Abstract
The growth dynamics of bacterial pathogens within infected hosts are a fundamental but poorly understood feature of most infections. We have focused on the in situ distribution and growth characteristics of two prevailing and transmissible Pseudomonas aeruginosa clones that have caused chronic lung infections in cystic fibrosis (CF) patients for more than 20 years. We used fluorescence in situ hybridization (FISH) directly on sputum specimens to examine the spatial distribution of the infecting P. aeruginosa cells. Mucoid variants were present in sputum as cell clusters surrounded by an extracellular matrix, whereas nonmucoid variants were present mainly as dispersed cells. To obtain estimates of the growth rates of P. aeruginosa in CF lungs, we used quantitative FISH to indirectly measure growth rates of bacteria in sputum samples (reflecting the in vivo lung conditions). The concentration of rRNA in bacteria isolated from sputa was measured and correlated with the rRNA contents of the same bacteria growing in vitro at defined rates. The results showed that most cells were actively growing with doubling times of between 100 and 200 min, with some growing even faster. Only a small stationary-phase subpopulation seemed to be present in sputa. This was found for both mucoid and nonmucoid variants despite their different organizations in sputum. The results suggest that the bacterial population may be confronted with selection forces that favor optimized growth activities. This scenario constitutes a new perspective on the adaptation and evolution of P. aeruginosa during chronic infections in CF patients in particular and on long-term infections in general.
An understanding of the growth dynamics of bacterial pathogens within infected hosts is a fundamental issue of general biological and medical relevance and with important consequences for how bacterial infections are understood and interfered with. In cases of lifelong persistent bacterial infections, within-host growth dynamics also become an important aspect specifically related to the mechanisms of pathogen adaptation and evolution.
Chronic lung infections by the opportunistic pathogen Pseudomonas aeruginosa in patients with the hereditary disease cystic fibrosis (CF) are one example of a persistent bacterial infection. Most CF patients acquire chronic P. aeruginosa infections, and eventually these infections cannot be eradicated, even with continuous intensive antibiotic treatment (18). The infection process in the CF airways is associated with extensive genetic adaptation and microevolution of the infecting bacteria (53). The accumulation of mutations results in strains with phenotypes of which many are not usually observed among environmental isolates. These phenotypes include loss of motility (32), loss of effector proteins of the type III secretion system (19), loss of O-antigen components of the lipopolysaccharide (12, 54), reduced virulence (27, 31), reduced capacity for in vitro biofilm formation (26), and increased antibiotic resistance (8). In some cases, specific genes have been found to be hot spots for mutations during CF infections. These common targets include lasR, which encodes a quorum-sensing regulator (53), and mucA, which results in the overproduction of alginate and conversion to the frequently found mucoid phenotype (34, 35). The occurrence of a range of genetic variants during chronic infections of the CF airways and the possibility of a conserved pattern of P. aeruginosa mutations suggest within-host, parallel evolution of the infecting bacteria that secures their persistence and long-term survival.
P. aeruginosa is located in both the respiratory zone and the conductive zone of the infected CF lung (3, 14, 16, 25, 58). Materials from both zones can be found in sputum, which is a mixture of airway mucus, factors of the innate immune system that are induced by the infections, and other bacteria and bacterial products. The CF airway mucus forms a stationary and thickened gel which is relatively hypoxic and adherent to the epithelial lining fluid of the airway surfaces (3, 58). The airways of CF patients may thus be regarded as a complex ecological niche in which P. aeruginosa must adapt to challenges imposed not only from the specific physical-chemical properties of the mucus but also from inflammatory cells and molecules of the innate and adaptive defense systems, frequent exposure to antibiotics, and the specific nutritional conditions in the different lung compartments.
Our previous molecular epidemiological studies of the P. aeruginosa population dynamics among CF patients in Copenhagen, Denmark, have shown that many of the long-term-infected CF patients are infected with the same, dominant clones because of extensive cross-infections (22). On the basis of pulsed-field gel electrophoresis and single-nucleotide polymorphism analysis of conserved genes, we identified two such dominant clones, which we designated “b” and “r”. In addition, we have shown that these two particular clones have been transmitted among different patients for more than 20 years (22). In order to understand better the processes that have secured the long-term persistence of these dominant and transmissible P. aeruginosa clones within the CF airway niche, we have focused on characterizing their in situ distribution and growth physiology using fluorescence in situ hybridization (FISH) analysis.
FISH using labeled oligonucleotide probes targeting rRNA has been a powerful technique for assessing both microbial identity (population structure) and spatial distributions in situ in complex environmental contexts (11, 15, 39, 47). Importantly, the hybridization of labeled probes to rRNA also yields a quantifiable fluorescent signal which is correlated to the cellular content of ribosomes. In several bacteria, the number of ribosomes is correlated with their growth rates (24, 38, 46, 51), and measurements of this parameter can therefore be used as an indicator of physiological activity (growth rate). The fundamental requirement for the quantification of ribosomes as a growth indicator is the establishment of a standard curve based on quantification of ribosome content in exponentially growing cultures with different growth rates (37, 49). Previous studies have used similar approaches to estimate the growth rates of bacteria colonizing different complex environments such as the mammalian gastrointestinal tract (30, 48), the rhizosphere of barley seedlings (50), and bean plant leaves (29). In the present study we have used FISH to examine the relationship between spatial organization and growth activities of both mucoid and nonmucoid cells within CF sputum specimens.
MATERIALS AND METHODS
CF patients.
Data for six long-term-infected CF patients included in this study are shown in Table 1 (22). All patients were attending the Danish CF Center, Rigshospitalet, Copenhagen, Denmark. The six patients have been chronically infected with P. aeruginosa for more than 13 years (median, 24 years; range, 13 to 30 years). Chronic P. aeruginosa infection was defined as the persistent presence of P. aeruginosa in sputum for 6 consecutive months, or for less time when persistence was combined with the presence of two or more precipitating antibodies against P. aeruginosa (23). One patient, not included in Table 1, was intermittently colonized by P. aeruginosa (strain B4-1 [Table 2]) (22).
TABLE 1.
Characteristics of the long-term chronically infected CF patients included in this study
| Patienta | Yr of birth | Gender | Yr chronic infection began (no. of yr chronically infected) | Genotypes (phenotypes) of infecting P. aeruginosa strainsb |
|---|---|---|---|---|
| p2 | 1966 | Female | 1976 (30) | r (M), r (NM) |
| p6 | 1975 | Female | 1984 (22) | Unique (NM), r (NM), b (NM)c |
| p7 | 1986 | Female | 1990 (16) | b (NM)c |
| p10 | 1963 | Male | 1980 (26) | r (NM)c |
| p11 | 1983 | Male | 1993 (13) | b (NM), 4 (M) |
| p16 | 1973 | Female | 1976 (30) | b (M), r (NM) |
The patients are identical to those studied in the work described in reference 22.
r, b, and 4 are different bacterial genotypes that have been identified in more than one patient. Genotypes found only in single patients or found only once in the data set are designated “unique.” M and NM refer to mucoid and nonmucoid isolates, respectively. For example, during the course of this study we have found mucoid and nonmucoid variants in patient p2. Both variants were of the same genotype.
The bacteriology records showed that mucoid variants have been found in occasional samples (but not stored) from the history of the patients.
TABLE 2.
P. aeruginosa strains used in this study
| Strain | Source | Yr of isolation | Genotypeb |
|---|---|---|---|
| p2 s1F6/05 | CF p2 | 2005 | r |
| p6 s1C3/05 | CF p6 | 2005 | Unique |
| p7 14429/91 | CF p7 | 1991 | b |
| p7 s1B1/05 | CF p7 | 2005 | b |
| p10 s1E9/05 | CF p10 | 2005 | r |
| p11 s1E9/05 | CF p11 | 2005 | b |
| p11 s3A1/05a | CF p11 | 2005 | 4 |
| p16 s1C7/05 | CF p16 | 2005 | r |
| B4-1 | CF B4 | 2005 | Unique |
| PAO1 | Laboratory | Unique | |
| PA14 | Laboratory | Unique |
Strain p11 s3A1/05 is a mucoid isolate.
r, b, and 4 are different bacterial genotypes that have been identified in more than one patient. Genotypes found only in single patients or found only once in the data set are designated “unique.”
Bacterial isolates.
The P. aeruginosa strain collection examined is listed in Table 2. Clinical P. aeruginosa strains from sputum samples were isolated on Pseudomonas isolation agar (Difco) containing ampicillin (100 μg/ml). All P. aeruginosa isolates were genotyped by single-nucleotide polymorphism typing using AT biochips (Clondiag Chip Technologies, Germany) (22).
Growth media and measurements of growth rates in laboratory media.
Pure-culture growth studies of P. aeruginosa strains were performed under aerobic conditions at 37°C in Luria-Bertani (LB) medium; in pig mucus medium; or in ABT minimal medium (9) supplemented with either 0.2% glucose plus 1% Casamino Acids, 0.5% glucose, 2% Casamino Acids, or 10 mM sodium citrate. The mucus medium was prepared by rinsing pig lungs with 0.9% NaCl. This solution was diluted (1:1, vol/vol) in ABT minimal medium and used for growth studies. CF sputum medium was prepared by dissolving sputum samples from patient p16 in ABT (1:20, vol/vol) by vortexing for 2 min, kept at 4°C for 30 min, and sterilized by filtering through a 0.45-μm filter. All bacteria in the sputa were efficiently removed as evaluated by plating aliquots of the CF sputum medium. Anaerobic growth was performed in LB medium plus 1% nitrate at 30°C with a constant nitrogen flow. Growth rates were measured by monitoring the optical density at 600 nm during growth in 50 ml medium in 250-ml flasks with shaking at 150 rpm. Growth rates are expressed as generation times in minutes or as specific growth rates (ln2/hour [i.e., reciprocal hours]).
Isolation of bacterial cells from sputum samples.
Sputum samples were processed within an hour after expectoration or stored at 4°C for later analysis. The sputum samples could be stored for more than 5 days without effects on growth rate determinations. To determine in situ growth rates, P. aeruginosa cells from sputum samples were extracted by dissolving the sample in 0.9% NaCl. For very thick sputa, Sputaosol (Oxoid, Hampshire, United Kingdom) was used to help dissolve samples. The samples were centrifuged at 800 × g for 5 min at 4°C to remove mucus and epithelial cells. The supernatant which contained bacteria was removed and fixed for rRNA hybridizations as described below.
Oligonucleotide probe.
Probe PSEUDAER (5′-GGACGTTATCCCCCACTAT-3′), specific to P. aeruginosa 16S rRNA (21), was labeled with Cy3 (Molecular Probes, Eugene, OR). The probe has previously been reported to have a sensitivity and specificity of 1.000 (21). Potential cross-reactivity of the PSEUDAER probe is of minor concern, as the probe did not hybridize to Pseudomonas putida cells under the conditions used in this work (data not shown).
Fixation of bacterial cells.
Sputum samples, isolated cells from sputum samples, and cells from laboratory cultures were fixed in 4% paraformaldehyde for 15 min and washed with phosphate-buffered saline (PBS) (pH 7.4). Fixed cells were stored at −20°C in storage buffer (50% ethanol, 10 mM Tris [pH 7.5], 0.1% Nonidet P-40) until use.
Whole-cell hybridization.
Fixed cells were applied homogenously on poly-l-lysine (Sigma Chemical, St. Louis, MO)-coated slides (36) and air dried. Hybridizations were carried out on the slides using 30 μl of solution I (30% formamide, 100 mM Tris [pH 7.5], 0.1% sodium dodecyl sulfate, 0.9 M NaCl) and 80 ng of the PSEUDAER-Cy3 probe. For FISH carried out directly on fresh sputum, hybridization solution was added directly to the sample. The slides were incubated in the dark in a humidified chamber for 3 h at 37°C, followed by washing of the slides with 45 μl of prewarmed (37°C) solution I for 30 min at 37°C and subsequently with 45 μl of prewarmed (37°C) solution II (100 mM Tris [pH 7.5], 0.9 M NaCl) for 40 min at 37°C. The slides were then quickly rinsed in distilled water and air dried. For sputum samples, 50 μl Calcofluor white (CFW) (fluorescent brightener 28; Sigma-Aldrich) was added for staining of alginate and incubated for 3 h in a humidified chamber before washing with distilled water. For FISH counterstained with the fluorescent nucleic acid dye DAPI (4′,6′-diamidino-2-phenylindole), the slides were then incubated in the dark with 14 mM DAPI in PBS at room temperature for 5 min and washed with PBS for 5 min. The slides were then rinsed and air dried.
Microscopy and image analysis.
Microscopic observations of cells within sputum samples were completed using a Zeiss LSM510 scanning confocal laser microscope (Carl Zeiss, Jena, Germany) equipped with an NeHe laser as well as an UV lamp and detectors and filter sets for simultaneous monitoring of red fluorescence emitted from the Cy3 probe (excitation, 543 nm; emission filter, 565 to 615 nm) and CVW fluorescence from fluorescent brightener 28 (UV lamp excitation; emission filter, 395 to 465 nm). Images were obtained using a 40×/1.3 Plan-Neofluar oil objective. Images were processed using the IMARIS software package (Bitplane AG, Zürich, Switzerland).
Visualization of hybridized cells isolated from sputum samples or from laboratory cultures was done using an Axioplan epifluorescence microscope (Carl Zeiss) equipped with a 100-W mercury lamp and a Cy3 filter. A 63×/1.25 Plan-Neofluar oil objective (Carl Zeiss) was used for inspection and image acquisition. Image analysis was done in 12 bits with PMIS-S200 software version 4.1.4 and the Unix-based CELLSTAT image analysis program (38). Hybridized cells were automatically identified by use of the CELLSTAT program, providing cell volume, fraction of dividing cells, and mean fluorescence intensity. In some cases, unfocused cells or images and extremely high-intensity signals from crystals were deleted manually before further analysis.
RESULTS
Organization of P. aeruginosa populations in CF sputum.
To characterize the growth physiology of P. aeruginosa in CF airways, we first combined FISH and CFW staining to visualize the distribution of both P. aeruginosa cells and exopolysaccharide materials (including alginate) directly within sputum samples (see Materials and Methods). In vitro control experiments using defined mucoid and nonmucoid derivatives of PAO1 showed that alginate was stained by CFW (data not shown). Figure 1 shows representative results from combined in situ CFW staining and hybridizations using a P. aeruginosa-specific 16S rRNA probe on sputum material from five CF patients. These patients have been diagnosed as being chronically infected for more than 13 years and are infected with P. aeruginosa clones of the r and/or b genotype (Table 1) (22). In sputum material from patients infected with both mucoid and nonmucoid clones (patients p2, p11, and p16), we observed clusters of P. aeruginosa cells surrounded by material stained by CFW as well as single cells in areas with only little CFW staining (Fig. 1). This finding is consistent with previous light microscopic and electron microscopic observations of cell clusters surrounded by a matrix of extracellular polymeric substances within different types of clinical specimens (including sputa) from CF patients (17, 25, 52).
FIG. 1.
Distribution of P. aeruginosa populations in CF sputum. Sputum samples either with exclusively nonmucoid variants or with both nonmucoid and mucoid variants from five CF patients were analyzed by CFW staining combined with FISH using a P. aeruginosa-specific rRNA probe. The bar in each photograph represents 15 μm.
In contrast, we observed mainly well-separated, single cells and little CFW staining in sputa from patients p7 and p10, who are infected with only nonmucoid clones (Fig. 1). These data suggest that clones of the r and b genotypes are organized as cell clusters within sputum only when they are present as mucoid variants.
To further show that the cell clusters are composed of mainly mucoid cells, we investigated a set of sputum samples from patient p11 before and after a 2-week intravenous antibiotic therapy course with tobramycin and meropenem. Patient p11 is chronically infected with two clones, i.e., nonmucoid cells of the b genotype and mucoid cells of the ′4′ genotype (Table 1) (22). Routine antibiotic resistance profiling performed prior to the antibiotic course showed that the mucoid cells were sensitive to both antibiotics, whereas the nonmucoid cells were resistant. Etests of individual strains isolated before therapy showed that nonmucoid isolates had >10-fold-higher MICs for both antibiotics than mucoid isolates (data not shown). In accordance with this observation, only few mucoid cells were found in sputum samples taken immediately after the antibiotic course as evaluated by plating (Fig. 2). Importantly, this reduction in the number of mucoid cells as a consequence of the antibiotic therapy was correlated with a clear reduction in the number of cell clusters in the sputum both during and at the end of the intensive treatment period as shown by FISH analysis (Fig. 2). We note that the antibiotic treatment did not eradicate the mucoid population, as it reappeared soon after the end of treatment (Fig. 2).
FIG. 2.
Distribution of P. aeruginosa populations in CF sputum during a course of intravenous antibiotic therapy. Visualization of P. aeruginosa in sputum samples from CF patient p11 obtained before (A), immediately following (B), and 2 months after (C) an intravenous antibiotic therapy course with tobramycin and meropenem is shown. The P. aeruginosa population was visualized directly within sputa by FISH (left column) or by plating (middle column). The genotypes of the infecting clones as well as the frequencies of nonmucoid and mucoid variants for each time point are shown on the right. The bars represent 10 μm.
Taken together, these data shows that P. aeruginosa cell clusters within sputum samples appear to represent mainly the mucoid population, as the clusters were not present in patients infected with only nonmucoid P. aeruginosa or in patients from whom the mucoid population was transiently removed by antibiotics.
The specific correlation between growth rate and cellular ribosome content varies between different clones.
To be able to measure in situ growth rates of the infecting bacteria, we first determined the specific relationship between bacterial growth rate and cellular ribosome content in a series of in vitro growth experiments. To this end, we analyzed eight P. aeruginosa strains isolated from sputum samples from six different long-term chronically infected CF patients (Table 1). These eight isolates were of four different genotypes, and one of the isolates was mucoid (Table 2). We also included an isolate from an intermittently colonized patient (strain B4-1) as well as reference strains PAO1 and PA14. The P. aeruginosa strains were cultivated in different media that supported a range of growth rates. Cells were also cultivated under anaerobic conditions with nitrate as an electron acceptor as well as in a mucus medium prepared from mucus isolated from pig lungs (see Materials and Methods). In these two media, we found the doubling times of strain p7 s1B1/05 to be 149 and 104 min, respectively. During exponential growth, the cellular ribosome content was determined by quantitative whole-cell hybridization with a fluorescently labeled 16S rRNA probe.
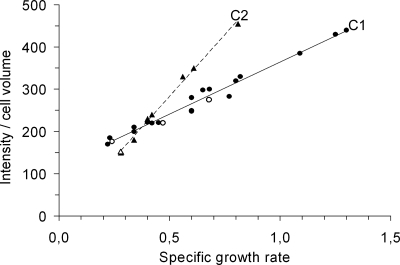
Figure 3 shows how the cellular content of ribosomes (measured as rRNA hybridization signal intensities per bacterial volume) increased approximately linearly with growth rates for all strains tested, as previously reported for other bacteria (30, 46, 48, 51). With the exception of strains of the b genotype (Table 2) isolated from patients p7 and p11, all other strains tested demonstrated the same specific correlation regardless of their genotype and their mucoid/nonmucoid phenotype (regression line C1 in Fig. 3). Interestingly, the linear correlation between ribosome content and growth rate for strains of the b genotype (regression line C2 in Fig. 3) was different, and the slope of the regression line was about 2 times greater than that for C1. These results show that there is a clone-specific relationship between ribosome content and growth rate and that the standard curves in Fig. 3 may be used for in situ determination of the growth activity of P. aeruginosa only if the genotype of the strain analyzed is known. The data used in Fig. 3 (including strain information, growth media, growth rate, and intensity per cell volume) are provided in the supplemental material.
FIG. 3.
Cellular content of ribosomes (fluorescence signal intensity per cell volume) inferred by whole-cell hybridization with a fluorescence-labeled 16S rRNA probe of balanced cultures grown in defined media supporting different specific growth rates. The strains and culture conditions used are described in Materials and Methods. The solid line labeled C1 and the dashed line labeled C2 are regression lines. Bacterial isolates of the b genotype (triangles) display the C2 correlation between ribosome content and specific growth rate (r2 = 0.995). All other isolates tested (circles) display the C1 correlation (r2 = 0.978). Key data points are highlighted by open symbols: anaerobic growth of isolate p7 s1B1/05 of the b genotype is shown by the open triangle, and open circles show growth of the mucoid isolate p11 s3A1/05 of the 4 genotype in minimal medium supplemented with (from left to right) glucose or glucose with Casamino Acids or in LB medium (fastest growth). Each measurement is the average value obtained from the analysis of the fluorescence signal from more than 100 cells. The standard error of the mean was less than 15% for all measurements. Further details related to the data set (strain information, growth media, growth rate, and intensity per cell volume) are provided in the supplemental material.
During the in vitro growth experiments, we also observed that P. aeruginosa isolates obtained from long-term chronically infected patients grow two- to threefold slower than the reference strains PAO1 and PA14 in the different growth conditions tested. For example, growth of PAO1 and PA14 in rich LB medium resulted in generation times of between 24 and 27 min. In contrast, the CF isolates grew with an average generation time of 64 min, with an range of 50 to 74 min. Likewise, growth in ABT minimal medium supplemented with 2% Casamino Acids resulted in generation times of between 33 and 36 min for the reference strains, whereas the CF isolates grew with an average generation time of 119 min with a range of 103 to 129 min. The slow-growth phenotype was not an intrinsic trait of clones capable of infecting CF patients, as the growth phenotypes of six P. aeruginosa strains isolated from four intermittently colonized CF patients were similar to those of the reference strains (data not shown). It therefore appears that selective pressures in the CF airway environment specifically enrich for slow-growing variants.
Detection of stationary-phase, nongrowing cells by rRNA hybridization.
Starved or slow-growing cells contain reduced numbers of ribosomes. To test whether fluorescence rRNA hybridization signals could be detected in starved nongrowing cells and to examine the effect of nutrient starvation on the cellular ribosome content, we transferred exponentially growing cells of the r and b genotypes as well as PA14 to ABT medium without a carbon source and measured fluorescence rRNA hybridization signals per cell volume at different time points on fixed cells counterstained with DAPI. As expected, a shift to nutrient-free medium resulted in a period of decline in ribosome content, after which the ribosome concentration remained approximately constant for at least 5 days at 50 to 40% of the level observed in exponentially growing cells (Fig. 4). Importantly, the observed significant reduction in the cellular ribosome content after only 2 h of starvation suggests that ribosomes are being degraded rapidly when cells are starved. The results also show that cells of both the b and r genotypes display similar regulation of rRNA in response to nutrient starvation, despite their genotype-specific relationship between ribosome concentration and growth rate.
FIG. 4.
Relative fluorescence signal intensity per cell volume during starvation. Exponentially growing cultures of PA14 (triangles), p2 s1F6/05 (squares), and p7 s1B1/05 (circles) in ABT plus 2% Casamino Acids were transferred to ABT medium without a carbon source. At the indicated time points, samples were taken and cellular ribosome content measured by whole-cell hybridization using a fluorescently labeled rRNA probe. The signal intensity measured prior to starvation was set to 100%. Each measurement is the average value obtained from the analysis of the fluorescence signal from more than 100 cells. The standard error of the mean was less than 15% for all measurements.
After 3 days of starvation, more than 90% of the DAPI-stained cells produced detectable rRNA hybridization signals. This result suggests that most starving nongrowing cells would indeed be detected in situ. As the data provide a measurement of the cellular ribosome content in nongrowing cells, it was possible to define a specific fluorescence signal intensity that clearly differentiated nongrowing, stationary-phase cells from growing cells. For each of the two genotypes analyzed, we used 120% of the fluorescence intensity measured after 20 h of starvation as the boundary separating growth from nongrowth. Cells showing fluorescence intensities lower than these values were thus considered to be stationary-phase, nongrowing cells.
Estimation of growth rates in sputum samples from CF patients.
To estimate in situ doubling times of P. aeruginosa in CF airways, we measured the ribosome contents of P. aeruginosa cells in sputa from CF patients p2, p7, and p11 (Table 1). We have previously shown that p2 and p7 are chronically infected with clones of either the r genotype (p2) or the b genotype (p7) and that these particular infections are entirely clonal (22). Both mucoid and nonmucoid variants are found in p2, whereas only nonmucoid cells are found in p7. Patient p11 is chronically infected with nonmucoid cells of the b genotype and mucoid cells of the 4 genotype (Table 2). For this particular patient, we analyzed only sputum samples taken after a 2-week intravenous antibiotic therapy that targeted the mucoid population and effectively reduced the number of mucoid cells to <1% as evaluated by plating (Fig. 2). As a result, the cellular ribosome content was measured only on the nonmucoid population of the b genotype, which was unaffected by the antibiotic treatment.
Total bacteria were extracted from fresh sputum samples, and the cellular ribosomal content was measured as fluorescence rRNA hybridization signals per cell volume as described in Materials and Methods. By use of the appropriate standard curves in Fig. 3 (C1 for p2 and C2 for p7 and p11), the fluorescence signal intensities were converted to apparent doubling times.
The distribution of growth rates of P. aeruginosa cells isolated from a sputum sample from patient p2 is shown in Fig. 5A. Interestingly, the vast majority of the cells analyzed were found to be actively growing, and only 15% of the population was found to be stationary-phase, nongrowing cells. Most of the growing cells showed generation times of between 100 and 200 min, but a significant fraction of cells (22%) were growing even faster. Some of these cells were approaching the in vitro growth rates measured in rich LB medium for the isolated clones from this patient. The average generation time calculated for all actively growing cells in vivo was 139 min, which is comparable to the in vitro generation time in ABT plus 2% Casamino Acids.
FIG. 5.
Distribution of generation times of P. aeruginosa cells isolated from sputum samples from CF patients p2 (A), p7 (B), and p11 (C). The cellular ribosome contents (fluorescence signal intensity per cell volume) of bacteria in the samples were measured by whole-cell hybridization using a fluorescently labeled rRNA probe. These measurements were converted into apparent doubling times using the appropriate standard correlations presented in Fig. 3.
Similar distributions of P. aeruginosa doubling times were observed in sputa from both patients p7 (Fig. 5B) and p11 (Fig. 5C). Again, only small fractions of the cell populations were found to be stationary-phase, nongrowing cells (8% for p7 and 11% for p11). The average generation time calculated for all actively growing cells was 115 min in p7 and 127 min in p11.
We further estimated in situ doubling times in two sputum samples with mixed populations of P. aeruginosa, i.e., where the samples contained more than one genotype. In a sample from p11 taken prior to the intravenous antibiotic course described above, both nonmucoid cells of the b genotype and mucoid cells of the 4 genotype were present, in an approximately 9:1 ratio. Fourteen percent of the cells analyzed were found to be stationary-phase cells, and the average doubling time for actively growing cells was found to be 134 min. In a sample from p16 which contained approximately 30% mucoid cells of the b genotype and 70% nonmucoid cells of the r genotype (Tables 1 and 2), we found that 13% of the cell population was in stationary phase and that the average doubling time for actively growing cells was 141 min. For both samples, the most conservative cutoff value for distinguishing between growing and nongrowing cells was used.
These results show that the majority of P. aeruginosa cells in sputum samples were actively growing with average doubling times ranging from 115 to 141 min, which is similar to pure-culture growth in ABT medium supplemented with 2% Casamino Acids. Importantly, these growth characteristics appeared to be independent of bacterial genotype, clonal versus mixed infections, mucoid/nonmucoid phenotype, and the patients from whom the sputum sample was obtained.
Pure-culture growth in CF sputum media.
In order to assess the relevance of the in situ growth rate measurements, the following in vitro experiment was performed. A sterile CF sputum growth medium was prepared from a sputum sample from p16 (see Materials and Methods). Pure-culture growth of PAO1 in this medium resulted in a doubling time of 66 min, which is significantly faster growth than observed in situ for the clinical strains in p16. Furthermore, pure-culture growth of strains p16 s1F4/05 and p7 s1B1/05 in the CF sputum medium resulted in generation times of 169 min and 112 min, respectively. Thus, the two- to threefold reduction in growth rate of the clinical isolates relative to reference strains observed in vitro in different growth media was retained in the CF sputum medium. Importantly, the agreement between these in vitro results and the in vivo measurements strongly indicates that the bacteria are indeed growing at their maximum potential in the sputum.
DISCUSSION
We have examined the distribution and growth characteristics of P. aeruginosa colonizing the airways of CF patients. We have previously shown that many of the long-term-infected CF patients at the CF Center in Copenhagen are infected with two prevailing clones because of extensive cross-infections among the patients (22). The patients and their infecting isolates studied here are therefore representatives of a large number of chronically infected patients.
To specifically visualize the P. aeruginosa populations colonizing the CF patients, we used FISH with a P. aeruginosa-specific rRNA probe directly on sputum samples (15). Compared to light and electron microscopy methods, FISH analysis provide specific, molecular detection of the particular bacteria studied. The spatial distribution of P. aeruginosa cells within sputum samples was found to be dependent on the mucoid/nonmucoid status of the infecting bacteria. Clusters of P. aeruginosa cells surrounded by an CFW-stainable exopolysaccharide matrix (presumably alginate) were found in patients with mucoid variants, whereas these clusters as well as CFW staining were absent in sputum material from patients infected with only nonmucoid variants. In these patients, the bacteria were found mainly as well-separated cells. In addition, cell clusters were also absent in patients from whom the mucoid population was transiently removed by use of antibiotics. These results indicate that mainly mucoid variants may represent the biofilm lifestyle, which has been associated with P. aeruginosa infections of the CF airways (25, 52), whereas the nonmucoid variants examined here may be found as free-living cells not enclosed in an extracellular matrix. It is possible that in patients colonized with mixed populations (i.e., both mucoid and nonmucoid variants), alginate produced by the mucoid subpopulation may trap or embed the nonmucoid population in such a way that the observed cell clusters are composed of both types of variants.
We have previously shown that nonmucoid variants of the r and b genotypes are revertants of mucoid parent cells, as they have mutations in the mucA gene (22). We speculate that these nonmucoid revertants (containing mucA mutations as well as second-site suppressor mutations) have evolved alternative adaptive solutions that can substitute for alginate overproduction in order to enhance survival in the CF lung. These solutions probably include preferential colonization of sputum in the conductive zone of the airways (14) and being more resistant to antibiotics than strains with mucoid phenotypes (8).
To obtain estimates of the growth rates of P. aeruginosa when present in the airways of CF patients, we used quantitative FISH to measure the concentration of rRNA in a large number of single cells from individual sputum samples. Based on standard correlations between growth rate and cellular ribosome contents obtained from controlled in vitro growth experiments, these measurements were converted to estimates of in situ growth rates of P. aeruginosa in CF sputum. Previous studies have used similar approaches to estimate the growth rates of Escherichia coli and Salmonella enterica serovar Typhimurium colonizing mouse intestines (30, 48) and Pseudomonas putida colonizing the rhizosphere of barley seedlings (50).
The growth experiments performed to establish the standard correlations between growth rate and ribosome content showed that the P. aeruginosa isolates of different genotypes obtained from long-term chronically infected CF patients grow two- to threefold slower than laboratory reference strains in a variety of standard laboratory media, such as complex LB medium and minimal medium with different carbon sources, as well as in CF sputum medium. The observed reduced growth rate of isolates from long-term-infected CF patients appears not to be a phenomenon specifically associated with the Copenhagen CF center, since some CF isolates collected at other CF clinics also showed reduced growth rates in LB medium compared with non-CF isolates (13). These results suggest that the slow-growing phenotype may be a common feature of prolonged CF airway colonization. Most likely, the slow-growth phenotype develops as a consequence of the stressful environment where the cells experience continuous exposure to antibiotics and contacts with the cells and molecules of the immune system. For example, it is well established that chromosomal mutations that confer antibiotic resistance often have growth rate-reducing effects (1, 5, 6, 33, 40). Indeed, antibiotic resistance is a common and serious clinical problem in the treatment of chronic P. aeruginosa lung infections, and many of the nonmucoid isolates from long-term chronically infected patients attending the Copenhagen CF center are resistant to several antibiotics (Fig. 2) (2, 8, 20). Attempts to select for fast-growing revertants by repetitive growth of slow-growing cells in rich laboratory media were unsuccessful, which suggests that multiple compensatory mutations are required to restore the normal growth rate.
When establishing the standard correlations between growth rate and ribosome content, it also became evident that the correlations were dependent on the clone analyzed. Although the majority of clones shared the same standard correlation (C1 in Fig. 3), clones of the b genotype isolated from two different patients exhibited a different correlation (C2 in Fig. 3). More specifically, the C2 correlation indicates that cells of the b genotype contain a higher level of rRNA at specific growth rates of >0.4 h−1 than the other clones that follow the C1 correlation. While the molecular basis for the clone-specific relationship between ribosome content and growth rate remains unknown, we note that strains of the b genotype displayed a pattern of regulation of rRNA in response to nutrient starvation that was similar to the pattern found for strains that followed the C1 correlation (Fig. 4), which suggests that the stringent response is fully functional in the b clone. In support of this finding, sequencing of the relA, spoT, and dksA genes in strains of the b genotype did not reveal any mutations that would alter the functions of the encoded proteins (data not shown). RelA, SpoT, and DksA are required for normal stringent regulation in many bacteria, including P. aeruginosa (42, 43, 45).
The in vitro growth experiments performed in order to establish the standard correlations between growth rate and ribosome content were designed to cover a wide range of growth conditions that supported different specific growth rates. These growth conditions included both aerobic and anaerobic growth in defined media as well as growth in complex media such as LB and in mucus purified from pig lungs. The important conclusion from quantitative FISH measurements of cells under these diverse growth conditions is that the ribosome content depends only on the actual growth rate (and the particular clone analyzed) and not on the specific medium used. In other words, all points related to the quantitative hybridizations fall on the standard curves presented in Fig. 3. For example, strain p7 s1B1/05 was found to grow with a doubling time of 149 min both during aerobic growth in ABT plus 10 mM sodium citrate and during anaerobic growth in LB medium. In these two cultures, we also measured similar ribosome contents (150 and 153 units of fluorescence intensity per cell volume, respectively).
The in vitro starvation experiments performed to measure the effect of nutrient starvation on the cellular ribosome content clearly showed that cells responded immediately (within 2 h) to starvation by rapid degradation of ribosomes. This means that the actual cellular ribosome concentrations reflect quite well the actual physiological states of the cells. If, in contrast, the ribosomes had turned out to be stable after the onset of starvation we would not have been able to distinguish between growing cells and stationary-phase cells with a nondegraded excess of ribosomes. Moreover, the data show that the reduced ribosome contents in cells subjected to prolonged starvation were still efficiently detected by FISH. The main conclusion from this experiment therefore is that the rapid reduction in ribosome content to a low but detectable level in response to starvation allowed determination of the fraction of stationary-phase, nongrowing cells in the sputum samples.
Our estimates of growth rates of P. aeruginosa in sputa by using quantitative FISH are, to the best of our knowledge, the first in situ description of the growth physiology of P. aeruginosa infecting CF airways. The experiments revealed remarkably similar results for three different CF patients infected by two distinct clones (Fig. 5) as well as for CF patients infected by both mucoid and nonmucoid strains of different genotypes. The observation that there were similar average doubling times in vivo for bacteria with different genotypes, different mucoid/nonmucoid phenotypes, and different correlations between growth rate and ribosome content, as well as for bacteria isolated from different patients with both clonal and mixed infections, suggests that the growth properties of the infecting strains have been optimized to the lung environment during long-term colonization and that these particular conditions are very similar in different patients. In this view, the reduced growth rates of the different clinical isolates may be regarded as an example of parallel evolution of an adaptive trait, analogous to the parallel adaptation of independent bacterial cultures to a defined laboratory environment during long-term evolution experiments (10, 28, 44).
The average doubling times calculated for actively growing cells in sputa from these patients were in all cases comparable to the doubling times measured in ABT minimal medium supplemented with 2% Casamino Acids in controlled in vitro growth experiments as well as to doubling times measured both in pig mucus media and in CF sputum media. High levels (15 to 20 mM) of amino acids have been observed in CF sputum (4, 56), and recently a transcriptome analysis of P. aeruginosa cells growing in medium made from CF sputum strongly suggested that amino acids within the sputum indeed are the likely candidates as carbon and nitrogen sources for the bacteria (41). Our data suggest that ABT minimal medium supplemented with 2% Casamino Acids, pig mucus medium, and CF sputum medium all may be appropriate growth substrates for in vitro studies of P. aeruginosa isolates from CF patients.
The distributions of in situ doubling times indicate that most cells are actively growing cells. Only 8 to 15% of the cell populations analyzed were characterized as being nongrowing cells (Fig. 5). We recognize that some inaccuracy exists in the discrimination between growing and nongrowing cells. For example, some nongrowing cells may have a hybridization signal too faint to be recorded. However, in control experiments with DAPI-counterstained starving cells, we found that a faint hybridization signal may result in only a slight underestimation of the number of stationary-phase cells. Also, the hybridization signal chosen for the differentiation between growing and nongrowing cells may contribute to some inaccuracies. We chose 120% of the hybridization signal value after 20 h of starvation as the cutoff value for growing cells, which is a highly conservative choice and may in fact result in significant overestimation of the number of nongrowing cells. Nevertheless, we consider the contribution of these potential errors to be insignificant in relation to our major conclusion that the clear majority of P. aeruginosa cells are actively growing in the airways of the CF patients.
The finding that most cells in vivo are actively growing at relatively high growth rates has implications for the understanding of the P. aeruginosa population dynamics and organization in chronic CF airway infections. First, the result suggests a high rate of turnover of bacterial biomass. This turnover is most likely caused by antibiotic treatment, immune system attacks, and removal of biomass by coughing. Second, the results indicate that the bacteria may not be organized in the same type of biofilms as described from in vitro investigations. Laboratory-based flow chambers and related setups (7) have been used to mimic and study the biofilm lifestyle, which for a long time has been linked to P. aeruginosa infections in CF patients (3, 14, 16, 25, 52). However, biofilm development in vitro most often results in spatial patterns of growth activity with the majority of the population being inactive, nongrowing cells located in the center of the biofilm and a subpopulation of active, rapidly growing cells near the nutrient- and oxygen-exposed surface (55, 57). We performed quantitative FISH analysis on cells of PAO1 and p7 s1B1/05 isolated from 3- and 8-day-old in vitro biofilms. For both strains, more than 45% of the cells were found to be nongrowing, stationary-phase cells using the same analysis as carried out on cells from sputum (data not shown). We therefore argue that the growth physiology of cells from in vitro biofilms may be incompatible with the in vivo growth activities measured here and that the absence of heterogeneity with respect to growth activities in sputum indicates a lifestyle characterized by growth of dispersed, single cells. In fact, we did observe that nonmucoid cells were organized mainly as dispersed, single cells in sputum (Fig. 1). On the other hand, mucoid variants were found to be organized in clusters surrounded by an extracellular matrix in sputum. However, as the majority of these cells were actively growing cells, we suggest that the cells are distributed within these clusters in such a manner that internal heterogeneities (e.g., gradients of nutrient availability) may not interfere with growth. We observed that the cells within in vitro biofilms formed by the mucoid isolate p11 s3A1/05 were not as densely organized as observed for PA01 in vitro biofilms (data not shown). This suggests that microenvironments that impose constraints on growth activities may not develop in situ, as the mucoid cells are essentially growing as well-dispersed cells within an extracellular matrix.
In conclusion, we propose that the complex CF airway environment impose two opposite selection forces on the P. aeruginosa populations that chronically infect CF patients. On one hand, there is a clear selection for clones with highly reduced growth rates. It remains to be seen if slow growth per se is the selected property or if slow growth is a consequence of other selected properties such as increased antibiotic tolerance. On the other hand, competition for nutrients favors relatively fast growth of the bacteria within the CF airways. This, in turn, may be incompatible with the biofilm mode of growth as defined from in vitro settings. This model constitutes a new perspective on both the adaptation and evolution of infecting P. aeruginosa populations but also on the within-host growth dynamics of the bacteria during chronic lung infections in CF patients. An understanding of both aspects has important consequences for the design of new preventive measures.
Supplementary Material
Acknowledgments
This work was supported by a cross-disciplinary research grant from the Danish Research Councils to S.M.
We thank Tove Johansen from BioCentrum-DTU and Jean Baptiste Rioux from INSA-Biosciences, Lyon, France, for excellent technical assistance.
Footnotes
Published ahead of print on 21 December 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andersson, D. I., and B. R. Levin. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2489-493. [DOI] [PubMed] [Google Scholar]
- 2.Bagge, N., O. Ciofu, M. Hentzer, J. I. Campbell, M. Givskov, and N. Høiby. 2002. Constitutive high expression of chromosomal beta-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob. Agents Chemother. 463406-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltimore, R. S., C. D. Christie, and G. J. Smith. 1989. Immunohistopathologic localization of Pseudomonas aeruginosa in lungs from patients with cystic fibrosis. Implications for the pathogenesis of progressive lung deterioration. Am. Rev. Respir. Dis. 1401650-1661. [DOI] [PubMed] [Google Scholar]
- 4.Barth, A. L., and T. L. Pitt. 1996. The high amino-acid content of sputum from cystic fibrosis patients promotes growth of auxotrophic Pseudomonas aeruginosa. J. Med. Microbiol. 45110-119. [DOI] [PubMed] [Google Scholar]
- 5.Bjorkman, J., D. Hughes, and D. I. Andersson. 1998. Virulence of antibiotic-resistant Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 953949-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorkman, J., I. Nagaev, O. G. Berg, D. Hughes, and D. I. Andersson. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 2871479-1482. [DOI] [PubMed] [Google Scholar]
- 7.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, Jr., A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 31020-42. [DOI] [PubMed] [Google Scholar]
- 8.Ciofu, O., V. Fussing, N. Bagge, C. Koch, and N. Høiby. 2001. Characterization of paired mucoid/non-mucoid Pseudomonas aeruginosa isolates from Danish cystic fibrosis patients: antibiotic resistance, beta-lactamase activity and RiboPrinting. J. Antimicrob. Chemother. 48391-396. [DOI] [PubMed] [Google Scholar]
- 9.Clark, J. D., and O. Maaloe. 1967. DNA replication and the cell cycle in Escherichia coli cells. J. Mol. Biol. 2399-112. [Google Scholar]
- 10.Cooper, T. F., D. E. Rozen, and R. E. Lenski. 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. USA 1001072-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 2431360-1363. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, R. E., L. M. Mutharia, L. Chan, R. P. Darveau, D. P. Speert, and G. B. Pier. 1983. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect. Immun. 42170-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Head, N. E., and H. Yu. 2004. Cross-sectional analysis of clinical and environmental isolates of Pseudomonas aeruginosa: biofilm formation, virulence, and genome diversity. Infect. Immun. 72133-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann, N., T. B. Rasmussen, P. O. Jensen, C. Stub, M. Hentzer, S. Molin, O. Ciofu, M. Givskov, H. K. Johansen, and N. Høiby. 2005. Novel mouse model of chronic Pseudomonas aeruginosa lung infection mimicking cystic fibrosis. Infect. Immun. 732504-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogardt, M., K. Trebesius, A. M. Geiger, M. Hornef, J. Rosenecker, and J. Heesemann. 2000. Specific and rapid detection by fluorescent in situ hybridization of bacteria in clinical samples obtained from cystic fibrosis patients. J. Clin. Microbiol. 38818-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Høiby, N. 2005. New insight into the pathogenesis and epidemiology of Pseudomonas aeruginosa infection in cystic fibrosis, p. 11-25. In H. Goossens, W. Peetermans, and M. Struelens (ed.), Proceedings of the 4th Elzenveld workshop on infectious diseases: non-fermenting Gram-negative bacilli: microbiological and clinical challenges. Bristol-Meyers Squibb, Antwerpen, Belgium.
- 17.Høiby, N. 1977. Pseudomonas aeruginosa infection in cystic fibrosis. Diagnostic and prognostic significance of pseudomonas aeruginosa precipitins determined by means of crossed immunoelectrophoresis. A survey. Acta Pathol. Microbiol. Scand. Suppl. 19771-96. [PubMed] [Google Scholar]
- 18.Høiby, N., and B. Frederiksen. 2000. Microbiology of cystic fibrosis, p. 83-107. In M. E. Hodson and D. M. Geddes (ed.), Cystic fibrosis, 2nd ed. Arnold, London, United Kingdom.
- 19.Jain, M., D. Ramirez, R. Seshadri, J. F. Cullina, C. A. Powers, G. S. Schulert, M. Bar-Meir, C. L. Sullivan, S. A. McColley, and A. R. Hauser. 2004. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J. Clin. Microbiol. 425229-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jalal, S., O. Ciofu, N. Høiby, N. Gotoh, and B. Wretlind. 2000. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 44710-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansen, G. J., M. Mooibroek, J. Idema, H. J. Harmsen, G. W. Welling, and J. E. Degener. 2000. Rapid identification of bacteria in blood cultures by using fluorescently labeled oligonucleotide probes. J. Clin. Microbiol. 38814-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jelsbak, L., H. K. Johansen, A. L. Frost, R. Thogersen, L. E. Thomsen, O. Ciofu, L. Yang, J. A. Haagensen, N. Høiby, and S. Molin. 2007. Molecular epidemiology and dynamics of Pseudomonas aeruginosa populations in lungs of cystic fibrosis patients. Infect. Immun. 752214-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansen, H. K., L. Norregaard, P. C. Gotzsche, T. Pressler, C. Koch, and N. Høiby. 2004. Antibody response to Pseudomonas aeruginosa in cystic fibrosis patients: a marker of therapeutic success? A 30-year cohort study of survival in Danish CF patients after onset of chronic P. aeruginosa lung infection. Pediatr. Pulmonol. 37427-432. [DOI] [PubMed] [Google Scholar]
- 24.Kjeldgaard, N. O., and C. G. Kurland. 1963. The distribution of soluble and ribosomal RNA as a function of growth rate. J. Mol. Biol. 6341-348. [Google Scholar]
- 25.Lam, J., R. Chan, K. Lam, and J. W. Costerton. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28546-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, B., J. A. Haagensen, O. Ciofu, J. B. Andersen, N. Høiby, and S. Molin. 2005. Heterogeneity of biofilms formed by nonmucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. J. Clin. Microbiol. 435247-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, V. T., R. S. Smith, B. Tummler, and S. Lory. 2005. Activities of Pseudomonas aeruginosa effectors secreted by the type III secretion system in vitro and during infection. Infect. Immun. 731695-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenski, R. E., and M. Travisano. 1994. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl. Acad. Sci. USA 916808-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leveau, J. H., and S. E. Lindow. 2001. Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. USA 983446-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Licht, T. R., K. A. Krogfelt, P. S. Cohen, L. K. Poulsen, J. Urbance, and S. Molin. 1996. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect. Immun. 643811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luzar, M. A., and T. C. Montie. 1985. Avirulence and altered physiological properties of cystic fibrosis strains of Pseudomonas aeruginosa. Infect. Immun. 50572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahenthiralingam, E., M. E. Campbell, and D. P. Speert. 1994. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect. Immun. 62596-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mariam, D. H., Y. Mengistu, S. E. Hoffner, and D. I. Andersson. 2004. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 481289-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin, D. W., M. J. Schurr, M. H. Mudd, J. R. Govan, B. W. Holloway, and V. Deretic. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 908377-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathee, K., O. Ciofu, C. Sternberg, P. W. Lindum, J. I. Campbell, P. Jensen, A. H. Johnsen, M. Givskov, D. E. Ohman, S. Molin, N. Høiby, and A. Kharazmi. 1999. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology 1451349-1357. [DOI] [PubMed] [Google Scholar]
- 36.Mazia, D., G. Schatten, and W. Sale. 1975. Adhesion of cells to surfaces coated with polylysine. Applications to electron microscopy. J. Cell Biol. 66198-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molin, S., and M. Givskov. 1999. Application of molecular tools for in situ monitoring of bacterial growth activity. Environ. Microbiol. 1383-391. [DOI] [PubMed] [Google Scholar]
- 38.Moller, S., C. S. Kristensen, L. K. Poulsen, J. M. Carstensen, and S. Molin. 1995. Bacterial growth on surfaces: automated image analysis for quantification of growth rate-related parameters. Appl. Environ. Microbiol. 61741-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moller, S., A. R. Pedersen, L. K. Poulsen, E. Arvin, and S. Molin. 1996. Activity and three-dimensional distribution of toluene-degrading Pseudomonas putida in a multispecies biofilm assessed by quantitative in situ hybridization and scanning confocal laser microscopy. Appl. Environ. Microbiol. 624632-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nilsson, A. I., O. G. Berg, O. Aspevall, G. Kahlmeter, and D. I. Andersson. 2003. Biological costs and mechanisms of fosfomycin resistance in Escherichia coli. Antimicrob. Agents Chemother. 472850-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer, K. L., L. M. Mashburn, P. K. Singh, and M. Whiteley. 2005. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 1875267-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paul, B. J., M. M. Barker, W. Ross, D. A. Schneider, C. Webb, J. W. Foster, and R. L. Gourse. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118311-322. [DOI] [PubMed] [Google Scholar]
- 43.Paul, B. J., W. Ross, T. Gaal, and R. L. Gourse. 2004. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 38749-770. [DOI] [PubMed] [Google Scholar]
- 44.Pelosi, L., L. Kuhn, D. Guetta, J. Garin, J. Geiselmann, R. E. Lenski, and D. Schneider. 2006. Parallel changes in global protein profiles during long-term experimental evolution in Escherichia coli. Genetics 1731851-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perron, K., R. Comte, and C. van Delden. 2005. DksA represses ribosomal gene transcription in Pseudomonas aeruginosa by interacting with RNA polymerase on ribosomal promoters. Mol. Microbiol. 561087-1102. [DOI] [PubMed] [Google Scholar]
- 46.Poulsen, L. K., G. Ballard, and D. A. Stahl. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 591354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poulsen, L. K., F. Lan, C. S. Kristensen, P. Hobolth, S. Molin, and K. A. Krogfelt. 1994. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect. Immun. 625191-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poulsen, L. K., T. R. Licht, C. Rang, K. A. Krogfelt, and S. Molin. 1995. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J. Bacteriol. 1775840-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramos, C., T. R. Licht, C. Sternberg, K. A. Krogfelt, and S. Molin. 2001. Monitoring bacterial growth activity in biofilms from laboratory flow chambers, plant rhizosphere, and animal intestine. Methods Enzymol. 33721-42. [DOI] [PubMed] [Google Scholar]
- 50.Ramos, C., L. Molbak, and S. Molin. 2000. Bacterial activity in the rhizosphere analyzed at the single-cell level by monitoring ribosome contents and synthesis rates. Appl. Environ. Microbiol. 66801-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaechter, M., O. Maaloe, and N. O. Kjeldgaard. 1958. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J. Gen. Microbiol. 19592-606. [DOI] [PubMed] [Google Scholar]
- 52.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407762-764. [DOI] [PubMed] [Google Scholar]
- 53.Smith, E. E., D. G. Buckley, Z. Wu, C. Saenphimmachak, L. R. Hoffman, D. A. D'Argenio, S. I. Miller, B. W. Ramsey, D. P. Speert, S. M. Moskowitz, J. L. Burns, R. Kaul, and M. V. Olson. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 1038487-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spencer, D. H., A. Kas, E. E. Smith, C. K. Raymond, E. H. Sims, M. Hastings, J. L. Burns, R. Kaul, and M. V. Olson. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 1851316-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sternberg, C., B. B. Christensen, T. Johansen, A. Toftgaard Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 654108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas, S. R., A. Ray, M. E. Hodson, and T. L. Pitt. 2000. Increased sputum amino acid concentrations and auxotrophy of Pseudomonas aeruginosa in severe cystic fibrosis lung disease. Thorax 55795-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Werner, E., F. Roe, A. Bugnicourt, M. J. Franklin, A. Heydorn, S. Molin, B. Pitts, and P. S. Stewart. 2004. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 706188-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Doring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.