from the hyperthermophilic M. jannaschii. The MJ1101 protein is 68% identical and 90% similar to its M. maripaludis ortholog (MMP1076). These genes are homologous to the E. coli glmU gene, which encodes a bifunctional protein catalyzing the acetylation of GlcN-1-P and the transfer of the acetamido sugar to UTP, producing UDP-GlcNAc and pyrophosphate (21). However, the more closely related Sulfolobus tokodaii homolog was only shown to catalyze UDP-GlcNAc formation from UTP and GlcNAc-1-P; no acetyltransferase activity was reported (45). The S. tokodaii enzyme also used a variety of nucleoside triphosphate substrates for the activation of Glc-1-P or GlcNAc-1-P. Another archaeal homolog, from P. furiosus, has nucleotidyltransferase activity with a broad range of sugar-1-phosphates (23); this enzyme is more similar to the M. jannaschii MJ1334 paralog (49% identical) than the MJ1101 protein (31% identical).
To test the specificity and catalytic activities of this enzyme, the amino-terminal decahistidine-tagged MJ1101 protein (His10-MJ1101) was heterologously expressed in E. coli and purified to homogeneity by affinity chromatography (Fig. 3). This thermostable protein had an apparent molecular mass of 51 kDa, close to its expected mass of 49 kDa. His10-MJ1101 eluted from an analytical size exclusion column with an apparent molecular mass of 203 kDa and a Stokes radius of 51 Å. This method provided no evidence for association of the MJ1100 and MJ1101 proteins.
In reaction mixtures containing Glc-1-P and UTP at 50°C, the enzyme catalyzed UDP-Glc formation. The MJ1101 enzyme was active across a wide pH range, demonstrating optimal activity at pH 7.5. The enzyme showed maximal activity from 50 to 60°C; the activity was 22% lower at 80°C and 60% lower at 100°C. Replacement of UTP with dUTP or dTTP resulted in a fourfold decrease in activity (Table 2), but no activity was observed using CTP, ATP, or GTP as the nucleotide substrate. The enzyme activity was fivefold higher with GlcN-1-P as the substrate. However, the addition of Ac-CoA to the reaction stimulated activity 30-fold, suggesting that acetylation precedes transfer to UTP, as observed for the bacterial enzyme. Reactions containing GlcN-6-P with His10-MMP1077, UTP, pyrophosphatase, Ac-CoA, and His10-MJ1101 produced UDP-GlcNAc and CoA (Fig. 5A). GlcN-1-P could replace GlcN-6-P and His10-MMP1077, simplifying the reaction. Neither galactose-1-phosphate nor galactosamine-1-phosphate could replace GlcN-1-P in this reaction, although Man-1-P could be activated to produce UDP-Man after extended incubation with excess Man-1-P.
TABLE 2.
Reactions catalyzed by MJ1101
| Substrates | Products | Sp act (μmol min−1 mg−1) |
|---|---|---|
| Forward reactiona | ||
| GlcN-1-P, UTP | UDP-GlcN, PPi | 0.25 ± 0.07 |
| GlcN-1-P, AcCoA, UTP | UDP-GlcNAc, PPi, CoA | 7.74 ± 1.37 |
| GlcN-1-P, AcCoA, dTTP | dTDP-GlcNAc, PPi, CoA | 2.64 ± 0.05 |
| GlcN-1-P, AcCoA, dUTP | dUDP-GlcNAc, PPi, CoA | 2.10 ± 0.14 |
| Glc-1-P, UTP | UDP-Glc, PPi | 1.38 ± 0.16 |
| Glc-1-P, dTTP | dTDP-Glc, PPi | 0.23 ± 0.02 |
| Reverse reactionb | ||
| UDP-GlcNAc, PPi | GlcNAc-1-P, UTP | 12.51 ± 0.18 |
| UDP-Glc, PPi | Glc-1-P, UTP | 8.40 ± 1.20 |
Thirty-microliter reaction mixtures containing saturating substrate concentrations (0.5 mM of nucleotide triphosphate and 0.5 mM Ac-CoA) with 1 mM DTT, 5 mM MgCl2 , 50 mM Tris-HCl (pH 7.5), 2 units of inorganic pyrophosphatase, and 50 ng of His10-MJ1101 were preincubated at 50°C for 2 min before the reaction was started by the addition of 0.5 mM sugar phosphate substrate. After 5 min of incubation at the same temperature, the reactions were stopped by the addition of 10 mM EDTA. Nucleotide products were detected by HPLC as described previously.
Assays were performed in a manner similar to forward reactions. The 30-μl reaction mixtures contained 0.5 mM UDP-GlcNAc, 1 mM pyrophosphate, 1 mM DTT, 5 mM MgCl2, 50 mM Tris-HCl (pH 7.5), and 50 ng His10-MJ1101. UTP product formation was determined using HPLC.
In reactions containing pyrophosphate and UDP-GlcNAc, the MJ1101 enzyme readily catalyzed the reverse reaction, forming GlcNAc-1-P and UTP. LC-MS analysis of the reaction products identified peaks at 606 and 628 m/z, corresponding to the UDP-GlcNAc substrate ions [M − H]− and [M − 3H + Mg]−, as well as peaks at 483 and 505 m/z, corresponding to UTP ions [M − H]− and [M − 3H + Mg]−, and a peak at 300 m/z, corresponding to GlcNAc-1-P [M—H]−.
UDP-GlcNAc 2-epimerase activity of MMP0705.
The M. maripaludis MMP0705 protein shares 36% amino acid identity with the E. coli UDP-GlcNAc 2-epimerase protein WecB, which is required for enterobacterial common-antigen biosynthesis (15, 25). These proteins belong to a family of phosphoglycosyl transferases (6). Carboxy-terminally hexahistidine-tagged MMP0705 (MMP0705-His6) was heterologously expressed as a soluble protein in E. coli with an apparent molecular mass of 40 kDa, close to its expected mass of 42.4 kDa. The affinity-purified protein was 99% pure, as judged by SDS-PAGE (Fig. 3). Analytical size exclusion chromatography of the MMP0705-His6 protein identified a single species with an apparent molecular mass of 101 kDa and a Stokes radius of 37 Å, which could be a dimeric form of the protein.
Reactions containing MMP0705-His6 protein with UDP- GlcNAc in D2O were monitored using 1H-NMR to detect UDP-GlcNAc epimerization (25). After 18 h of incubation at 37°C, these reactions showed a 21% decrease in intensity for the resonance of the UDP-GlcNAc anomeric proton at 5.53 ppm (dd, J = 7.2, 3.3 Hz) and the appearance of a new low-intensity peak at 5.47 ppm (d, J = 7.4 Hz), characteristic of the anomeric proton of [2"-2H]-UDP-ManNAc (25). A peak at 5.52 ppm (d, J = 7.1 Hz) also indicated the presence of [2"-2H]-UDP-GlcNAc. These results confirm that MMP0705 catalyzes the reversible epimerization of UDP-GlcNAc to produce UDP-ManNAc. The UDP-ManNAc product could not be separated from UDP-GlcNAc substrate by our HPLC method. However, incubation of MMP0705-His6 protein with UDP-GlcNAc also produced UDP and a new product that eluted at 2.6 min with a maximal absorbance at 242 nm (Fig. 5B). This compound may be 2-acetamidoglucal, which is an intermediate in the proposed elimination reaction (25). Insufficient amounts of this early-eluting compound were obtained for further characterization.
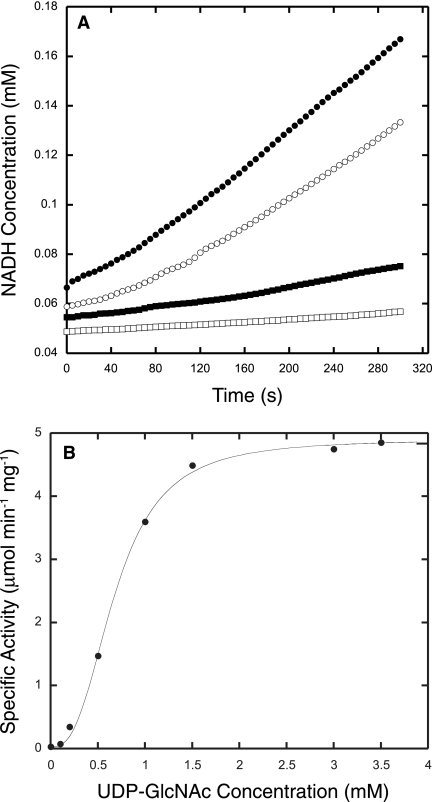
The rate of UDP-GlcNAc epimerase activity catalyzed by MMP0705 was measured by adding limiting amounts of MMP0705-His6 and an excess of MMP0706 dehydrogenase (described below) in a coupled assay that measured the production of NADH (Fig. 6A) (15). The enzyme showed maximal activity at 50°C, although standard assays were done at 37°C. The protein was active over a wide pH range from 6 to 10; therefore, the coupled assays were performed at pH 8.5. Epimerase activity was unaffected by the metal ions, but the addition of 200 mM KCl stimulated activity twofold. A plot of initial rates at various substrate concentrations showed a sigmoidal relationship, so the steady-state parameters were obtained by fitting the data to the Hill equation (Fig. 6B). Such an analysis resulted in a Hill coefficient of 2.7, indicating positive cooperativity. The KM for UDP-GlcNAc was 0.36 mM, and the kcat was 3.4 s−1. These parameters are similar to those reported for the E. coli protein: a Hill coefficient of 2.3, a KM of 0.73 mM, and a kcat of 4.8 s−1 (25). M. maripaludis encodes a paralog of MMP0705 at locus MMP0357; however, purified His10-MMP0357 protein had no detectable UDP-GlcNAc 2-epimerase activity (unpublished data).
FIG. 6.
A coupled assay measured the UDP-GlcNAc 2-epimerase activity of MMP0705. (A) Reaction progress curves show activities for coupled epimerase and dehydrogenase reactions containing 3 mM UDP-GlcNAc (filled circles) and 1.5 mM UDP-GlcNAc (open circles). Control reactions without MMP0705 enzyme (filled squares) or without UDP-GlcNAc (open squares) show background levels of activity. The reaction mixtures contained 3 to 5 mM NAD+, 0.4 μg MMP0705-His6, and 150 μg His6-MMP0706 with buffer salts, as described in the text. The reduction of NAD+ to NADH was monitored by following the increase in absorbance at 340 nm. (B) Hill plot of the rate of MMP0705-catalyzed epimerization of UDP-GlcNAc. Initial rate data were fitted to the Hill equation by nonlinear regression. The Hill coefficient was 2.7, the KM for UDP-GlcNAc was 0.36 mM, and the kcat was 3.4 s−1.
UDP-ManNAc dehydrogenase activity of MMP0706.
The MMP0706 gene lies adjacent to the MMP0705 UDP-GlcNAc 2-epimerase gene in M. maripaludis. The MMP0706 protein is homologous to the UDP-ManNAc dehydrogenase (WecC) from E. coli (15, 25), as well as the UDP-Glc dehydrogenase (Ugd in E. coli), the UDP-N-acetylgalactosamine dehydrogenase, and the GDP-mannose dehydrogenase. To identify the substrate specificity of the MMP0706 protein and to distinguish it from the paralogous MMP0353 protein, we assayed the dehydrogenase activities of purified protein.
The amino-terminally hexahistidine-tagged MMP0706 protein (His6-MMP0706) was expressed in E. coli as a soluble protein with an apparent molecular mass of 50 kDa, close to its expected mass of 51.4 kDa. Protein purified by affinity chromatography was substantially pure (Fig. 3). This protein had an apparent native molecular mass of 193 kDa and a Stokes radius of 46 Å, consistent with a tetrameric form. A mixture of the MMP0705 and MMP0706 proteins showed no evidence of association between the two proteins during chromatography. Reactions containing MMP0705-His6, His6-MMP0706, UDP-GlcNAc, and NAD+ produced NADH and a new, late-eluting nucleotide sugar (Fig. 5C). This compound was purified using preparative anion-exchange chromatography. Analytical ion-exchange chromatography indicated that the product was 92% pure. ESI-LC-MS analysis of the compound identified a 620 m/z base peak, corresponding to the UDP-ManNAcA molecular ion (M−), and a 403 m/z peak, corresponding to the UDP fragment. Tandem MS of the ion with 620 m/z identified the following peaks: 523 m/z (UMP-ManNAcA), 403 m/z (UDP), and 385 m/z (UDP-H2O). The loss of an internal phosphate from UDP-ManNAcA due to a rearrangement of the phosphate-N-acetylmannosamine moiety is consistent with previous studies of UDP-GlcNAc (20). High-resolution ESI-MS analysis of the molecular ion was consistent with the structural assignment of UDP-ManNAcA: expected, 620.05356 m/z for C17H24N3O18P2−; observed, 620.0517 m/z. The 1H-NMR spectrum was consistent with a low-resolution spectrum of UDP-ManNAcA published previously (see the supplemental material) (42).
To determine the ratio of NAD+ molecules reduced per molecule of UDP-ManNAcA produced in the dehydrogenase reaction, MMP0705 and MMP0706 proteins were incubated with UDP-GlcNAc and excess NAD+. The products were analyzed by anion-exchange chromatography, and NAD+ and UDP-ManNAcA were quantified by the method of standard addition. A ratio of 1.95 NAD+ molecules consumed per UDP-ManNAcA molecule produced confirmed that two equivalents of NAD+ are required for the four-electron oxidation. UDP-ManNAc dehydrogenase activity was measured in coupled reactions containing an excess of MMP0705 and limiting amounts of MMP0706. Dehydrogenase activity was 76% lower in the absence of any reducing agent than that in the presence of 1 mM TCEP. Salts or divalent cations were not required for the dehydrogenase activity. No appreciable activity was detected when NAD+ was replaced with NADP+ in the coupled reaction. Also, UDP-glucose or N-acetyl-d-mannosamine could not replace UDP-GlcNAc in the coupled assay. In contrast to the epimerase activities, initial rates of NADH production at various NAD+ concentrations fit the hyperbolic Michaelis-Menten-Henri equation. The kcat was 0.4 s−1, and the apparent KM for NAD+ was 0.57 mM for this reaction.
The dehydrogenase activity of MMP0706 in reactions containing a 1 mM mixture of UDP-GlcNAc and UDP-ManNAc was reduced 50% by the following compounds: 0.07 mM ManNAc, 0.5 mM UDP, 0.6 mM UDP-glucose, 1 mM UTP, and 1.3 mM GlcNAc. This inhibition profile supports the specificity of the MMP0706 protein: it binds the separate ManNAc and UDP components in preference to GlcNAc or UTP. For MMP0705 epimerase inhibition assays, the reaction mixtures contained limiting amounts of MMP0705-His6, an excess of His6-MMP0706, and 0.4 mM UDP-GlcNAc in the coupled assay. The compounds that reduced activity in this assay by 50% included 0.3 mM UDP, 0.6 mM UTP, 0.8 mM GlcNAc, 0.8 mM UDP-glucose, and 3 mM d-glucosamine. Other nucleotides, including UMP, ATP, CTP, GTP, GDP, and the sugars d-glucose, d-mannose, and ManNAc, did not cause any noteworthy inhibition. Therefore, both the epimerase and dehydrogenase enzymes specifically bind their UDP sugar substrates. They are only moderately inhibited by substrate fragments, and they discriminate against epimers of these fragments.
DISCUSSION
We have established that hexosamine biosynthesis in the euryarchaea follows the bacterial rather than the eukaryotic pathway. The two pathways differ in the order of acetylation, and the more modular eukaryotic pathway could have evolved to incorporate exogenous GlcNAc. From the results presented here, we can identify orthologous genes required for UDP-acetamido sugar biosynthesis in most methanogens, halobacteria, and Thermococcales (Table 3). These organisms have several homologs of each gene, which made biochemical characterization of the proteins essential for pathway assignment. Although euryarchaea and bacteria share the biosynthetic pathway to UDP-GlcNAc and UDP-ManNAcA shown in Fig. 2, their respective genes evolved separately.
TABLE 3.
Homologs of identified acetamido sugar biosynthesis proteins
| Reaction no.a | Reaction | Protein
|
||||
|---|---|---|---|---|---|---|
| E. coli | M. maripaludis | M. jannaschii | Halobacterium sp. strain NRC-1 | P. furiosus | ||
| 1 | Glutamine-fructose-6-phosphate transaminase | GlmS | MMP1680b | MJ1420c | VNG0006G | PF0157 |
| 2 | Phosphoglucosamine mutase | GlmM | MMP1077b | MJ1100 | VNG2276G | PF0861 |
| 3 | Glucosamine-1-phosphate uridylyltransferase/ acetyltransferase | GlmU | MMP1076 | MJ1101b | VNG0009G | PF0235 |
| 4 | UDP-GlcNAc 2-epimerase | WecB | MMP0705b | MJ1504 | VNG2639G | PF0794 |
| 5 | UDP-ManNAc 6-dehydrogenase | WecC | MMP0706b | MJ0468 | VNG0046G | PF1631 |
| VNG1048G | ||||||
Numbers correspond to reactions shown in Fig. 2.
Activity confirmed in this work.
The MJ1420 protein contains a 499-amino-acid intein.
The glmS glucosamine 6-phosphate synthase gene appears to have been vertically inherited in almost all archaea, as it is in bacteria and eukaryotes. The M. jannaschii GlmS protein contains an intein sequence of about 500 amino acids; all 18 inteins in this organism sit in highly conserved and apparently essential genes. Therefore, amino sugar biosynthesis may be essential in archaea, as it is in E. coli and Saccharomyces cerevisiae. The absence of hexokinase or phosphotransferase system homologs in methanogens implies that they cannot use exogenous GlcNAc. On the other hand, the euryarchaeon T. kodakaraensis contains both a full complement of UDP-GlcNAc biosynthesis genes and a gene cluster for chitin degradation. This cluster includes an exo-β-d-glucosaminidase gene adjacent to a glucosamine-6-phosphate deaminase gene (glmD), a paralog of the glmS glucosamine 6-phosphate synthase gene (36).
In addition to glmS, most euryarchaea have homologs of glmM, which encodes GlcN-6-P phosphomutase. The crenarchaeon S. solfataricus has one copy of glmS in a gene cluster with glmU; however, the only known phosphohexomutase in the organism fails to isomerize GlcN-6-P (32). Therefore, there is a missing step in the S. solfataricus pathway that remains to be identified. GlcN-6-P phosphomutase activity appears to have evolved independently several times. The MMP1077 protein sequence contains the GXEXS amino acid signature motif of GlcN-6-P phosphomutase sugar-binding loops, and it was predicted to have this catalytic activity (34). However, the mode of substrate binding is still poorly understood in this subgroup of phosphohexomutases. Phylogenetic analysis suggests that the archaeal GlcN-6-P phosphomutases are more closely related to archaeal phosphopentomutases and archaeal glucose/mannose phosphomutases, rather than to bacterial GlcN-6-P phosphomutases (see Fig. S1 in the supplemental material). Both the M. jannaschii and M. maripaludis GlcN-6-P phosphomutases formed soluble aggregates of purified protein. The E. coli GlmM was also shown to oligomerize at neutral pH (14). Additional studies will be required to determine the effect of aggregation on activity.
The phosphohexomutases require serine phosphorylation for activation. Hexose 1,6-bisphosphates, which represent the reaction intermediate, are usually added to enzyme preparations to form the active phosphoprotein. Whereas E. coli GlmM was activated with glucose-1,6-bisphosphate in both the forward and reverse reactions, the M. maripaludis protein was better activated by fructose-1,6-bisphosphate in the reverse reaction. Glucose-1,6-bisphosphate is not believed to form in methanogens, and the physiologically relevant activator could be a serine kinase protein, as observed in Streptococcus pneumoniae (27). The amino-terminal domain of the S. pneumoniae protein kinase is distantly related to the carboxy-terminal domain of a euryarchaeal protein (the MMP0004 protein in M. maripaludis), so future studies will test the ability of this protein to activate and regulate GlmM.
The bifunctional GlcN-1-P acetyltransferase/uridylyltransferase (GlmU) protein contains an amino-terminal nucleotidyltransferase domain and a smaller carboxy-terminal acetyltransferase domain (3, 28). Because of its modular structure, GlmU is often underannotated in sequence databases as solely a nucleotidyltransferase. We showed that the M. jannaschii homolog catalyzes both reactions, completing the pathway to UDP-GlcNAc biosynthesis. Based on its 90% sequence similarity, the M. maripaludis ortholog likely has the same function. Previous results suggested that acetylation of GlcN-1-P precedes uridylyltransferase activity, and the data shown here are consistent with that model. Both the M. jannaschii and M. maripaludis genomes contain a cluster of genes that are transcribed in the same orientation and that may constitute an operon: MJ1100 (GlmM), MJ1101 (GlmU), and MJ1102 (dUTP diphosphatase). The dUTP diphosphatase is an important scavenger of nucleotides produced by spontaneous dCTP deamination (19). Although the MJ1101 enzyme can activate GlcNAc-1-P with dUTP or dTTP, the activity is fourfold higher with UTP. The MJ1102 protein has no detectable hydrolytic activity on UDP-GlcNAc or dUDP-GlcNAc.
The UDP-GlcNAc 2-epimerase and dehydrogenase genes extend acetamido sugar biosynthesis in euryarchaea to produce the acidic sugar donor UDP-ManNAcA. Steady-state kinetic analysis showed that MMP0705 epimerase activity is cooperative and specific for UDP-GlcNAc. The specificity constant for the MMP0705 protein (5.7 × 105 M−1 min−1) is comparable to the value reported for the E. coli WecB homolog (3.9 × 105 M−1 min−1) (25). While a full kinetic analysis of the MMP0706 UDP-ManNAc dehydrogenase could not be performed due to a lack of pure UDP-ManNAc, the enzyme's apparent KM for NAD+ is similar to that reported for the E. coli WecC homolog (15). The lower specific activity and turnover of our MMP0706 protein may be due to the presence of inhibitory UDP-GlcNAc in the reaction mixtures. These results support the identification of ManNAcA in methanogen flagellin and coenzyme B. These two genes are broadly, but erratically, distributed among bacterial genomes and may be frequently acquired by horizontal gene transfer. This widespread distribution suggests that ManNAcA is a common sugar on bacterial and archaeal surfaces.
Besides illuminating the evolutionary history of acetamido sugar biosynthesis, our biochemical evidence for the functions of five proteins will improve genome sequence annotations and metabolic reconstruction projects. The results complement genetic studies on archaeal posttranslational glycosylation. Future manipulations of the archaeal surface layer and flagellar glycoproteins could identify the role of oligosaccharides in stabilizing these proteins and engineer new epitope expression systems for glycopeptides.
Supplementary Material
Acknowledgments
This work was supported in part by grants from NIH (AI06444-01), the Petroleum Research Foundation (44382-G4), and the Welch Foundation (F-1576).
We thank Mehdi Moini and Lara Mahal for helpful discussions.
Footnotes
Published ahead of print on 8 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abu-Qarn, M., and J. Eichler. 2006. Protein N-glycosylation in Archaea: defining Haloferax volcanii genes involved in S-layer glycoprotein glycosylation. Mol. Microbiol. 61511-525. [DOI] [PubMed] [Google Scholar]
- 2.Akutsu, J.-I., Z. Zhang, M. Tsujimura, M. Sasaki, M. Yohda, and Y. Kawarabayasi. 2005. Characterization of a thermostable enzyme with phosphomannomutase/phosphoglucomutase activities from the hyperthermophilic archaeon Pyrococcus horikoshii OT3. J. Biochem. 138159-166. [DOI] [PubMed] [Google Scholar]
- 3.Brown, K., F. Pompeo, S. Dixon, D. Mengin-Lecreulx, C. Cambillau, and Y. Bourne. 1999. Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily. EMBO J. 184096-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J.-F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. M. Geoghagen, H. O. Smith, C. R. Woese, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 2731017-1140. [DOI] [PubMed] [Google Scholar]
- 5.Burda, P., and M. Aebi. 1999. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426239-257. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, R. E., S. C. Mosimann, M. E. Tanner, and N. C. J. Strynadka. 2000. The structure of UDP-N-acetylglucosamine 2-epimerase reveals homology to phosphoglycosyl transferases. Biochemistry 3914993-15001. [DOI] [PubMed] [Google Scholar]
- 7.Chaban, B., S. Voisin, J. Kelly, S. M. Logan, and K. F. Jarrell. 2006. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol. Microbiol. 61259-268. [DOI] [PubMed] [Google Scholar]
- 8.Drevland, R. M., A. Waheed, and D. E. Graham. 2007. Enzymology and evolution of the pyruvate pathway to 2-oxobutyrate in Methanocaldococcus jannaschii. J. Bacteriol. 1894391-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichler, J., and M. W. W. Adams. 2005. Posttranslational protein modification in Archaea. Microbiol. Mol. Biol. Rev. 69393-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrante, G., I. Ekiel, and G. D. Sprott. 1986. Structural characterization of the lipids of Methanococcus voltae, including a novel N-acetylglucosamine 1-phosphate diether. J. Biol. Chem. 26117062-17066. [PubMed] [Google Scholar]
- 11.Ghuysen, J.-M., and G. D. Shockman. 1973. Biosynthesis of peptidoglycan, p. 37-130. In L. Leive (ed.), Bacterial membranes and walls. Marcel Dekker, New York, NY.
- 12.Helgadóttir, S., G. Rosas-Sandoval, D. Söll, and D. E. Graham. 2007. Biosynthesis of phosphoserine in the Methanococcales. J. Bacteriol. 189575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrickson, E. L., R. Kaul, Y. Zhou, D. Bovee, P. Chapman, J. Chung, E. Conway de Macario, J. A. Dodsworth, W. Gillett, D. E. Graham, M. Hackett, A. K. Haydock, A. Kang, M. L. Land, R. Levy, T. J. Lie, T. A. Major, B. C. Moore, I. Porat, A. Palmeiri, G. Rouse, C. Saenphimmachak, D. Söll, S. Van Dien, T. Wang, W. B. Whitman, Q. Xia, Y. Zhang, F. W. Larimer, M. V. Olson, and J. A. Leigh. 2004. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 1866956-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jolly, L., P. Ferrari, D. Blanot, J. van Heijenoort, F. Fassy, and D. Mengin-Lecreulx. 1999. Reaction mechanism of phosphoglucosamine mutase from Escherichia coli. Eur. J. Biochem. 262202-210. [DOI] [PubMed] [Google Scholar]
- 15.Kawamura, T., N. Ishimoto, and E. Ito. 1979. Enzymatic synthesis of uridine diphosphate N-acetyl-d-mannosaminuronic acid. J. Biol. Chem. 2548457-8465. [PubMed] [Google Scholar]
- 16.Kawamura, T., N. Ishimoto, and E. Ito. 1982. UDP-N-acetyl-d-glucosamine 2′-epimerase from Escherichia coli. Methods Enzymol. 83515-519. [DOI] [PubMed] [Google Scholar]
- 17.Kuntz, C., J. Sonnenbichler, I. Sonnenbichler, M. Sumper, and R. Zeitler. 1997. Isolation and characterization of dolichol-linked oligosaccharides from Haloferax volcanii. Glycobiology 7897-904. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 19.Li, H., H. Xu, D. E. Graham, and R. H. White. 2003. The Methanococcus jannaschii dCTP deaminase is a bifunctional deaminase and diphosphatase. J. Biol. Chem. 27811100-11106. [DOI] [PubMed] [Google Scholar]
- 20.Liu, H.-D., Y.-M. Li, J.-T. Du, J. Hu, and Y.-F. Zhao. 2006. Novel acetylation-aided migrating rearrangement of uridine-diphosphate-N-acetylglucosamine in electrospray ionization multistage tandem mass spectrometry. J. Mass Spectrom. 41208-215. [DOI] [PubMed] [Google Scholar]
- 21.Mengin-Lecreulx, D., and J. van Heijenoort. 1994. Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: characterization of the glmU gene product as a bifunctional enzyme catalyzing two subsequent steps in the pathway for UDP-N-acetylglucosamine synthesis. J. Bacteriol. 1765788-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milewski, S., I. Gabriel, and J. Olchowy. 2006. Enzymes of UDP-GlcNAc biosynthesis in yeast. Yeast 231-14. [DOI] [PubMed] [Google Scholar]
- 23.Mizanur, R. M., C. J. Zea, and N. L. Pohl. 2004. Unusually broad substrate tolerance of a heat-stable archaeal sugar nucleotidyltransferase for the synthesis of sugar nucleotides. J. Am. Chem. Soc. 12615993-15998. [DOI] [PubMed] [Google Scholar]
- 24.Moore, B. C., and J. A. Leigh. 2005. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J. Bacteriol. 187972-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan, P. M., R. F. Sala, and M. E. Tanner. 1997. Eliminations in the reactions catalyzed by UDP-N-acetylglucosamine 2-epimerase. J. Am. Chem. Soc. 11910269-10277. [Google Scholar]
- 26.Mukhopadhyay, B., E. F. Johnson, and R. S. Wolfe. 1999. Reactor-scale cultivation of the hyperthermophilic methanarchaeon Methanococcus jannaschii to high cell densities. Appl. Environ. Microbiol. 655059-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nováková, L., L. Sasková, P. Pallová, J. Janeček, J. Novotná, A. Ulrych, J. Echenique, M.-C. Trombe, and P. Branny. 2005. Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates. FEBS J. 2721243-1254. [DOI] [PubMed] [Google Scholar]
- 28.Pompeo, F., Y. Bourne, J. van Heijenoort, F. Fassy, and D. Mengin-Lecreulx. 2001. Dissection of the bifunctional Escherichia coli N-acetylglucosamine-1-phosphate uridyltransferase enzyme into autonomously functional domains and evidence that trimerization is absolutely required for glucosamine-1-phosphate acetyltransferase activity and cell growth. J. Biol. Chem. 2763833-3839. [DOI] [PubMed] [Google Scholar]
- 29.Ramakrishnan, V., Q. Teng, and M. W. Adams. 1997. Characterization of UDP amino sugars as major phosphocompounds in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 1791505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rashid, N., H. Imanaka, T. Fukui, H. Atomi, and T. Imanaka. 2004. Presence of a novel phosphopentomutase and a 2-deoxyribose 5-phosphate aldolase reveals a metabolic link between pentoses and central carbon metabolism in the hyperthermophilic archaeon Thermococcus kodakaraensis. J. Bacteriol. 1864185-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rashid, N., T. Kanai, H. Atomi, and T. Imanaka. 2004. Among multiple phosphomannomutase gene orthologues, only one gene encodes a protein with phosphoglucomutase and phosphomannomutase activities in Thermococcus kodakaraensis. J. Bacteriol. 1866070-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray, W. K., S. M. Keith, A. M. DeSantis, J. P. Hunt, T. J. Larson, R. F. Helm, and P. J. Kennelly. 2005. A phosphohexomutase from the archaeon Sulfolobus solfataricus is covalently modified by phosphorylation on serine. J. Bacteriol. 1874270-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauer, F. D., B. A. Blackwell, J. K. G. Kramer, and B. J. Marsden. 1990. Structure of a novel cofactor containing N-(7-mercaptoheptanoyl)-O-3-phosphothreonine. Biochemistry 297593-7600. [DOI] [PubMed] [Google Scholar]
- 34.Shackelford, G. S., C. A. Regni, and L. J. Beamer. 2004. Evolutionary trace analysis of the α-d-phosphohexomutase superfamily. Protein Sci. 132130-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shams-Eldin, H., B. Chaban, S. Niehus, R. T. Schwarz, and K. F. Jarrell. 2008. Identification of the archaeal alg7 gene homolog (encoding N-acetylglucosamine-1-phosphate transferase) of the N-linked glycosylation system by cross-domain complementation in Saccharomyces cerevisiae. J. Bacteriol. 1902217-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka, T., F. Takahashi, T. Fukui, S. Fujiwara, H. Atomi, and T. Imanaka. 2005. Characterization of a novel glucosamine-6-phosphate deaminase from a hyperthermophilic archaeon. J. Bacteriol. 1877038-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tavares, I. M., L. Jolly, F. Pompeo, J. H. Leitao, A. M. Fialho, I. Sa-Correia, and D. Mengin-Lecreulx. 2000. Identification of the Pseudomonas aeruginosa glmM gene, encoding phosphoglucosamine mutase. J. Bacteriol. 1824453-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teplyakov, A., C. Leriche, G. Obmolova, B. Badet, and M.-A. Badet-Denisot. 2002. From Lobry de Bruyn to enzyme-catalyzed ammonia channelling: molecular studies of d-glucosamine-6P synthase. Nat. Prod. Rep. 1960-69. [DOI] [PubMed] [Google Scholar]
- 39.Varki, A., R. Cummings, J. Esko, H. Freeze, G. Hart, and J. Marth (ed.). 1999. Essentials of glycobiology. Cold Spring Harbor Laboratory Press, New York, NY. [PubMed]
- 40.White, R. H. 1993. Structures of the modified folates in the thermophilic archaebacteria Pyrococcus furiosus. Biochemistry 32745-753. [DOI] [PubMed] [Google Scholar]
- 41.White, R. H., and H. Xu. 2006. Methylglyoxal is an intermediate in the biosynthesis of 6-deoxy-5-ketofructose-1-phosphate: a precursor for aromatic amino acid biosynthesis in Methanocaldococcus jannaschii. Biochemistry 4512366-12379. [DOI] [PubMed] [Google Scholar]
- 42.Yamazaki, T., C. D. Warren, A. Herscovics, and R. W. Jeanloz. 1980. A convenient synthesis of uridine 5′-(2-acetamido-2-deoxy-α-d-manno-pyranosyluronic acid pyrophosphate). Carbohydr. Res. 79C9-C12. [Google Scholar]
- 43.Zachara, N. E., and G. W. Hart. 2006. Cell signaling, the essential role of O-GlcNAc! Biochim. Biophys. Acta 1761599-617. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, H., Y. Zhou, H. Bao, and H.-W. Liu. 2006. Vi antigen biosynthesis in Salmonella typhi: characterization of UDP-N-acetylglucosamine C-6 dehydrogenase (TviB) and UDP-N-acetylglucosaminuronic acid C-4 epimerase (TviC). Biochemistry 458163-8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, Z., M. Tsujimura, J.-I. Akutsu, M. Sasaki, H. Tajima, and Y. Kawarabayasi. 2005. Identification of an extremely thermostable enzyme with dual sugar-1-phosphate nucleotidylyltransferase activities from an acidothermophilic archaeon, Sulfolobus tokodaii strain 7. J. Biol. Chem. 2809698-9705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.