Abstract
Using Streptococcus thermophilus phages, plasmid transduction in Lactococcus lactis was demonstrated. The transduction frequencies were 4 orders of magnitude lower in L. lactis than in S. thermophilus. These results are the first evidence that there is phage-mediated direct transfer of DNA from S. thermophilus to L. lactis. The implications of these results for phage evolution are discussed.
Bacteriophages of Streptococcus thermophilus and Lactococcus lactis, two important starter strains in the dairy industry (5, 25), share significant sequence homology (3, 6). Therefore, horizontal gene transfer between the two genera has to be supposed. S. thermophilus has recently been shown to be capable of natural transformation (1), but L. lactis lacks natural competence for DNA uptake (29). Conjugal transfer of plasmids from L. lactis to S. thermophilus has been demonstrated (14); however, no native conjugative plasmids have been found in S. thermophilus yet. Transduction of several plasmids with different phages of S. thermophilus within this species has been shown to occur (17; A. Ammann, H. Neve, A. Geis, and K. J. Heller; unpublished); however, transduction between the two species has not been reported previously, nor have phages of one species been demonstrated to be able to infect the other species.
In this paper, we describe for the first time plasmid transduction with three S. thermophilus phages beyond the genus into L. lactis. Three virulent cos-type S. thermophilus phages were selected for this study (Table 1), which had different host ranges for three S. thermophilus strains (strains a10, 55n, and St11) (18, 21). Phage P1109 could infect all three hosts, while phage P53 could be propagated in strains a10 and 55n. Phage a10/J9 had a narrow host range (only strain a10). For transduction assays, plasmid pAG106AE was introduced into the plasmid-free host strains a10 and St11 either by electroporation or by transduction. This plasmid construct is based on a native S. thermophilus plasmid (8) that has been cloned in a charomid vector conferring erythromycin resistance (Table 1). For preparation of plasmid-transducing lysates, 0.1 to 0.3 ml of phage lysate was added at low multiplicities of infection (MOI) (between 10−4 and 1) to 0.1- to 0.3-ml portions of overnight cultures of plasmid-bearing S. thermophilus strains in the presence of 10 mM CaCl2. After 10 min of adsorption at 40°C, each mixture was diluted with 10 ml of prewarmed thM17 medium (containing 10 mM CaCl2) (15). The infected cultures were incubated at 40°C aerobically without shaking until cell lysis occurred. In cases of premature lysis at low optical densities at 620 nm (OD620) (OD620, <0.2), the lysates were inoculated again with 1 ml of the same host culture grown to the early logarithmic phase (OD620, approximately 0.5). Residual bacterial cells and cell debris were finally removed by membrane filtration (pore size, 0.45 μm).
TABLE 1.
Strains, phages, and plasmid used
| Strain(s), phage, or plasmid | Propertiesa | Source or reference |
|---|---|---|
| Strains | ||
| S. thermophilus a10 | Host for phages P1109, P53, and a10/J9 | FRCNF-Kiel strain collection |
| S. thermophilus 55n | Host for phages P1109 and P53 | FRCNF-Kiel strain collection |
| S. thermophilus St11 | Host for phage P1109 | 18 |
| L. lactis Bu2-60 | Plasmid-cured derivative of L. lactis Bu2, Smr Rmr | 20 |
| L. lactis Bu2-60-STD1, Bu2-60-STD2, and Bu2-60-STD3 | Bu2-60 transductants containing pAG106AE obtained by transduction with S. thermophilus phage P1109, Smr Rmr Emr | This study |
| Phages | ||
| P1109 | Virulent S. thermophilus phage, cos type | FRCNF-Kiel phage collection |
| P53 | Virulent S. thermophilus phage, cos type | 21 |
| a10/J9 | Virulent S. thermophilus phage, cos type | 21 |
| P008 | Virulent L. lactis phage, cos type, 936 species | 12 |
| Plasmid | ||
| pAG106AE | 9.2 kb, pSt106(AatII)::charomid 9-36, Δspacer and Δcos of charomid 9-36, Emr | 8 |
Smr, streptomycin resistance; Rmr, rifampin resistance; Emr, erythromycin resistance.
For transduction assays with plasmid-free strains S. thermophilus a10 and L. lactis Bu2-60 (20), cultures grown in thM17 (strain a10) or in GM17 (28) (strain Bu2-60) were harvested in the logarithmic phase (OD620, 0.5) and resuspended in 0.2 volume of 10 mM ice-cold MgSO4 (approximately 1 × 109 CFU/ml). Phage lysates (150 μl) were added at an MOI of 0.01 (S. thermophilus) or 0.1 (L. lactis) to 300 μl of bacteria. CaCl2 was added at a final concentration of 10 mM (S. thermophilus) or 100 mM (L. lactis) before the sample volume was increased to 1 ml by addition of fresh thM17 or GM17 medium. Phage adsorption was performed for 10 min at 42°C (S. thermophilus) or for 30 min at 30°C (L. lactis). Phage adsorption was stopped on ice by addition of 1 ml of ice-cold thM17 or GM17 broth containing 20 mM (S. thermophilus) or 200 mM (L. lactis) trisodium citrate. Transduction mixtures were subsequently incubated for 50 min at 42 or 30°C. All transduction assay mixtures were finally plated on GM17 agar (L. lactis) or thLM17 agar (S. thermophilus) supplemented with 10 μg/ml erythromycin. Control assays either without phage or without cells were also included.
Plasmid pAG106AE was transduced in L. lactis Bu2-60 using phage P1109 grown on S. thermophilus St11 (harboring pAG106AE) and phages P53 and a10/J9 grown on S. thermophilus a10 (harboring pAG106AE) (Table 1). For all phages tested, the transduction frequencies (calculated by determining the number of erythromycin-resistant CFU per added PFU) were ca. 4 orders of magnitude lower in L. lactis Bu2-60 than in S. thermophilus a10 (Table 2).
TABLE 2.
Transduction of plasmid pAG106AE in S. thermophilus and L. lactis with S. thermophilus phages
| Phage | S. thermophilus host strain | Phage neutralization by P53-specific antiserum (%)a |
S. thermophilus a10a
|
L. lactis Bu2-60b
|
||||
|---|---|---|---|---|---|---|---|---|
| Transduction frequency (CFU/PFU) | Transduction frequency after phage neutralization (CFU/PFU) | Reduction in transduction frequency (%) | Transduction frequency (CFU/PFU) | Transduction frequency after phage neutralization (CFU/PFU) | Reduction in transduction frequency (%) | |||
| P1109 | St11(pAG106AE) | 53 ± 8 | 1.9 × 10−2 ± 0.6 × 10−2 | 1.4 × 10−2 ± 0.1 × 10−2 | ∼26 | 3.4 × 10−5 ± 1.5 × 10−5 | 1.9 × 10−5 ± 0.6 × 10−5 | ∼44 |
| P53 | a10(pAG106AE) | 79 ± 14 | 6.8 × 10−3 ± 3.1 × 10−3 | 9.8 × 10−4 ± 0.3 × 10−4 | ∼86 | 9.5 × 10−6 ± 1.6 × 10−6 | 1.4 × 10−6 ± 0.3 × 10−6 | ∼85 |
| a10/J9 | a10(pAG106AE) | 99 ± 0 | 6.3 × 10−5 ± 3.2 × 10−5 | ND (<3.1 × 10−5)c | NAd | 5.6 × 10−8 ± 2.6 × 10−8 | ND (<3.2 × 10−8)e | NA |
Data from two experiments.
Data from four experiments.
ND, not detectable (0.2 ml from a 2-ml transduction mixture was plated).
NA, not applicable.
ND, not detectable (1 ml from a 2-ml transduction mixture was plated).
L. lactis Bu2-60 transductants were easily identified and distinguished from S. thermophilus host cells phenotypically by their growth characteristics (e.g., no growth at 45°C; resistance to streptomycin and rifampin [20]) and by their sensitivity to lactococcal virulent phage P008 (26) (data not shown). Hence, contamination of S. thermophilus plasmid donor cells could be ruled out.
L. lactis has recently been shown to harbor an incomplete set of genes involved in natural competence (2). To exclude the possibility that natural transformation by contaminating plasmid DNA was the cause of the gene transfer observed, we used polyclonal phage antiserum raised against phage P53 for neutralization of the phage particles (19). Phage suspensions were mixed with the same volume of a 1:100 dilution of antibodies in phosphate-buffered saline and incubated for 10 min at room temperature before the transduction experiments were carried out. Inactivation of phage particles was determined by spotting dilutions of phage-antibody suspensions on soft agar plates inoculated with S. thermophilus a10. A reduction in the transduction frequency corresponded with a reduction in phage infectivity (Table 2), showing that intact phage particles and not naked DNA was responsible for plasmid transfer. As a second control, S. thermophilus phage lysates were replaced by 0.5 μg of intact plasmid pAG106AE DNA in the transduction assays. No transformants were detected (data not shown).
For another demonstration of the efficient transductional gene transfer of plasmid pAG106AE from S. thermophilus to L. lactis, phage P1109 was propagated in its plasmid-bearing S. thermophilus host strain and purified by CsCl buoyant density gradient centrifugation (24). The phage-containing fraction was used for transduction assays with the lactococcal recipient strain Bu2-60 at different MOI (0.1, 0.01, and 0.001). The number of L. lactis transductants obtained correlated well with the serial dilutions of the phages used (634, 41, and 5 transductants per ml), while the calculated transduction frequency remained remarkably stable (2 × 10−5 to 4 × 10−5) (Table 3). When the purified phages were neutralized by using phage P53-specific antiserum (99.9% inactivation), no transductants were obtained (data not shown). Preincubation of these purified phages with DNase (0.2 to 1 mg/ml) did not affect the transduction frequency in L. lactis Bu2-60 (data not shown). DNase activity was confirmed by spiking transduction assay mixtures with purified pAG106AE DNA and then performing agarose gel electrophoresis and demonstrating hydrolysis of pAG106AE DNA (not shown).
TABLE 3.
Transduction of plasmid pAG106AE in L. lactis Bu2-60 with S. thermophilus phage P1109 purified by CsCl density gradient centrifugationa
| P1109 titer (PFU/ml) | MOI | Transductants/ml | Transduction frequency (CFU/PFU) |
|---|---|---|---|
| 1.8 × 107 | 0.1 | 634 ± 10 | 3.5 × 10−5 ± 0.1 × 10−5 |
| 1.8 × 106 | 0.01 | 41 ± 3 | 2.3 × 10−5 ± 0.1 × 10−5 |
| 1.8 × 105 | 0.001 | 5 ± 4 | 2.5 × 10−5 ± 1.9 × 10−5 |
Data from two experiments.
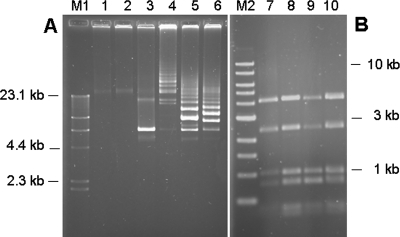
Plasmids were extracted from recipient cells to confirm transduction events. Plasmid pAG106AE was isolated from L. lactis Bu2-60 transductants in a high-molecular-weight concatemeric form (Fig. 1A). This was most likely due to reduced expression in L. lactis of the resolvase gene pSt106, the plasmid on which the theta-replicating S. thermophilus plasmid pAG106AE was based (8). Concatemers were also observed when pAG106AE was transferred via electroporation (data not shown). The StyI restriction enzyme profiles of the plasmid DNA extracted from the L. lactis transductants Bu2-60-STD1, -STD2, and -STD3 (obtained by P1109-mediated transduction) were in most cases identical to the corresponding pattern of the plasmid extracted from the S. thermophilus a10(pAG106AE) donor strain (Fig. 1B). However, we sometimes observed restriction fragments that did not fit the control profile. These fragments may have been due either to incomplete hydrolysis with StyI or to inhomogeneities in the plasmids created by incorrect resolution of concatemers. It should be noted that the plasmid was packaged into the phage as concatemers, as demonstrated by restriction analysis of packaged DNA followed by Southern blot hybridization with plasmid DNA as the probe (data not shown).
FIG. 1.
(A) Agarose gel electrophoresis of intact plasmid DNA extracted from plasmid-free recipient strains S. thermophilus a10 (lane 1) and L. lactis Bu2-60 (lane 2), from S. thermophilus donor strain a10(pAG106AE) (lane 3), and from three L. lactis Bu2-60 transductants, Bu2-60-STD1, Bu2-60-STD2, and Bu2-60-STD3 (lanes 4 to 6). (B) Agarose gel electrophoresis of StyI digests of plasmid DNA extracted from S. thermophilus donor strain a10(pAG106AE) (lane 7) and from the three L. lactis Bu2-60 transductants (lanes 8 to 10). Phage lambda cut with HindIII (lane M1) and a 1-kb ladder (lane M2) were used as size references.
In order to elucidate the efficiency of adsorption of phages to S. thermophilus or L. lactis, we analyzed reversible and irreversible binding to the recipient cells. As has been demonstrated previously mostly for phages of gram-negative bacteria, reversible binding may considerably enhance adsorption (9) and may thus increase the efficiency of infection when there is poor irreversible binding (10). Cells were harvested from GM17 or thM17 early-log-phase broth cultures (OD620, 0.5) and resuspended in 0.02 volume of 10 mM MgSO4. The adsorption assays were performed as described by Østergaard Breum et al. (23) at a low MOI (0.01). After 20 min of adsorption at either 30°C (L. lactis assays) or 40°C (S. thermophilus assays), the samples were diluted 1:100 in ice-cold 0.25× Ringer buffer (supplemented with 1 M NaCl) for determination of irreversible adsorption. For determination of reversible adsorption, assay mixtures were centrifuged immediately after 20 min of adsorption, followed by dilution (1:100) of the supernatant with 0.25× Ringer buffer-1 M NaCl and determination of the number of PFU. The three S. thermophilus phages adsorbed to S. thermophilus a10 cells equally well with high efficiency (98 to 99% reversible adsorption and 96 to 98% irreversible adsorption). Furthermore, phage P1109, which exhibited the highest transduction efficiency in L. lactis Bu2-60 (Table 2), adsorbed significantly (46% reversible adsorption and 43% irreversible adsorption) to L. lactis Bu2-60, while adsorption of the two remaining phages was either not detectable or close to the limit of detection (Table 4). This correlated well with their low transduction efficiencies in L. lactis. The fact that there was basically no difference between reversible adsorption and irreversible adsorption indicates that binding of S. thermophilus phages to the S. thermophilus or L. lactis cell surface either is a single-step process with no reversible binding or is a two-step process with irreversible binding that occurs so fast that differentiation between reversible binding and irreversible binding is not possible.
TABLE 4.
Reversible and irreversible adsorption of S. thermophilus phages to S. thermophilus a10 and L. lactis Bu2-60 cells at a low MOI (0.01)a
| Strain | Adsorption of S. thermophilus phages (%)
|
|||||
|---|---|---|---|---|---|---|
| P1109
|
P53
|
a10/J9
|
||||
| Reversible | Irreversible | Reversible | Irreversible | Reversible | Irreversible | |
| S. thermophilus a10b | 99 | 98 | 99 | 98 | 98 | 96 |
| L. lactis subsp. lactis Bu2-60c | 46 ± 2 | 43 ± 4 | NDd | 4.1 ± 0.2 | 2.8 ± 0.2 | 4.0 ± 0.9 |
Adsorption was performed for 20 min at 30°C (L. lactis) or at 40°C (S. thermophilus).
Data from one experiment.
Means ± standard deviations for two experiments.
ND, not detectable.
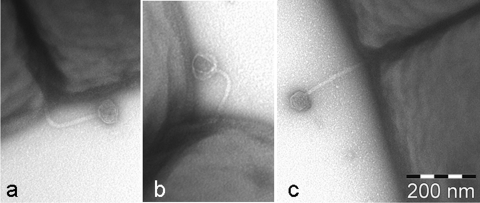
In order to visualize adsorption of P1109 phage particles to L. lactis Bu2-60 cells, transduction assays were performed at higher MOI (approximately 1 to 10). Samples were taken for negative staining with 1% (wt/vol) uranyl acetate and analyzed with a Tecnai 10 transmission electron microscope (FEI Company, Eindhoven, The Netherlands) as described previously (23, 26). Figure 2 shows specific adsorption of intact S. thermophilus P1109 phage particles (Fig. 2a and c) and of a phage ghost (Fig. 2b) to the L. lactis Bu2-60 cell surface; no free phages were detected in the space between cells. Most of the adsorbed phage particles analyzed appeared to be intact, indicating that DNA injection had not occurred yet. It is notable that adsorption did not occur randomly but occurred preferably at regions involved in cell division.
FIG. 2.
Transmission electron micrographs of S. thermophilus P1109 phage particles adsorbed to L. lactis Bu2-60 cells.
Our experiments provide evidence that plasmids can be transferred via transduction from S. thermophilus to L. lactis. We demonstrate that the S. thermophilus phages tested are able to adsorb to L. lactis cells and to inject their DNA into these cells, albeit at a low frequency. So far, no S. thermophilus phage able to propagate on L. lactis has been described. Our experiments indicate that the failure of S. thermophilus phages to propagate on L. lactis may be due to their low efficiency of adsorption, as shown for phages P53 and a10/J9 (Table 3). If we assume, on the basis of the reduced transduction efficiency, that the adsorption rate is 103-fold lower for L. lactis than for S. thermophilus, this would (for a phage producing approximately 102 progeny phage per infected cell, as is the case for the S. thermophilus phages described here) result in the failure of the phage to form plaques. However, we cannot rule out the possibility that, in addition, intracellular multiplication proceeds inefficiently, if at all. An in-depth analysis of host range mutants of two S. thermophilus phages revealed previously that at least three structural tail proteins may be involved in the phage-host interactions in S. thermophilus (7). The mutants were obtained at a low frequency (10−6), and host range extensions were shown to be due to single nucleotide mutations in the structural genes. Host range phage mutants had expanded host ranges and were still able to adsorb to the original host cells. On the other hand, two prophages of L. lactis IL-1403 (phages bIL286 and bIL309) possess structural proteins with high levels of similarity to proteins of S. thermophilus phages (portal protein, capsid protein, tail protein, tail host specificity protein). These data also indicate that S. thermophilus phages can interact with the surface of lactococcal cells.
The high levels of similarity of genes of S. thermophilus and L. lactis phages support the hypothesis that horizontal gene transfer occurs between these species belonging to two different genera (4). Exchange of genetic modules between S. thermophilus and L. lactis phages appears to be one of the motors of the evolution of these organisms (16, 22). In principle, recombination between two phage genomes can occur by two mechanisms following phage infection: (i) recombination may take place between the genome of an infecting phage and a prophage residing in the genome of the infected cell; and (ii) recombination may take place between the genomes of two phages simultaneously infecting a host cell. Since we have shown that transduction by S. thermophilus phages occurs in L. lactis, this implies that at least for the S. thermophilus phages, which are capable of transduction in L. lactis, infection of L. lactis is possible. Thus, recombination and exchange of genetic modules by either of the two mechanisms indicated above appear to be a realistic option. As a consequence, a mixture of recombined lactococcal and streptococcal progeny phages (with respect to host specificity) is produced, which carry newly acquired genetic modules. The phages with S. thermophilus host specificity are able to transfer the newly acquired genetic modules back into S. thermophilus. It remains to be determined whether genetic exchange involving L. lactis phages and S. thermophilus recipient cells is also possible. Comparison of the genome sequences of two S. thermophilus yoghurt strains clearly showed that their genomes contain more than 100 “nonstreptococcal” genes potentially acquired by horizontal gene transfer. Approximately 30 of these genes exhibit the highest levels of similarity to L. lactis genes (11). It is also notable that a number of these L. lactis-related genes code for putative membrane proteins. Two findings have indicated that phage-related traits are functional in both genera. First, the AbiA lactococcal abortive infection mechanism, when introduced into an S. thermophilus host, has been shown to be effective at 30°C in S. thermophilus cells against six S. thermophilus phages. As observed with lactococcal phages, AbiA affected S. thermophilus phages by also interfering with DNA replication (27). Second, the S. thermophilus TP-J34 phage protein Ltp, involved in superinfection exclusion, has been shown to be functionally active in L. lactis against phage of this host. In both hosts, Ltp interfered with the process of DNA injection (26).
In conclusion, we present for the first time evidence that phage-mediated horizontal gene transfer occurs in gram-positive dairy starter cultures from S. thermophilus to L. lactis. To our knowledge, horizontal transductional gene transfer by broad-host-range phages infecting different species has been demonstrated previously only for gram-negative phages (13). Our results support the hypothesis that some S. thermophilus and L. lactis bacteriophages have evolved by direct exchange of genetic modules.
Acknowledgments
The expert technical assistance of Inka Lammertz is gratefully acknowledged. We also thank Yahya Ali for classification of phage P1109 as a cos-type phage by an unpublished multiplex PCR.
Footnotes
Published ahead of print on 8 February 2008.
REFERENCES
- 1.Blomqvist, T., H. Steinmoen, and L. S. Havarstein. 2006. Natural genetic transformation: a novel tool for efficient genetic engineering of the dairy bacterium Streptococcus thermophilus. Appl. Environ. Microbiol. 726751-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brüssow, H., A. Bruttin, F. Desiere, S. Lucchini, and S. Foley. 1998. Molecular ecology and evolution of Streptococcus thermophilus bacteriophages—a review. Virus Genes 1695-109. [DOI] [PubMed] [Google Scholar]
- 4.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M.-C. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daly, C. 1983. The use of mesophilic cultures in the dairy industry. Antonie van Leeuwenhoek 49297-312. [DOI] [PubMed] [Google Scholar]
- 6.Desiere, F., W. M. McShan, D. van Sinderen, J. J. Ferretti, and H. Brüssow. 2001. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic streptococci: evolutionary implications for prophage-host interactions. Virology 288325-341. [DOI] [PubMed] [Google Scholar]
- 7.Duplessis, M., C. M. Levesque, and S. Moineau. 2006. Characterization of Streptococcus thermophilus host range phage mutants. Appl. Environ. Microbiol. 723036-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geis, A., H. A. M. El Demerdash, and K. J. Heller. 2003. Sequence analysis and characterization of plasmids from Streptococcus thermophilus. Plasmid 5053-69. [DOI] [PubMed] [Google Scholar]
- 9.Heller, K. J. 1992. Molecular interaction between bacteriophage and the Gram-negative cell envelope. Arch. Microbiol. 158235-248. [DOI] [PubMed] [Google Scholar]
- 10.Heller, K. J., and D. Bryniok. 1984. O-antigen-dependent mutant of bacteriophage T5. J. Virol. 4920-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hols, P., F. Hancy, L. Fontaine, B. Grossiord, D. Prozzi, N. Leblond-Bourget, B. Decaris, A. Bolotin, C. Delorme, S. D. Ehrlich, E. Guédon, V. Monnet, P. Renault, and M. Kleerebezem. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29435-463. [DOI] [PubMed] [Google Scholar]
- 12.Jarvis, A. W., G. F. Fitzgerald, M. Mata, A. Mercenier, H. Neve, I. B. Powell, C. Ronda, M. Saxelin, and M. Teuber. 1991. Species and type phages of lactococcal bacteriophages. Intervirology 322-9. [DOI] [PubMed] [Google Scholar]
- 13.Jensen, E. C., H. S. Schrader, B. Rieland, T. L. Thomson, K. W. Lee, K. W. Nickerson, and T. A. Kokjohn. 1998. Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl. Environ. Microbiol. 64575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleinschmidt, J., B. Soeding, M. Teuber, and H. Neve. 1993. Evaluation of horizontal and vertical gene-transfer and stability of heterologous DNA in Streptococcus thermophilus isolated from yogurt and yogurt starter cultures. Syst. Appl. Microbiol. 16287-295. [Google Scholar]
- 15.Krusch, U., H. Neve, B. Luschei, and M. Teuber. 1987. Characterization of virulent bacteriophages of Streptococcus salivarius subsp. thermophilus by host specificity and electron microscopy. Kiel. Milchwirtsch. Forschungsber. 39155-167. [Google Scholar]
- 16.Lucchini, S., F. Desiere, and H. Brüssow. 1999. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J. Virol. 738647-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mercenier, A., P. Slos, M. Faelen, and J. P. Lecocq. 1988. Plasmid transduction in Streptococcus thermophilus. Mol. Gen. Genet. 212386-389. [DOI] [PubMed] [Google Scholar]
- 18.Mollet, B., J. Knol, B. Poolman, O. Marciset, and M. Delley. 1993. Directed genomic integration, gene replacement, and integrative gene expression in Streptococcus thermophilus. J. Bacteriol. 1754315-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neve, H., W. Freudenberg, F. Diestel-Feddersen, R. Ehlert, and K. J. Heller. 2003. Biology of the temperate Streptococcus thermophilus bacteriophage TP-J34 and physical characterization of the phage genome. Virology 315184-194. [DOI] [PubMed] [Google Scholar]
- 20.Neve, H., A. Geis, and M. Teuber. 1984. Conjugal transfer and characterization of bacteriocin plasmids in group N (lactic acid) streptococci. J. Bacteriol. 157833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neve, H., U. Krusch, and M. Teuber. 1989. Classification of virulent bacteriophages of Streptococcus salivarius subsp. thermophilus isolated from yoghurt and Swiss-type cheese. Appl. Microbiol. Biotechnol. 30624-629. [Google Scholar]
- 22.Neve, H., K. I. Zenz, F. Desiere, A. Koch, K. J. Heller, and H. Brüssow. 1998. Comparison of the lysogeny modules from the temperate Streptococcus thermophilus bacteriophages TP-J34 and Sfi21: implications for the modular theory of phage evolution. Virology 24161-72. [DOI] [PubMed] [Google Scholar]
- 23.Østergaard Breum, S., H. Neve, K. J. Heller, and F. K. Vogensen. 2007. Temperate phages TP901-1 and φLC3, belonging to the P335 species, apparently use different pathways for DNA injection in Lactococcus lactis subsp. cremoris 3107. FEMS Microbiol. Lett. 276156-164. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Stiles, M. E., and W. H. Holzapfel. 1997. Lactic acid bacteria of foods and their current taxonomy. Int. J. Food Microbiol. 361-29. [DOI] [PubMed] [Google Scholar]
- 26.Sun, X., K. J. Heller, and H. Neve. 2006. The ltp gene of temperate Streptococcus thermophilus phage TP-J34 encodes a lipoprotein which is expressed during lysogeny. Virology 350146-157. [DOI] [PubMed] [Google Scholar]
- 27.Tangney, M., and G. F. Fitzgerald. 2002. AbiA, a lactococcal abortive infection mechanism functioning in Streptococcus thermophilus. Appl. Environ. Microbiol. 686388-6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wydau, S., R. Dervyn, J. Anba, D. Ehrlich, and E. Maguin. 2006. Conservation of key elements of natural competence in Lactococcus lactis ssp. FEMS Microbiol. Lett. 25732-42. [DOI] [PubMed] [Google Scholar]